Summary
To find links between the biotic characteristics and abiotic process parameters in anaerobic digestion systems, the microbial communities of nine full‐scale biogas plants in South Tyrol (Italy) and Vorarlberg (Austria) were investigated using molecular techniques and the physical and chemical properties were monitored. DNA from sludge samples was subjected to microarray hybridization with the ANAEROCHIP microarray and results indicated that sludge samples grouped into two main clusters, dominated either by Methanosarcina or by Methanosaeta, both aceticlastic methanogens. Hydrogenotrophic methanogens were hardly detected or if detected, gave low hybridization signals. Results obtained using denaturing gradient gel electrophoresis (DGGE) supported the findings of microarray hybridization. Real‐time PCR targeting Methanosarcina and Methanosaeta was conducted to provide quantitative data on the dominating methanogens. Correlation analysis to determine any links between the microbial communities found by microarray analysis, and the physicochemical parameters investigated was conducted. It was shown that the sludge samples dominated by the genus Methanosarcina were positively correlated with higher concentrations of acetate, whereas sludge samples dominated by representatives of the genus Methanosaeta had lower acetate concentrations. No other correlations between biotic characteristics and abiotic parameters were found. Methanogenic communities in each reactor were highly stable and resilient over the whole year.
Introduction
In the face of climate change and global warming, the rapid depletion of fossil fuel reserves and the accumulation of waste in our ‘throw‐away’ society, the production of clean bioenergy has undergone a rebirth in recent years. The production of biogas from the anaerobic digestion of organic wastes is one example. In fact, biogas technology is considered to be an excellent tool to avoid negative influences on the environment and climate (Insam and Wett, 2008).
A number of anaerobic digesters have been developed and installed in Austria during the last 20 years and the number is continuously rising. In 2002, there were 97 plants in Austria, while by the end of 2010, there were 360 plants in operation (E‐CONTROL, 2010). In South Tyrol (Italy), 30 plants are in operation and several others are in the process of planning or under construction (INBIMO, 2011) due to the incentives currently being offered for the operation of anaerobic digestion plants.
The anaerobic digestion process itself requires specific environmental conditions and is dependent on the microorganisms involved to cooperate in a close and efficient syntrophism (Schink, 1997). Digestion occurs in four major stages (hydrolysis, acidogenesis, acetogenesis and methanogenesis) and complex polymers are degraded in a stepwise manner to yield CO2 and CH4. In the first step, hydrolysing and fermenting microbes degrade the organic macromolecules, such as proteins, carbohydrates and fats, into amino acids, sugars and fatty acids. Acidogenic bacteria convert the sugars, amino acids and fatty acids to organic acids, alcohols and ketones, acetate, CO2 and H2. Acetogenic bacteria convert the fatty acids and alcohols into acetate, H2 and CO2, products used by methanogenic archaea to form methane (Ahring, 2003). Methanogens thus hold the key position in the anaerobic digestion process.
Since anaerobic digestion is a very complex process involving biotic and abiotic factors, improvements in operation can be difficult to achieve. There are however different approaches to increasing the biogas potential of a particular reactor, such as optimizing the reactor configuration, increasing the digestibility of the input material, optimizing process control and stability and improving the microbial processes and their efficiency (Ahring, 2003). Improvements in anaerobic biotechnology and an increased/stable biogas production require a better understanding of reactor functioning and the microbial communities involved. The aim of this study was to reveal and link anaerobic digester process functioning and environmental parameters with the microbial communities present.
Results
Data regarding reactor volume, substrate composition and the results of physicochemical parameter measurements are presented in Table 1. Only two of the physicochemical parameters measured were found to exceed the thresholds for stable reactor conditions quoted (BMVIT, 2007). These were the pH in A sp and the dry matter in B sp.
Table 1.
Operational and physicochemical parameters of the biogas reactors in spring, summer and autumn
| Biogas reactor | Season | Province/country | Main substrate | Percentage of co‐substrates | Reactor volume | Temperature | pH | EC | Dry matter | Loss on ignition | NH4‐N | NH3‐N | CH4 | CO2 | H2S | H2 | Acetate | Propionic acid |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (%) | (m3) | (°C) | (mS cm−1) | (%) | (%) | (mg l−1) | (mg l−1) | (%) | (%) | (ppm) | (mmol l−1) | (mmol l−1) | (mmol l−1) | |||||
| A | Spring | AU/Vb | Cow manure | 7 (Waste vegetable oil) | 380 | 40.0 | 8.2 | 16.8 | 7.2 | 75.0 | 2144 | 366 | 55.0 | 43.5 | 380 | nm | 1.8 | nd |
| A | Summer | AU/Vb | Cow manure | 7 (Waste vegetable oil) | 380 | 40.0 | 7.6 | 17.6 | 7.1 | 73.4 | 1840 | 106 | 56.4 | 42.8 | 2579 | 13 139 | 0.7 | nd |
| A | Autumn | AU/Vb | Cow manure | 7 (Waste vegetable oil) | 380 | 40.0 | 7.5 | 18.5 | 6.8 | 73.0 | 1680 | 78.0 | 55.0 | 44.0 | 90.0 | 11 630 | nd | nd |
| B | Spring | I/ST | Cow manure | 18 (Pomace) | 1 000 | 42.0 | 7.8 | 15.6 | 9.5 | 74.0 | 1768 | 172 | 57.0 | 39.0 | 40.0 | nm | 4.0 | nd |
| B | Summer | I/ST | Cow manure | 18 (Pomace) | 1 000 | 42.0 | 7.8 | 17.0 | 7.6 | 66.0 | 1520 | 157 | 53.3 | 40.3 | 4000 | 12 075 | 0.1 | nd |
