Abstract
Alternative splicing is a post-transcriptional mechanism that can substantially change the pattern of gene expression. Up to 95% of human genes have multi-exon alternative spliced forms, suggesting that alternative splicing is one of the most significant components of the functional complexity of the human genome. Nevertheless, alternative splicing regulation has received comparatively little attention in the study of cardiac diseases. When investigating SCN5A splicing abnormalities in heart failure, we found 47 of 181 known splicing regulators were upregulated in HF when compared to controls, which indicate that splicing regulation may play a key role in heart failure. Our results shows that AngII and hypoxia, signals common to HF, result in increased LUC7L3 and RBM25 splicing regulators, increased binding of RBM25 to SCN5A mRNA, increased SCN5A splice variant abundances, decreased full-length SCN5A mRNA and protein, and decreased Na+ current. These observations could shed light on a mechanism whereby cardiac function and arrhythmic risk are associated and allow for refined predictions of which patients may be at highest arrhythmic risk or suffer from Na+ channel blocking anti-arrhythmic drug complications.
Despite extensive research and novel treatments, human systolic heart failure (HF) remains a substantial clinical problem affecting millions of Americans, and HF-associated arrhythmia still remains a cause of morbidity and mortality. (Bristow et al., 2004) Currently, drugs with Na+ channel blocking activity, such as amiodarone, are still used in HF-associated arrhythmia, especially as adjuvant therapy to reduce implanted cardiac defibrillator (ICD) shock risk. (Dorian et al., 2008; Kamath and Mittal, 2008; Singh and Murawski, 2007) They suppress premature ventricular contractions and should reduce the sudden death risk. Nevertheless, a landmark clinical trial, the Cardiac Arrhythmia Suppression Trial (CAST), showed that use of sodium channel blocking drugs worsens mortality after myocardial infarction, despite an open label titration phase demonstrating the efficacy of the drugs to suppress asymptomatic ventricular arrhythmias. (Akhtar et al., 1990) Subsequent analysis showed that increased mortality was inversely associated with cardiac function. (Akhtar et al., 1990; Morganroth et al., 1985) This finding was confirmed in later reports. (Bardy et al., 2005; Bristow et al., 2004; Moss et al., 2002) The mechanism whereby these drugs worsen outcomes in HF, despite the fact that they reduce premature ventricular contractions and nonsustained ventricular tachycardia, remains undetermined.
Voltage-gated sodium channels are responsible for generating sodium current propagation of most electrically excitable cells, such as cardiac myocytes (Shibata et al., 2006) and neurons. (Abriel and Kass, 2005) The cardiac sodium channel is a transmembrane protein, composed of four homologous domains, each containing six transmembrane segments. The cardiac Na+ channel consists of the main pore-forming α-subunit and auxiliary β1, β2 subunits. α-Subunit alone is sufficient to produce a functional channel. β-Subunit co-expression can increase the level of Na+ channel expression and alter the voltage-dependent inactivation. (Abriel and Kass, 2005) SCN5A, encoding the α-subunit of the sodium channel, was cloned by Gellens et al in 1992 and mapped on chromosome 3p21 by George et al in 1995. (Gellens et al., 1992) (George, Jr. et al., 1995) Since the SCN5A gene was cloned, more than one hundred mutation sites have been found on SCN5A. (Ye et al., 2003) They are responsible for several inherited sodium channel disease (Brugada syndrome, (Chung et al., 2007) LTQ3, (Otagiri et al., 2008) and sudden death of infants (Hong et al., 2005)). These data indicate that even subtle alterations of SCN5A may underline cardiac disease.
