Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) has a modular domain architecture that couples DNA damage detection to poly(ADP-ribosyl)ation activity through a poorly understood mechanism. Here we report the crystal structure of a DNA double-strand break in complex with human PARP-1 domains essential for activation (Zn1, Zn3, WGR-CAT). PARP-1 engages DNA as a monomer, and the interaction with DNA damage organizes PARP-1 domains into a collapsed conformation that can explain the strong preference for automodification. The Zn1, Zn3, and WGR domains collectively bind to DNA, forming a network of interdomain contacts that links the DNA damage interface to the catalytic domain (CAT). The DNA damage-induced conformation of PARP-1 results in structural distortions that destabilize the CAT. Our results suggest that an increase in CAT protein dynamics underlies the DNA-dependent activation mechanism of PARP-1.
PARP-1 creates poly(ADP-ribose) (PAR) covalently attached onto target proteins that mediate gene transcription, DNA damage repair, and cell death signaling (1, 2), and it has emerged as a promising drug target for the treatment of cancer due to its role in maintaining genome stability (3, 4). The primary target for PARP-1 mediated poly(ADP-ribosyl)ation is PARP-1 itself (1). PARP-1 binding to DNA strand breaks dramatically elevates PAR synthesis activity over a low basal level (1). The modular architecture of PARP-1 contains six domains (Fig. 1A). Two zinc-binding domains, Zn1 and Zn2, enable PARP-1 to recognize particular DNA structures (5–9). A third zinc-binding domain, Zn3, has a distinct structure and function from that of Zn1 and Zn2 (10, 11). The automodification domain (AD) bears the major sites of automodification and contains a BRCT (BRCA1 C–terminus) fold (1). The WGR is an essential domain of unknown function (12). The CAT is composed of two subdomains, the helical subdomain (HD) and the ART subdomain, which is conserved in other ADP-ribosyl transferases (ARTs), and includes the amino acids involved in catalysis and binding of NAD+ (13, 14).
Fig. 1. Overview of the PARP-1/DNA crystal structure.
(A) Modular domain architecture of human PARP-1. (B) Colorimetric assay of PARP-1 DNA-dependent automodification using the indicated domain combinations (see also Fig. S1 A,B). (C) Surface representation of the PARP-1/DNA structure. (D) A 90° rotation was applied to the view in C. (E) The Zn2 and BRCT domains (light grey) were manually positioned on the PARP-1/DNA structure using the structures of Zn2 and BRCT (PDB codes 3odc 2cok, respectively). The arrow accents the close proximity of AD and CAT.
We determined the x-ray crystal structure of human PARP-1 domains Zn1, Zn3, and WGR-CAT bound to a DNA double-strand break. These domains are essential for PARP-1 activity in the presence of DNA double-strand breaks (5, 10–12), and represent the minimal assembly of domains that supports DNA-dependent PARP-1 activity. Indeed, the Zn1 and Zn3 domains stimulate PAR synthesis of the WGR-CAT fragment in the presence of DNA (Fig. 1B). The Zn2 and BRCT domains are dispensable for PARP-1 activity in the presence of DNA double-strand breaks (Fig. S1)(5, 12, 15). Crystals of the PARP-1/DNA complex diffract anisotropically to 3.25 Å and the structure was determined by molecular replacement (Table S1, Fig. S2).
The PARP-1/DNA structure reveals the assembly of PARP-1 domains on DNA damage. The WGR domain is a central component of the complex, interacting with Zn1, Zn3, CAT, and the DNA (Fig. 1C, D). Zn1 and Zn3 rest next to each other on the DNA, both domains interacting with one face of WGR. The opposite face of WGR contacts the HD of the CAT. PARP-1 contacts with the DNA break are transmitted to the CAT through a network of interdomain communication that acts on the HD. In contrast, the ART subdomain is not involved in direct contacts with DNA or PARP-1 regulatory domains.
