Summary
Evidence for the involvement of the nonhomologous end joining (NHEJ) pathway in Agrobacterium-mediated transferred DNA (T-DNA) integration into the genome of the model plant Arabidopsis remains inconclusive.
Having established a rapid and highly efficient Agrobacterium-mediated transformation system in rice (Oryza sativa) using scutellum-derived calli, we examined here the involvement of the NHEJ pathway in Agrobacterium-mediated stable transformation in rice. Rice calli from OsKu70,OsKu80 and OsLig4 knockdown (KD) plants were infected with Agrobacterium harboring a sensitive emerald luciferase (LUC) reporter construct to evaluate stable expression and a green fluorescent protein (GFP) construct to monitor transient expression of T-DNA.
Transient expression was not suppressed, but stable expression was reduced significantly, in KD plants. Furthermore, KD-Ku70 and KD-Lig4 calli exhibited an increase in the frequency of homologous recombination (HR) compared with control calli. In addition, suppression of OsKu70,OsKu80 and OsLig4 induced the expression of HR-related genes on treatment with DNA-damaging agents.
Our findings suggest strongly that NHEJ is involved in Agrobacterium-mediated stable transformation in rice, and that there is a competitive and complementary relationship between the NHEJ and HR pathways for DNA double-strand break repair in rice.
Keywords: DNA double-strand breaks (DSBs), DNA ligase 4 (Lig4), homologous recombination (HR), Ku70, Ku80, nonhomologous end joining (NHEJ), Oryza sativa (rice), transferred DNA (T-DNA)
Introduction
Plant cells are continually exposed to endogenous and exogenous genotoxic stresses, such as reactive oxygen species and UV light, which lead to the accumulation of numerous types of DNA damage, including cross-linking of DNA, base oxidation or alkylation, mismatch of bases, DNA single-strand breaks and DNA double-strand breaks (DSBs). DSBs are amongst the most serious types of DNA damage in living cells and can lead to cell death if not repaired. There are at least two repair pathways for DSB repair: nonhomologous end joining (NHEJ), which involves rejoining of the broken DNA ends; and homologous recombination (HR), which is an accurate pathway that uses homologous DNA sequences from the sister chromatid as a template. NHEJ is used preferentially to deal with DSBs in higher eukaryotes, including higher plants (Mladenov & Iliakis, 2011; Symington & Gautier, 2011; Waterworth et al., 2011), whereas HR is the main DSB repair pathway in bacteria and yeast (Pâques & Haber, 1999; Aylon & Kupiec, 2004).
The NHEJ pathway in vertebrates is thought to be as follows. A Ku70/Ku80 complex binds initially to two DNA ends at the DSB site, and then recruits a DNA-dependent protein kinase (DNA-PKcs), which has not been identified in plants. DNA-PKcs phosphorylates and activates many proteins, including nuclease and itself. Ultimately, the Lig4–Xrcc4 complex rejoins the two DNA ends of the break (Mladenov & Iliakis, 2011; Symington & Gautier, 2011). Many proteins involved in NHEJ found in mammalian cells have also been identified in Arabidopsis and rice (reviewed by Singh et al., 2010; Edlinger & Schlögelhofer, 2011), including Arabidopsis Ku70 (Tamura et al., 2002), rice Ku70 (Hong et al., 2010), Arabidopsis Ku80 (Tamura et al., 2002), Arabidopsis DNA ligase 4 (Lig4) (West et al., 2000) and Arabidopsis Xrcc4 (West et al., 2000).
In Arabidopsis, mutants of the ku70, ku80 and lig4 genes have been shown to display hypersensitivity to DSB-inducing agents, including γ-irradiation, methyl methanesulfonate, ionizing radiation and bleomycin (van Attikum et al., 2003; Friesner & Britt, 2003; Gallego et al., 2003; Li et al., 2005; Hong et al., 2010; Wang et al., 2010), indicating that Ku70/Ku80 and Lig4 proteins play an important role in DSB repair in Arabidopsis. In addition, recent studies in mammalian cells have shown that DSBs can be rejoined in the presence of chemical inhibition or mutation of key NHEJ factors, such as DNA-PKcs, Ku70, Ku80, Lig4 and XRCC4, suggesting the existence of backup pathways for NHEJ (Mladenov & Iliakis, 2011; Symington & Gautier, 2011). Such a backup NHEJ pathway may utilize poly(ADP-ribose) polymerase-1, MRN, histone H, DNA ligase III and XRCC1 (Cheng et al., 2011; Mladenov & Iliakis, 2011; Symington & Gautier, 2011). In Arabidopsis, several studies have also provided evidence for the existence of Ku- and Lig4-independent pathways, and have identified proteins, such as AtLig1 and AtXRCC1, that function in these pathways (Charbonnel et al., 2010, 2011; Waterworth et al., 2009, 2011).
Agrobacterium-mediated plant genetic transformation uses the transferred DNA (T-DNA) region on a binary plasmid harbored by Agrobacterium tumefaciens as a vector for genetic engineering. DNA repair pathways have been thought to be involved in the integration of T-DNA into the plant genome, and two major models for this integration have been proposed. The first – the DSB repair model – hypothesizes that single-strand T-DNAs imported into the plant cell nucleus by the virulence protein complex are replicated to a double-stranded form and are subsequently integrated into DSBs in the host genome (Gelvin, 2010; Pitzschke & Hirt, 2010; Magori & Citovsky, 2011). By contrast, the second – the strand-invasion model – assumes that the 3′ end of single-stranded T-DNA finds a microhomology to plant DNA and invades the target site host DNA. The VirD2-attached 5′ end of the T-DNA binds to a nick in the plant DNA and is ligated. The complementary strand of the T-DNA is synthesized, resulting in integration of a double-strand copy of the T-DNA into the plant genome (Gelvin, 2010; Pitzschke & Hirt, 2010; Magori & Citovsky, 2011).
In Arabidopsis, mutation of either the AtKu80 or AtLig4 gene caused a decrease in the frequency of stable T-DNA integration following an in planta floral dip transformation assay (Friesner & Britt, 2003). Involvement of AtKu80 in T-DNA integration was also observed in a root tumorigenesis assay (Li et al., 2005). By contrast, no decrease in T-DNA integration was observed in AtKu80 mutants using the in planta floral dip transformation assay (Gallego et al., 2003). Furthermore, the AtLig4 mutant was not impaired in T-DNA integration using either the in planta floral dip or tumorigenesis assay (van Attikum et al., 2003). The discrepancy between these previous reports might be attributable to differences in the transformation mechanisms between germline and somatic cells, and could also depend on the physiological condition of the plant material used for the experiments. Thus, evidence for the involvement of the NHEJ pathway in Agrobacterium-mediated T-DNA integration into the plant genome remains inconclusive.
The choice of NHEJ and HR pathways for DSB repair in eukaryotes depends on the cell type, cell cycle stage and complexity of the DNA end (Shrivastav et al., 2008; Heyer et al., 2010; Symington & Gautier, 2011). In addition, disruption of the NHEJ pathway has been shown to lead to an increased frequency of HR in fungi (Ninomiya et al., 2004; Villalba et al., 2008), mammals (Liang et al., 1996; Pierce et al., 2001; Allen et al., 2003) and plants (Gallego et al., 2003), suggesting that the NHEJ pathway competes with the HR pathway for DSB repair. However, the AtKu80 mutant did not show an enhanced frequency of intrachromosomal HR (Gallego et al., 2003).
We have developed a stable and efficient Agrobacterium-mediated transformation system for rice (Toki et al., 2006). Most recently, we have further constructed a sequential monitoring system for stable transformation by visualizing cells in which T-DNA is successfully integrated into the rice genome using a nondestructive and highly sensitive visible marker in rice (H. Saika et al., unpublished). In this report, we investigated the involvement of the NHEJ pathway in an Agrobacterium-mediated stable transformation system, and the choice between NHEJ and HR for DSB repair in rice plants. The evidence presented here indicates that the NHEJ pathway participates in Agrobacterium-mediated stable transformation in rice, and that this pathway possibly competes with the HR pathway for DSB repair. Alternatively, an increase in the HR pathway might result from genome instability and the up-regulation of HR genes derived from the suppression of the NHEJ pathway in rice somatic cells.
Materials and Methods
Plant materials
Oryza sativa L. cv Nipponbare (genetic background of KD-Ku70, KD-Ku80 and KD-Lig4) and O. sativa cv Dongjin (genetic background of OsKu70 T-DNA insertional line) were used in this study. The OsKu70 T-DNA insertional line was obtained from the Rice T-DNA Insertion Sequence Database (http://signal.salk.edu/cgi-bin/RiceGE). Plant genotypes were determined by PCR using the T-DNA right border primer pGA2715 RB (5′-ttggggtttctacaggacgtaac-3′) and OsKu70 gene-specific primers (5′-ccaaccttagtttcactcttgttacgtg-3′ and 5′-ggaaagcctaagtgacatcactggaa-3′).