Recently, we report three cardiac sodium channel (SCN5A) mRNA alternative splicing variants are upregulated in human HF tissue. (Shang et al., 2007) By using RNA ligase-mediated rapid amplification of cDNA ends (RACE) assay, we reported three new human cardiac Na+ channel C-terminal splicing variants in exon 28 designated as E28B (27 bp), E28C (39bp), and E28D (114 bp) (GenBank accession nos. EF092292, EF092293, and EF092294, respectively)(Shang et al., 2007) Our studies demonstrated that HF is associated with an increase in these splicing variants resulting from splicing at cryptic splice sequences in the terminal exon of SCN5A (i.e., exon 28). These variants encode cardiac Na+ channels truncated before the pore-forming segment of domain IV. Variant levels reach greater that >50% of the total SCN5A mRNA. As expected, these variants do not form functional channels. Moreover, the presence of the variants causes reduced abundance of the full-length SCN5A mRNA. A mouse model in which one allele of the SCN5A gene was substituted by a truncation variant was embryonic lethal, showing a >80% reduction in cardiac Na+ current and a significant reduction in conduction velocity in syncytial embryonic stem cell-derived cardiac myocytes. (Shang et al., 2007) These observations may explain the reduction in Na+ channels and Na+ current known to accompany human HF. Moreover, our results may help explain, for the first time, the physiological basis for the CAST trial observations that pro-arrhythmic risk from Na+ channel blocking drugs is inversely related to the severity of structural heart disease. Moreover, our data show that SCN5A abnormalities are not only responsible for inherited sodium channel disease, but also could play an important role in acquired cardiac disease, such as HF, by alternative splicing regulation. (Shang et al., 2007)
RBM25/LUC7L3complex mediates abnormal SCN5A mRNA regulation
Alternative splicing is a mechanism used to generate protein isoforms from a single gene. (Maniatis and Tasic, 2002) The spliceosome, a multi-protein complex, is responsible for excision of introns and conjoining exons to make mature mRNA. (Jurica and Moore, 2003) Splicing can be modulated by cis elements and trans factors, leading to splicing variation. (Matlin et al., 2005) A number of RNA binding proteins are known to act as splicing regulators. When bound, these proteins tend to influence nearby splicing site selection. When splicing sites cannot be recognized by canonical sequences, they are known as cryptic sites. Splice selection has been shown to be a function of the amount of splicing regulator present (Matlin et al., 2005).
RNA binding motif protein 25 (RBM25) localizes to the nuclear speckles and associates with multiple splicing components such as splicing cofactors SRm160/300, U1 small nuclear (sn) RNAs, assembled splicing complexes, and spliced mRNAs. Characterization of RBM25 strongly suggests that it functions in precursor mRNA (pre-mRNA) processing and that this regulation is gene specific. (Zhou et al., 2008) LUC7L3, a human homolog of yeast U1 snRNP-associated factor, is also a nuclear protein with a role in pre-mRNA splicing. LUC7L3 has two zinc finger motifs. (Nishii et al., 2000) The first cross-links the pre-mRNA and is required for LUC7L3 splicing activity. LUC7L3 acts as a bridge between the pre-mRNA and the U1 snRNP through its second zinc finger. (Nishii et al., 2000) According to a recent report, RBM25 associates selectively with the LUC7L3 and activates proapoptotic Bcl-xS 5′ splicing via its interaction with the exonic splicing enhancer cis-element, CGGGCA.(Zhou et al., 2008) In conjunction with LUC7L3, another spliceosome-associated factor, it shifts the ratio of Bcl-x long and short alternatively spliced mRNA forms leading to an increase in apoptosis when RBM25 is upregulated. The amount of RBM25 is related to the amount of the pro-apoptotic Bcl-xs isoform in HEK293 or HeLa cells. (Zhou et al., 2008) The effect of RBM25 is mediated by LUC7L3, which bridges RBM25 and the U1 spliceosome ribonuclear protein. LUC7L3 is known as an acidosis- and hypoxia-sensitive splicing factor. Microarray analysis of human heart samples from patients with and without HF was undertaken to look at changes in mRNA splicing factor abundance. From this analysis, LUC7L3 and RBM25 were upregulated by 1.7 and 1.5 fold, respectively. The changes were confirmed by RT-PCR and Western Blot assay on human HF tissue samples. Scanning the entire SCN5A RNA sequence revealed only a single binding site for RBM25 at the place where SCN5A splicing variants were detected. (Shang et al., 2007) (Gao et al., 2011) Gel mobility shift assays showed that RMB25 bound to the canonical sequence, CGGGCA, in SCN5A exon 28, and up and downregulation of the splicing regulators confirmed that they were necessary and sufficient to cause abnormal SCN5A mRNA splicing.