A model of full-length PARP-1 (including Zn2 and BRCT) was constructed based on the relative locations of the N- and C-termini of domains present in the structure (Fig. 1E) [Supplementary Online Material (SOM) text]. In the model, the AD is anchored between Zn3 and WGR in close proximity to the CAT and available for in cis modification, explaining the strong preference of PARP-1 to perform automodification over heteromodification of target proteins (16). The PARP-1/DNA complex contains a single copy of each domain, and the model for full-length PARP-1 indicates that a monomeric PARP-1 polypeptide can assume the active conformation. A monomeric mode of DNA binding has been reported recently for PARP-1 (5, 6, 17, 18); however, an earlier study reported that the Zn1-Zn2 fragment binds to DNA as a dimer under certain conditions (8) (SOM text). Our sedimentation analysis indicates that full-length PARP-1 exists as a highly extended monomer in solution in the absence of DNA, and that PARP-1 domains compact upon binding to DNA as a monomer with a 1:1 stoichiometry (Fig. S3), consistent with the PARP-1/DNA structure.
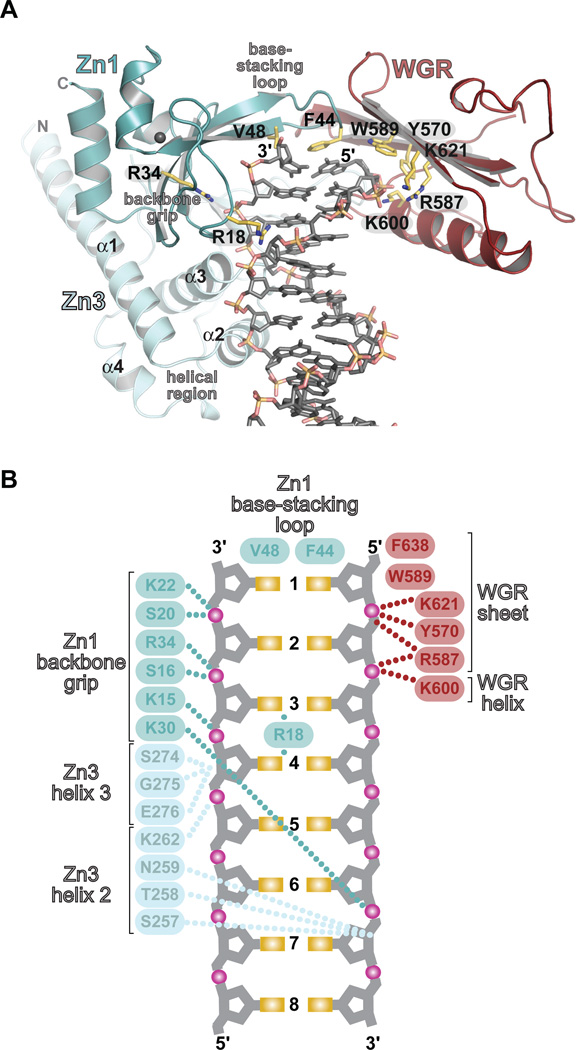
The PARP-1/DNA structure and biochemical experiments demonstrate that the Zn1, Zn3, and WGR domains collaboratively bind to DNA damage (Fig. 2, Fig S4, and Table S2). PARP-1 domains primarily contact the ribose-phosphate backbone of the DNA and therefore mediate sequence-independent interactions. PARP-1 engages the break in the DNA through hydrophobic contacts with exposed nucleotide bases (Fig. 2A, B), a common feature of damaged DNA structures (Fig. S5). Zn1 binds to DNA using two conserved regions termed the backbone grip and the base-stacking loop (Fig. 2A, B), as observed previously (5). Zn3 binds to DNA adjacent to Zn1 using its N-terminal α-helical region to span the minor groove (Fig. 2A). WGR binds to the 5'-terminus of one DNA strand, holding the DNA backbone between the central β-sheet and the α-helix of WGR (Fig. 2A). WGR residue Trp589 stacks against the ribose sugars of nucleotides located at the end of the 5'-terminated DNA strand (Fig. 2A, B).
Fig. 2. PARP-1 forms a multi-domain DNA binding interface.
Zn1, Zn3, and WGR collectively form an interface with DNA. (A) Key Zn1–, Zn3–, and WGR–DNA contacts are highlighted. (B) Schematic representation of PARP-1 contacts with DNA.
