Abstract
Bacteria in the gut microbiome shed microbial-associated molecule patterns (MAMPs) into the portal venous circulation, where they augment various aspects of systemic immunity via low-level stimulation. Because the liver is immediately downstream of the intestines, we proposed that gut-derived MAMPs shape liver immunity and affect Kupffer cell (KC) phenotype. Germ-free (GF), antibiotic-treated (AVMN), and conventional (CL) mice were used to study KC development, function, and response to the significant stress of cold storage, reperfusion, and orthotopic transplantation. We found that a cocktail of physiologically active MAMPs translocate into the portal circulation, with flagellin (Toll-like receptor 5 ligand) being the most plentiful and capable of promoting hepatic monocyte influx in GF mice. In MAMP-deficient GF or AVMN livers, KCs are lower in numbers, have higher phagocytic activity, and have lower major histocompatibility complex II expression. MAMP-containing CL livers harbor significantly increased KC numbers via induction of intercellular adhesion molecule 1 on liver sinusoidal endothelium. These CL KCs have a primed yet expected phenotype, with increased major histocompatibility complex class II and lower phagocytic activity that increases susceptibility to liver preservation/reperfusion injury after orthotopic transplantation. The KC number, functional activity, and maturational status are directly related to the concentration of gut-derived MAMPs and can be significantly reduced by broad-spectrum antibiotics, thereby affecting susceptibility to injury.
More than 100 trillion, largely colon-restricted, autochthonous bacteria comprise the gut microbiome.1 They not only help shape gut morphologic features and mucosal immunity2 but also contribute to the development of the extraintestinal immune system. For example, germ-free (GF) rodents exhibit smaller, less cellular spleens and lower systemic antibody levels,3 and the gut-derived microbe-associated molecular pattern (MAMP) peptidoglycan (PDG) can prime the systemic innate immunity.4 Extraintestinal effects of the gut microbiome are thought to be mediated by MAMPs, which are recognized by germline encoded pattern recognition receptors (PRRs) expressed on cells throughout the body, including Kupffer cells (KCs), hepatocytes, and liver sinusoidal endothelial cells (LSECs).5
Gut-derived MAMPs reach the liver via blood from the portal vein and first encounter PRR-bearing KCs, the most abundant of all tissue macrophage populations, and sinusoidal endothelium. Despite a valid assumption that MAMPs reach the liver via the portal venous blood, the relative physiologic composition of various portal venous MAMPs under homeostatic conditions and their effects, if any, on LSEC and KC populations, including their physiologic activation/maturation state, are poorly understood. Interactions among gut-derived MAMPs, LSECs, and KCs, however, have the potential to control KC activation status and signaling pathways and trigger protein secretion that can profoundly influence all nearby cells, including hepatocytes, stellate cells, LSECs, and intrahepatic leukocytes. Consequently, gut-derived MAMP interactions with LSECs and KCs can significantly influence susceptibility to injury and hepatic physiologic and pathophysiologic responses to environmental challenges. Included are host defense, regeneration, fibrogenesis, carcinogenesis, toxin exposure, ischemia/reperfusion injury, and immunologic tolerance.6
Particular bacterial species (ie, Clostridium species, segmented filamentous bacteria) and specific MAMPs (ie, polysaccharide A, PDG) can induce the development of lymphoid tissue and immune cell subsets within the gut,2 and MAMPs translocate to the liver via portal circulation under homeostatic conditions. Therefore, we tested the hypothesis that gut bacteria orchestrate the development of the normal KC population of the liver and, in turn, help determine the susceptibility to injury and response to environmental challenges. Results indicate that the number and activation/maturational status of KCs directly correlate with density of gut bacteria and susceptibility to injury, as indicated by the extent of cold preservation/reperfusion injury after orthotopic liver transplantation. This is mediated via gut-derived MAMPs that increase LSEC intercellular adhesion molecule 1 (ICAM-1) and lymphatic vessel endothelial receptor 1 (LYVE-1) expression, which in turn recruit KCs from circulating KC progenitors. Most importantly, broad-spectrum antibiotic therapy can decrease both the gut microbiome and liver KC population, which in turn influences a wide variety of hepatic immunology, drug reactions, physiologic processes, and susceptibility to injury.