Generation of transgenic plants
Vectors for the generation of OsKu70-, OsKu80- and OsLig4-suppressed rice plants using the RNA interference (RNAi) method were constructed with the vector pANDA (Miki & Shimamoto, 2004). The 3′ end of OsKu70, OsKu80 or OsLig4 cDNA as an RNAi trigger was amplified from first-strand cDNA by PCR using the following primer sets, OsKu70 RNAi (forward 5′-cacccggtggtggacttgaaatct-3′ and reverse 5′-ctctgcagactggagtgacatt-3′), OsKu80 RNAi (forward 5′-cacccttctgtctgaaacccgagc-3′ and reverse 5′-cagagcttctggaggtgagg-3′), OsLig4 RNAi (forward 5′-caccacaccgctgaaacaacgagta-3′ and reverse 5′-ggcgacgtccttgtaactgac-3′), and was cloned into the vector pENTR/D-TOPO using directional TOPO cloning methods (Life Technologies, Carlsbad, CA, USA) to yield an entry vector. The RNAi trigger fragments of OsKu70 (325 bp), OsKu80 (339 bp) and OsLig4 (341 bp) were re-cloned into the RNA silencing binary pANDA vector using a Gateway LR clonase reaction (Life Technologies).
The LU-UC recombination substrate was constructed as follows. The 120-bp artificial synthesized fragment containing a multi-cloning site and two I-SceI recognition sites (AscI-SacI-I-SceI-AvrII-AatII-I-SceI-Csp45I-PacI) was cloned into the AscI/PacI site of pZAmI, which is a derivative of pPZP201 (Hajdukiewicz et al., 1994), with a herbicide bispyribac sodium resistance cassette (rice acetolactate synthase (ALS) promoter 3 kb + mutant ALS gene (W548L/S627I) (Shimizu et al., 2005) + ALS terminator 0.7 kb). The full length and 5′ region of the luciferase (LUC) cDNA were amplified by PCR using pSP-luc+NF vector (Promega) as a template and the following primer pairs: full length (LUC-F 5′-tctagaatggtcaccgacgccaaaaacat-3′ (XbaI site in italics) and LUC-R 5′-gagctcttacacggcgatctttccgcc-3′ (SacI site in italics)) and 5′ region (LUC-F and LUC-R 891 5′-gagctcggcgaagaaggagaatagggtt-3′ (SacI site in italics)). The full length and 5′ region of LUC cDNA were cloned into the XbaI/SacI site between the maize ubiquitin1 promoter + first intron and the transcription terminator of the Arabidopsis ribulose-bisphosphate carboxylase small subunit (rbcS) gene in pENTR (Life Technologies), yielding the vectors pPubi:LUC and pPubi:LU, respectively. The fragment containing the maize ubiquitin1 promoter + first intron and 5′ region of LUC cDNA in the pPubi:LUC vector was digested with AscI/SacI and cloned into pZAmI, yielding pZAmLU. A 3.2-kb fragment containing the promoter sequence of rice elongation factor 1, mutant codA gene (D314A) (Mahan et al., 2004) and transcription terminator of the rice glycerol 3-phosphate dehydrogenase gene was amplified from pE_Pef:codAm:Tg3p vector (K. Osakabe et al., unpublished) using primers 5′-cctaggaagctttataatcccgtgcg-3′ (AvrII site in italics) and 5′-gacgtcgaattccaagcaccaccgcga-3′ (AatII site in italics). The amplified fragment was integrated into the AvrII/AatII site of pZAmLU, yielding pZAmLUcodA. A fragment containing the 3′ region of LUC cDNA (1485 bp) and the transcription terminator of the rbcS gene in pPubi:LUC was digested with Csp45I (internal restriction site)/PacI and integrated into pZAmLUcodA, yielding pZAmLUcodAUC. For construction of the I-SceI expression plasmid, the yeast I-SceI gene was cloned into the SpeI/SacI site between the 2 × CaMV35 promoter + Ω and the transcription terminator of rice heat shock protein 17.3 of pZDB (K. Osakabe et al., unpublished).
These plant binary vectors were transferred into Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) by electroporation. Agrobacterium-mediated transformation of rice (O. sativa cv Nipponbare) was performed as described previously (Toki, 1997; Toki et al., 2006).
RNA extraction and quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted from calli and seedlings of rice plants using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). Quantitative RT-PCR was performed with a Power SYBR Green PCR Master Mix (Life Technologies) and an ABI7300 (Life Technologies) according to the manufacturer's protocols. Primer pairs for quantitative RT-PCR were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and are as follows: OsAct1 (5′-cattgctgacaggatgagcaa-3′ and 5′-gggcgaccaccttgatctt-3′); OsKu70 (5′-acgtgcaagagatgcacaag-3′ and 5′-aactcctcatcgggcctact-3′); OsKu70 3′ (5′-cggtggtggacttgaaatct-3′ and 5′-gctgacgagtgcctctttct-3′); OsKu80 (5′-agctcaacgtgggttcagac-3′ and 5′-ctccagtgccttttggtgat-3′); OsLig4 (5′-ttggtgaatgcggactacaa-3′ and 5′-aatgctgcacacttgaccac-3′); OsRad51A2 (5′-tggtggacgcttggattgat-3′ and 5′-caggattccagggcgctat-3′); OsBRCA1 (5′-tgcaatctgcaacctctttg-3′ and 5′-agccctgtgcatcttagatttc-3′); OsPARP2A (5′-gcggtacgttctccatgttt-3′ and 5′-tacggtttcatctccgtgct-3′).
Agrobacterium-mediated stable transformation assay
The Agrobacterium-mediated stable transformation assay was performed according to the method established by H. Saika et al. (unpublished). The reporter constructs used (Fig. 2a, p35Smini:Eluc vector) contain the gfbsd2 gene (Ochiai-Fukuda et al., 2006) encoding a fusion gene of gfp and bsd (herbicide Blasticidin S resistance) under the control of the constitutive rice elongation factor 1 promoter (Pef) and emerald luciferase (Eluc) gene under the control of the CaMV35S minimal promoter (m), which can express the Eluc gene only after stable integration into an enhancer-like sequence (H. Saika et al., unpublished). Rice calli from control plants transformed with pANDA empty vector and KD-OsKu70, KD-OsKu80 and KD-OsLig4 plants were induced and grown on N6D medium containing 50 mg l−1 hygromycin for 7 d at 33°C, and were infected with Agrobacterium harboring p35Smini:Eluc vector. After 3 d of co-cultivation with Agrobacterium at 22–25°C under continuous darkness on 2N6-AS medium, calli were washed and cultured on N6D medium containing 50 mg l−1 hygromycin (Wako Pure Chemical Industries, Osaka, Japan), 10 mg l−1 blasticidin S (Wako Pure Chemical Industries) and 25 mg l−1 meropenem (Wako Pure Chemical Industries). Green fluorescent protein (GFP) fluorescence and LUC luminescence were then detected and quantified sequentially in rice callus transformed with the p35Smini:Eluc reporter construct.
Observation of GFP fluorescence
GFP fluorescence images were taken using a Molecular Imager FX (Bio-Rad, USA) with an excitation wavelength of 488 nm and an emission wavelength of 530 nm.
Observation of LUC luminescence
Rice calli were treated with 0.2 mM Beetle Luciferin potassium salt (Promega). LUC luminescence images were taken using a high-resolution photon counting camera (C2400–700 VIM camera, Hamamatsu Photonics, Hamamatsu, Japan) with a 15 min exposure time, and processed using Aquacosmos (Hamamatsu Photonics).
Genomic DNA extraction and Southern blot analysis
Genomic DNA was extracted from rice calli transformed with the LU-UC recombination substrate using Nucleon PhytoPure (GE Healthcare, Little Chalfont, Buckingham, UK) according to the manufacturer's protocol, digested with HindIII and fractionated in a 1.0% agarose gel. Southern blot analysis was performed according to a standard protocol. Specific DNA probes for the codA gene were synthesized with a PCR digoxigenin (DIG) probe synthesis kit (Roche Diagnostics) according to the manufacturer's protocol, using the primers 5′-ggcccatggtgtcgaataacgctttacaaac-3′ and 5′-cccgagctctcaacgtttgtaatcgatggct-3′.
Genotoxic stress treatment
Seedlings of control, KD-Ku70, KD-Ku80 and KD-Lig4 plants were grown on Murashige and Skoog medium for 7 d at 30°C under continuous light in a growth chamber, transferred to water containing 5 μM bleomycin and incubated for 12 h in the growth chamber. The aerial parts of plants were collected and immediately frozen in liquid N2 and stored at −80°C for quantitative RT-PCR analysis and microarray analysis.
Microarray analysis
Total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen), labeled using a Quick-Amp Labeling Kit (Agilent Technologies, Santa Clara, CA, USA) and hybridized to a rice 4 × 44 K custom oligoDNA microarray (Agilent Technologies) according to the manufacturer's instructions. Hybridization microarray slides were scanned with an Agilent Microarray Scanner (Agilent Technologies). The images generated were analyzed using Feature Extraction software (Agilent Technologies), applying standard normalization procedures.
Results
Suppression of NHEJ-related gene expression causes inhibition of Agrobacterium-mediated stable transformation in rice calli
We searched for genes with high sequence homology to AtKu80 and AtLig4 in the rice genome annotation project database (http://rice.plantbiology.msu.edu/), and identified the rice homologs of Ku80 (LOC_Os03g63920.1) and Lig4 (LOC_Os04g51700.1). We generated rice plants transformed with RNAi constructs using the 3′ untranslated region (UTR) of OsKu70, OsKu80 or OsLig4 mRNA. Among a number of T2 lines generated, two independent lines of each transgenic rice plant with a single T-DNA insertion and reduced expression of the targeted gene were selected and named KD-OsKu70 (-1 and -4), KD-OsKu80 (-1 and -4) and KD-OsLig4 (-8 and -14), respectively (Fig.a–c). T0 plants were self-pollinated and T1 seeds were used for further analysis.