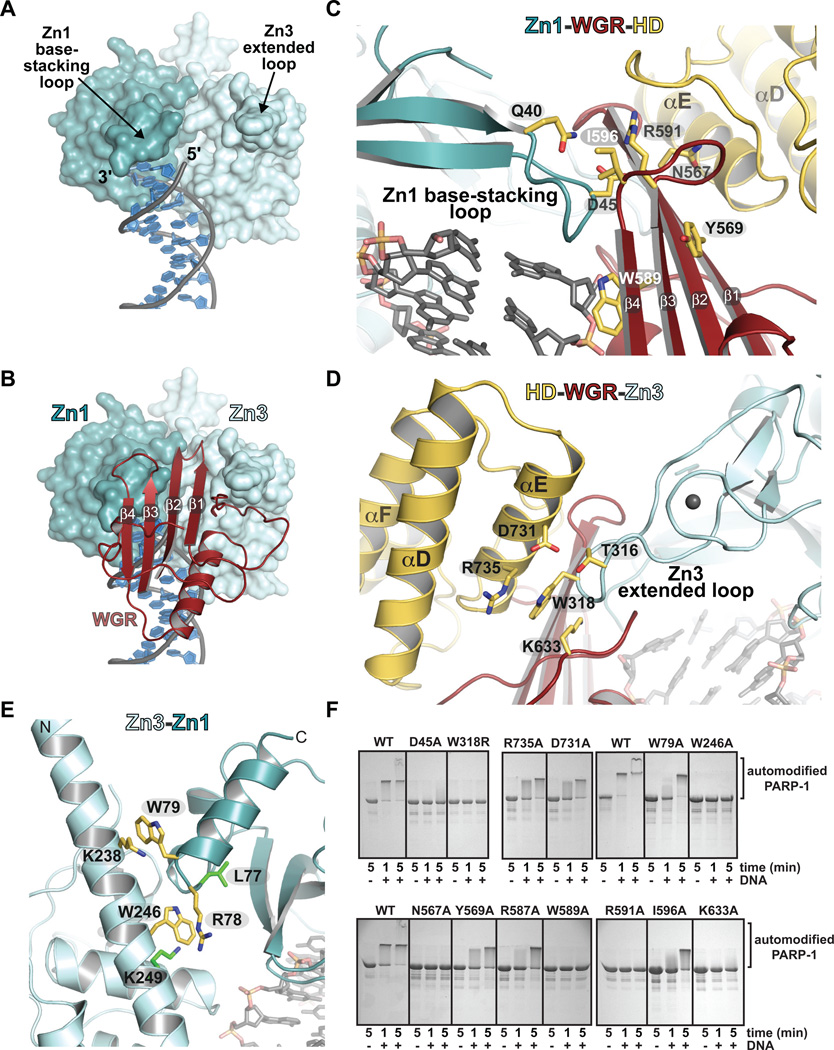
The PARP-1/DNA structure reveals a network of interdomain contacts formed upon DNA binding. The Zn1 base-stacking loop and the Zn3 extended loop provide a two-point contact surface with the appropriate orientation and spacing for engaging WGR (Fig. 3A, B). A key Zn1-WGR contact is a salt bridge formed between Asp45 of Zn1 and Arg591 of WGR (Fig. 3C). Arg591 also interacts with HD; thus WGR provides a bridge between the Zn1 DNA damage interface and the CAT. Zn3 contacts WGR and HD using the extended loop of its zinc ribbon fold, with residue Trp318 occupying a central location at this interface (Fig. 3D). Zn3 forms a second interface in the PARP-1/DNA complex, using its N-terminal helical region to interact with Zn1, near their points of contact with the DNA duplex (Fig. 3E). Mutation of full-length PARP-1 at residues located at domain interfaces resulted in severe to moderate defects in DNA-dependent activity relative to wild-type (WT) PARP-1 (Fig. 3F) (5, 19, 20). These biochemical results confirm that the interdomain contacts observed in the PARP-1/DNA structure are critical for DNA-dependent activation of full-length PARP-1.
Fig. 3. DNA-induced interdomain contacts are critical for DNA-dependent PARP-1 activation.