| B | Autumn | I/ST | Cow manure | 18 (Pomace) | 1 000 | 41.0 | 7.5 | 13.1 | 6.9 | 65.0 | 800 | 33.8 | 55.8 | 43.8 | 150 | 7 195 | 0.1 | nd |
| C | Spring | AU/Vb | Cow manure | 0 | 560 | 40.0 | 7.9 | 11.6 | 9.0 | 79.0 | 1792 | 176 | 53.4 | 46.5 | 100 | nm | 0.6 | 7.5 |
| C | Summer | AU/Vb | Cow manure | 0 | 560 | 40.0 | 7.5 | 14.8 | 6.2 | 76.0 | 1360 | 59.1 | 55.0 | 44.9 | 391 | 9 965 | 0.5 | nd |
| C | Autumn | AU/Vb | Cow manure | 0 | 560 | 39.5 | 7.5 | 12.9 | 6.8 | 75.0 | 1200 | 48.4 | 51.5 | 39.6 | nd | 8 865 | 0.1 | nd |
| D | Spring | AU/Vb | Sewage sludge | 0 | 10 000 | 38.0 | 7.2 | 9.8 | 2.6 | 55.0 | 985 | 20.2 | 61.4 | 38.3 | 297 | 94.5 | nd | nd |
| D | Summer | AU/Vb | Sewage sludge | 0 | 10 000 | 38.0 | 7.1 | 5.6 | 2.3 | 56.0 | 560 | 9.6 | 64.0 | 36.0 | 1400 | 3 191 | nd | nd |
| D | Autumn | AU/Vb | Sewage sludge | 0 | 10 000 | 37.5 | 7.1 | 6.0 | nm | nm | 640 | 9.5 | 60.4 | 39.3 | nd | 3 530 | 0.3 | nd |
| E | Spring | AU/Vb | Cow manure | 0 | 850 | 40.0 | 8.1 | 17.3 | 5.2 | 71.0 | 1336 | 207 | 43.2 | 38.7 | 100 | 146 | nd | nd |
| E | Summer | AU/Vb | Cow manure | 0 | 850 | 40.0 | 7.7 | 17.6 | 5.6 | 69.0 | 1600 | 105 | 51.1 | 33.5 | 480 | 11 604 | 0.8 | nd |
| E | Autumn | AU/Vb | Cow manure | 0 | 850 | 38.0 | 7.6 | 17.2 | 5.0 | 67.0 | 1520 | 68.1 | 50.0 | 37.0 | nd | 11 112 | 0.3 | nd |
| F | Spring | I/ST | Pomace | 23 (Pomace) | 1 000 | 48.0 | 8.0 | 15.5 | 9.0 | 72.0 | 1224 | 237 | 55.0 | 44.5 | 1055 | nm | 6.8 | 1.1 |
| F | Summer | I/ST | Pomace | 23 (Pomace) | 1 000 | 48.0 | 7.5 | 15.2 | nm | nm | 880 | 63.9 | nm | nm | nm | 7 769 | 9.2 | 1.3 |
| F | Autumn | I/ST | Pomace | 23 (Pomace) | 1 000 | 48.0 | 7.4 | 15.8 | nm | nm | 800 | 43.0 | 54.3 | 42.8 | 650 | 7 069 | 5.6 | 0.8 |
| G | Spring | I/ST | Cow manure | < 1 (Biowaste) | 130 | 40.0 | 7.9 | 18.7 | 8.2 | 70.0 | 2008 | 197 | 54.0 | 45.0 | 1040 | nm | 1.6 | nd |
| G | Summer | I/ST | Cow manure | < 1 (Biowaste) | 130 | 40.0 | 7.7 | 25.0 | 7.2 | 66.0 | 1840 | 137 | 57.0 | 41.2 | 100 | 303 | 2.1 | nd |
| G | Autumn | I/ST | Cow manure | < 1 (Biowaste) | 130 | 41.0 | 7.8 | 22.5 | 7.4 | 69.0 | 1920 | 158 | 58.0 | 41.3 | 100 | 311 | 2.2 | nd |
| H | Spring | I/ST | Cow manure | 2 (Waste vegetable oil) | 670 | 38.5 | 7.8 | 19.4 | 7.2 | 72.0 | 2000 | 162 | 55.0 | 44.0 | 1050 | nm | 4.0 | nd |
| H | Summer | I/ST | Cow manure | 2 (Waste vegetable oil) | 670 | 38.5 | 7.8 | 18.7 | 6.3 | 71.0 | 1760 | 136 | 57.0 | 42.3 | 4000 | 210 | 1.5 | 0.1 |
| H | Autumn | I/ST | Cow manure | 2 (Waste vegetable oil) | 670 | 38.5 | 7.7 | 19.0 | 4.8 | 67.0 | 1760 | 98.9 | 56.5 | 43.3 | nm | 188 | 1.5 | 0.1 |
| I | Spring | AU/Vb | Cow, swine and chicken manure | 21 (Waste vegetable oil, pomace, garden waste, biowaste) | 750 | 42.0 | 7.9 | 19.4 | 5.9 | 75.0 | 2280 | 288 | 57.6 | 42.2 | 44.0 | 555 | 1.8 | nd |
| I | Summer | AU/Vb | Cow, swine and chicken manure | 21 (Waste vegetable oil, pomace, garden waste, biowaste) | 750 | 41.9 | 7.6 | 16.8 | 6.5 | 79.0 | 2160 | 143 | 56.7 | 42.2 | 148 | 227 | 1.7 | 0.1 |
| I | Autumn | AU/Vb | Cow, swine and chicken manure | 21 (Waste vegetable oil, pomace, garden waste, biowaste) | 750 | 41.9 | 7.7 | 22.0 | 7.3 | 73.0 | 2640 | 211 | 60.1 | 39.2 | 200 | 459 | 6.2 | 0.2 |
AU, Austria; I, Italy; Vb, Vorarlberg; ST, South Tirol; nd, not detected (detection limit was 0.1 mmol l−1); nm, not measured.
Temperature within most reactors ranged between 37.5°C and 42°C and did not exceed the quoted maximum temperatures for mesophilic digestion processes of 42°C. The only thermophilic reactor investigated was reactor F. At the times of sampling, it was operated at 48°C, below the optimum range for thermophilic reactors of 50–57°C, according to Fachagentur für Nachwachsende Rohstoffe (2006).
pH was very similar in all plants (pH 7.4–8.2) and within good operation conditions, according to BMVIT (2007), with the exception of reactor D, where values of 7.1–7.2 were recorded. DM (dry matter) ranged from 2.3% (D su) to 9.5% (B sp), loss on ignition from 55% (D sp) to 79% (C sp and I su). Conductivity varied slightly among all manure digesters with values from 11.6 to 25 mS cm−1. Again, the sewage sludge AD plant (D) was found to differ, with values of 5.6–9.8 mS cm−1 recorded.
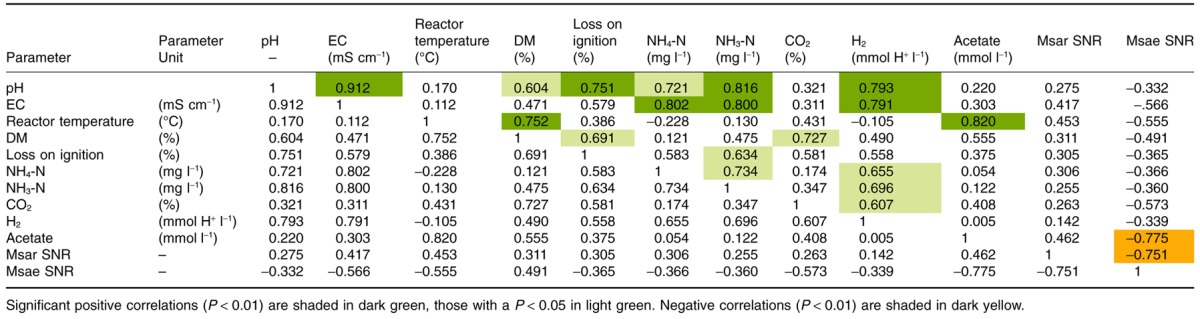
NH4‐N and NH3‐N concentrations varied from digester to digester, as shown in Table 1. Reactor D however, had considerably lower values than those found in the other reactors (560 mg l−1 NH4‐N in D su and 9.5 mg l−1 NH3‐N in D au). Generally, the highest methane contents were found in reactor D (60.4%, 64%, 61.4%), while methane contents were found to range from 43.2% to 58% in the other reactors (exception of I au with 60.1%). CO2 concentration ranged from 33.5% in E su to 46.5% in C sp. A huge range of H2 concentrations with values as low as 94.5 mmol l−1 (D sp) to values as high as 13139 mmol l−1 (A su) were measured. Similarly, acetate levels presented a broad heterogeneity over the year in each of the reactors, as shown in Fig. 1. The concentrations resulted in an obvious clustering: reactors A to E with concentrations below 1.8 mmol l−1 (with the exception of B sp) and reactors F to I with concentrations between 1.5 and 9.2 mmol l−1. Other volatile fatty acids (VFAs) were measured (i‐butyrate, butyric acid, valeric acid, isovaleric acid) but values either did not exceed the detection limit of 0.1 mmol l−1 or were found at very low concentration (data not shown in Table 1). The results of correlation analyses (according to Pearson) between physicochemical parameters and the different sludge samples are presented in Table 2. Significant positive correlations (P < 0.01) are shaded in dark green, those with a lower significance (P < 0.05) in light green. Significant negative correlations (P < 0.01) are shaded in dark yellow.
Figure 1.