Materials and Methods
Animals
All mice received humane care according to the guidelines in the Guide for the Care and Use of Laboratory Animals. Procedures performed on animals were approved by the University of Pittsburgh’s Institutional Animal Care and Use Committee protocols 1001281B-4 and 1002448. GF and conventional (CL) Swiss Webster male mice aged 3 to 4 weeks or 6 to 9 weeks were purchased from Taconic (New York, NY) and the Center for Gastrointestinal Biology and Disease Core at the University of North Carolina (Chapel Hill, NC). GF mice were maintained inside of a sterile vinyl isolator for 48 hours with sterilized food and water. CL mice were maintained under specific pathogen-free conditions. To generate the antibiotic-treated (AVMN) experimental groups for various experiments, antibiotics (ampicillin, 1 g/L; vancomycin, 0.5 g/L; metronidazole, 1 g/L; and neomycin, 1 g/L) were delivered to CL mice (4 to 10 weeks old) in drinking water (changed every 3 days) for 2.4 to 7 weeks, depending on body weight minimums needed for transplantation. All mice, however, were 11 to 13 weeks old at the time of transplantation.
The second group was given antibiotics later so treatment time could be reduced to 2 weeks (not to risk bacterial overgrowth by resistant strains and still reach optimal body weight for successful transplantation).
Cell Preparations
Livers were perfused with cold Hanks' balanced salt solution and digested with collagenase (2 mg). Perfused livers were placed in a 1:1 solution of cold PBS (Invitrogen, San Diego, CA) and RPMI 1640 (Sigma-Aldrich, The Woodlands, TX) supplemented with 5% fetal bovine serum, nonessential amino acids, sodium pyruvate, HEPES, l-glutamine, β-mercaptoethanol, and penicillin/streptomycin (Invitrogen). For in vivo challenge experiments, nonparenchymal cells (NPCs) were suspended in cold RPMI only. Livers were minced through a cell dissociation sieve (size 40; Sigma-Aldrich) for single cell suspension, separated into hepatocytes and NPC by centrifugation (60 × g), filtered three times (40 μm cell strainer; BD Falcon, Franklin Lakes, NJ), and stained for flow cytometry (FC). More than 85% of isolated NPCs were CD45+ (CD45-Alexafluor700; Biolegend, San Diego, CA).
Peripheral blood mononuclear cells (PBMCs) were obtained from systemic blood suspended in 2 mL of Hanks' balanced salt solution for Ficoll (Invitrogen) gradient separation. The buffy coat was removed, washed, filtered (40-μm cell strainer; BD Falcon), and stained for FC.
Bone marrow was flushed using a 1:1 solution of PBS and RPMI Complete Media. Bone marrow derived cells (BMDCs) were washed, filtered (40-μm cell strainer; BD Falcon), and stained for FC.
Flow Cytometry
Cells were resuspended in FACS buffer (1% bovine serum albumin, 0.1% sodium azide in PBS). Cells were analyzed on a BD LSR II with FACS Diva software version 6.1.3. FC data were analyzed with Flow Jo (Tree Star Inc., Ashland, OR). The antibodies used are listed in Table 1.
Table 1.
