Abstract
HACE1 is an E3 ubiquitin ligase located in 6q21, the genomic region frequently deleted in natural killer (NK) cell malignancies. Here, we report HACE1 as a candidate tumor suppressor gene silenced through a combination of deletion and cytosine phosphate guanine island hypermethylation. We detected deletion of HACE1 in malignant NK cell lines (6 of 9, 67%) and primary biopsies (5 of 15, 33%) by quantitative PCR, with most of the specimen showing cytosine phosphate guanine island hypermethylation in the remaining allele, leading to low mRNA transcription. The ectopic expression of HACE1 in an HACE1-null NK cell line led to apoptosis and G2/M cell cycle arrest. Moreover, HACE1 expression was up-regulated in IL-2-activated normal NK cells and NK cells cocultured with an engineered NK cell target, K562 Clone 9.mbIL21, suggesting its role in the regulation of NK cell homeostasis. In conclusion, HACE1 is another potent tumor suppressor gene located within the 6q21 region, and loss of function of multiple tumor suppressor genes within 6q21 may be a critical determinant of NK cell lymphomagenesis.
HACE1 belongs to the HECT family of ubiquitin ligases (HECT E3), which have the unique capability of intrinsic catalytic activity and specificity for substrates involved in the regulation of growth and apoptosis such as p53, p73, and phosphatase and tensin homologue.1 HECT E3s have been shown to be involved in cancer development because of their capability of targeting specific genes for proteosomal degradation associated with cellular growth and survival.1 A comprehensive study reported HACE1 as a tumor suppressor gene, which was involved in the spontaneous tumorigenesis in several cancers in vivo.2 Knock-down of HACE1 via small-interfering RNA converted nontumorigenic cell lines to tumorigenic ones and HACE1 cooperated with p53 in tumor suppression.2 HACE1 was shown to be silenced in Wilms tumor cases,2,3 and HACE1 silencing was shown to be mediated through hypermethylation of the two cytosine phosphate guanine (CpG) islands, CpG-29 and CpG-177, located upstream of the transcription start site (TSS).2 CpG-177 hypermethylation of HACE1 was frequently observed in colorectal and gastric carcinomas,4,5 and the association of HACE1 hypermethylation with the clinicopathologic findings, especially lymph node metastasis, has been shown for colorectal carcinomas.4 HACE1 was reported to be located in the frequently deleted 6q21 locus by array comparative genomic hybridization (aCGH), and HACE1 expression was down-regulated in natural killer cell lymphoma/leukemia (NKCL) samples.6,7 However, the role of CpG island methylation on HACE1 silencing was not evaluated in those two studies, and the frequency of hemizygous deletion of HACE1 detected by the aCGH platforms (30% to 40% of the cases) was not sufficient enough to account for the down-regulation of HACE1 in NKCLs.
HACE1 was shown to inhibit the tumor suppressor gene RARβ,8 to ubiquitylate Rac19—a gene involved in cell proliferation and G2/M cell cycle progression,10 and to regulate Golgi biogenesis during cell cycle.11 It was shown to target and degrade cyclinD1 in HEK293T cells.2 Those studies suggest that loss of function of HACE1 in NKCLs may be associated with the deregulation of its target genes associated with cell cycle and/or apoptosis in NK cells that contribute to the neoplastic transformation of NK cells.
Here, we report the silencing of HACE1 in NK cell malignancies through a combination of deletion and CpG island hypermethylation and show the tumor suppressive role of HACE1 in NK cell lines through functional assays.
Materials and Methods
Patient and Cell Line Material
The characteristics of NK cell tumor cases and NK cell lines have been reported earlier12 and are summarized in Supplemental Table S1. DNA and RNA were isolated with AllPrep DNA/RNA mini kit (Qiagen Inc., Valencia, CA). All NK cell lines were cultured in RPMI 1640 (Gibco-Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum, penicillin G (100 U/mL) and streptomycin (100 μg/mL), and 5 to 7 ng/mL IL-2 (R&D Bioscience, San Diego, CA) at 37°C in 5% CO2.
HACE1 Copy Number Analysis
Copy number analysis of HACE1 was performed with quantitative real-time PCR (qPCR) with the use of primers designed against the HACE1 genomic DNA by applying the same qPCR-based method used earlier for the detection of monoallelic deletion of PRDM1 in diffuse large B-cell lymphomas and NKCLs, respectively.12,13 Briefly, the copy number of HACE1 is normalized to a reference gene, and the normalized copy number was compared with a control sample [ie, freshly isolated human peripheral blood (PB) NK cells] that was considered to have no genomic abnormality. If the normalized numeric value of the sample was less than the cutoff value (0.75-fold of the control sample), the sample was considered to have the deletion. Genomic DNA (20 ng) was used as the template for qPCR. RPL13A was used as the reference gene to normalize the copy number.12 The HACE1 primers used for copy number analysis were as follows: forward, 5′-AACTCTTAGTTCCAGGGTCCCACA-3′, and reverse, 5′-TTGGAGTATATGGCACAGCAGCGA-3′.