Fig 1.
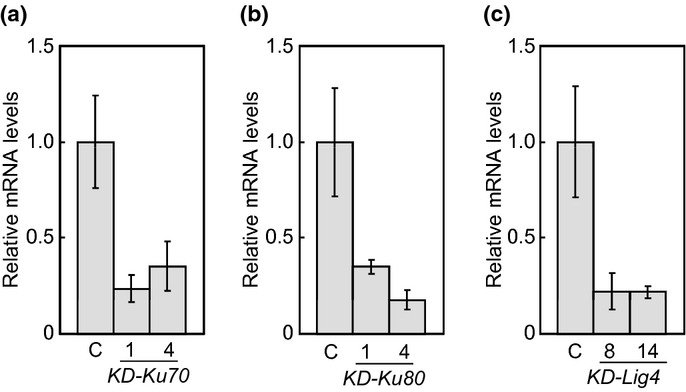
Expression of OsKu70, OsKu80 and OsLig4 in control, KD-Ku70, KD-Ku80 and KD-Lig4 transgenic rice (Oryza sativa) calli. Total RNA was extracted from 1-wk-old control, KD-Ku70, KD-Ku80 and KD-Lig4 transgenic rice calli. A quantitative PCR analysis was carried out to determine the expression levels of OsKu70 (a), OsKu80 (b) and OsLig4 (c). Relative transcript levels were normalized to OsAct1 mRNA. Error bars represent ± SD of three individual experiments.
There were no significant differences between wild-type and KD-OsKu70, KD-OsKu80 and KD-OsLig4 plants in the frequency of callus induction, callus proliferation, callus shape, vegetative growth or fertility (data not shown).
In previous reports of Agrobacterium-mediated transformation in rice calli, two patterns of expression derived from the foreign gene on the T-DNA have been detected: transient expression of the foreign gene from the unintegrated double-stranded T-DNA, and stable expression following integration of the foreign gene into the plant genome (Toki et al., 2006). To distinguish between transient and stable expression more clearly and sensitively, we constructed a new T-DNA vector, p35Smini:Eluc, containing the GFP gene under the control of a constitutive promoter and the emerald LUC gene under the control of a minimal promoter (Fig.a; H. Saika et al., unpublished). This system allows us to evaluate Agrobacterium-mediated transformation events by visualizing stable transgene expression in rice. The GFP signal observed in rice calli at the early transformation stage (3–4 d after Agrobacterium infection) was derived from transient expression of T-DNA that was not integrated into the rice genome. The emerald LUC luminescence seen in rice calli at later transformation stages (7–9 d after Agrobacterium infection) was derived from stable expression of T-DNA that had integrated successfully into the rice genome and was expressed because of a nearby enhancer.
Fig 2.
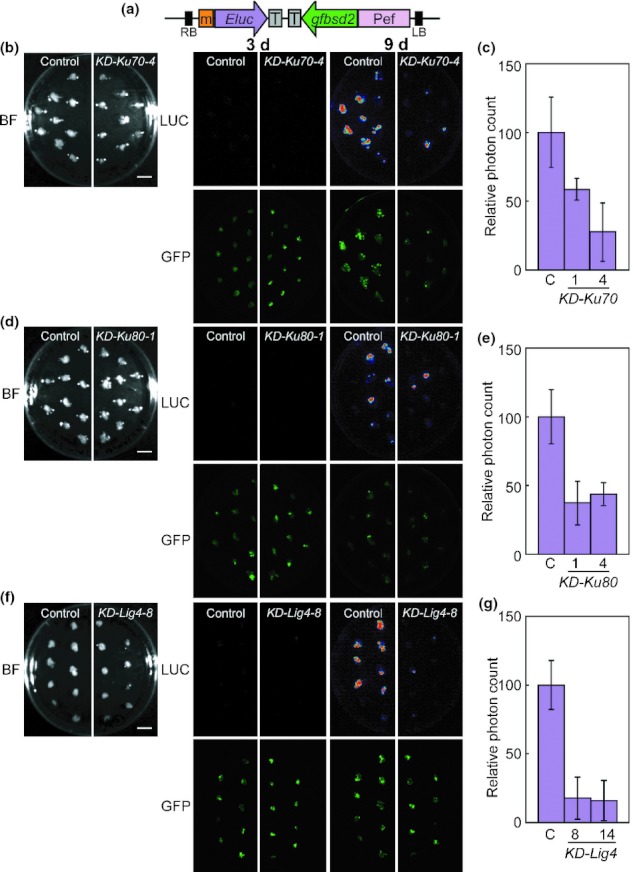
Efficiency of stable transformation in control and KD-OsKu70 (b, c), KD-OsKu80 (d, e) and KD-OsLig4 (f, g) transgenic rice (Oryza sativa) calli. (a) Schematic diagram of the reporter constructs (p35Smini:Eluc) for the Agrobacterium-mediated stable transformation assay. The reporter constructs (p35Smini:Eluc vector) contain the gfbsd2 gene (Ochiai-Fukuda et al., 2006) encoding a fusion gene of green fluorescent protein (GFP) and bsd (herbicide Blasticidin S resistance) under the control of the constitutive rice elongation factor 1 promoter (Pef) and emerald luciferase (LUC) gene under the control of the CaMV35S minimal promoter (m), which can express the LUC gene only after stable integration into an enhancer-like sequence. T, transcription terminator; LB, left border; RB, right border. (b, d, f) Representative bright-field image (BF, left panel), LUC luminescence image (LUC, upper right panel) and GFP fluorescence image (GFP, lower right panel) of control, and KD-OsKu70 (b), KD-OsKu80 (d) and KD-OsLig4 (f) transgenic rice calli. The GFP signal observed on rice calli at an early transformation stage (3 d after Agrobacterium infection) was derived from the transient expression of T-DNA which was not integrated into the rice genome. The LUC luminescence, as represented by the false color scale, on rice calli at the late transformation stage (after 9 d of Agrobacterium infection) was derived from the stable expression of T-DNA which was then successfully integrated into the rice genome. Bars, 1 cm. (c, e, g) Graphical representation of LUC luminescence intensity on control, and KD-OsKu70 (c), KD-OsKu80 (e) and KD-OsLig4 (g) transgenic rice calli at 9 d after infection. Relative LUC luminescence levels were normalized to LUC luminescence levels of control calli (= 100). Agrobacterium-mediated stable transformation assays were performed using three independent batches. Error bars represent ±SD.
To test the efficiency of Agrobacterium-mediated stable transformation, calli from control plants transformed with the empty pANDA vector and from KD-OsKu70, KD-OsKu80 and KD-OsLig4 plants were induced and grown on N6D medium for 7 d. Calli were infected with Agrobacterium harboring the p35S mini:Eluc vector. At 3 d after infection, transient expression of GFP derived from nonintegrated T-DNA in KD-OsKu70, KD-OsKu80 and KD-OsLig4 calli was comparable with that in control calli (Fig.b,d,f), whereas LUC signals, monitoring stable expression of the transgene in KD-OsKu70-1, KD-OsKu70-4, KD-OsKu80-1, KD-OsKu80-4, KD-OsLig4-8 and KD-OsLig4-14 calli, were decreased to 60, 30, 40, 45, 20 and 20% of that in control calli at 9 d after infection (Fig.b–g).
Furthermore, a mutant line (3A-01546; Hong et al., 2010; Fig. 3a), containing a T-DNA insertion in the OsKu70 gene (KO-Ku70), was acquired from the Rice T-DNA Insertion Sequence Database (http://www.postech.ac.kr/life/pfg/risd/) and self-fertilized to obtain a pure homozygous line. However, progeny derived from homozygous T-DNA insertion lines for the OsKu70 gene (KO-Ku70−/−) were not obtained. It has been reported that plants homozygous for the OsKu70 T-DNA insertion mutation display a sterile phenotype and severely retarded vegetative organ growth (Hong et al., 2010). Thus, mature seeds of the heterozygous KO-Ku70 mutant (KO-Ku70+/−) were analyzed for segregation of the mutant allele and transcript levels of the OsKu70 gene (Fig.d). The seeds were then inoculated on N6D medium to induce calli. As 7-d-old calli of KO-Ku70−/− showed severely reduced growth compared with calli of wild-type or knockdown (KD) plants (Fig.b), we could not use these calli for the Agrobacterium-mediated stable transformation assay. Seven-day-old rice calli of the control and KO-Ku70+/− plants were infected with Agrobacterium harboring p35S mini:Eluc vector, and GFP fluorescence and LUC luminescence were analyzed after 3 d of co-culture and elimination of the bacteria. Consistent with results from KD-Ku70, no significant differences in the transient expression of GFP between control and KO-Ku70+/− plants were observed (Fig.c). However, LUC luminescence was suppressed slightly in KO-Ku70+/− calli when compared with control calli at 9 d after infection (Fig.c,e). These results suggest that the suppression of NHEJ-related gene expression causes an inhibition of Agrobacterium-mediated stable transformation, but not of import of T-DNA into the nuclei of infected cells.
Fig 3.