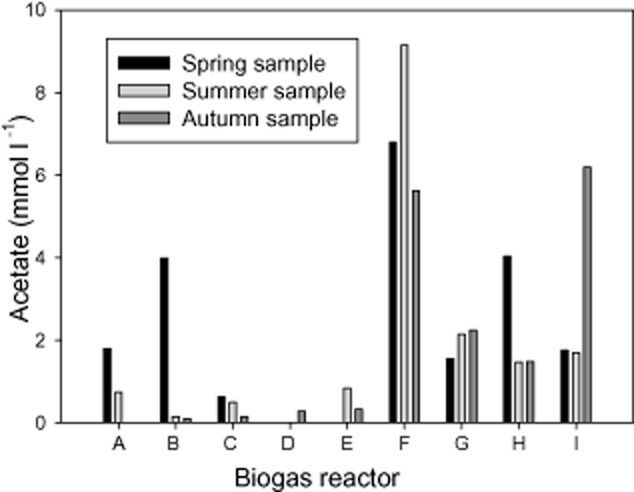
Acetate concentrations (mmol l−1) in the different biogas reactor sludges as determined by HPLC.
Table 2.
Correlation analysis of physicochemical parameters according to Pearson
Microarray hybridization
The signal‐to‐noise ratios (SNRs) obtained after hybridization of the ANAEROCHIP microarray were determined, and are presented in Table 3. A statistical evaluation of microarray SNR results was performed using CANOCO 4.5 (ter Braak and Šmilauer, 2002) combined with principal component analysis (PCA). Figure 1 shows a canonical analysis loading plot; the two axes explaining 76.2% of the variance, the first and second axis representing 59.8% and 16.4% of the variance respectively. Reactors from different sampling time points are represented by circles, the oligonucleotide probes of the different genera and the acetate concentration by vectors. The lengths of the vectors indicate the significance for sample differentiation, and vectors point in the direction of samples with above average signal. Probes with similar vector directions have high covariance, meaning they tend to occur jointly on the microarrays.
Table 3.
Microarray results showing SNR values obtained for the different probes upon hybridization of the sludge samples
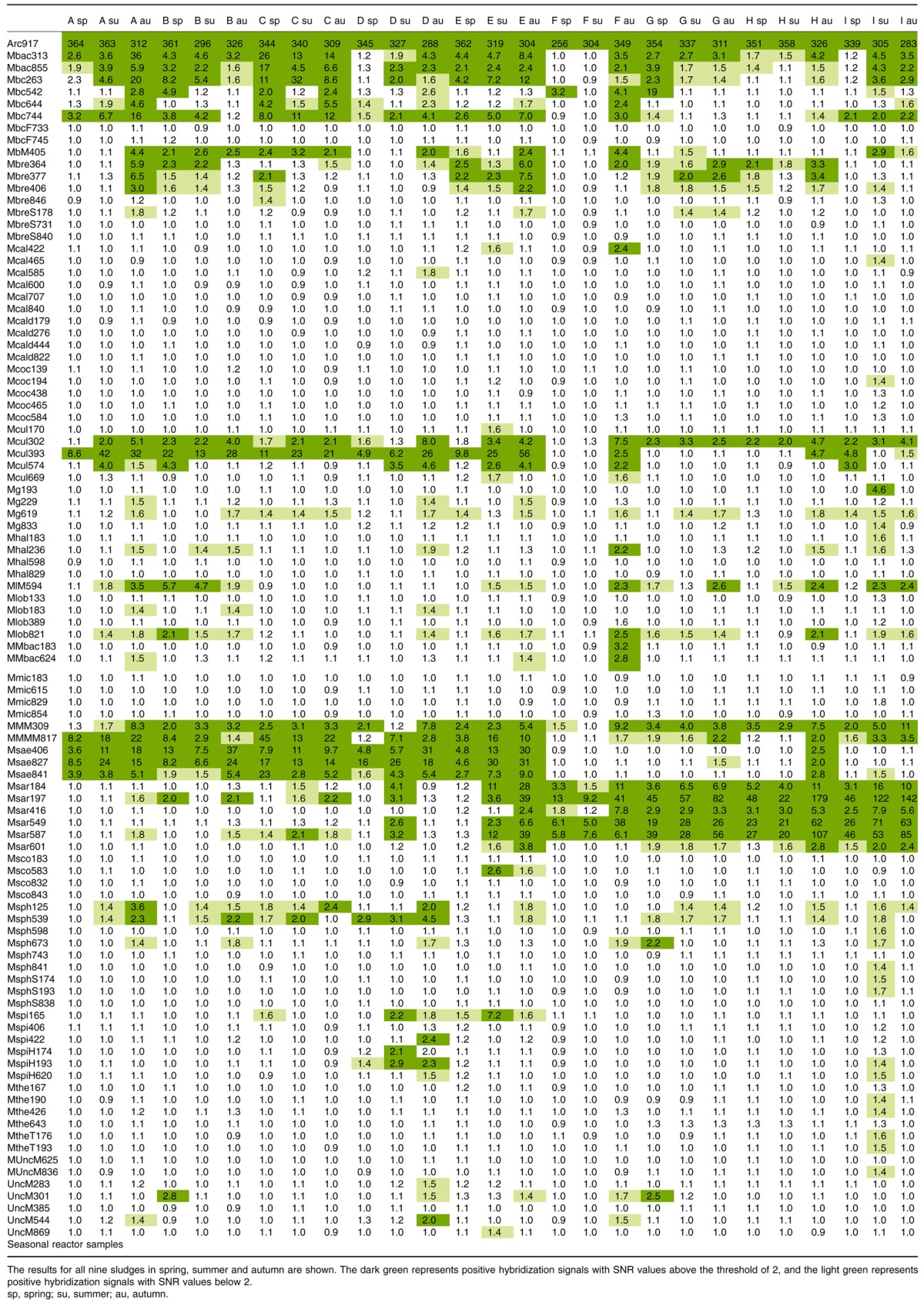
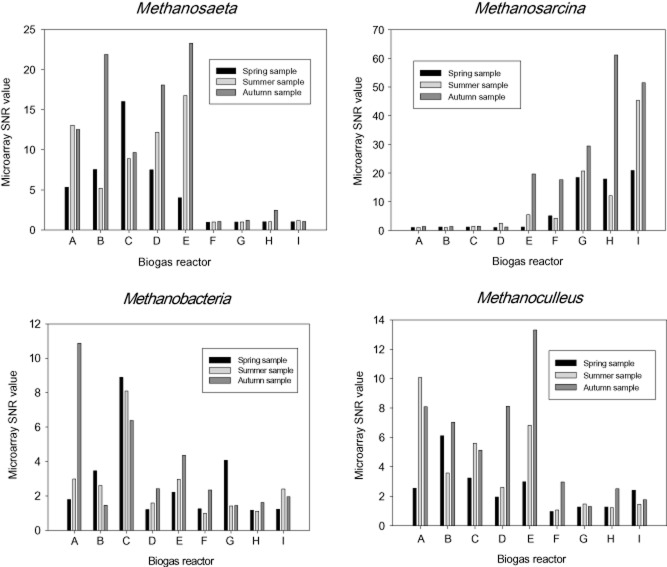
Multivariate analysis showed that the archaeal communities of the sludges were divided into two clusters (Fig. 2). Samples from reactors A to E grouped together, and were dominated by Methanosaeta (Msae), with SNR values ranging from 4 to 23.3 for the probes Msae841, Msae827 and Msae406 (see Franke‐Whittle et al., 2009a for probe details). Reactors F to I, on the other hand, were dominated by Methanosarcina, with SNR values of 4.3–61.2 being obtained for the different Methanosarcina probes (Msar549, Msar197, Msar416, Msar587, Msar184, Msar601). PASW‐SPSS 17.0 (SPSS, Chicago, IL, USA) analysis corroborated this finding (Table 2), revealing a significant negative correlation of −0.775 between acetate concentration and Methanosaeta.
Figure 2.