FISH Analysis of NK Cell Lines
Standard interphase fluorescent in situ hybridization FISH study was performed on NK92 and KAI3 cell suspensions with the use of direct-labeled centromere probes for chromosome 6 (Abbott/Vysis, Inc., Abbott Park, IL) and the HACE1 gene region (6q21; Empire Genomics, Buffalo, NY). FISH was performed by co-denaturation on a ThermoBrite instrument (Abbott-Vysis, Inc.) at a denaturation temperature of 75°C for 1 minute, followed by an overnight hybridization at 37°C. The slides were then washed with 0.4× standard saline citrate/0.3% NP-40 at 72°C for 2 minutes, followed by a 1-minute wash in 2× standard saline citrate/0.1% NP-40 at room temperature. The cells were counterstained with DAPI II and viewed on a Leica DM5500B microscope (Leica, Wetzlar, Germany) equipped with appropriate filters. One hundred interphase nuclei were examined for each probe.
Quantification of the HACE1 mRNA
HACE1 mRNA expression was determined with quantitative RT-PCR (RT-qPCR) in malignant NK cell lines, NKCL cases, IL-2-activated peripheral blood NK cells, or human peripheral blood NK cells activated by coculturing with an engineered K562 cell line (K562 Clone 9.mbIL21) kindly provided by Dr. Dean A. Lee (MD Anderson Cancer Center, Houston, TX). The K562 Clone 9.mbIL21 cell line was derived from the K562 cells, an erthyroleukemia cell line originated from a patient with chronic malignant leukemia with blast crisis,14 engineered to express additional activation molecules (ie, 4-1BBL, CD86, and mbIL21) on the cell surface so that robust signals of NK cell activation can be obtained.15 The rationale of choosing the engineered K562 cell line was to address whether HACE1 expression is up-regulated during NK cell activation, suggesting a possible functional role for HACE1 during NK cell activation. NK cells activated by K562 Clone 9.mbIL21 cells can be expanded, allowing longer periods of observation time. In addition, this model provided us with the opportunity to observe HACE1 expression in the later stages of activation of NK cells, which can be determined by the time period at which the engineered K562 cells are cleared by the cytotoxic action of NK cells. Melting curve analysis was performed to ensure PCR-band specificity. ΔΔCt method was used for the RT-qPCR data analysis. RPL13A16 was used as the housekeeping gene to calibrate HACE1 expression. The HACE1 RT-qPCR primers used were as follows: forward, 5′-CCAGTTATTCAGTGGTTCTGGGAAG-3′, and reverse, 5′-CCACCCATGATATTAGCAAACCCACC-3′.
Methylation Analysis of HACE1
HACE1 promoter methylation was determined with the methylation-specific cell counting (MSCC) procedure as described earlier17 in NKCL cases (n = 11) and with the use of quantitative Methyl-specific PCR (qMSP) in NK cell lines (n = 7). We specifically concentrated on the CpG islands close to the promoter of HACE1, whereas the complete methylation profile by MSCC will be published separately. The library preparation and high-throughput sequencing was performed at the epigenetic core laboratory facility by Illumina Genome Analyzer II X. To determine the significance of change in count data, an analysis for fit that was based on the Poisson distribution at the P = 0.05 level was used. On the basis of this analysis, a significant change was determined to be a fourfold or greater difference between two samples. qMSP has been performed on seven NK cell lines with the use of previously reported MSP primers2 with the EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA) and 250 ng of genomic DNA as the template. DyNAmo HS SYBR Green qPCR kit (Finnzymes, Waltham, MA) was used for qPCR. Resting PB NK cells were used as the unmethylated control, M. SssI (New England Biolabs, Ipswich, MA) methylated human lymphocyte genomic DNA (Roche Diagnostic Corporation, Indianapolis, IN) was used as the methylated control for qMSP, and 48-hour IL-2-activated PB NK cells were used as the control for MSCC. The differential methylation in NKCL cases compared with the control was calculated according to the count data. The primer sequences used for qMSP are as follows: CpG-177 forward, 5′-GAATGGAAGGTTTAATTTCGC-3′, and CpG-177 reverse, 5′-CTAAAACCCTACGTCAACCG-3′.