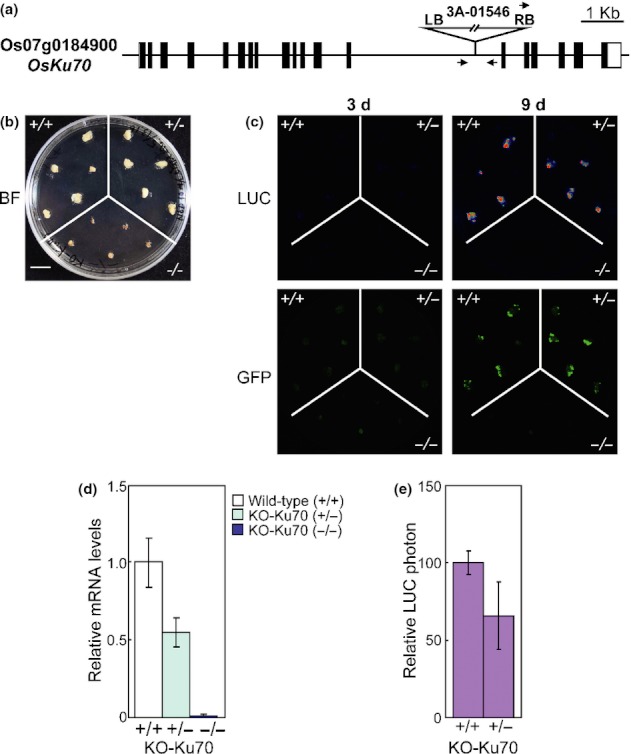
Efficiency of stable transformation in wild-type and KO-OsKu70 mutant rice (Oryza sativa) calli. (a) Schematic diagram of the OsKu70 (Os07g0184900) gene showing the position of the inserted T-DNA (3A-01546). Open and solid bars show the untranslated regions and coding region, respectively. The primer sets used in genotyping are represented by arrows. (b,c) Bright-field image (BF, left panel), luciferase luminescence image (LUC, upper right panel) and green fluorescent protein fluorescence image (GFP, lower right panel) of wild-type (+/+), KO-OsKu70 heterozygous mutant (+/−) and KO-OsKu70 homozygous mutant (−/−) rice calli. Bar, 1 cm. The Green fluorescent protein (GFP) signal observed on rice calli at an early transformation stage (3 d after Agrobacterium infection) was derived from the transient expression of T-DNA which was not integrated into the rice genome. Luciferase luminescence (LUC), as represented by the false color scale, on rice calli at the late transformation stage (9 d after Agrobacterium infection) was derived from the stable expression of T-DNA which was integrated successfully into the rice genome. Bar, 1 cm. (d) Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of OsKu70 transcripts in wild-type (+/+), KO-OsKu70 heterozygous mutant (+/−) and KO-OsKu70 homozygous mutant (−/−)rice calli. Error bars represent ± SD of three individual experiments. (e) Graphical representation of LUC luminescence intensity on wild-type (+/+), KO-OsKu70 heterozygous mutant (+/−) and KO-OsKu70 homozygous mutant (−/−) rice calli at 9 d after infection. Relative LUC luminescence levels were normalized to LUC luminescence levels of control calli (= 100). Agrobacterium-mediated stable transformation assays were performed using three independent batches. Error bars represent ± SD.
Suppression of NHEJ-related gene expression leads to enhanced frequency of HR
To investigate whether suppression of the NHEJ pathway causes an increase in HR frequency, we used an HR assay system that permits recombination events to be visualized as luminescence from a reconstituted recombination substrate locus. This recombination substrate consists of two partially duplicated fragments of the LUC gene (LU-UC) interrupted by a cytosine deaminase (codA) expression cassette and two recognition sites for meganuclease, I-SceI (Fig.a). These fragments of the LUC gene share a duplicated 720-bp region in tandem arrangement. We generated rice plants transformed with recombination substrate (LU-UC), and evaluated the number of integration loci of the transgene. Southern blot analysis with a CodA probe showed that almost all transformants contained a single copy or two copies of the T-DNA (data not shown). Transgenic lines with a single copy (LU-UC lines 8) and two copies (LU-UC lines 9) of the LU-UC recombination substrate were selected (Fig.b), and were transformed with Agrobacterium to introduce pANDA empty vector (as a control) or the KD constructs of OsKu70 or OsLig4 (Fig.c).
Fig 4.
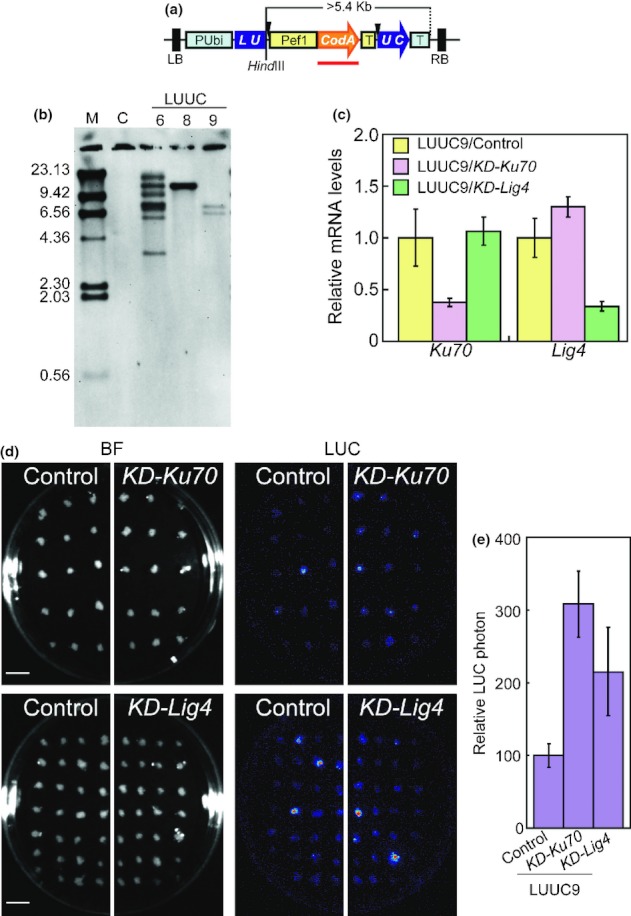
Luciferase (LUC) luminescence of recombination events by spontaneous double-strand breaks (DSBs) in control, KD-OsKu70 and KD-OsLig4 rice (Oryza sativa) calli. (a) Schematic diagram of the LU-UC recombination substrate. This recombination substrate consists of two partially duplicated fragments of the LUC gene interrupted by a cytosine deaminase (codA) expression cassette and two recognition sites for the meganuclease I-SceI (arrowheads). PUbi, maize ubiquitin1 promoter; Pef1, rice elongation factor 1 promoter; T, transcription terminator; LB, left border; RB, right border. Red bar represents DNA probe for Southern blot analysis. (b) Southern blot analysis using a codA probe (shown by the red bar in a) and 1 μg of genomic DNA extracted from wild-type and LU-UC recombination substrate transgenic rice calli and digested with HindIII. (c) Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of OsKu70 (left) and OsLig4 (right) transcripts in LU-UC-9 transgenic lines transformed with pANDA empty vector (as a control), KD-Ku70 or KD-Lig4. Relative transcript levels were normalized to OsAct1 mRNA. Error bars represent ± SD of three individual experiments. (d) Bright-field image (BF) and LUC luminescence image (LUC) of control, KD-Ku70 (upper panel) and KD-Lig4 (lower panel) rice calli. LUC luminescence derived from recombination events of recombination substrate by spontaneous DSB were detected in 4-wk-old transgenic rice calli without induction of I-SceI expression. Bar, 1 cm. (e) Graphical representation of the LUC luminescence intensity on control and KD-Ku70 and KD-Lig4 rice calli. Relative LUC luminescence levels were normalized to LUC luminescence levels of control calli (= 100). Error bars represent ± SD.
Mature seeds of T1 control, KD-Ku70 and KD-Lig4 transgenic (T2 LU-UC transgenic) plants were grown and selected on N6D medium containing hygromycin and bispyribac without 5-fluorocytosine, which is converted to toxic 5-fluorouracil by codA. The negative selection by codA results in the preferential growth of calli in which the codA expression cassette has been removed from the recombination substrate locus. After 4 wk of selection, LUC luminescence derived from reconstituted recombination substrate was analyzed on transgenic calli. KD-Ku70 and KD-Lig4 transgenic calli containing two copies of the recombination substrate showed a two- to three-fold increase in the frequency of HR compared with control calli (Fig.d,e). However, LUC luminescence was not detected on transgenic calli containing a single copy of LU-UC (data not shown).
Next, we investigated the effect of suppression of NHEJ on the frequency of DSB-inducible HR using transient I-SceI expression. To introduce DSBs at the two I-SceI sites flanking the codA expression cassette of LU-UC recombination substrates by inducing transient I-SceI expression, 4-wk-old control, KD-OsKu70 and KD-OsLig4 transgenic calli containing LU-UC recombination substrates were infected with Agrobacterium harboring the I-SceI expression vector (Supporting Information Fig. S1a). LUC luminescence derived from DSB-inducible reconstituted recombination substrate was analyzed 5 d after infection, at which point T-DNA is largely unintegrated into the rice genome (Toki et al., 2006). We found no difference in LUC luminescence between control, KD-OsKu70 and KD-OsLig4 calli (Fig. S1b).