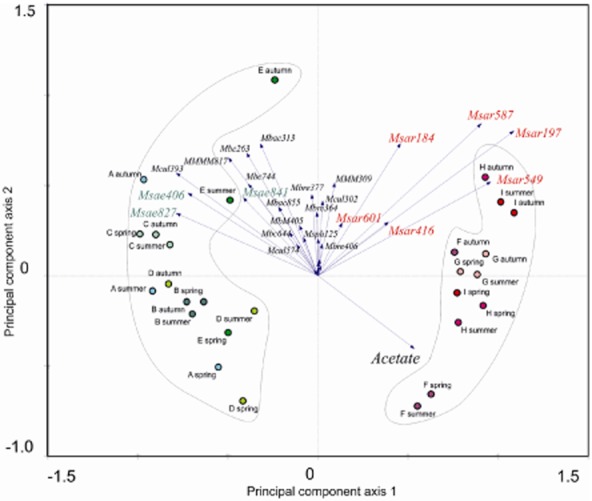
Loading plot obtained by redundancy analysis, depicting the clustering of sludge samples. The vectors represent the different probes.
No significant changes in archaeal community composition could be found in the samples collected in the different seasons of the year. The average SNR values of all probes (which were printed in triplicate on the array) for the genera Methanosarcina, Methanosaeta, Methanoculleus and Methanobacteria were generated in order to compare the relative abundance of the organisms in the different sludge samples. Figure 3 shows clearly the abundance of Methanosaeta in the A–E sludges, and of Methanosarcina in the F–I sludges. Methanosaeta dominated sludges also appeared to be correlated with higher numbers of the hydrogenotrophic Methanobacteria and Methanoculleus than the Methanosarcina dominated sludges (Fig. 3).
Figure 3.
Overview of important genera determined by microarray hybridization. The x‐axes denote the biogas reactors A to I, the y‐axes the SNR values of spring, summer and autumn samples.
Real‐time PCR
Dominant genera in the biogas reactors determined by the results of hybridization of sludges with the ANAEROCHIP microarray prompted a real‐time quantitative PCR (RT‐PCR) analysis. Figures 4 and 5 show the results of this analysis. Supporting the results of the microarrays, Methanosaeta dominated the sludges of reactors A–E with gene copy numbers of 9.6 × 104–7.43 × 107 gene copies ml−1 reactor sample while Methanosarcina dominated in the reactors F–I with 1.14 × 107–5.83 × 108 gene copies ml−1 reactor sample (Fig. 4). Methanosaeta concilii DSM 2139 was used as a positive control, and the standard curve parameters for RT‐PCR were: slope −3.522, intercept 30.99 and R2 > 0.999. Methanosarcina barkeri (DSM 800) was the positive control in the other RT‐PCR, and the standard curve parameters for this RT‐PCR were: slope −4.40, intercept 40.47 and R2 value 0.988. In the Methanosaeta dominated reactors, higher gene copy numbers of Methanosarcina were found, whereas only low levels of Methanosaeta appeared in the Methanosarcina dominated reactors.
Figure 4.
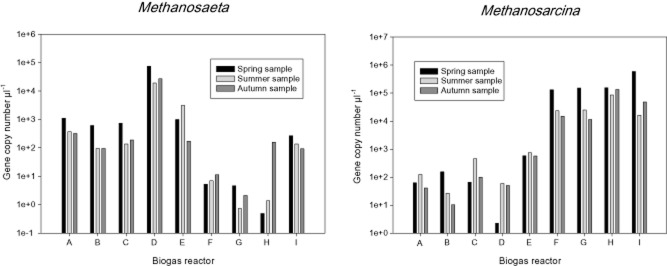
RT quantification of the genera Methanosaeta and Methanosarcina in the biogas reactors A to I during spring, summer and autumn.
Figure 5.
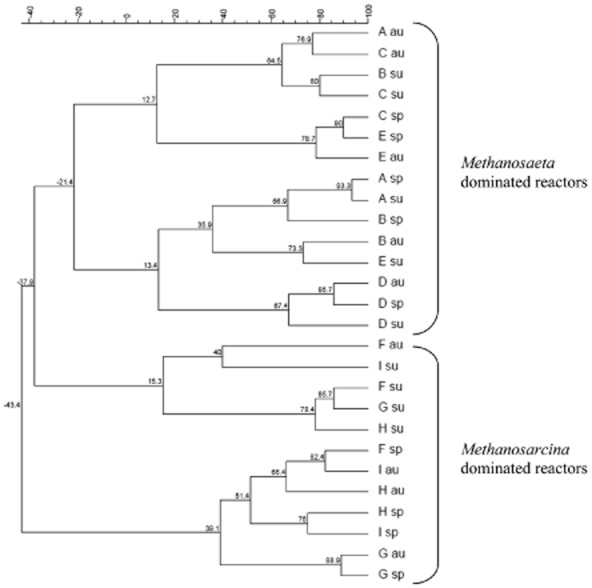
Cluster analysis of DGGE fingerprints based on the PCR of archaeal 16S rRNA genes extracted from the sludge samples of nine biogas plants in spring, summer and autumn.
Denaturing gradient gel electrophoresis (DGGE)
Analyses of DGGE banding patterns supported the findings from microarray and RT‐PCR, whereby the Methanosaeta‐dominated sludges grouped distinctly from the Methanosarcina‐dominated sludges (Fig. 5). The Methanosaeta‐dominated sludges were found to form two clusters, as were the Methanosarcina‐dominated sludges. Interestingly, the summer samples (as well as one autumn sample) of the Methanosarcina‐dominated sludges were found to cluster together, and separately from the spring and remaining autumn samples. It would seem that some particular seasonal change must have caused this clustering. However, this finding was not able to be verified by any of the other results obtained in this study.
Discussion
Anaerobic digestion is a renewable energy technology that is gaining increasing interest and importance worldwide. In this study, nine different anaerobic digestion plants were monitored and investigated to determine any links between the abiotic and biotic factors involved. All nine reactors monitored were found to be operating under stable conditions with good biogas yields, even when some values exceeded the critical value for good operation conditions (BMVIT, 2007). Correlation analysis indicated links between pH, conductivity, NH4‐N, NH3‐N and H2. This was not unexpected, since the pH approximates the negative logarithm (base 10) of the molar concentration of dissolved hydronium ions and thus regulates the balance of NH4‐N and NH3‐N (Fachagentur für Nachwachsende Rohstoffe, 2006). The thermophilic reactor F was found to have the highest acetate level of all reactors investigated. This correlation between acetate and reactor temperature has also been reported in previous studies (Van Lier et al., 1996; Ahring et al., 2001).
The archaeal community structure was investigated using a variety of molecular‐biological approaches. Initially, 16S rRNA gene‐based microarrays and DGGE were used to obtain an overview of the archaeal community composition present in the different sludge samples. The dominating genera were then analysed by RT‐PCR to obtain quantitative data regarding the genera in the reactor samples. Microarray analysis with the ANAEROCHIP microarray as well as fingerprinting patterns as revealed by DGGE showed the microbial communities of the sludge samples to cluster into two main groups, dominated either by Methanosarcina or by Methanosaeta (Fig. 2). Both genera are comprised of acetoclastic methanogens, which have been reported to be responsible for approximately 70% of the methane produced in biogas reactors (Jetten et al., 1992; Ahring et al., 1995). Microarray data can however only be interpreted semi‐quantitatively, because of the variation in binding efficiency of different oligonucleotide probes with the various regions on the 16S rRNA gene. RT‐PCR is thus a perfect tool to counteract this disadvantage and results obtained in this study supported those obtained by microarray analysis (Fig. 4), whereby Methanosaeta dominated in the reactors A–E while Methanosarcina dominated in the reactors F–I. De Vrieze and colleagues (2012) stated that the ratio of Methanosarcina to Methanosaeta in reactors appears to be even more important than total archaeal numbers in monitoring reactor operational stability.