Mutation Analysis of HACE1 in Malignant NKCL Cases
RNA-seq has been performed on eight NKCL cases. In this study, we concentrated on HACE1, and the complete transcriptomic profile will be published later. The RNA-seq data of the HACE1 coding sequence and intron/exon junctions were analyzed with Integrative Genomics Viewer software version 2.0 (http://www.broadinstitute.org/igv) to screen for possible mutations. If suspected for a splice site mutation, DNA from the corresponding NKCL case was PCR-amplified with primers spanning the HACE1 intron/exon splice junctions. The primers used in the mutation analysis are listed Table 1.
Table 1.
HACE1 Mutation Analysis Primer Sets (NC_000006.11)
| Location | Forward | Reverse |
|---|---|---|
| 68169-68760 | 5′-TGAAACAGATGGTCAGTGGGATGA-3′ | 5′-AGCATGGTGGCTGCATATTACA-3′ |
| 74341-75180 | 5′-GGCATTACTATCACGTTAACTTGGGAAGAT-3′ | 5′-ACAAGGTGACATTCACCATTCTACA-3′ |
| 74401-74860 | 5′-GGCATTACTATCACGTTAACTTGGGAAGAT-3′ | 5′-ACATCCTGACAATCTGCACTGGCT-3′ |
| 75541-76140 | 5′-TCAGTGTCAGTGTATCTGAATTTGC-3′ | 5′-GCTCCTTTCAGGAAATGTGGCA-3′ |
| 82381-82980 | 5′-GGTAGTCAAACTATTTCAGCTAAGTTCTG-3′ | 5′-ATGCCTTCTTCTCCATGGAACCGT-3′ |
Reconstitution of HACE1 in Malignant NK Cell Lines
HACE1-pCMV6-XL4 (Origene, Rockville, MD) was used as the HACE1 source sequence during cloning of expression plasmids. The National Center for Biotechnology Information reference sequence used for HACE1 is NM_020771.2. HACE1 coding sequences were PCR-cloned with the high-fidelity PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies, Palo Alto, CA) upstream of the internal ribosome entry site with the use of the NotI and SalI sites into PMIG. Sequence validation of all of the inserts was performed by Sanger sequencing. HACE1 retroviral construct generated and used in this study are shown (Supplemental Figure S1). The retroviral transduction was performed as described previously.12
Apoptosis and Cell Cycle Assays
Apoptosis of retrovirally transduced cells was quantified with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) after staining the cells with Annexin V-PE (Apoptosis Detection Kit; BD Pharmingen, San Diego, CA) according to the manufacturer’s instructions. The cell cycle profile of retrovirally transduced cells was analyzed with Hoechst 33342 staining (H3570; Invitrogen, Carlsbad, CA). The cells were washed once with phosphate-buffered saline (PBS) and resuspended in buffer containing 1× PBS plus 0.1% bovine serum albumin at 0.5 × 106 cells/mL. Hoechst dye was added to the cells at a final concentration of 10 μg/mL. The cells were incubated at 37°C for 15 minutes and then analyzed with a BD LSR II flow cytometer.
Isolation of Primary NK Cells from the Peripheral Blood
Normal NK cells were isolated from the peripheral blood of healthy donors with negative selection with the use of the NK Cell Isolation Kit (Miltenyi Biotech, Auburn, CA). The purity of NK cells was determined with CD56-APC/CD3-PE double staining and FACSCalibur flow cytometry (Supplemental Figure S2A).
NK Cell Activation by Coculturing Peripheral Blood Lymphocytes with the Engineered K562 Cells
Derivation of primary NK cells from the coculture of peripheral blood lymphocytes and K562 Clone 9.mbIL21 was performed as described earlier.15 Briefly, 1.5 × 106 peripheral blood lymphocytes were mixed with 100-Gy irradiated 3 × 106 K562 Clone 9.mbIL21 cells (1:2 ratio). Seven days after stimulation, second stimulation was performed by combining cocultured cells with fresh irradiated engineered K562 cells (1:1 ratio). The proportion of NK cells in the cocultured population was determined with flow cytometry with the use of CD56 and CD3 surface staining 7, 12, and 14 days after the coculture (Supplemental Figure S2, B and D), and CD56+/CD3− cells were considered as human NK cells as described earlier.18
Statistical Analysis
Statistical analysis was performed with Student’s t-test. P < 0.05 was considered significant.