Effect of suppression of NHEJ-related gene expression on induction of DSB-inducible genes
The enhancement of HR frequency observed in KD-OsKu70 and KD-OsLig4 calli could be explained by competition for DNA repair between HR and NHEJ at the sites of DSBs. However, it could also be explained by the induction of HR-related gene expression. We analyzed the effect of suppression of NHEJ-related gene expression on the transcript levels of OsRad51A2, which plays a crucial role in HR repair of DSBs (Rajanikant et al., 2008), and OsBRCA1, which colocalizes with Rad51 at DSB sites and activates HR via Rad51 (Lafarge & Montané, 2003). Seven-day-old control, KD-OsKu70, KD-OsKu80 and KD-OsLig4 rice plants were either treated with 5 μM bleomycin, which generates DSBs and DNA single-strand breaks, or left untreated for 12 h. No significant differences were observed in the levels of OsRad51A2 and OsBRCA1 transcripts between controls and transgenic plants without bleomycin treatment (Fig.a,b), whereas, on treatment with 5 μM bleomycin for 6 and 12 h, transcripts of both genes increased markedly in transgenic relative to control plants (Fig.a,b). Suppression of OsKu70 expression showed the highest transcript levels of HR-related genes at 12 h after treatment with bleomycin.
Fig 5.
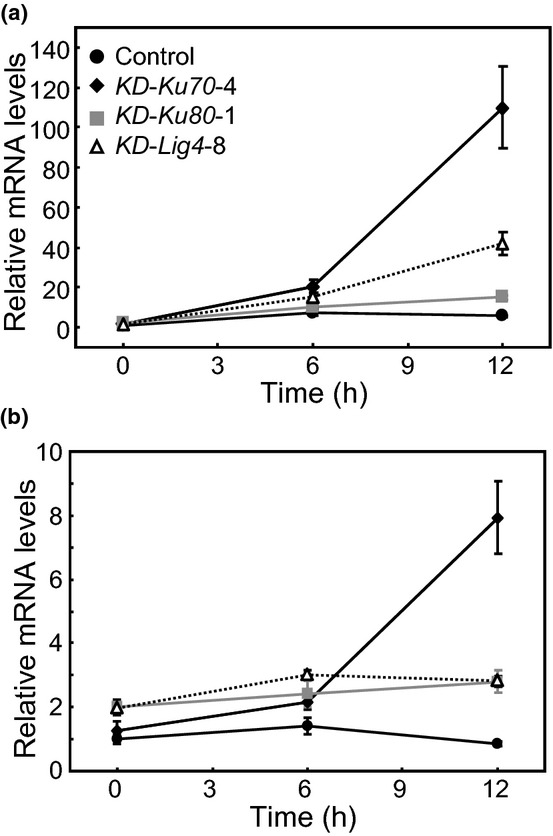
Effect of suppression of the nonhomologous end joining (NHEJ) pathway on the expression of genes involved in homologous recombination (HR). Seven-day-old seedlings of rice (Oryza sativa) wild-type, KD-Ku70, KD-Ku80 and KD-Lig4 seedlings were treated with water containing 5 μM bleomycin and incubated for 12 h in the growth chamber. Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of OsRad51A2 (a) and OsBRCA1 (b) transcripts in control, KD-Ku70, KD-Ku80 and KD-Lig4. Relative transcript levels were normalized to OsAct1 mRNA. Error bars represent ± SD of three individual experiments.
We further analyzed the transcriptional responses to bleomycin treatment of OsKu70/80 and OsLig4 suppression plants using an Agilent rice 44K microarray. The expression levels of HR-related genes are shown in Table 1. Transcript levels of not only OsBRCA1 and OsRad51A2, but also OsDMC1A and OsDMC1B, the meiotic Rad51 paralog (Metkar et al., 2004), were induced in KD-OsKu70 and KD-OsLig4 rice plants relative to control plants under treatment with bleomycin (Table 1). In addition, under DNA-damaging conditions, the expression of OsRad54, one of the key proteins necessary for HR and DNA repair (Osakabe et al., 2006), and OsTOP3 (putative topoisomerase 3) (Singh et al., 2010) was increased in KD-OsKu70 plants. Putative OsBARD1 – a BRCA1-associated RING domain protein (Singh et al., 2010) – was up-regulated in KD-OsLig4 plants, but not in control plants, whereas no differences in the expression of HR-related genes other than OsBRCA1 were noted between KD-OsKu80 and control plants.
Table 1.
Expression data of homologous recombination (HR)-related genes in control, KD-Ku70, KD-Ku80 and KD-Lig4 rice (Oryza sativa) plants with (12 h) or without (0 h) 5 μM bleomycin treatment
| Accessions | Description | Control (12 h) | KD-Ku70 (0 h) | KD-Ku70 (12 h) | KD-Ku80 (0 h) | KD-Ku80 (12 h) | KD-Lig4 (0 h) | KD-Lig4 (12 h) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log10 ratio | P | Log10 ratio | P | Log10 ratio | P | Log10 ratio | P | Log10 ratio | P | Log10 ratio | P | Log10 ratio | P | ||
| Os05g0512000 | OsBRCA1 | −0.44 | 0.19 | −0.02 | 0.92 | 0.68 | 0.00 | 0.01 | 0.93 | 0.32 | 0.03 | 0.39 | 0.00 | 0.52 | 0.00 |
| Os01g0164900 | OsBRCA2 | 0.00 | 1.00 | 0.00 | 1.00 | 0.41 | 0.21 | −0.13 | 0.83 | 0.00 | 1.00 | 0.15 | 0.76 | 0.00 | 1.00 |
| Os04g0635900 | MRE11 | −0.06 | 0.30 | 0.01 | 0.91 | −0.10 | 0.12 | 0.00 | 0.98 | −0.12 | 0.05 | −0.04 | 0.50 | −0.08 | 0.20 |
| Os01g0948100 | OsMUS81 | −0.13 | 0.04 | −0.15 | 0.02 | −0.10 | 0.12 | −0.14 | 0.03 | −0.13 | 0.04 | 0.00 | 0.96 | −0.04 | 0.56 |
| Os02g0497500 | Rad50 | −0.02 | 0.72 | 0.03 | 0.66 | 0.09 | 0.17 | −0.07 | 0.27 | −0.06 | 0.36 | −0.04 | 0.55 | 0.04 | 0.58 |
| Os11g0615800 | OsRad51A1 | −0.12 | 0.07 | 0.11 | 0.08 | 0.26 | 0.00 | 0.25 | 0.00 | 0.17 | 0.01 | −0.05 | 0.41 | −0.20 | 0.00 |
| Os12g0497300 | OsRad51A2 | 0.84 | 0.00 | 0.00 | 1.00 | 1.86 | 0.00 | −0.19 | 0.01 | 0.57 | 0.00 | 0.50 | 0.10 | 1.45 | 0.00 |
| Os05g0121700 | OsRad51B | −0.43 | 0.00 | 0.05 | 0.73 | −0.01 | 0.90 | −0.10 | 0.39 | −0.23 | 0.09 | −0.09 | 0.51 | 0.09 | 0.48 |
| Os01g0578000 | OsRad51C | −0.09 | 0.17 | −0.04 | 0.51 | −0.07 | 0.26 | 0.01 | 0.90 | −0.02 | 0.70 | −0.07 | 0.28 | −0.09 | 0.17 |
| Os09g0104200 | OsRad51D | −0.02 | 0.72 | −0.09 | 0.15 | −0.07 | 0.25 | 0.03 | 0.67 | −0.02 | 0.72 | −0.02 | 0.79 | −0.04 | 0.56 |
| Os02g0762800 | OsRad54 | 0.00 | 1.00 | −0.26 | 0.65 | 0.72 | 0.00 | 0.00 | 1.00 | 0.05 | 0.93 | −0.05 | 0.93 | 0.26 | 0.04 |
| Os05g0392400 | OsRad54-like | −0.17 | 0.01 | 0.02 | 0.77 | −0.10 | 0.11 | −0.16 | 0.02 | −0.18 | 0.01 | −0.03 | 0.63 | 0.01 | 0.85 |
| Os10g0487300 | OsNBS1 | −0.16 | 0.01 | −0.09 | 0.16 | −0.21 | 0.00 | 0.01 | 0.81 | −0.12 | 0.06 | −0.06 | 0.36 | −0.15 | 0.02 |
| Os04g0433800 | OsRecQl4 | −0.36 | 0.00 | 0.04 | 0.59 | 0.20 | 0.00 | 0.20 | 0.00 | 0.01 | 0.94 | 0.39 | 0.00 | 0.16 | 0.02 |
| Os01g0273000 | DSS1/SEM1 | 0.05 | 0.46 | 0.04 | 0.55 | 0.06 | 0.33 | 0.20 | 0.00 | 0.15 | 0.02 | 0.14 | 0.02 | 0.11 | 0.09 |
| Os02g0562100 | XRCC3 | −0.09 | 0.31 | 0.07 | 0.50 | 0.17 | 0.03 | 0.17 | 0.05 | 0.12 | 0.15 | 0.25 | 0.00 | 0.18 | 0.04 |
| Os03g0165000 | TOP3 | −0.40 | 0.01 | 0.10 | 0.49 | 0.38 | 0.00 | 0.14 | 0.19 | 0.06 | 0.61 | 0.36 | 0.00 | 0.25 | 0.03 |
| Os09g0500600 | TOP3 | −0.01 | 0.83 | 0.01 | 0.93 | −0.04 | 0.56 | −0.08 | 0.22 | −0.06 | 0.38 | −0.09 | 0.16 | −0.09 | 0.21 |
| Os05g0509700 | SSB | −0.11 | 0.07 | −0.08 | 0.21 | 0.02 | 0.78 | 0.07 | 0.23 | 0.02 | 0.75 | 0.15 | 0.02 | 0.06 | 0.37 |
| Os01g0642900 | SSB | −0.06 | 0.35 | 0.05 | 0.40 | 0.20 | 0.00 | 0.10 | 0.12 | 0.00 | 0.95 | 0.14 | 0.03 | 0.10 | 0.09 |
| Os04g0648700 | EME1 | 0.16 | 0.01 | 0.16 | 0.01 | 0.23 | 0.00 | 0.04 | 0.51 | 0.08 | 0.19 | 0.09 | 0.17 | 0.09 | 0.15 |
| Os11g0146800 | OsDMC1A | 0.26 | 0.00 | 0.00 | 0.98 | 0.72 | 0.00 | −0.13 | 0.04 | 0.20 | 0.00 | −0.12 | 0.07 | 0.66 | 0.00 |
| Os12g0143800 | OsDMC1B | 0.50 | 0.00 | 0.21 | 0.20 | 1.32 | 0.00 | 0.05 | 0.71 | 0.40 | 0.00 | 0.06 | 0.62 | 1.64 | 0.00 |
| Os09g0280600 | OsMND1 | 0.09 | 0.14 | 0.11 | 0.08 | 0.17 | 0.01 | 0.20 | 0.00 | 0.18 | 0.00 | −0.03 | 0.64 | −0.05 | 0.46 |
| Os01g0901200 | RecA | −0.17 | 0.01 | −0.08 | 0.22 | 0.08 | 0.22 | 0.01 | 0.92 | −0.06 | 0.35 | −0.02 | 0.77 | −0.07 | 0.26 |
| Os03g0639700 | RecA | −0.01 | 0.86 | −0.02 | 0.80 | −0.07 | 0.28 | 0.01 | 0.82 | −0.03 | 0.63 | 0.00 | 0.97 | 0.01 | 0.83 |
| Os11g0302700 | RecA | −0.06 | 0.36 | −0.10 | 0.14 | −0.13 | 0.05 | −0.07 | 0.26 | −0.11 | 0.09 | −0.19 | 0.00 | −0.19 | 0.01 |
| Os02g0710800 | RecG | −0.12 | 0.06 | −0.06 | 0.32 | −0.12 | 0.06 | −0.04 | 0.54 | −0.13 | 0.04 | −0.02 | 0.75 | −0.05 | 0.41 |
| Os05g0486600 | BARD1 | 0.16 | 0.03 | 0.08 | 0.31 | 0.03 | 0.71 | −0.02 | 0.77 | −0.21 | 0.01 | 0.32 | 0.00 | 0.39 | 0.00 |