All in all, microorganisms assumed to be acetotrophic were more abundant than hydrogenotrophic methanogens in the reactors studied, although cattle manure as a main substrate (except in reactor D) served as a constant inoculum of mainly hydrogenotrophic methanogens. The most frequently observed hydrogenotroph was Methanobacterium, which was detected in all investigated reactors using the ANAEROCHIP microarray. The hydrogenotrophic genera Methanoculleus and Methanobrevibacter were also detected, but yielded lower SNR signals. Furthermore, the number of hydrogenotrophic methanogens appeared to be higher in the Methanosaeta dominated reactors than in the Methanosarcina dominated reactors. Possibly, Methanosarcina, which is able to use the acetoclastic and the hydrogenotrophic methanogenesis pathways by utilizing H2 and CO2 (Jetten et al., 1992; Kendall and Boone, 2006), was able to outcompete the hydrogen utilizing archaea.
In this study we were able to confirm that acetate, which is considered to be the most important precursor during mesophilic anaerobic digestion and accounts for 60–80% of the CH4 produced (Jeris and McCarty, 1965; Smith and Mah, 1966; Van den Berg et al., 1974; Mountfort and Asher, 1978), was primarily responsibly for the clustering of the dominating archaea in the anaerobic digester sludges. Under low acetate concentrations (reactors A to E), Methanosaeta holds a competitive advantage over Methanosarcina spp., because of its 5–10 times higher substrate affinity. This higher substrate affinity is based on a high energy input in the activation of acetate by an acetyl‐CoA synthetase (Zinder, 1990; Jetten et al., 1992; Kendall and Boone, 2006). Methanosarcina spp. cannot, reportedly, successfully compete under such limiting conditions. Raskin and colleagues (1996) reported acetate values of 0.1–0.18 mmol l−1 to be a minimum threshold for Methanosarcina growth. Min and Zinder (1989) and Westermann and colleagues (1989) also reported concentrations of 0.4–1.2 mmol l−1 acetate to be a minimum threshold for Methanosarcina growth. These acetate concentrations perfectly support the findings in this study, whereby Methanosaeta was able to dominate at low acetate levels (≤ 0.8 mmol l−1, with the exception of two samples, where the acetate concentration was 1.8 mmol l−1 and 4 mmol l−1). In contrast, Methanosarcina outcompeted Methanosaeta in reactors F to I, where the acetate levels were mostly higher. According to the literature (Liu et al., 1985; Schmidt et al., 2000; Conklin et al., 2006; Shin et al., 2011) high changeover rates, low generation times (i.e. doubling times of 1.0–1.2 days), tolerance to sudden changes in pH (around 0.8–1.0 units) and higher ammonia and VFA tolerance ensure the dominance of Methanosarcina. While Methanosaeta sp. has a low maximum specific growth rate (μmax) of 0.20 day−1 and a half saturation constant (Ks) of 10–50 mg chemical oxygen demand (COD) l−1, Methanosarcina sp. is characterized by a high μmax of 0.60 day−1 and a Ks of 200–280 mg COD l−1 (Gujer and Zehnder, 1983; McMahon et al., 2004; Conklin et al., 2006; Yu et al., 2006; Qu et al., 2009).
Furthermore, Methanosarcina cells have a coccoidal form and grow in irregular flocks, protecting the cell against many harmful chemical agents (Demirel and Scherer, 2008). Methanogens like Methanosaeta, which occur as large filaments or non‐motile rods (Patel and Sprott, 1990) appear to be more sensitive to high ammonia concentrations than Methanosarcina cells (Zhilina, 1976). According to Calli et al. (2005) and Goberna and colleagues (2010) this resistance of Methanosarcina is attributed to its ability to form cell clusters and flocs. The size and form of Methanosarcina thus corresponds to a higher volume‐to‐surface ratio (four to seven times higher than for Methanosaeta sp.) correlating to a lower ammonia diffusion rate per unit of cell mass, compared with filamentous methanogens. However, we found no significant correlations between ammonium ions (NH4+) or free ammonia (NH3+) and the archaeal community in this study, since toxic concentrations as postulated in the literature (> 2.7 g l−1 NH4+ and > 0.15 g l−1 NH3+; Fachagentur für Nachwachsende Rohstoffe, 2006) were not exceeded.
All the physicochemical parameters measured in this study were found to correlate positively (although not significantly) with Methanosarcina (Table 2), indicating a higher stress tolerance than Methanosaeta, for which the correlations were all negative.
Syntrophic acetate oxidation (SAO), a two step process where acetate is first oxidized to CO2 and H2 (often through Clostridium sp.) and subsequently converted to methane by hydrogenotrophic methanogens, is reportedly favoured under thermophilic conditions (Zinder and Koch, 1984; Petersen and Ahring, 1991), at very low levels of H2 partial pressure (i.e. between 2.6 and 74 Pa according to Hattori, 2008) and in the presence of very high inhibitor concentrations, particularly ammonium and VFAs (Schnurer et al., 1999; Schnurer and Nordberg, 2008). Under such conditions, especially in Methanosaeta dominated reactors, which are very sensitive to stress, an increase in the organic loading rate can cause a shift from direct acetate cleavage towards syntrophic acetate oxidation coupled with hydrogenotrophic methanogenesis (Sasaki et al., 2011). According to Schnurer and Nordberg (2008), Nettmann and colleagues (2010) and Sasaki and colleagues (2011) such community changes were reported in ammonium ranges of 3000 mg total ammonia nitrogen (TAN) l−1. Also high acetate concentrations can result in a shift to syntrophic acetate oxidation (Hao et al., 2011). Methanosarcina species, on the other hand were found in over 90% of the methanogenic population analysed in biogas reactors with SAO as the main methanogenic pathway and seem to act as hydrogen‐utilizing methanogens in reactors with SAO (Karakashev et al., 2006; Karlsson et al., 2012). According to these results, an interaction between SAO and Methanosarcina in reactors F–I is possible, but not proven, since the focus of this study was limited to methanogenic 16S rDNA.
The archaeal communities appeared to be highly stable and resilient in all biogas reactors. As already known (Wittebolle et al., 2009) community evenness corresponds with functional stability, by having a higher capacity to use redundant functional pathways. Traversi and colleagues (2011) detected a positive correlation between biogas production efficiency and the genera Methanosarcina and Methanosaeta. It seems that these organisms, primarily Methanosarcina, can act as an indicator for reliable reactor functioning, and help to diagnose imbalances in the microbial community (De Vrieze et al., 2012).
Experimental procedures
Sampling
As a part of a greater biogas monitoring project, samples from nine anaerobic digestion plants in South Tyrol (Italy) and Vorarlberg (Austria) were analysed in the year 2009. Spring samples (sp) were taken in March/April, summer samples (su) in June/July and autumn samples (au) in September.
With the exception of two reactors which treated sewage sludge (reactor D) and a mixture of cow, pig and chicken manure (reactor I), anaerobic digesters were fed predominantly cow manure. Reactors A and H co‐digested small amounts (< 7%) of waste vegetable oil, reactors B and F co‐digested small amounts (∼ 20%) of pomace and reactor I a composition (21%) of waste vegetable oil, pomace, garden waste and biowaste (Table 1). Overpressure vents, paddle cases or manholes were used to collect sludge samples (about 0.5 l), which were stored on ice until they reached the laboratory. Following the completion of DNA extraction and the investigation of physical and chemical parameters, samples were stored at −20°C to preserve microbial community structure and chemical composition.