Results
Deletion and CpG Island Methylation Is Frequent and Cooperates in Silencing of HACE1 in NKCL Cases
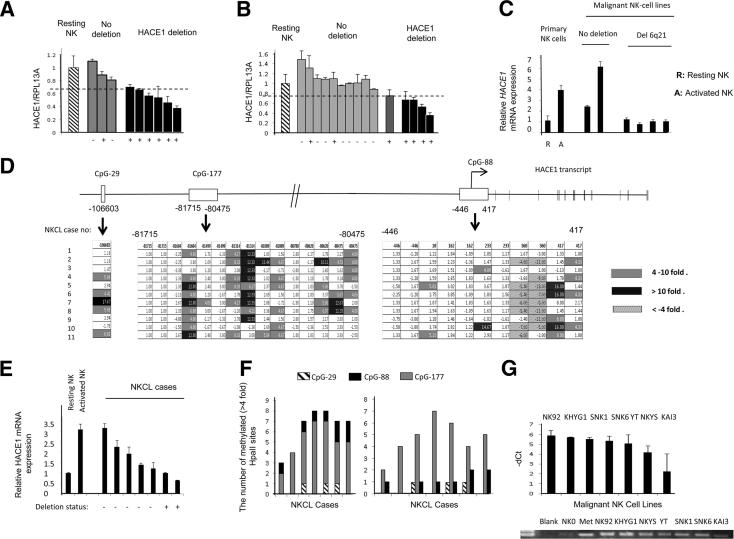
Previously, hemizygous deletion of HACE1 was observed only in ∼37.5% of the NKCL cases.6 Consistent with these observations, we observed monoallelic deletion of HACE1 in 6 of 9 (67%) NK cell lines (Figure 1A) and 5 of 15 (33%) NKCL cases (Figure 1B) with the use of the qPCR method for monoallelic deletion reported recently.12,13 In general, monoallelic deletion of HACE1 detected by qPCR in NK cell lines and NKCL cases (8 of 9, 89%, for the NK cell lines and 13 of 15, 87%, for the NKCL cases) were present in the samples that have the monoallelic deletion of the 6q21 minimal common region locus detected in previous studies by bacterial artificial chromosome aCGH16 or Cartes d’Identité des Tumeurs aCGH.19 We evaluated HACE1 copy number status with FISH on NK92 and KAI3 cell lines that have no deletion or HACE1 deletion detected by qPCR, respectively. Consistent with the findings with qPCR assay, we observed monoallelic deletion of HACE1 in KAI3 but not in the NK92 cell line (Supplemental Figure S3). To address whether monoallelic deletion of HACE1 decreased mRNA expression, we performed RT-qPCR on six NK cell lines and observed lower HACE1 expression in four NK cell lines with deletion (Figure 1C).
Figure 1.
HACE1 is silenced through a combination of hemizygous deletion and CpG island hypermethylation in NKCL samples. A and B: NK cell lines and NKCL cases with or without deletion were determined by the HACE1/RPL13A ratio for each sample. NKCL cases with HACE1/RPL13A <0.75 that of human resting primary PB NK cell DNA were defined to have deletion. HACE1/RPL13A for each sample represented the average of two independent experiments for both HACE1 and RPL13A. The deletion status of the NKCL cases by aCGH was shown with the ± signs at the bottom of the figure. The plus sign indicates a hemizygous deletion in the minimal common region of 6q21 according to the two previous reports that applied bacterial artificial chromosome-aCGH on NK cell lines and NKCL cases16 or Cartes d’Identité des Tumeurs CGH on NKCL cases.19 The dashed horizontal bar represents the cutoff used to determine the deletion status of the NK cell lines and NKCL cases. C: Quantification of HACE1 mRNA in NK cell lines. The HACE1 mRNA expression in NK cell lines was determined with RT-qPCR. Relative expression values were normalized to the values of the resting NK cells. Resting and 4-day IL-2–activated primary PB NK cells were used as control. Data are means ± SDs of two experiments. D: Hypermethylation of the three CpG islands, CpG-29, CpG-177, and CpG-88, in NKCL cases was determined with MSCC. The differential methylation of the HpaII sites was shown after normalization of the HpaII count sites to the count sites of the 48-hour IL-2–activated peripheral blood NK cells. Gray (4- to 10-fold) and black (>10-fold) boxes show the hypermethylated HpaII sites. E: HACE1 mRNA expression in seven NKCL cases. HACE1 mRNA expression in NKCL cases was normalized to the HACE1 expression in resting PB NK cells. Resting and 4-day IL-2–activated PB NK cells were used as the controls. The HACE1 deletion status was marked with plus and minus signs on the x axis. F: The total number of hypermethylated HpaII sites (ie, hypermethylated HpaII sites in CpG-177, CpG-88, and CpG-29 altogether) (left panel) and the number of hypermethylated HpaII sites in each CpG island compared with normal 48-hour IL-2–activated NK cells (right panel) were indicated for the same NKCL cases with the same sample order as in E. Fourfold count was set as the threshold for hypermethylation. G: MSP-qPCR results of the seven malignant NK cell lines are shown. Resting NK cells (NK0) were used as the negative, and M.SssI methylated lymphocyte DNA (Met) was used as the positive control, respectively. −ΔCt values are calculated as follows: −(CtNK cell line − CtNK0).