| Os04g0512400 | BARD1 | −0.49 | 0.00 | 0.00 | 0.95 | 0.23 | 0.00 | 0.23 | 0.00 | 0.20 | 0.00 | 0.47 | 0.00 | 0.37 | 0.00 |
The log10 ratios of HR-related genes in control, KD-Ku70, KD-Ku80 and KD-Lig4 plants with (12 h) or without (0 h) 5 μM bleomycin treatment compared with those in control plants without bleomycin treatment are presented. Bold characters represent genes whose expression is significantly induced (log10 ratio > 0.3 and P < 0.01).
Discussion
Here, we have demonstrated that the suppression of NHEJ-related gene expression causes decreased Agrobacterium-mediated stable transformation of the rice genome, especially in KD-OsLig4 (Figs 2, 3). Lig4 proteins interact with XRCC4 and catalyze the ligation step of DSBs in the NHEJ pathway (Mladenov & Iliakis, 2011; Symington & Gautier, 2011). The most generally accepted mechanisms of T-DNA integration into the plant genome are the strand-invasion model and the DSB repair model depending on the NHEJ pathway. The results presented here suggest that NHEJ is the major pathway of T-DNA integration into the rice genome, at least when rice scutellum-derived calli are used for transformation, and that the OsLig4 protein may play an important role in this process.
The decreased stable transformation observed in KD-OsKu70 and KD-OsKu80 plants and OsKu70+/− plants is in accordance with a previous report (Li et al., 2005), in which a decreased frequency of T-DNA integration was observed in atku80 knockout mutants of Arabidopsis by root tumorigenesis assay. In addition, mRNA levels of OsKu70 and OsRad51A2, which are well-known genes associated with DNA damage, are higher in calli than in leaves, roots and anthers (Yang et al., 2010). Yadav et al. (2009) have revealed an enrichment of DNA repair gene expression in Arabidopsis shoot apical meristem stem cells. Furthermore, consistent with previous observations (Hong et al., 2010), we found that knockout of the OsKu70 gene resulted in a growth defect of calli (Fig.b), but had no effect on the growth of intact plants. Summarizing these results, DSBs arise constantly in rice callus as a result of active cell division. These DSBs are repaired mainly by the NHEJ pathway. DSBs are toxic for rice callus if not repaired correctly by the NHEJ pathway. T-DNA has been shown in several cases to be inserted into DSB sites (Salomon & Puchta, 1998). Thus, T-DNA is most likely to be integrated into the rice genome via NHEJ using endogenously induced DSBs.
There was no correlation between the transcript levels of Ku80 and LUC luminescence derived from T-DNA stable transformation in KD-OsKu80 rice calli (Figs 1b, 2d,e). It has been shown that the Ku70/80 protein functions as a heterodimer to repair DSBs, with the two constituents stabilizing each other (Smider et al., 1994). Accordingly, these results may be attributed to a decrease in protein levels of both OsKu80 and OsKu70 caused by the down-regulation of the expression of OsKu80 mRNA in KD-Ku80 rice calli.
We also detected decreased Agrobacterium-mediated stable transformation in KD-OsLig4 rice callus. However, it has also been reported that the absence of AtLig4 does not affect the frequency of T-DNA integration in an in vitro root tumorigenesis assay (van Attikum et al., 2003). Although we cannot rule out the possibility that AtLig4 and OsLig4 act differently in each species in the process of Agrobacterium-mediated stable transformation, differences in the assay system itself could lead to different results. In the root tumorigenesis assay, the efficiency of T-DNA integration was evaluated by tumor formation in roots that were grown via cell division and cell expansion for 4–5 wk after Agrobacterium infection; many environmental factors are involved in this process. Our Agrobacterium-mediated stable transformation assay system allows transient and stable expression to be visualized using a sensitive reporter gene, and thus stable transformation events can be detected rapidly (within 7–10 d after Agrobacterium infection).
It has been shown recently in Arabidopsis that the epidermal cells of the root tip are enlarged and show increased nuclear DNA content in response to DSBs derived from treatment with the genotoxic agent zeocin, indicating that DSBs induce the onset of the endocycle (Adachi et al., 2011). In other words, DNA-damaged cells in Arabidopsis could select the repair of DNA damage, the induction of cell death or, alternatively, the induction of the endocycle to avoid cell death. We speculated that a fraction of cells with integrated T-DNA could enter the endocycle as a result of DSB signals, as DSBs have been reported to be major integration sites of T-DNA (Salomon & Puchta, 1998). In contrast with Arabidopsis, the endocycle has never been reported in rice except in the endosperm. We have recently confirmed that this is true, even after genotoxic stress treatment inducing DSBs (Endo et al., 2012). According to this concept, the frequency of T-DNA integration could be underestimated in Arabidopsis, as AtKu70/80 or AtLig4 mutants accumulate DSBs as a result of insufficient NHEJ repair.
Decreased T-DNA integration has also been reported in atku80 and atlig4 mutants when analyzed using an in planta floral dip transformation assay (Friesner & Britt, 2003), whereas no differences were detected between atku80 mutants and wild-type plants (Gallego et al., 2003). Furthermore, Ziemienowicz et al. (2008) have reported that Arabidopsis type I DNA ligase (Lig1) can function as a ligase for T-DNA in vitro, suggesting that T-DNA integration may be catalyzed by Lig1 via the strand-invasion model in the case of in planta floral dip transformation. The presence of a micro-homology sequence in the T-DNA integration site identified via in planta floral dip transformation (Brunaud et al., 2002) also supports this idea. Micro-homology-dependent integration of T-DNA into the rice genome was also reported when scutellum-derived calli were used for transformation (Sha et al., 2004). To better understand the mechanism of T-DNA integration in plants, the effect of the suppression of Arabidopsis or rice Lig1 on the efficiency of T-DNA integration via in planta floral dip or callus transformation should be investigated.
We observed a sterile phenotype and delayed callus proliferation in OsKu70 mutants. By contrast, Arabidopsis ku70, ku80 and lig4 mutant plants are viable and exhibit a fertile phenotype (Bundock et al., 2002; van Attikum et al., 2003; Friesner & Britt, 2003). In addition, no significant difference in callus formation and callus regeneration from roots was observed between Arabidopsis ku80 mutant and wild-type plants (Li et al., 2005). These differences suggest that the defect in the NHEJ pathway that depends on Ku70, Ku80 and Lig4 is more severely affected in rice than in Arabidopsis. Furthermore, it has been reported that cells with DNA damage enter into the endocycle to prevent their proliferation and cell death in Arabidopsis (Adachi et al., 2011). Consequently, mutations of Arabidopsis ku70, ku80 and lig4 may have little direct effect on viability and productivity.