List of Antibodies Used in the Study
| Antigen | Host (isotype) | Clone | Source |
|---|---|---|---|
| Flow cytometry | |||
| Gr1 (Ly6G) | Rat (IgG2b, κ) | RB6-8C5 | eBioscience (San Diego, CA) |
| F4/80 | Rat (IgG2a, κ) | BM8 | eBioscience |
| CD11b | Rat (IgG2b, κ) | M1/70 | Biolegend |
| MHCII (I-A/I-E) | Rat (IgG2b, κ) | M5/114.15.2 | eBioscience |
| CD80 (B7-1) | Armenian Hamster (IgG) | 16-10A1 | eBioscience |
| CD86 (B7-2) | Rat (IgG2a, κ) | GL1 | eBioscience |
| Ly6C | Rat (IgG2c, κ) | HK1.4 | Biolegend |
| CD281 (TLR1) | Rat (IgG2a, κ) | eBioTR23 | eBioscience |
| CD282 (TLR2) | Rat (IgG2b, κ) | 6C2 | eBioscience |
| TLR4/MD2 complex | Rat (IgG2a, κ) | MTS510 | eBioscience |
| CD289 (TLR9) | Rat (IgG2a) | M9.D6 | eBioscience |
| TLR5 | Mouse (IgG2a, κ) | 19D759.2 | Imgenex (San Diego, CA) |
| CD16/CD32 (Fc block) | Rat (IgG2a, λ) | 93 | eBioscience |
| Tissue staining | |||
| F4/80 | Rat (IgG2b) | CI:A3-1 | AbD Serotec (Raleigh, NC) |
| ICAM-1 (CD54) | Armenian Hamster (IgG1, κ) | 3E2 | BD Pharmingen (Franklin Lakes, NJ) |
| CD44 (LYVE1) | Rat (IgG1) | ALY7 | eBioscience |
| Ki67 | Rabbit | SP6 | GeneTex (Irvine, CA) |
Stool Isolation and Bacterial Detection
Total DNA was extracted from 80 mg of cecal stool using a QIAmp DNA stool mini-kit (Qiagen, Santa Clarita, CA). Extracted DNA was submitted to real-time PCR of the conserved region of the bacterial 16S rRNA gene using universal primers (forward 5′-TCCTACGGGAGGCAGCAGT-3′ and reverse 5′-GGACTACCAGGGTATCTAATCCTGTT-3′) as described by Nadkarni et al.7 To calibrate the quantitative real-time PCR (qPCR) reaction, DNA from Escherichia coli (maximum efficiency DH5α competent cells; Invitrogen) was used. Amplification and detection of DNA by qPCR were performed with the 7500 Fast Real-Time PCR Instrument (Applied Biosystems, Foster City, CA). Enumeration of bacteria was also performed using the Bacteria Counting Kit for Flow Cytometry (B-7277; Invitrogen) according to the manufacturer instructions.
Tissue Staining and Image Analysis
Formalin-fixed, paraffin-embedded, and OCT embedded tissue was sectioned (4 μm) and stained with the antibodies listed in Table 1. For immunofluorescence, Qdot streptavidin conjugates (Invitrogen) were used after application of biotinylated hamster secondary (BD Pharmingen, Franklin Lakes, NJ) or biotinylated rat and rabbit secondary (Vector Laboratories, Burlingame, CA) antibodies. Images were captured using the Mirax MIDI Whole Slide Scanner (Zeiss, Thornwood, NY). Quantitative image analysis was completed using internally developed NearCYTE software (http://www.nearcyte.org) with KCs defined as nucleated F4/80+ cells and normalized to a standard tissue area in 4-μm sections.
Western Blotting
Protein was isolated from CL, GF, and AVMN livers by tissue homogenization in RIPA buffer containing complete protease inhibitors (Roche Applied Science, Indianapolis, IN). Protein quantification was assessed with a BCA Protein Assay Kit (Pierce, Chicago, IL). Seventy-five micrograms of protein was run on SDS-PAGE. After electrotransfer to a nitrocellulose membrane, 5% milk in Tris buffered saline Tween was used for blocking, and monoclonal β-actin (clone AC-15; Sigma-Aldrich) and ICAM-1 (clone M19; Santa Cruz Biotechnology, Santa Cruz, CA) were used to probe the membrane. ECL Western blotting substrate (Thermo Scientific, Ashville, NC) was used to visualize the protein.
RNA Isolation, RT-PCR, and PCR for Murine Genes
Total RNA was extracted from homogenized liver tissue using Trizol (Invitrogen). Reverse transcription was performed using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems) according to the manufacturer instructions. To determine expression of Csf1 and Csf2 relative to the Hprt internal control, 25 ng of cDNA was used in a 10-μL reaction using the Taqman Fast Universal PCR Master Mix (Applied Biosystems) and Taqman array 96 well fast plates for mouse immune response (Applied Biosystems). Analysis was performed on a 7500 Fast Real-Time PCR Instrument (Applied Biosystems).
In Vivo Phagocytosis
Mice were injected (tail vein) with sphero carboxyl magnetic particles (1.22 μm) once (200 μL, ∼5.4 × 108 particles; Spherotech Inc., Libertyville, IL). NPCs and active liver phagocytes were isolated by magnetic separation. Phagocytes were then counted and analyzed by FC.