(A) Surface representation of Zn1 and Zn3 bound to DNA. WGR and CAT have been omitted for clarity. (B) The Zn1 base-stacking loop, the Zn3 extended loop, and the 5'-terminated DNA strand form a binding site for WGR. (C) The Zn1–WGR–HD interface. (D) The HD-WGR–Zn3 interface. (E) The Zn3–Zn1 interface. Residues targeted for mutagenesis are labeled (yellow). Residues identified in a random screen for inactive mutants (20) are labeled (green). (F) SDS-PAGE assay of DNA-dependent PARP-1 automodification activity. WT and the indicated mutants were monitored for a shift in migration due to the covalent addition of PAR.
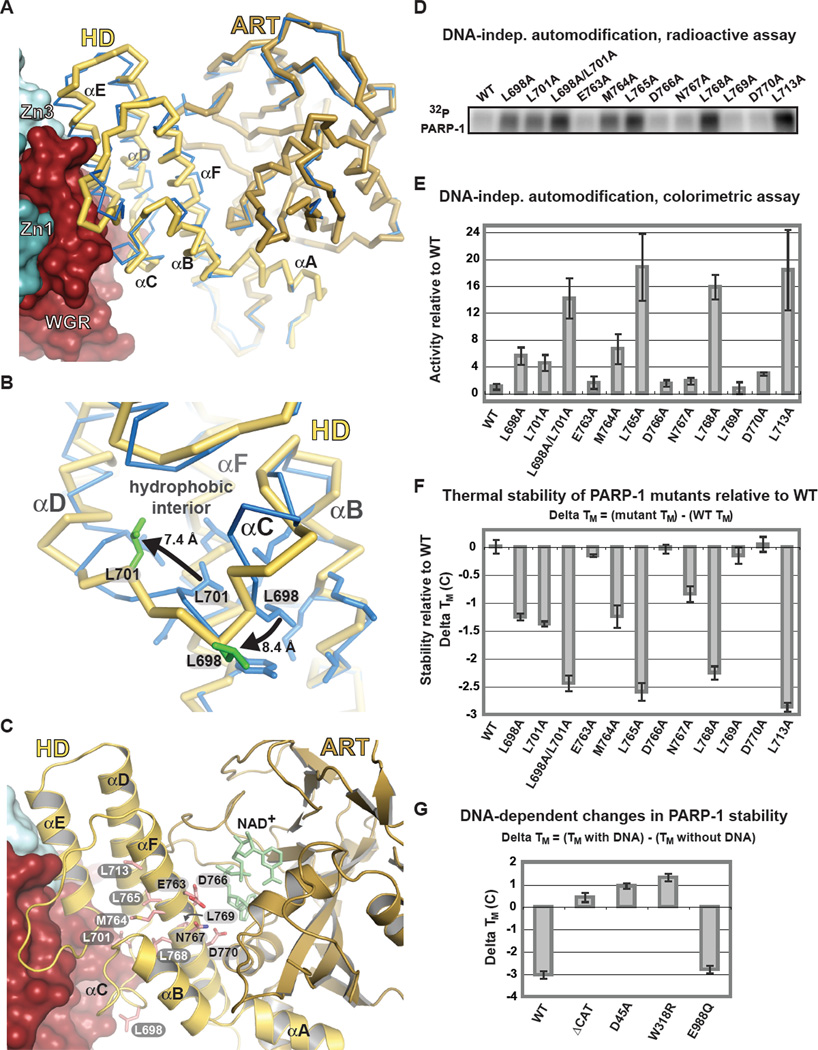
The PARP-1/DNA structure reveals that the HD is distorted when compared to structures of isolated CAT domains (Fig. 4A, B, Fig. S6). The HD region surrounding αC is remodeled and moved away from αF and αB and toward the WGR interface, displacing conserved residues Leu698 and Leu701 from the HD hydrophobic core (Fig. 4B). In contrast, the ART is similar to isolated CAT domain structures (Fig. 4A and Fig. S6). Thus, the DNA-dependent regulatory domains of PARP-1 act to distort the HD hydrophobic core, suggesting that HD distortion underlies DNA-dependent activation. We tested whether mutagenesis of the HD could simulate the observed HD distortions and thereby mimic the effect of PARP-1 interaction with DNA. Indeed, an increase in DNA-independent PARP-1 activity was observed for mutations that targeted the HD hydrophobic core (Fig. 4C–E, Fig. S7). In contrast, mutation of HD residues facing away from the hydrophobic interior had no effect on DNA-independent activity (Fig. 4C–E, Fig. S7). Random mutagenesis identified a hydrophobic core mutant L713F that activated PARP-1 in the absence of DNA (21), consistent with our biochemical analysis. HD hydrophobic core mutants mimic the effect of DNA-damage induced HD distortions, increasing PARP-1 DNA-independent activity up to ~20-fold, and elevating the efficiency of poly(ADP-ribosyl)ation (VMAX) while not affecting affinity for NAD+ (KM) (Fig. S8, S9).