During our studies, we identified angiotensin II (AngII) and hypoxia as signals for upregulation of the LUC7L3/RBM25 complex and abnormal SCN5A mRNA splicing. In human embryonic stem cell-derived cardiomyocytes (hESC-CMs), hypoxia increased the expression of SCN5A variants E28C and E28D by 3.7- and 6.4-fold, respectively whereas the expression of the full-length SCN5A transcript is decreased by 0.7-fold. With AngII, the expression of SCN5A variants E28C and E28D was increased by 2.9- and 4.3-fold, respectively, whereas the expression of the full-length SCN5A transcript was decreased by 0.8-fold. Ang II treatment of hESC-CMs showed current reductions in the range known to contribute to arrhythmic risk. These results are consistent with clinical data suggesting that renin-angiotensin system (RAS) inhibition and revascularization have antiarrhythmic effects. (Gao et al., 2011)
The abnormal RNA splicing effects may be additive to Na+ channel promoter downregulation with RAS activation. We have shown that AngII can downregulate the cardiac Na+ channel through an H2O2-dependent pathway that involves NFκB activation. (Shang et al., 2008) The data indicate that AngII acts through an NADPH oxidase-dependent oxidative species and NFκB to reduce Na+ channel transcription The fact that Na+ channel transcriptional regulation was affected by NFκB activation is consistent with both the human and mouse Na+ channel promoters having NFκB consensus binding sites (GenBank accession numbers AY313163 and AY769981). Based on the ChIP assays and NFκB subunit overexpression, Na+ channel downregulation seemed to be mediated by p50/p65 subunit binding to the SCN5A promoter. The chronic effects seen here may be additive with previous reported acute effects of AngII or oxidative stress to enhance Na+ channel dysfunction. Therefore, it is likely that multiple acute and chronic deleterious effects on Na+ channels occur during pathophysiological conditions associated with oxidative stress and RAS activation. (Shang et al., 2008)
Aside from the direct effect of NFκB on the Na+ channel promoter, it is possible that this transcription factor may play a role in alternative splicing. Gene array comparisons of splicing factor expression between normal and heart failure tissue has demonstrated changes in hypoxia inducing factor-1α (HIF1α). HIF1α is a key transcriptional regulatory molecule elevated in hypoxia and inflammation. The HIF1α promoter contains an NFκB binding element, and NFκB is an upstream regulator during HIF1α activation. Among other things, HIF1α regulates mRNA splicing regulators, such as LUC7L3 and RBM25, suggesting NFκB activation may be upstream of the alternative splicing of this channel during cardiac oxidative stress, inflammation, or hypoxia. (Gao et al., 2011; Gao and Dudley, Jr., 2009)
The possibility of a blood test that helps predict arrhythmic risk in HF
In addition to heart, SCN5A Na+ channels have been described in lymphocytes, macrophages, and skeletal muscle. (Carrithers et al., 2007; Fraser et al., 2004; Roselli et al., 2006) In a monocytic cell line, SCN5A seems to be involved in providing a countercurrent to allow endosomal acidification. The predominant Na+ channel in Jurkat immortalized T lymphocytes is SCN5A. (Fraser et al., 2004) All four variants SCN5A splice variants were detected in the human lymphoblast. (Shang et al., 2007) The identical SCN5A splice variants regulation is found in Jurkat cells compare to hESC-derived cardiomyocytes, suggesting that white blood cells might serve as readily accessible surrogates for the status of Na+ channel splicing in the myocardium. If this proves correct, then it may be possible to develop a blood test that helps predict arrhythmic risk in heart failure. This possibility is being tested in a human clinical trial known as SOCS-HEFT (Sodium Channel Splicing in Heart Failure Trial, NCT01185587). The preliminary data from 50 patients for each group showed that abnormal Na+ channel mRNA splicing is associated with increased appropriate ICD discharge, and splicing in heart correlates to that in leukocytes.