KC Depletion and in Vivo Antibody Blockade
CL mice were treated as follows: day 0: clodronate liposome injection (300 μL i.p.); day 3: blocking antibody injection i.p. using 50 μg (100 μL) of anti–ICAM-1 (YN1/1.7.4; Abcam, Cambridge, MA) and/or 20 μg (100 μL) of anti-LYVE1 (CD44, IM7.8.1; Cedarlane, Burlington, NC). Rat IgG2b (50 μg, 100 μL) (BD Biosciences, San Jose, CA) was used as an isotype control; and day 6: KC repopulation and monocyte frequency assessment by FC. Cl2MDP (or clodronate) was a gift from Roche Diagnostics GmbH, Mannheim, Germany. Other reagents include phosphatidylcholine (LIPOID E PC; Lipoid GmbH, Ludwigshafen, Germany), cholesterol (Sigma Chemical Co., St. Louis, MO), and CL2MDP (laboratory of Dr. Nico van Rooijen, Amsterdam, The Netherlands).
PRR Stimulation
Flagellin (FLA-ST; Invitrogen) (dose of 100 ng, 100 μL) was injected via tail vein into CL and GF mice. Controls were injected with 100 μL of PBS. NPCs were isolated 24 hours after challenge.
PRR Ligand Screening
Portal and systemic blood were collected from adult CL Swiss Webster mice. Blood plasma was collected, and alkaline phosphatase was heat inactivated by 30-minute incubation at 56°C. Briefly, Invitrogen performed PRR ligand screening by assessing NF-κB activation in HEK293 cells expressing a single PRR. NF-κB activation was monitored by measuring secreted alkaline phosphatase, a reporter gene in HEK293 cells induced by NF-κB. Plasma from CL mice was tested on nonobese diabetic (NOD) 1, NOD2, and Toll-like receptors (TLRs) 2, 3, 4, 5, and 9 mice. Plasma was tested in duplicate using a 1:10 dilution and compared with control ligands. The following were used as positive controls: heat-killed Listeria monocytogenes (TLR-2), poly (I:C) (TLR-3), E. coli K12 lipopolysaccharide (LPS) (TLR-4), Salmonella typhimurium flagellin (TLR-5), CpG ODN 1826 (TLR-9), C12-iEDAP (NOD1), L18-MDP (NOD2), and tumor necrosis factor α (NF-κB control cells). NF-κB control cells (no PRR) and unstimulated PRR-expressing HEK293 cells were used as negative controls. The results are provided as mean OD values. For semiquantification of ligands, plasma was concentrated twofold for testing (Amicon Ultra 3K Centrifugal Filters; Millipore, Marlborough, MA). Then 10-fold dilutions of plasma and positive control ligands were tested in triplicate for activation of TLR-2, TLR-5, and TLR-9. The upper and lower limits of detection for TLR screening using HEK293 cells are as follows: 106 to 109 cells/mL of TLR-2 agonist heat-killed L. monocytogenes or 0.01 to 10 ng/mL of Pam3CSK4, 100 pg/mL to 1 μg/mL of TLR-4 agonist LPS, and 1 ng/mL to 1 μg/mL of TLR-9 agonist ODN 1826.
Mouse Orthotopic Liver Transplantation
Techniques of liver harvesting and orthotopic liver transplantation without hepatic artery reconstruction were based on the method of Qian et al,8 with minor modifications.9,10 Liver grafts from AVMN or untreated C57Bl/6J mice (Jackson Labs, Bar Harbor, ME) were perfused with 5.0 mL of University of Wisconsin solution via the inferior vena cava in a retrograde fashion, stored for 24 hours at 4°C, and then implanted orthotopically into untreated B6 mice by anastomosis of the suprahepatic vena cava with a running 10-0 suture and anastomosis of the portal vein and inferior vena cava using a cuff technique. Mean ± SD anhepatic time was 22.2 ± 2.3 minutes. The bile duct was connected via ligation over the stent. The recipient animals were euthanized 6 hours after reperfusion for serum alanine aminotransferase and aspartate aminotransferase determinations and graft histopathologic analysis.