Physicochemical parameters
Chemical and physical parameters were measured, documented and compared with the results of other studies or technical literature (Fachagentur für Nachwachsende Rohstoffe, 2006; BMVIT, 2007). The parameters investigated included: reactor temperature, pH, electrical conductivity (EC), dry matter (DM), volatile solids (VS), and ionized ammonia (NH4‐N), free ammonia (NH3‐N), hydrogen (H2), hydrogen sulfide (H2S), acetate, carbon dioxide (CO2) and methane (CH4) concentrations. Physical and chemical parameters were monitored either directly at the plants (pH, EC, temperature and gas measurements) or in the laboratory (DM, VOS, NH4‐N, NH3‐N, H2). A portable Biogas Check BM 2000 instrument (Geotechnical Instruments, Warwickshire, UK) was used to measure the proportion of CH4, CO2, O2 and H2S in the biogas.
Sample preparation for HPLC analysis was performed by dialysis. Dialysis tubing was filled with 10 ml of distilled water, closed and submerged into the liquid sample immediately after sample collection. The bottle was shaken three times and stored overnight at 4°C to reach a total equilibrium in the dialysate. Tubing was then removed, washed with distilled water and opened. Dialysate (0.5 ml) was subjected to high‐performance liquid chromatography (HPLC) analysis on an Aminex HPX‐87H column (Bio‐Rad, Hercules, USA). A 5 mM H2SO4 mobile phase was used at 0.7 ml min−1 and the detection wavelength was set at 210 nm. The detection limit ranged at 1 mmol l−1.
To determine dry matter (DM), approximately 100 g of fresh sludge were dried at 105°C for 24 h and weighed after cooling in a desiccator. VS were calculated as the loss of weight after igniting 5 g of the oven‐dried residue at 550°C in a muffle furnace (Schinner et al., 1993). Ammonium was determined using the colorimetric tube test (Macherey‐Nagel, Düren, Germany). The PASW‐SPSS 17.0 software (SPSS, Chicago, IL, USA) was used to determine correlations between parameters.
DNA extraction
The PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA) was used to extract DNA according to the instructions provided. One modification that was employed however, was that the supernatant obtained after centrifugation of the horizontally shaken sample was exposed to three freeze–thaw cycles (30 min at −80°C followed by 5 min at 65°C). DNA extracts were stored at −20°C.
Preparation of fluorescently labelled target DNAs by PCR
The universal archaeal primers 109f and 934r (Grosskopf et al., 1998) were used to amplify the 16S rRNA gene of methanogens in the sludge samples. PCR amplifications were conducted in 25 μl standard reaction mixes containing a final concentration of 1× reaction buffer [16 mM (NH4)2SO4, 67 mM Tris–HCl, pH 8.8, 1.5 mM MgCl2, 0.01% Tween 20] (GeneCraft, Münster, Germany), 200 μM of each dNTP, 0.8 μM of forward primer (Cy5‐labelled at the 5′ end), 0.2 μM of reverse primer (PO4‐group at the 5′ end), 0.625 U DNA polymerase (GeneCraft, Munster, Germany) and sterile water. In addition, 10 mM tetramethylammonium chloride and 0.4 mg ml−1 bovine serum albumin (BSA) was included in the reactions to enhance the specificity. Amplifications were performed using a FlexCycler Thermal cycler (Analytikjena, Jena, Germany). After an initial denaturation at 95°C for 5 min, amplification reactions were subjected to 1 min at 80°C, 1 min at 55°C and 2 min at 72°C. Thermal cycling then proceeded with 33 cycles of 95°C for 1 min, 55°C for 1 min and 72°C for 2 min. Temperature cycling was followed by 1 min at 95°C, 1 min at 55°C and a final extension at 72°C for 10 min.
The GenElute PCR clean‐up kit (Sigma, Missouri, USA) was used to purify pooled amplification products. For the subsequent preparation of fluorescently labelled single‐stranded DNA targets, the phosphorylated DNA strand was removed, using Lambda exonuclease enzyme (Epicentre, Wisconsin, USA). One thousand ng of the amplified double‐stranded DNA (dsDNA) was incubated with 18 U Lambda exonuclease, 1× Lambda exonuclease buffer and sterile distilled water in a total volume of 50 μl at 37°C for 3 h. Products were then vacuum dried and stored at −20°C until hybridization.
Microarray hybridization
For the in situ hybridization on microarray slides, single‐stranded Cy5‐labelled PCR product was resuspended in 19 μl of a hybridization buffer consisting of 5× SSC, 1% blocking reagent (Roche, Mannheim, Germany), 0.02% SDS, 0.1% n‐laurylsarcosine and 5% formamide (Loy et al., 2002). One microlitres of a 100 nM Cy5‐labelled control oligonucleotide (5′‐AGGAAGGAAGGAAGGAAG‐3′) was added to each tube as a hybridization control. After a denaturation step of 10 min at 95°C, DNA was placed directly on ice, and then cooled DNA was transferred onto a pre‐chilled ANAEROCHIP microarray (Franke‐Whittle et al., 2009a) and covered with a glass coverslip to guarantee a uniform moistening of the array surface. Slides were placed into the hybridization chamber of a Hybex® microsample incubator (SciGene Corporation, Sunnyvale, USA) for 4 h at 55°C.
After hybridization, slides were washed immediately at room temperature, each for 3 min in buffer 1 (1× SSC, 0.2% SDS), followed by buffer 2 (0.1× SSC, 0.2% SDS) and buffer 3 (0.05× SSC). Finally, arrays were briefly submerged into distilled water, air‐dried and scanned using a ScanArray Gx microarray scanner (Perkin Elmer, MA, USA) as described by Franke‐Whittle and colleagues (2009a). Scan power was set to 90% and PMT gain to 500 at 633 nm. Quantification of fluorescence was conducted by superimposing a grid of circles onto the image using the ScanArray Gx software (Perkin Elmer, MA, USA). The signal to noise ratio (SNR) for all spots was calculated according to the following calculation, as described by Loy and colleagues (2002):
where F635P and F635NB denote the median fluorescent signals measured at 635 nm for each sample (P) and the non‐binding control (NB), and B635P and B635NB the corresponding background signals. Signals were assumed to be positive if an SNR value of ≥ 2 was obtained (Loy et al., 2002). Statistical analyses of data were performed using the program Canoco 4.5 (ter Braak and Šmilauer, 2002) and PASW‐SPSS 17.0 (SPSS, Chicago, IL).
Real‐time PCR
Quantification of the dominant genera Methanosaeta and Methanosarcina from the DNA of sludge samples was performed, using RT‐PCR. RT‐PCR amplifications were conducted using the Quantimix Easy SYG kit (Biotools, Spain) and performed in a Rotor‐GeneTM 6000 (Corbett Life Sciences, Sydney, Australia) in 20 μl volumes. Each standard reaction mix contained a final concentration of 1× Quantimix Easy SYG, 100 nM each primer, 0.4 mg ml−1 BSA and distilled water (Franke‐Whittle et al., 2009b; Goberna et al., 2010). Primer sequences are listed in Goberna and colleagues (2010). Two microlitres of sludge DNA was used as the template in each reaction. After an initial denaturation at 95°C for 5 min, thermal cycling comprised of 40 cycles of 20 s at 95°C, 20 s at 60°C (Methanosaeta) or 64°C (Methanosarcina) and 20 s at 72°C. Thermal cycling was completed with a melting analysis (65–95°C, ramp 0.5°C min−1) to check for primer dimer formation and product specificity. Standard curves were constructed with PCR‐amplified 16S rRNA from pure cultures as described in Franke‐Whittle and colleagues (2009b). All standards and samples were run in duplicate.