The importance of splicing regulation in heart failure
Alternative splicing is a post-transcriptional mechanism that can substantially change the pattern of gene expression. Up to 95% of human genes have multi-exon alternative spliced forms, suggesting that alternative splicing is one of the most significant components of the functional complexity of the human genome. (Modrek et al., 2001; Pan et al., 2008) Proper regulation of alternative splicing is important for cell physiology, and aberrant splicing may lead to cellular dysfunction and clinical manifestations. Although our understanding of the role of alternative mRNA splicing is elemental, a growing list of human diseases, such as cancer, (Ryan et al., 2010) neurodegenerative disorders (Du et al., 2010) and autoimmune diseases (Novak et al., 2009) are associated with alternative splicing. Alternative splicing events in cancer allow for ‘splicing signatures’ associated with different tumor subgroups. Nevertheless, alternative splicing regulation has received comparatively little attention in the study of cardiac diseases. When investigating SCN5A splicing abnormalities in heart failure, we found 47 of 181 known splicing regulators were upregulated in HF when compared to controls. Interestingly, no significant downregulation of splicing regulators was observed. (Gao et al., 2011) These splicing regulators were grouped according to known pathogenic processes such as hypoxia, (Stearman et al., 2005) inflammation, (Ricco and Kanduc, 2010) wall tension, (Inuzuka et al., 2009) or hormonal factors (Claus et al., 2008) involved in HF. Our results indicate that abnormal SCN5A splicing in HF is likely just the tip of the iceberg of expression changes in HF.
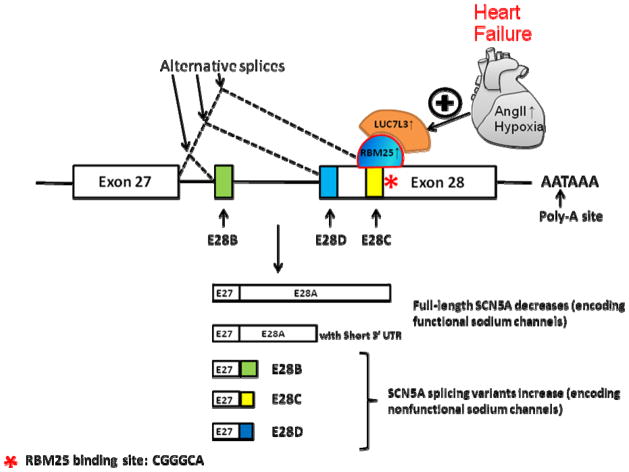
In summary, AngII and hypoxia, signals common to HF, result in increased hLuc7A and RBM25 protein, increased binding of RBM25 to SCN5A mRNA, increased SCN5A splice variant abundances, decreased full-length SCN5A mRNA and protein, and decreased Na+ current. These observations could shed light on a mechanism whereby cardiac function and arrhythmic risk are associated, allow for refined predictions of which patients may be at highest arrhythmic risk or suffer from Na+ channel blocking anti-arrhythmic drug complications. In addition, this work could lead to development of therapies to reduced sudden death risk in HF (Figure 1).
Figure 1.
Illustration of the mechanism of SCN5A splicing regulation during HF. The top line represents the genomic structure of SCN5A (showing exon 27 and the terminal codon, exon 28). Untranslated, translated, and nontranscribed sequences are shown as open bars, closed bars, and lines, respectively. The arrows and dotted lines indicate SCN5A splicing variants. Splicing patterns of SCN5A C-terminal variants are identified as E28A, B, C, and D respectively. * indicate RBM25 binding site and sequence.
Acknowledgments
This work was supported by R01 HL106592, R01 HL104025, P01 HL058000 (SCD), and a VA MERIT grant.