Next, we evaluated the influence of the CpG island methylation on HACE1 expression as an epigenetic mechanism of gene silencing. Three CpG islands were reported around or upstream of the TSS for HACE1.3 The hypermethylation-associated silencing of the upstream CpG islands, CpG-177 and CpG-29, was shown in Wilms tumor,2 and hypermethylation of CpG-177 was detected in a variety of solid tumors.2–5 Consistent with the previous reports, we observed hypermethylation of at least one HpaII (CCGG) site for the CpG-177 (−81715 to −80475 of TSS) in all NKCL cases compared with the activated NK cells by MSCC (Figure 1D). CpG-88, the CpG island around the HACE1 TSS, had hypermethylated sites; however, the frequency of hypermethylation was lower than that observed for the CpG-177, and there was one hypomethylated site (360 of TSS) in 9 of 11 (82%) NKCL cases. We had one MSCC site from CpG-29, and 5 of 11 (45%) of the NKCL cases had hypermethylation at this site. Next, we determined HACE1 expression in seven NKCL cases by RT-qPCR (Figure 1E). We observed lower HACE1 expression in NKCL cases having higher numbers of hypermethylated HpaII sites in NKCL cases, and the cases with deletion and high quantity of hypermethylated sites had the lowest mRNA expression (Figure 1, E and F). The association between CpG island hypermethylation and low HACE1 expression was more obvious in CpG-177 than in CpG-88 and CpG-29 (Figure 1F). We determined CpG-177 methylation in 6 of 7 (86%) NK cell lines with MSP (Figure 1G).
Mutation Analysis of HACE1 in the Malignant NKCL Cases
RNA-seq showed retention of introns 11 and 15 in two of eight NKCL cases; however, no mutations were detected in the coding region. Insufficient splicing may lead to loss of function of HACE1 that can cooperate with deletion and CpG island methylation. The retention of the introns may be due to insufficient splicing that may be related to the mutations in the intron/exon junctions. To evaluate whether there was any mutation in the intron/exon junctions leading to aberrant splicing, we designed primers targeting the genomic DNA that spanned these junctions. Sanger sequencing of the intron/exon junctions spanning the exons 11 to 12 and 14 to 15 for the corresponding NKCL case did not show any single nucleotide variations in the intron/exon junctions screened.
Ectopic Expression of HACE1 in an NK Cell Line Leads to Cell Cycle Arrest and Apoptosis
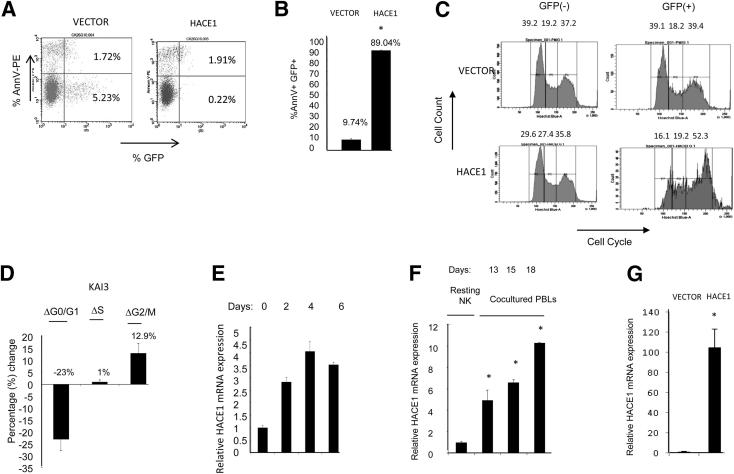
We tested the functional consequence(s) of the ectopic expression of HACE1 in the KAI3 cell line, an NK cell line with del6q21 and low HACE1 expression. We directly tested cell cycle and apoptosis in green fluorescent protein (GFP)+, unsorted cells with Hoechst and Annexin V staining. We observed increase in apoptosis (Figure 2, A and B) and induction of G2/M cell cycle arrest in KAI3 cells transduced with HACE1 compared with the vector-only control 2 days after transduction (Figure 2, C and D).
Figure 2.