Of all rice tissues, HR events occur with highest frequency in callus tissue under normal conditions (Yang et al., 2010). Consistent with this observation, we detected LUC luminescence derived from reconstituted recombination substrate in both control and transgenic calli; HR activity in KD-Ku70 and KD-Lig4 transgenic calli was two- to three-fold higher than in control calli (Fig.d,e). Thus, the frequency of HR seemed to be enhanced by suppression of the NHEJ pathway in rice plants, whereas there was no difference in the frequency of DSB-inducible HR between control and transgenic calli (Fig. S1b). In this study, to eliminate the effect of the efficiency of T-DNA integration on I-SceI expression levels, we analyzed LUC luminescence by monitoring DSB-inducible HR at 5 d after infection with Agrobacterium harboring an I-SceI expression vector. It was assumed that LUC luminescence derived from DSB-inducible HR might not be detected correctly because I-SceI expression levels derived from free T-DNA may be very low. Alternatively, it is possible that no differences were observed in the frequency of DSB-inducible HR between control and transgenic calli because DSBs induced by I-SceI expression at the two I-SceI sites flanking the codA expression cassette of LU-UC recombination substrates were repaired via the single-strand annealing (SSA) pathway. It has been reported that DSBs flanking directly repeated sequences are repaired mainly by the SSA pathway using homologous sequences (Siebert & Puchta, 2002). In future, evaluation of the frequency of DSB-inducible HR should be performed with KD-Ku70/80 or KD-Lig4 harboring a recombination substrate and a stably integrated I-SceI expression cassette driven by a chemically inducible system. To assess HR frequency in KD-Ku70/80 or KD-Lig4 in detail, the establishment of an HR assay system in rice is also required to detect the reconstruction of the recombination substrate only via HR, for example, intrachromosomal HR reporter with indirect repeats (Gherbi et al., 2001) or interchromosomal HR reporter (Molinier et al., 2004).
A large number of genes, including HR-related genes, are up-regulated in response to ionizing radiation (Nagata et al., 2005; Culligan et al., 2006; Ricaud et al., 2007). In addition, previous studies of Arabidopsis ku80 mutants have shown that several genes, including DSB repair genes, display transcriptional induction (West et al., 2004). Consistent with these findings, we demonstrate here that the transcript levels of OsRad51A2 and OsBRCA1 are higher in KD-Ku70, KD-Ku80 and KD-Lig4 plants than in control plants under conditions of DNA damage (Fig.). Considering the transcriptional induction of HR-related genes in KD-Ku70, KD-Ku80 and KD-Lig4, suppression of the NHEJ pathway causes the accumulation of DNA damage under stressful conditions, such as active cell division and treatment with DNA-damaging agents, and the HR pathway is then activated and DNA damage repaired as a result of the induction of HR-related gene expression. Recently, it has been shown that Ku proteins interact with a number of proteins and function not only in the NHEJ pathway for DSB repair, but also in transcriptional regulation and signal transduction in mammals (Bertinato et al., 2003; Brenkman et al., 2010; Zhang et al., 2011; Fell & Schild-Poulter, 2012). Wang et al. (2010) have identified Arabidopsis ovate family protein 1 (AtOFP1) as a novel AtKu70-interacting protein using yeast two-hybrid screening and pull-down assay. In this report, suppression of OsKu70 expression has a major effect on HR-related genes, such as OsRad51A2 and BRCA1 (Fig., Table 1), therefore suggesting that OsKu70 may also be involved in the signaling pathway of the DSB response.
In conclusion, our results clearly show the involvement of the NHEJ pathway in the Agrobacterium-mediated stable transformation in rice and the presence of the competitive and complementary relationship between the NHEJ and HR pathways for DSB repair in rice. Although it takes a relatively long time to set up an assay system in rice relative to Arabidopsis because of its longer life-cycle, a well-established evaluation system in rice could be applied to other important crops in Gramineae.
In contrast with random T-DNA integration, sequence-specific integration of T-DNA into the endogenous homologous sequence – gene targeting (GT) – occurs through the HR pathway (Smithies et al., 1985; Paszkowski et al., 1988; Müller, 1999; Iida & Terada, 2004). As a result, it has been reported that GT events of endogenous genes by HR occur on the order of 0.01–0.1% compared with random integration in higher plants (Paszkowski et al., 1988; Offringa et al., 1990; Puchta et al., 1996). Our data indicate that suppression of the NHEJ pathway is expected to cause an increase in the occurrence of sequence-specific integration by HR and inhibition of random integration by NHEJ, resulting in a synergistic effect that will improve GT experiments. In fact, it has been reported that deletion of the Ku or Lig4 gene can increase the frequency of GT in several organisms, such as fungi (de Boer et al., 2010; Ushimaru et al., 2010; Nakazawa et al., 2011), bacteria (Zhang et al., 2012) , birds (Iiizumi et al., 2008), mammals (Bertolini et al., 2009; Fattah et al., 2008; Iiizumi et al., 2008) and plants (Tanaka et al., 2010; Jia et al., 2012). We are progressing towards the establishment of a high-efficiency GT procedure using KD rice plants targeting NHEJ-related genes.
Acknowledgments
We thank Dr Nagamura and R. Motoyama (National Institute of Agrobiological Sciences) for data analysis and technical help with microarray analysis, Dr Kimura (RIKEN) for providing the gfbsd2 gene, Dr Rothnie for English editing, Dr An (Postech) for providing rice T-DNA insertion mutant lines and K. Amagai, A. Nagashii and F. Suzuki for general experimental technical support. This research was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation GMC-0001) and KAKENHI (23658012 and 23310142).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Luciferase (LUC) luminescence of DNA double-strand break (DSB)-inducible homologous recombination (HR) events by transient I-SceI expression in control, KD-OsKu70 and KD-OsLig4 rice calli.
References
- Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2011;108:10004–10009. doi: 10.1073/pnas.1103584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C, Halbrook J, Nickoloff JA. Interactive competition between homologous recombination and non-homologous end joining. Molecular Cancer Research. 2003;1:913–920. [PubMed] [Google Scholar]
- van Attikum H, Bundock P, Overmeer RM, Lee LY, Gelvin SB, Hooykaas PJ. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Research. 2003;31:4247–4255. doi: 10.1093/nar/gkg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Kupiec M. New insights into the mechanism of homologous recombination in yeast. Mutation Research. 2004;566:231–248. doi: 10.1016/j.mrrev.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Bertinato J, Tomlinson JJ, Schild-Poulter C, Haché RJ. Evidence implicating Ku antigen as a structural factor in RNA polymerase II-mediated transcription. Gene. 2003;302:53–64. doi: 10.1016/s0378111902010892. [DOI] [PubMed] [Google Scholar]
- Bertolini LR, Bertolini M, Maga EA, Madden KR, Murray JD. Increased gene targeting in Ku70 and Xrcc4 transiently deficient human somatic cells. Molecular Biotechnology. 2009;41:106–114. doi: 10.1007/s12033-008-9098-8. [DOI] [PubMed] [Google Scholar]
- de Boer P, Bastiaans J, Touw H, Kerkman R, Bronkhof J, van den Berg M, Offringa R. Highly efficient gene targeting in Penicillium chrysogenum using the bi-partite approach in deltalig4 or deltaku70 mutants. Fungal Genetics and Biology. 2010;47:839–846. doi: 10.1016/j.fgb.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Brenkman AB, van den Broek NJ, de Keizer PL, van Gent DC, Burgering BM. The DNA damage repair protein Ku70 interacts with FOXO4 to coordinate a conserved cellular stress response. FASEB Journal. 2010;24:4271–4280. doi: 10.1096/fj.10-158717. [DOI] [PubMed] [Google Scholar]
- Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, et al. T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Reports. 2002;3:1152–1157. doi: 10.1093/embo-reports/kvf237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, van Attikum H, Hooykaas P. Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Research. 2002;30:3395–3400. doi: 10.1093/nar/gkf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnel C, Allain E, Gallego ME, White CI. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair. 2011;10:611–619. doi: 10.1016/j.dnarep.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Charbonnel C, Gallego ME, White CI. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant Journal. 2010;64:280–290. doi: 10.1111/j.1365-313X.2010.04331.x. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Barboule N, Frit P, Gomez D, Bombarde O, Couderc B, Ren GS, Salles B, Calsou P. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Research. 2011;39:9605–9619. doi: 10.1093/nar/gkr656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant Journal. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- Edlinger B, Schlögelhofer P. Have a break: determinants of meiotic DNA double strand break (DSB) formation and processing in plants. Journal of Experimental Botany. 2011;62:1545–1563. doi: 10.1093/jxb/erq421. [DOI] [PubMed] [Google Scholar]
- Endo M, Nakayama S, Umeda-Hara C, Ohtsuki N, Saika H, Umeda M, Toki S. CDKB2 is involved in mitosis and DNA damage response in rice. Plant Journal. 2012;69:967–977. doi: 10.1111/j.1365-313X.2011.04847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proceedings of the National Academy of Sciences, USA. 2008;105:8703–8708. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell VL, Schild-Poulter C. Ku regulates signaling to DNA damage response pathways through the Ku70 von Willebrand A domain. Molecular and Cellular Biology. 2012;32:76–87. doi: 10.1128/MCB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner J, Britt AB. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant Journal. 2003;34:427–440. doi: 10.1046/j.1365-313x.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- Gallego ME, Bleuyard JY, Daoudal-Cotterell S, Jallut N, White CI. Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant Journal. 2003;35:557–565. doi: 10.1046/j.1365-313x.2003.01827.x. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Plant proteins involved in Agrobacterium-mediated genetic transformation. Annual Review of Phytopathology. 2010;48:45–68. doi: 10.1146/annurev-phyto-080508-081852. [DOI] [PubMed] [Google Scholar]