Table of Abbreviations
- HF
Human systolic heart failure
- ICD
Implanted cardiac defibrillator
- CAST
Cardiac Arrhythmia Suppression Trial
- SCN5A
Voltage-gated cardiac sodium channels, α-subunit
- Brugada Syndrome
An inherited sudden death syndrome that can be caused by heterozygous mutations in the SCN5A gene (Brugada syndrome-1)
- LTQ3
An inherited sudden death syndrome caused by mutations in the SCN5A gene
- E28A
The full length SCN5A transcript
- E28B (27 bp)
The type B SCN5A transcript variant
- E28C (39 bp)
The type C SCN5A transcript variant
- E28D (114 bp)
The type C SCN5A transcript variant
- RBM25
RNA binding motif protein 25
- LUC7L3
A spliceosome protein containing an arginine and glutamate rich domain (RE domain) and an arginine and serine rich domain (RS domain)
- U1 snRNA
The small nuclear RNA (snRNA) component of U1 snRNP (small nuclear ribonucleoprotein)
- spliceosome
a complex of small nuclear RNA and protein subunits that removes introns from a transcribed pre-mRNA segment
- AngII
angiotensin II
- hESC-CMs
embryonic stem cell-derived cardiomyocytes
- RAS
renin-angiotensin system
- NFκB
the transcription factor, nuclear factor κB, a key factor in the immune response
- ChIP assays
Chromatin immunoprecipitation, a powerful tool for identifying proteins by binding to nucleic acids
- HIF1α
Hypoxia inducing factor-1α
- Jurkat cells
An immortalized line of T lymphocyte cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abriel H, Kass RS. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc Med. 2005;15(1):35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Akhtar M, et al. CAST and beyond. Implications of the Cardiac Arrhythmia Suppression Trial. Task Force of the Working Group on Arrhythmias of the European Society of Cardiology. Circulation. 1990;81(3):1123–1127. doi: 10.1161/01.cir.81.3.1123. [DOI] [PubMed] [Google Scholar]
- Bardy GH, et al. Amiodarone or an implantable cardioverter-defibrillator for congestive heart failure. N Engl J Med. 2005;352(3):225–237. doi: 10.1056/NEJMoa043399. [DOI] [PubMed] [Google Scholar]
- Bristow MR, et al. Cardiac-resynchronization therapy with or without an implantable defibrillator in advanced chronic heart failure. N Engl J Med. 2004;350(21):2140–2150. doi: 10.1056/NEJMoa032423. [DOI] [PubMed] [Google Scholar]
- Carrithers MD, Dib-Hajj S, Carrithers LM, Tokmoulina G, Pypaert M, Jonas EA, Waxman SG. Expression of the voltage-gated sodium channel NaV1.5 in the macrophage late endosome regulates endosomal acidification. J Immunol. 2007;178(12):7822–7832. doi: 10.4049/jimmunol.178.12.7822. [DOI] [PubMed] [Google Scholar]
- Chung SK, et al. Long QT and Brugada syndrome gene mutations in New Zealand. Heart Rhythm. 2007;4(10):1306–1314. doi: 10.1016/j.hrthm.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Claus EB, Park PJ, Carroll R, Chan J, Black PM. Specific genes expressed in association with progesterone receptors in meningioma. Cancer Res. 2008;68(1):314–322. doi: 10.1158/0008-5472.CAN-07-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorian P, Al-Khalidi HR, Hohnloser SH, Brum JM, Dunnmon PM, Pratt CM, Holroyde MJ, Kowey P. Azimilide reduces emergency department visits and hospitalizations in patients with an implantable cardioverter-defibrillator in a placebo-controlled clinical trial. J Am Coll Cardiol. 2008;52(13):1076–1083. doi: 10.1016/j.jacc.2008.05.055. [DOI] [PubMed] [Google Scholar]
- Du H, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17(2):187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser SP, Diss JK, Lloyd LJ, Pani F, Chioni AM, George AJ, Djamgoz MB. T-lymphocyte invasiveness. control by voltage-gated Na+ channel activity. FEBS Lett. 2004;569(1–3):191–194. doi: 10.1016/j.febslet.2004.05.063. [DOI] [PubMed] [Google Scholar]
- Gao G, Dudley SC., Jr Redox regulation, NF-κB, and atrial fibrillation. Antioxid Redox Signal. 2009;11(9):2265–2277. doi: 10.1089/ars.2009.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, et al. Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation. 2011;124(10):1124–1131. doi: 10.1161/CIRCULATIONAHA.111.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellens ME, George AL, Jr, Chen LQ, Chahine M, Horn R, Barchi RL, Kallen RG. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci USA. 1992;89(2):554–558. doi: 10.1073/pnas.89.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George AL, Jr, Varkony TA, Drabkin HA, Han J, Knops JF, Finley WH, Brown GB, Ward DC, Haas M. Assignment of the human heart tetrodotoxin-resistant voltage-gated Na+ channel α-subunit gene (SCN5A) to band 3p21. Cytogenet Cell Genet. 1995;68(1–2):67–70. doi: 10.1159/000133892. [DOI] [PubMed] [Google Scholar]