Denaturing gradient gel electrophoresis (DGGE)
For the amplification of methanogenic communities for DGGE, a nested PCR was applied. The primers 109f and 934r (Grosskopf et al., 1998) were used in the first round of PCR, and PCR products from the first amplification were used as a template for the second PCR. The primer pair 0357fGC and 0691r (Watanabe et al., 2004) were used in the second PCR. Each PCR mixture contained 0.2 μM of each primer, 0.625 U Bio Therm™ DNA Polymerase (Gene Craft, Germany), 1× DNA polymerase buffer, 2.5 μg of BSA, 200 μM each dNTP, 2.5 mM MgCl2 and 1 μl of extracted DNA or PCR product in a final volume of 25 μl. The cycling programme for the first‐round PCR was as follows: After an initial denaturation at 95°C for 5 min, 30 cycles at 94°C for 1 min, 55°C for 1 min and 72°C for 3 min were performed. An elongation step at 72°C for 15 min completed DNA amplification. The second‐round PCR was comprised of an initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 49°C for 1 min and 72°C for 2 min, with a final elongation step at 72°C for 15 min.
Denaturing gradient gel electrophoresis was performed using an Ingeny PhorU2 system (Ingeny International BV, the Netherlands). Two microlitres of PCR product (either sludge DNA or pure culture of M. barkeri DSMZ 800, M. concilii DSMZ 2139 or Methanobacterium formicicum DSMZ 1535) was loaded onto a 7% to 8% (w/v) polyacrylamide gel with a denaturing gradient of 40% to 65% (100% denaturant consists of 7 M urea plus 40% formamide in 1× TAE buffer) and was run for 16 h at 100 V, at a constant temperature of 60°C in 1× TAE buffer (pH 7.4).
After electrophoresis, gels were stained with silver nitrate using an automated gel stainer (Amersham Pharmacia Biotech, Germany), photographed and air dried for storage. DGGE banding patterns were normalized and analysed using the GelCompar II software package, version 4.0 (Applied Maths, Ghent, Belgium). Calculation of the pairwise similarities was based on the Dice correlation coefficient. Dendrograms were created using the algorithm of Ward (Legendre and Legendre, 1998).
Conclusions
Although the control of anaerobic digestion processes has received much attention in the past few years, science is still far from understanding all of the process interactions occurring between biotic and abiotic parameters in this complex system. In this study, acetotrophic archaea dominated all reactors and revealed significant correlations with acetate levels. Archaeal communities remained highly stable over a whole year of operation, showing that seasonal changes in input materials had little effect on the apparently resilient microbiota in all biogas plants. This study has focused only on the archaea, but considering the close and efficient syntrophisms known to exist between bacteria and archaea, further research will target both phylogenetic groups.
Acknowledgments
We would like to thank biogas plant operators for their cooperation, and the Tirol and South Tirol provinces for financial assistance. Further thanks go to Sieglinde Farbmacher and Pamela Vrabl for conducting the HPLC measurements of VFAs. Ingrid Franke‐Whittle and Andreas Walter were funded by the Fonds zur Förderung der wissenschaftlichen Forschung (FWF) Austria, P‐200010.
References
- Ahring B.K. Berlin, Heidelberg, New York: Springer; 2003. [Google Scholar]
- Ahring B.K., Sandberg M., Angelidaki I. Volatile fatty acids as indicators of process imbalance in anaerobic digestors. Appl Microbiol Biotechnol. 1995;43:559–565. , and . [Google Scholar]
- Ahring B.K., Ibrahim A.A., Mladenovska Z. Effect of temperature increase from 55 to 65°C on performance and microbial population dynamics of an anaerobic reactor treating cattle manure. Water Res. 2001;35:2446–2452. doi: 10.1016/s0043-1354(00)00526-1. , and . [DOI] [PubMed] [Google Scholar]
- BMVIT (Bundesministeriums für Verkehr, Innovation und Technologie) 2007. ) Aufbau eines Bewertungssystems für Biogasanlagen ‘Gütesiegel Biogas’, Projektnummer 807742 [WWW document]. URL http://www.eonerc.rwth‐aachen.de/global/show_document.asp?id=aaaaaaaaaabsint.
- ter Braak C.J.F., Šmilauer P. Ithaca, NY, USA: Microcomputer Power; 2002. [Google Scholar]
- Calli B., Mertoglu B., Inanc B., Yenigun O. Community changes during start‐up in methanogenic bioreactors exposed to increasing levels of ammonia. Environ Technol. 2005;26:85–91. doi: 10.1080/09593332608618585. , and . [DOI] [PubMed] [Google Scholar]
- Conklin A., Stensel H.D., Ferguson J. Growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion. Water Environ Res. 2006;78:486–496. doi: 10.2175/106143006x95393. , and . [DOI] [PubMed] [Google Scholar]
- De Vrieze J., Hennebel T., Boon N., Verstraete W. Methanosarcina: the rediscovered methanogen for heavy duty biomethanation. Bioresour Technol. 2012;112:1–9. doi: 10.1016/j.biortech.2012.02.079. , and . [DOI] [PubMed] [Google Scholar]
- Demirel B., Scherer P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review. Rev Environ Sci Biotechnol. 2008;7:173–190. , and . [Google Scholar]
- E‐CONTROL. 2010. ) Tabelle anerkannte Ökoanlagen 20022010 [WWW document]. URL http://www.e‐control.at/portal/page/portal/medienbibliothek/oeko‐energie/dokumente/pdfs/Tabelle%20anerkannte%20%C3%96koanlagen%202002‐2010_korr_mai2011.pdf.
- Fachagentur für Nachwachsende Rohstoffe. 2006.
- Franke‐Whittle I.H., Goberna M., Pfister V., Insam H. Design and development of the ANAEROCHIP microarray for investigation of methanogenic communities. J Microbiol Methods. 2009a;79:279–288. doi: 10.1016/j.mimet.2009.09.017. , and . [DOI] [PubMed] [Google Scholar]
- Franke‐Whittle I.H., Goberna M., Insam H. Design, testing and application of real‐time PCR primers for the detection of MethanoculleusMethanosarcinaMethanothermobacter and uncultured methanogens. Can J Microbiol. 2009b;55:611–616. doi: 10.1139/w08-157. , and . [DOI] [PubMed] [Google Scholar]
- Goberna M., Gadermaier M., García C., Wett B., Insam H. Adaptation of methanogenic communities to the cofermentation of cattle excreta and olive mill wastes at 37°C and 55°C. Appl Environ Microbiol. 2010;76:6564–6571. doi: 10.1128/AEM.00961-10. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf R., Janssen P.H., Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujer W., Zehnder A.J.B. Conversion processes in anaerobic‐digestion. Water Sci Technol. 1983;15:127–167. , and . [Google Scholar]
- Hao L.P., Lu F., He P.J., Li L., Shao L.M. Predominant contribution of syntrophic acetate oxidation to thermophilic methane formation at high acetate concentrations. Environ Sci Technol. 2011;45:508–513. doi: 10.1021/es102228v. , and . [DOI] [PubMed] [Google Scholar]
- Hattori S. Syntrophic acetate‐oxidizing microbes in methanogenic environments. Microbes Environ. 2008;23:118–127. doi: 10.1264/jsme2.23.118. [DOI] [PubMed] [Google Scholar]
- INBIMO. 2011. ) INBIMO Workshop 21.1.2011: Stand Biogas in Südtirol [WWW document]. URL http://alpibiogas.com/de/projekte/WORKSHOPBiogas/03_Steger.pdf.
- Insam H., Wett B. Control of GHG emission at the microbial community level. Waste Manag. 2008;28:699–706. doi: 10.1016/j.wasman.2007.09.036. , and . [DOI] [PubMed] [Google Scholar]
- Jeris J.S., McCarty P.L. The biochemistry of methane fermentation using C14 tracers. J Water Pollut Control Fed. 1965;37:178–192. , and . [Google Scholar]
- Jetten M.S.M., Stams A.J.M., Zehnder A.J.B. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol Rev. 1992;88:181–198. , and . [Google Scholar]
- Karakashev D., Batstone D.J., Trably E., Angelidaki I. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl Environ Microbiol. 2006;72:5138–5141. doi: 10.1128/AEM.00489-06. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A., Einarsson P., Schnürer A., Eljertsson J., Svensson B.H. 2012.