- Gherbi H, Gallego ME, Jalut N, Lucht JM, Hohn B, White CI. Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Reports. 2001;2:287–291. doi: 10.1093/embo-reports/kve069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliqa P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Molecular Biology. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual Review of Genetics. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JP, Byun MY, An K, Yang SJ, An G, Kim WT. OsKu70 is associated with developmental growth and genome stability in rice. Plant Physiology. 2010;152:374–387. doi: 10.1104/pp.109.150391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene-transfer to plants. Transgenic Research. 1993;2:208–218. [Google Scholar]
- Iida S, Terada R. A tale of two integrations, transgene and T-DNA: gene targeting by homologous recombination in rice. Current Opinion in Biotechnology. 2004;15:132–138. doi: 10.1016/j.copbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Iiizumi S, Kurosawa A, So S, Ishii Y, Chikaraishi Y, Ishii A, Koyama H, Adachi N. Impact of non-homologous end-joining deficiency on random and targeted DNA integration: implications for gene targeting. Nucleic Acids Research. 2008;36:6333–6342. doi: 10.1093/nar/gkn649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Bundock P, Hooykaas P, de Pater S. Agrobacterium tumefaciens T-DNA integration and gene targeting in Arabidopsis thaliana non-homologous end-joining mutants. Journal of Botany. 2012;2012:ID 989272. [Google Scholar]
- Lafarge S, Montané MH. Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Research. 2003;31:1148–1155. doi: 10.1093/nar/gkg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Vaidya M, White C, Vainstein A, Citovsky V, Tzfira T. Involvement of KU80 in T-DNA integration in plant cells. Proceedings of the National Academy of Sciences, USA. 2005;102:19231–19236. doi: 10.1073/pnas.0506437103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M. Chromosomal double-strand break repair in Ku80-deficient cells. Proceedings of the National Academy of Sciences, USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magori S, Citovsky V. Epigenetic control of Agrobacterium T-DNA integration. Biochimica et Biophysica Acta. 2011;1809:388–394. doi: 10.1016/j.bbagrm.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan SD, Ireton GC, Stoddard BL, Black ME. Alanine-scanning mutagenesis reveals a cytosine deaminase mutant with altered substrate preference. Biochemistry. 2004;43:8957–8964. doi: 10.1021/bi049720z. [DOI] [PubMed] [Google Scholar]
- Metkar SS, Sainis JK, Mahajan SK. Cloning and characterization of the DMC1 genes in Oryza sativa. Current Science. 2004;87:353–357. [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiology. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutation Research. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Molinier J, Ries G, Bonhoeffer S, Hohn B. Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell. 2004;16:342–352. doi: 10.1105/tpc.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U. Ten years of gene targeting: targeted mouse mutants, from vector design to phenotype analysis. Mechanisms of Development. 1999;82:3–21. doi: 10.1016/s0925-4773(99)00021-0. [DOI] [PubMed] [Google Scholar]
- Nagata T, Yamada H, Du Z, Todoriki S, Kikuchi S. Microarray analysis of genes that respond to gamma-irradiation in Arabidopsis. Journal of Agriculture and Food Chemistry. 2005;53:1022–1103. doi: 10.1021/jf0486895. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Ando Y, Kitaaki K, Nakahori K, Kamada T. Efficient gene targeting in ΔCc.ku70 or ΔCc.lig4 mutants of the agaricomycete Coprinopsis cinerea. Fungal Genetics and Biology. 2011;48:939–946. doi: 10.1016/j.fgb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proceedings of the National Academy of Sciences, USA. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai-Fukuda T, Takahashi-Ando N, Ohsato S, Igawa T, Kadokura K, Hamamoto H, Nakasako M, Kudo T, Shibata T, Yamaguchi I, et al. A fluorescent antibiotic resistance marker for rapid production of transgenic rice plants. Journal of Biotechnology. 2006;122:521–527. doi: 10.1016/j.jbiotec.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Offringa R, de Groot MJ, Haagsman HJ, Does MP, van den Elzen PJ, Hooykaas P. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO Journal. 1990;9:3077–3084. doi: 10.1002/j.1460-2075.1990.tb07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yoshioka T, Osakabe Y, Todoriki S, Ichikawa H, Hohn B, Toki S. Isolation and characterization of the RAD54 gene from Arabidopsis thaliana. Plant Journal. 2006;48:827–842. doi: 10.1111/j.1365-313X.2006.02927.x. [DOI] [PubMed] [Google Scholar]
- Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J, Baur M, Bogucki A, Potrykus I. Gene targeting in plants. EMBO Journal. 1988;7:4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes & Development. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO Journal. 2010;29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proceedings of the National Academy of Sciences, USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanikant C, Melzer M, Rao BJ, Sainis JK. Homologous recombination properties of OsRad51, a recombinase from rice. Plant Molecular Biology. 2008;68:479–491. doi: 10.1007/s11103-008-9385-6. [DOI] [PubMed] [Google Scholar]
- Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montané MH. ATM-mediated transcriptional and developmental responses to gamma-rays in Arabidopsis. PLoS ONE. 2007;2:e430. doi: 10.1371/journal.pone.0000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO Journal. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Y, Li S, Pei Z, Luo L, Tian Y, He C. Generation and flanking sequence analysis of a rice T-DNA tagged population. Theoretical and Applied Genetics. 2004;108:306–314. doi: 10.1007/s00122-003-1423-9. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Kaku K, Kawai K, Miyazawa T, Tanaka Y. Molecular characterization of acetolactate synthase in resistant weeds and crops. In: Clark JM, Ohkawa H, editors. ACS Symposium Series 899: Environmental Fate and Safety Management of Agrochemicals. Washington, DC, USA: American Chemical Society; 2005. pp. 255–271. [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Research. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Siebert R, Puchta H. Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell. 2002;14:1121–1131. doi: 10.1105/tpc.001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Roy S, Choudhury SR, Sengupta DN. DNA repair and recombination in higher plants: insights from comparative genomics of Arabidopsis and rice. BMC Genomics. 2010;11:443. doi: 10.1186/1471-2164-11-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smider V, Rathmell WK, Lieber MR, Chu G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annual Review of Genetics. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H. Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant Journal. 2002;29:771–781. doi: 10.1046/j.1365-313x.2002.01258.x. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Ishii C, Hatakeyama S, Inoue H. High efficient gene targeting on the AGAMOUS gene in an ArabidopsisAtLIG4 mutant. Biochemical and Biophysical Research Communications. 2010;396:289–293. doi: 10.1016/j.bbrc.2010.04.082. [DOI] [PubMed] [Google Scholar]
- Toki S. Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Molecular Biology Reporter. 1997;15:16–21. [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant Journal. 2006;47:69–76. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Ushimaru T, Terada H, Tsuboi K, Kogou Y, Sakaguchi A, Tsuji G, Kubo Y. Development of an efficient gene targeting system in Colletotrichum higginsianum using a non-homologous end-joining mutant and Agrobacterium tumefaciens-mediated gene transfer. Molecular Genetics and Genomics. 2010;284:357–371. doi: 10.1007/s00438-010-0572-1. [DOI] [PubMed] [Google Scholar]
- Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, Cirer B, Morin D, Bruel C, Beffa R, Lebrun MH. Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genetics and Biology. 2008;45:68–75. doi: 10.1016/j.fgb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Wang YK, Chang WC, Liu PF, Hsiao MK, Lin CT, Lin SM, Pan RL. Ovate family protein 1 as a plant Ku70 interacting protein involved in DNA double-strand break repair. Plant Molecular Biology. 2010;74:453–466. doi: 10.1007/s11103-010-9685-5. [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Drury GE, Bray CM, West CE. Repairing breaks in the plant genome: the importance of keeping it together. New Phytologist. 2011;192:805–822. doi: 10.1111/j.1469-8137.2011.03926.x. [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Kozak J, Provost CM, Bray CM, Angelis KJ, West CE. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biology. 2009;9:79. doi: 10.1186/1471-2229-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C, Waterworth WM, Sunderland PA, Bray CM. Arabidopsis DNA double-strand break repair pathway. Biochemical Society Transactions. 2004;32:964–966. doi: 10.1042/BST0320964. [DOI] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Jiang Q, Bray CM. Arabidopsis DNA ligase IV is induced by gamma-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant Journal. 2000;24:67–78. doi: 10.1046/j.1365-313x.2000.00856.x. [DOI] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proceedings of the National Academy of Sciences, USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tang L, Li M, Chen L, Xu J, Wu G, Li H. Monitoring homologous recombination in rice (Oryza sativa L) Mutation Research. 2010;691:55–63. doi: 10.1016/j.mrfmmm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. Journal of Immunology. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen W, Zhang Y, Jiang L, Chen Z, Wen Y, Li J. Deletion of ku homologs increases gene targeting frequency in Streptomyces avermitilis. Journal of Industrial Microbiology and Biotechnology. 2012;39:917–925. doi: 10.1007/s10295-012-1097-x. [DOI] [PubMed] [Google Scholar]
- Ziemienowicz A, Tzfira T, Hohn B. Mechanisms of T-DNA integration. In: Tzfira T, Citovsky V, editors. Agrobacterium: from biology to biotechnology. New York, NY, USA: Springer; 2008. pp. 395–440. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Luciferase (LUC) luminescence of DNA double-strand break (DSB)-inducible homologous recombination (HR) events by transient I-SceI expression in control, KD-OsKu70 and KD-OsLig4 rice calli.