- Hong K, et al. Cryptic 5′ splice site activation in SCN5A associated with Brugada syndrome. J Mol Cell Cardiol. 2005;38(4):555–560. doi: 10.1016/j.yjmcc.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Inuzuka Y, et al. Suppression of phosphoinositide 3-kinase prevents cardiac aging in mice. Circulation. 2009;120(17):1695–1703. doi: 10.1161/CIRCULATIONAHA.109.871137. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing. awash in a sea of proteins. Mol Cell. 2003;12(1):5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kamath GS, Mittal S. The role of antiarrhythmic drug therapy for the prevention of sudden cardiac death. Prog Cardiovasc Dis. 2008;50(6):439–448. doi: 10.1016/j.pcad.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418(6894):236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing. towards a cellular code. Nat Rev Mol Cell Biol. 2005;6(5):386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29(13):2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganroth J, Nestico PF, Horowitz LN. A review of the uses and limitations of tocainide--a class IB antiarrhythmic agent. Am Heart J. 1985;110(4):856–863. doi: 10.1016/0002-8703(85)90470-3. [DOI] [PubMed] [Google Scholar]
- Moss AJ, et al. Prophylactic implantation of a defibrillator in patients with myocardial infarction and reduced ejection fraction. N Engl J Med. 2002;346(12):877–883. doi: 10.1056/NEJMoa013474. [DOI] [PubMed] [Google Scholar]
- Nishii Y, Morishima M, Kakehi Y, Umehara K, Kioka N, Terano Y, Amachi T, Ueda K. CROP/Luc7A, a novel serine/arginine-rich nuclear protein, isolated from cisplatin-resistant cell line. FEBS Lett. 2000;465(2–3):153–156. doi: 10.1016/s0014-5793(99)01744-5. [DOI] [PubMed] [Google Scholar]
- Novak AJ, et al. Genetic variation in B-cell-activating factor is associated with an increased risk of developing B-cell non-Hodgkin lymphoma. Cancer Res. 2009;69(10):4217–4224. doi: 10.1158/0008-5472.CAN-08-4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otagiri T, Kijima K, Osawa M, Ishii K, Makita N, Matoba R, Umetsu K, Hayasaka K. Cardiac ion channel gene mutations in sudden infant death syndrome. Pediatr Res. 2008;64(5):482–487. doi: 10.1203/PDR.0b013e3181841eca. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Ricco R, Kanduc D. Hepatitis B virus and Homo sapiens proteome-wide analysis: A profusion of viral peptide overlaps in neuron-specific human proteins: Biologics. 2010;4:75–81. doi: 10.2147/btt.s8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselli F, Livrea P, Jirillo E. Voltage-gated sodium channel blockers as immunomodulators: Recent Pat CNS. Drug Discov. 2006;1(1):83–91. doi: 10.2174/157488906775245255. [DOI] [PubMed] [Google Scholar]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks. the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang LL, et al. Human heart failure is associated with abnormal C-terminal splicing variants in the cardiac sodium channel. Circ Res. 2007;101(11):1146–1154. doi: 10.1161/CIRCRESAHA.107.152918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang LL, Sanyal S, Pfahnl AE, Jiao Z, Allen J, Liu H, Dudley SC., Jr NF-κB-dependent transcriptional regulation of the cardiac scn5a sodium channel by angiotensin II. Am J Physiol Cell Physiol. 2008;294(1):C372–C379. doi: 10.1152/ajpcell.00186.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata EF, Brown TL, Washburn ZW, Bai J, Revak TJ, Butters CA. Autonomic regulation of voltage-gated cardiac ion channels. J Cardiovasc Electrophysiol. 2006;17(Suppl 1):S34–S42. doi: 10.1111/j.1540-8167.2006.00387.x. [DOI] [PubMed] [Google Scholar]
- Singh S, Murawski MM. Implantable cardioverter defibrillator therapy and the need for concomitant antiarrhythmic drugs. J Cardiovasc Pharmacol Ther. 2007;12(3):175–180. doi: 10.1177/1074248407305608. [DOI] [PubMed] [Google Scholar]
- Stearman RS, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167(6):1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Valdivia CR, Ackerman MJ, Makielski JC. A common human SCN5A polymorphism modifies expression of an arrhythmia causing mutation. Physiol Genomics. 2003;12(3):187–193. doi: 10.1152/physiolgenomics.00117.2002. [DOI] [PubMed] [Google Scholar]
- Zhou A, Ou AC, Cho A, Benz EJ, Jr, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol Cell Biol. 2008;28(19):5924–5936. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]