- Kendall M.M., Boone D.R. The order Methanosarcinales. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.H., Stackebrandt E., editors. Vol. 3. New York, USA: Springer; 2006. pp. 244–256. , and . In The Prokaryotes. A Handbook on the Biology of Bacteria, Vol. , and (eds). , pp. [Google Scholar]
- Legendre P., Legendre L. 2nd English edn. Amsterdam, the Netherlands: Elsevier; 1998. [Google Scholar]
- Liu Y., Boone D.R., Sleat R., Mah R.A. Methanosarcina mazei lyc, a new methanogenic isolate which produces a disaggregating enzyme. Appl Environ Microbiol. 1985;49:608–613. doi: 10.1128/aem.49.3.608-613.1985. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A., Lehner A., Lee N., Adamczyk J., Meier H., Ernst J. Oligonucleotide microarray for 16S rRNA gene‐based detection of all recognized lineages of sulfate‐reducing prokaryotes in the environment. Appl Environ Microbiol. 2002;68:5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon K.D., Zheng D.D., Stams A.J.M., Mackie R.I., Raskin L. Microbial population dynamics during start‐up and overload conditions of anaerobic digesters treating municipal solid waste and sewage sludge. Biotechnol Bioeng. 2004;87:823–834. doi: 10.1002/bit.20192. , and . [DOI] [PubMed] [Google Scholar]
- Min H., Zinder S.H. Kinetics of acetate utilization by two thermophilic acetotrophic methanogens: Methanosarcina sp. strain CALS‐1 and Methanothrix sp. strain CALS‐1. Appl Environ Microbiol. 1989;55:488–491. doi: 10.1128/aem.55.2.488-491.1989. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountfort D.O., Asher R.A. Changes in proportions of acetate and carbon dioxide used as methane precursors during the anaerobic digestion of bovine waste. Appl Environ Microbiol. 1978;35:648–654. doi: 10.1128/aem.35.4.648-654.1978. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettmann E., Bergmann I., Pramschufer S., Mundt K., Plogsties V., Herrmann C., Klocke M. Polyphasic analyses of methanogenic archaeal communities in agricultural biogas plants. Appl Environ Microbiol. 2010;76:2540–2548. doi: 10.1128/AEM.01423-09. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B., Sprott D. Methanosaeta concilii gen. nov., sp. nov. (‘Methanothrix concilii’) and Methanosaeta thermoacetophila nom. rev., comb. nov. Int J Syst Bacteriol. 1990;40:79–82. , and . [Google Scholar]
- Petersen P.S., Ahring B.K. Acetate oxidation in a thermophilic anaerobic sewage‐sludge digestor: the importance of non‐aceticlastic methanogenesis from acetate. FEMS Microbiol Lett. 1991;86:149–152. , and . [Google Scholar]
- Qu X., Vavilin V.A., Mazeas L., Lemunier M., Duquennoi C., He P.J., Bouchez T. Anaerobic biodegradation of cellulosic material: batch experiments and modelling based on isotopic data and focusing on aceticlastic and nonaceticlastic methanogenesis. Waste Manag. 2009;29:1828–1837. doi: 10.1016/j.wasman.2008.12.008. , and . [DOI] [PubMed] [Google Scholar]
- Raskin L., Rittmann B.E., Stahl D.A. Competition and coexistence of sulfate‐reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62:3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki D., Hori T., Haruta S., Ueno Y., Ishii M., Igarashi Y. Methanogenic pathway and community structure in a thermophilic anaerobic digestion process of organic solid waste. J Biosci Bioeng. 2011;111:41–46. doi: 10.1016/j.jbiosc.2010.08.011. , and . [DOI] [PubMed] [Google Scholar]
- Schink B. Energetics of syntrophic cooperation in methane degradation. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinner F., Öhlinger R., Kandeler E., Margesin R. Berlin, Heidelberg, New York: Springer Verl.; 1993. [Google Scholar]
- Schmidt J.E., Mladenovska Z., Lange M., Ahring B.K. Acetate conversion in anaerobic biogas reactors: traditional and molecular tools for studying this important group of anaerobic microorganisms. Biodegradation. 2000;11:359–364. doi: 10.1023/a:1011695409308. , and . [DOI] [PubMed] [Google Scholar]
- Schnurer A., Nordberg A. Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Sci Technol. 2008;57:735–740. doi: 10.2166/wst.2008.097. , and . [DOI] [PubMed] [Google Scholar]
- Schnurer A.G., Zellner G., Svensson B. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol Ecol. 1999;29:249–261. , and . [Google Scholar]
- Shin S.G., Zhou B.W., Lee S., Kim W., Hwang S. Variations in methanogenic population structure under overloading of pre‐acidified high‐strength organic wastewaters. Process Biochem. 2011;46:1035–1038. , and . [Google Scholar]
- Smith P.H., Mah R.A. Kinetics of acetate metabolism during sludge digestion. Appl Microbiol. 1966;14:368–371. doi: 10.1128/am.14.3.368-371.1966. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traversi D., Villa S., Acri M., Pietrangeli B., Degan R., Gilli G. The role of different methanogen groups evaluated by Real‐Time qPCR as high‐efficiency bioindicators of wet anaerobic co‐digestion of organic waste. AMB Express. 2011;1:1–7. doi: 10.1186/2191-0855-1-28. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg L., Lentz C.P., Athey R.J., Rooke E.A. Assessment of methanogenic activity in anaerobic digestion: apparatus and method. Biotechnol Bioeng. 1974;16:1459–1469. , and . [Google Scholar]
- Van Lier J.B., Sanz Martin J.L., Lettinga G. Effect of temperature on the anaerobic thermophilic conversion of volatile fatty acids by dispersed and granular sludge. Water Res. 1996;30:199–207. , and . [Google Scholar]
- Watanabe T., Asakawa S., Nakamura A., Nagaoka K., Kimura M. DGGE method for analyzing 16S rDNA of methanogenic archaeal community in paddy field soil. FEMS Microbiol Lett. 2004;232:153–163. doi: 10.1016/S0378-1097(04)00045-X. , and . [DOI] [PubMed] [Google Scholar]
- Westermann P., Ahring B.K., Mah R.A. Temperature compensation in Methanosarcina barkeri by modulation of hydrogen and acetate affinity. Appl Environ Microbiol. 1989;55:1262–1266. doi: 10.1128/aem.55.5.1262-1266.1989. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittebolle L., Marzorati M., Clement L., Balloi A., Daffonchio D., Heylen K. Initial community evenness favours functionality under selective stress. Nature. 2009;458:623–626. doi: 10.1038/nature07840. et al. [DOI] [PubMed] [Google Scholar]
- Yu Y., Kim J., Hwang S. Use of real‐time PCR for group‐specific quantification of aceticlastic methanogens in anaerobic processes: population dynamics and community structures. Biotechnol Bioeng. 2006;93:424–433. doi: 10.1002/bit.20724. , and . [DOI] [PubMed] [Google Scholar]
- Zhilina T.N. Biotypes of Methanosarcina. Microbiology (USSR) 1976;45:414–421. [PubMed] [Google Scholar]
- Zinder S.H. Conversion of acetic acid to methane by thermophiles. FEMS Microbiol Rev. 1990;75:125–138. [Google Scholar]
- Zinder S.H., Koch M. Non‐aceticlastic methanogenesis from acetate: acetate oxidation by a thermophilic syntrophic coculture. Arch Microbiol. 1984;138:263–272. , and . [Google Scholar]