Reconstitution of HACE1 in a HACE1-null NK cell line induces apoptosis and cell cycle arrest. A: Vector or HACE1-transduced KAI3 cells were stained with Annexin V-PE and tested with fluorescence-activated cell sorting. B: Ectopic expression of HACE1 caused apoptosis in the KAI3 cell line (vector: 9.74% ± 0.92%, n = 2; HACE1: 89.04% ± 0.21%, n = 2). The rate of apoptosis was determined by measuring the proportion of Annexin V+ cells in the GFP+ population 2 days after transduction. Data are means ± SDs of two independent experiments. *P < 0.001 versus vector. C: Vector- or HACE1-transduced KAI3 cells were stained with Hoechst 33342, and the cell cycle profile on the GFP-gated population of vector- or HACE1-transduced KAI3 cells was determined with fluorescence-activated cell sorting. D: Ectopic HACE1 expression causes G2/M cell cycle arrest in the KAI3 cell line [(%HACE1 − %Vector); (ΔG0/G1 = −23%, ΔS = 1%, ΔG2/M = 12.9%)]. Data are means ± SDs of two independent experiments. E: HACE1 mRNA expression was determined in IL-2–activated PB NK cells. RPL13A was used as the housekeeping gene for normalization. F: HACE1 mRNA expression in cocultured primary NK cells is shown. *P < 0.05 versus Resting NK cells. G: Ectopic expression of HACE1 in KAI3 cells. RT-qPCR was applied on GFP+ KAI3 cells sorted 42 hours after transduction with vector or HACE1. RPL13A was used as the housekeeping gene for normalization. Data are means ± SDs of two independent experiments. *P < 0.001 versus vector.
HACE1 Is Up-Regulated during Normal NK Cell Activation
Next, we asked whether HACE1 expression was altered on NK cell activation. To address this question, we tested HACE1 expression in IL-2-activated human peripheral blood NK cells and observed up to a fourfold increase in a 6-day period (Figure 2E). We observed a similar progressive up-regulation of HACE1 between days 13 and 18 in NK cells cocultured with engineered K562 cells, which reached 10-fold compared with the levels in resting NK cells, suggesting a possible role of HACE1 in the regulation of NK cell activation and homeostasis (Figure 2F). Ectopic expression of HACE1 has been shown with RT-qPCR on transduced KAI3 cells (Figure 2G).
Discussion
Monoallelic deletion of the 6q21 locus (del6q21) is one of the most recurrent genomic aberrations in a variety of cancers, including lymphomas/leukemias.20–22 High frequency of del6q21 suggests the presence of tumor suppressor gene(s) in this locus and loss of function that may contribute to the neoplastic transformation of NK cells and the pathogenesis of NKCLs. With the application of the high-resolution aCGH technique, the candidate tumor suppressor genes in del6q21 have been identified.16 Indeed, reconstitution and knock-down experiments showed that PRDM1 is a tumor suppressor gene in NKCLs.7,12 However, genomic deletion may affect multiple genes simultaneously in a genomic locus that may harbor multiple tumor suppressor genes. In fact, haploinsufficiency or loss of function of multiple tumor suppressor genes may provide additive or synergistic growth advantage during the neoplastic transformation of NK cells. Haploinsufficiency can account for the neoplastic transformation through decreased expression23; however, in most instances, both copies of a tumor suppressor gene are inactivated with loss of function. Inactivation of the remaining alleles can occur through a genetic or epigenetic mechanism,24 and promoter or CpG island hypermethylation is one of the mechanisms for silencing tumor suppressor genes.25 SHP1, p73, and PRDM1 have been reported to be silenced through promoter methylation in NKCLs.16,26,27 In this study, we observed a combination of deletion and CpG island hypermethylation to be responsible for the silencing of HACE1 in NKCLs. HACE1 mutations in NKCL cases may be uncommon or not present according to our analysis. Intronic sequences detected with RNA-seq may be an artifact of the genomic DNA contamination because no mutation was observed in the intron/exon junctions by Sanger sequencing, consistent with a previous report that showed no mutation in the exons and intron/exon junctions in Wilms tumor cases.3 Ectopic expression of HACE1 was much less efficient compared with vector-alone transduction. This phenomenon was frequently observed during the ectopic expression of the tumor suppressor genes with the use of unregulated systems, possibly because of early elimination of transduced cells by the cytotoxic action of these genes before the experimental endpoints can be tested. This phenomenon was also observed during the ectopic expression PRDM1 (BLIMP1), another tumor suppressor gene in the deleted 6q21 locus, in B- and NK-cell lines, respectively.28,29
In conclusion, we provided strong evidence that HACE1 may be a tumor suppressor gene in NK cell malignancies by using genetic, epigenetic, and functional approaches. Further studies are needed to elucidate the putative targets and the functional role of HACE1 in primary NK cells and NK cell malignancies.