Fig. 4. Distortion of the HD structure modulates PARP-1 catalytic activity.
(A) Cα trace of CAT structure in the absence of DNA and regulatory domains (PDB code 1a26; blue), and CAT in the PARP-1/DNA structure (yellow/brown). (B) Detailed view of Leu698 and Leu701 contributions to the HD hydrophobic interior of the isolated CAT (blue), and their re-positioning in the PARP-1/DNA structure (yellow with green side chains). (C) Ribbon representation of CAT in the PARP-1/DNA complex. Residues mutated are drawn in pink and labeled: HD interior hydrophobic core (white), HD exterior (black). The position of NAD+ was modeled for reference. (D) Radioactive assay of DNA-independent PARP-1 automodification using 32P-NAD+. (E) Colorimetric assay of DNA-independent PARP-1 automodification. Values represent quantification of the 90-minute time point of a time course experiment (Fig. S7) and are an average of three independent experiments with associated standard deviations. (F) Relative thermal stability of PARP-1 mutants obtained by differential scanning fluorimetry. Changes in thermal stability (ΔTM) were calculated as shown. (G) Relative change in thermal stability upon DNA binding. ΔTM were calculated as shown.
A strong anticorrelation exists between catalytic activity and relative thermal stability of HD mutants. HD hydrophobic core mutants exhibit increased DNA-independent activity and decreased thermal stability relative to WT PARP-1, while mutants that face the exterior of the HD hydrophobic core have the same activity and stability as WT (Fig. 4E, F). The HD mutations that elevate DNA-independent activity act solely through alterations to CAT, and do not depend on the regulatory domains. Indeed, the isolated CAT bearing HD hydrophobic core mutation L713F exhibited elevated DNA-independent activity and decreased thermal stability relative to WT CAT (Fig. S10). Consistent with the mutational analysis, full-length PARP-1 in the presence of DNA exhibited a decrease in thermal stability that depended on the presence of the CAT domain, and residues mediating interdomain contacts in the PARP-1/DNA structure (Fig. 4G). As a control, the catalytic ART mutant E988Q (22, 23), expected to form all interdomain contacts observed in the PARP-1/DNA structure, showed the same decrease in thermal stability as WT (Fig. 4G). Collectively, our results demonstrate that the stability of the CAT decreases upon PARP-1 binding to DNA damage, and the change in stability depends on the interdomain contacts that mediate HD distortion in the PARP-1/DNA structure.
PARP-1 binding to DNA damage organizes the Zn1, Zn3, and WGR domains into a conformation that distorts the hydrophobic core of the HD, and thereby decreases the thermal stability of CAT (Fig. S11). The observed HD distortions are unlikely to have a significant influence on active site access to NAD+ substrate or target protein substrate (SOM text). Rather, we propose that distortions to the structure and stability of HD are linked to the stability of ART, and that changes in the stability and conformational dynamics of ART underlie the DNA-dependent activation mechanism of PARP-1 (Fig. S11). The positioning of the AD in close proximity to the CAT could also contribute to PARP-1 DNA-dependent activity by increasing CAT domain exposure to protein substrate. Our study illustrates a DNA damage-dependent intramolecular mechanism for rapidly elevating the low basal level of PARP-1 activity, and suggests the structural basis for PARP-1 preference for automodification. The critical requirement of domain interfaces for PARP-1 DNA-dependent activity suggests that these domain interfaces represent targets for the design of novel PARP-1 inhibitors.
Supplementary Material
Acknowledgments
Supported by funds from the National Institutes of Health (R01087282), the American Cancer Society (RSG0918301), and the Kimmel Cancer Center X-ray Crystallography and Molecular Characterization Facility at Thomas Jefferson University (P30CA56036). We thank NSLS staff for access and assistance with data collection (beamlines X6A and X29A, Brookhaven National Laboratory); G. Cingolani, Y.-M. Hou, J.D. Steffen for helpful discussions, and S. Carney for cloning the Zn1–Zn3 interface mutants. The structure of the complex has been deposited under accession no. 4dqy in the Protein Data Bank.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/
Materials and Methods
SOM text
Figs. S1 to S11
Tables S1 to S2
References (25–48)
REFERENCES
- 1.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Biochem J. 1999 Sep 1;342(Pt 2):249. [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnakumar R, Kraus WL. Mol Cell. 2010 Jul 9;39:8. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fong PC, et al. N Engl J Med. 2009 Jul 9;361:123. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 4.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. Nat Rev Cancer. 2010 Apr;10:293. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langelier MF, Planck JL, Roy S, Pascal JM. J Biol Chem. 2011 Mar 25;286:10690. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eustermann S, et al. J Mol Biol. 2011 Mar 18;407:149. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lonskaya I, et al. J Biol Chem. 2005 Apr 29;280:17076. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 8.Pion E, et al. Biochemistry. 2003 Oct 28;42:12409. doi: 10.1021/bi0344432. [DOI] [PubMed] [Google Scholar]
- 9.D'Silva I, et al. Biochim Biophys Acta. 1999 Feb 10;1430:119. doi: 10.1016/s0167-4838(98)00278-7. [DOI] [PubMed] [Google Scholar]
- 10.Langelier MF, Servent KM, Rogers EE, Pascal JM. J Biol Chem. 2008 Feb 15;283:4105. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 11.Tao Z, Gao P, Hoffman DW, Liu HW. Biochemistry. 2008 May 2; doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- 12.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Nucleic Acids Res. 2009 Jun;37:3723. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruf A, de Murcia G, Schulz GE. Biochemistry. 1998 Mar 17;37:3893. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- 14.Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Proc Natl Acad Sci U S A. 1996 Jul 23;93:7481. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wacker DA, et al. Mol Cell Biol. 2007 Nov;27:7475. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogata N, Ueda K, Kawaichi M, Hayaishi O. J Biol Chem. 1981 May 10;256:4135. [PubMed] [Google Scholar]
- 17.Lilyestrom W, van der Woerd MJ, Clark N, Luger K. J Mol Biol. 2010 Feb 5;395:983. doi: 10.1016/j.jmb.2009.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spagnolo L, Barbeau J, Curtin NJ, Morris EP, Pearl LH. Nucleic Acids Res. 2012 Jan 5; doi: 10.1093/nar/gkr1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. J Biol Chem. 2010 Apr 13; doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trucco C, Flatter E, Fribourg S, de Murcia G, Menissier-de Murcia J. FEBS Lett. 1996 Dec 16;399:313. doi: 10.1016/s0014-5793(96)01347-6. [DOI] [PubMed] [Google Scholar]
- 21.Miranda EA, Dantzer F, O'Farrell M, de Murcia G, de Murcia JM. Biochem Biophys Res Commun. 1995 Jul 17;212:317. doi: 10.1006/bbrc.1995.1972. [DOI] [PubMed] [Google Scholar]
- 22.Marsischky GT, Wilson BA, Collier RJ. J Biol Chem. 1995 Feb 17;270:3247. doi: 10.1074/jbc.270.7.3247. [DOI] [PubMed] [Google Scholar]
- 23.Ruf A, Rolli V, de Murcia G, Schulz GE. J Mol Biol. 1998 Apr 24;278:57. doi: 10.1006/jmbi.1998.1673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.