Footnotes
Supported by Lymphoma SPOREP50CA136411-01(NCI) (W.C.C. and J.I.) and a grant from the National Cancer Institute (5U01/CA114778).
C.K. and X.H. contributed equally to this work.
Supplemental Data
HACE1-CDS cDNA was cloned inside MSCV-IRES-GFP (PMIG) as indicated.
A: The purity of primary PB NK cells was determined with CD56 and CD3 staining after isolation with the Miltenyi NK cell isolation kit. The proportion of NK cells was determined with CD56-APC and/or CD3-PE staining 7 days (B), 12 days (C), and 14 days (D) after the initiation of coculture of PB lymphocytes and the engineered K562 cells.
Interphase-FISH analysis of KAI3 and NK92 cell lines. HACE1 probe was labeled with red; and the chromosome 6 centromeric probe (CEP 6) was labeled with green. One representative cell was shown for each KAI3 (A) and NK92 (B) cell line. Both cell lines show tetraploidy for chromosome 6.
Supplemental Data
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2012.09.012.
References
- 1.Bernassola F., Karin M., Ciechanover A., Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L., Anglesio M.S., O’Sullivan M., Zhang F., Yang G., Sarao R., Mai P.N., Cronin S., Hara H., Melnyk N., Li L., Wada T., Liu P.P., Farrar J., Arceci R.J., Sorensen P.H., Penninger J.M. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat Med. 2007;13:1060–1069. doi: 10.1038/nm1621. [DOI] [PubMed] [Google Scholar]
- 3.Anglesio M.S., Evdokimova V., Melnyk N., Zhang L., Fernandez C.V., Grundy P.E., Leach S., Marra M.A., Brooks-Wilson A.R., Penninger J., Sorensen P.H. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms’ tumor versus normal kidney. Hum Mol Genet. 2004;13:2061–2074. doi: 10.1093/hmg/ddh215. [DOI] [PubMed] [Google Scholar]
- 4.Hibi K., Sakata M., Sakuraba K., Shirahata A., Goto T., Mizukami H., Saito M., Ishibashi K., Kigawa G., Nemoto H., Sanada Y. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2008;28:1581–1584. [PubMed] [Google Scholar]
- 5.Sakata M., Kitamura Y.H., Sakuraba K., Goto T., Mizukami H., Saito M., Ishibashi K., Kigawa G., Nemoto H., Sanada Y., Hibi K. Methylation of HACE1 in gastric carcinoma. Anticancer Res. 2009;29:2231–2233. [PubMed] [Google Scholar]
- 6.Huang Y., de Reynies A., de Leval L., Ghazi B., Martin-Garcia N., Travert M., Bosq J., Briere J., Petit B., Thomas E., Coppo P., Marafioti T., Emile J.F., Delfau-Larue M.H., Schmitt C., Gaulard P. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood. 2010;115:1226–1237. doi: 10.1182/blood-2009-05-221275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karube K., Nakagawa M., Tsuzuki S., Takeuchi I., Honma K., Nakashima Y., Shimizu N., Ko Y.H., Morishima Y., Ohshima K., Nakamura S., Seto M. Identification of FOXO3 and PRDM1 as tumor-suppressor gene candidates in NK cell neoplasms by genomic and functional analyses. Blood. 2011;118:3195–3204. doi: 10.1182/blood-2011-04-346890. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J., Zhang Z., Vucetic Z., Soprano K.J., Soprano D.R. HACE1: a novel repressor of RAR transcriptional activity. J Cell Biochem. 2009;107:482–493. doi: 10.1002/jcb.22146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torrino S., Visvikis O., Doye A., Boyer L., Stefani C., Munro P., Bertoglio J., Gacon G., Mettouchi A., Lemichez E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell. 2011;21:959–965. doi: 10.1016/j.devcel.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Moore K.A., Sethi R., Doanes A.M., Johnson T.M., Pracyk J.B., Kirby M., Irani K., Goldschmidt-Clermont P.J., Finkel T. Rac1 is required for cell proliferation and G2/M progression. Biochem J. 1997;326(Pt 1):17–20. doi: 10.1042/bj3260017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang D., Xiang Y., De Renzis S., Rink J., Zheng G., Zerial M., Wang Y. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nat Commun. 2011;2:501. doi: 10.1038/ncomms1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kucuk C., Iqbal J., Hu X., Gaulard P., De Leval L., Srivastava G., Au W.Y., McKeithan T.W., Chan W.C. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Natl Acad Sci U S A. 2011;108:20119–20124. doi: 10.1073/pnas.1115128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tam W., Gomez M., Chadburn A., Lee J.W., Chan W.C., Knowles D.M. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 14.Lozzio C.B., Lozzio B.B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 15.Somanchi S.S., Senyukov V.V., Denman C.J., Lee D.A. Expansion, purification, and functional assessment of human peripheral blood NK cells. J Vis Exp. 2011;(48) doi: 10.3791/2540. pii: 2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iqbal J., Kucuk C., Deleeuw R.J., Srivastava G., Tam W., Geng H., Klinkebiel D., Christman J.K., Patel K., Cao K., Shen L., Dybkaer K., Tsui I.F., Ali H., Shimizu N., Au W.Y., Lam W.L., Chan W.C. Genomic analyses reveal global functional alterations that promote tumor growth and novel tumor suppressor genes in natural killer-cell malignancies. Leukemia. 2009;23:1139–1151. doi: 10.1038/leu.2009.3. [DOI] [PubMed] [Google Scholar]
- 17.Ball M.P., Li J.B., Gao Y., Lee J.H., LeProust E.M., Park I.H., Xie B., Daley G.Q., Church G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nature Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Wetering D., de Paus R.A., van Dissel J.T., van de Vosse E. IL-23 modulates CD56+/CD3- NK cell and CD56+/CD3+ NK-like T cell function differentially from IL-12. Int Immunol. 2009;21:145–153. doi: 10.1093/intimm/dxn132. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y., de Reynies A., de Leval L., Ghazi B., Martin-Garcia N., Travert M., Bosq J., Briere J., Petit B., Thomas E., Coppo P., Marafioti T., Emile J.F., Delfau-Larue M.H., Schmitt C., Gaulard P. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood. 2010;115:1226–1237. doi: 10.1182/blood-2009-05-221275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyytinen E.R., Saadut R., Chen C., Paull L., Koivisto P.A., Vessella R.L., Frierson H.F., Jr., Dong J.T. Defining the region(s) of deletion at 6q16-q22 in human prostate cancer. Genes Chromosomes Cancer. 2002;34:306–312. doi: 10.1002/gcc.10065. [DOI] [PubMed] [Google Scholar]
- 21.Inoue M., Marx A., Zettl A., Strobel P., Muller-Hermelink H.K., Starostik P. Chromosome 6 suffers frequent and multiple aberrations in thymoma. Am J Pathol. 2002;161:1507–1513. doi: 10.1016/S0002-9440(10)64426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Matthiesen P., Harder S., Siebert R., Castoldi G., Calasanz M.J., Wong K.F., Rosenwald A., Ott G., Atkin N.B., Schlegelberger B. A 3-cM commonly deleted region in 6q21 in leukemias and lymphomas delineated by fluorescence in situ hybridization. Genes Chromosomes Cancer. 2000;27:52–58. doi: 10.1002/(sici)1098-2264(200001)27:1<52::aid-gcc7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 23.Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 25.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 26.Oka T., Ouchida M., Koyama M., Ogama Y., Takada S., Nakatani Y., Tanaka T., Yoshino T., Hayashi K., Ohara N., Kondo E., Takahashi K., Tsuchiyama J., Tanimoto M., Shimizu K., Akagi T. Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res. 2002;62:6390–6394. [PubMed] [Google Scholar]
- 27.Siu L.L., Chan J.K., Wong K.F., Kwong Y.L. Specific patterns of gene methylation in natural killer cell lymphomas: p73 is consistently involved. Am J Pathol. 2002;160:59–66. doi: 10.1016/s0002-9440(10)64349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kucuk C., Iqbal J., Hu X., Gaulard P., De Leval L., Srivastava G., Au W.Y., McKeithan T.W., Chan W.C. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Natl Acad Sci U S A. 2011;108:20119–20124. doi: 10.1073/pnas.1115128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K., Staudt L.M. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HACE1-CDS cDNA was cloned inside MSCV-IRES-GFP (PMIG) as indicated.
A: The purity of primary PB NK cells was determined with CD56 and CD3 staining after isolation with the Miltenyi NK cell isolation kit. The proportion of NK cells was determined with CD56-APC and/or CD3-PE staining 7 days (B), 12 days (C), and 14 days (D) after the initiation of coculture of PB lymphocytes and the engineered K562 cells.
Interphase-FISH analysis of KAI3 and NK92 cell lines. HACE1 probe was labeled with red; and the chromosome 6 centromeric probe (CEP 6) was labeled with green. One representative cell was shown for each KAI3 (A) and NK92 (B) cell line. Both cell lines show tetraploidy for chromosome 6.