Statistical Analysis
Student’s t-test or U-test was used to determine significance between two groups. In vivo analyses were performed using three or more animals per group. Analysis of variance was used when comparing more than two groups. Data are presented as the arithmetic mean. Error bars represent SEM. All data are representative of two or more independent experiments. For all, P < 0.05 was considered statistically significant.
Results
Gut Bacteria Are a Major Source of Systemic MAMPs
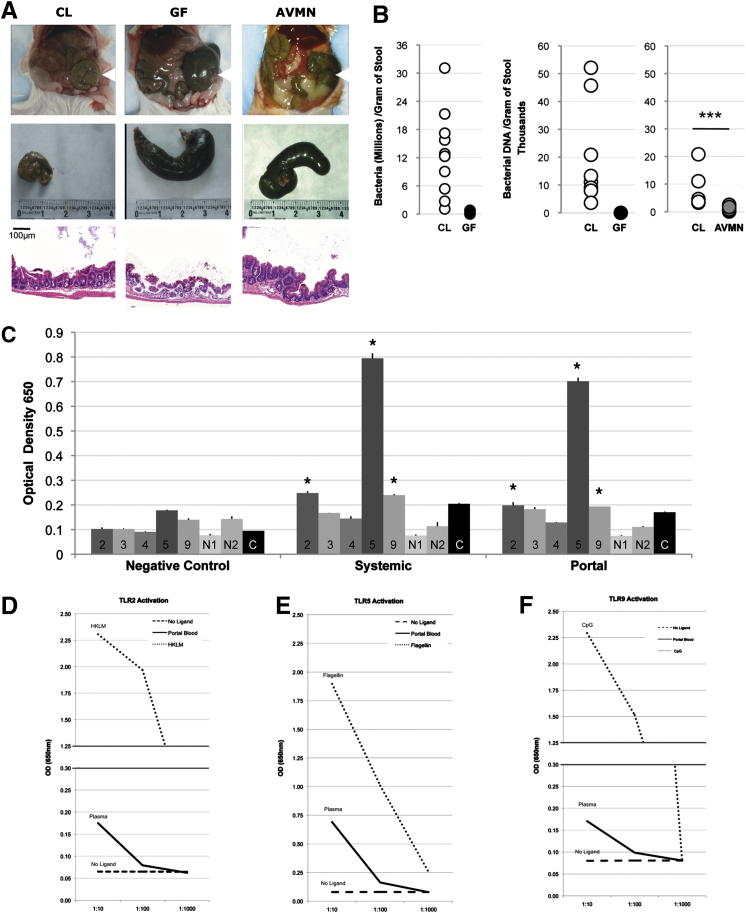
Gut bacteria help shape normal gut morphologic features as evidenced by cecal dilation in GF mice, which has been previously attributed to water and mucous accumulation and a thinner cecal muscularis propria described by Smith et al3 (Figure 1A).
Figure 1.
Gut bacteria and intestinal permeability. A: Top row: Representative images of intestines reveal cecal dilation in GF and AVMN but not in CL mice (arrowheads). Middle row: Representative images of the isolated cecum highlight dilation in GF and AVMN mice. Bottom row: Longitudinal H&E sections of cecal wall reveal thinned muscularis propria in GF and AVMN mice. Scale bar = 100 μm. ***P < 0.01. B: Bacterial density of cecal stool assessed by a Flow Cytometry Bacterial Counting Kit (see Materials and Methods) confirms the gut bacterial status of CL and GF mice and marked variability in CL mice (left). Bacterial DNA (in nanograms) in cecal stool assessed by real-time PCR amplification of 16S rRNA gene again reveals absence (GF) or marked reduction (AVMN) and considerable variability of gut bacteria in CL mice (n = 10) (P = 0.05) (right). ***P < 0.005. C: PRR ligand detection in CL portal and systemic blood assessed by NF-κB activation in HEK293 cells expressing mouse TLR-2, mouse TLR-3, mouse TLR-4/CD14/MD2, mouse TLR-5, mouse NOD1 (N1), mouse NOD2 (N2), or no PRR on NF-κB control cells. Blood pooled from 10 CL mice; means are representative of two experiments performed in duplicate, revealing significantly above background levels for TLR-2, -5, and -9. *P < 0.05 compared to control (C) levels. D–F: Estimation of TLR ligand concentrations for TLR-2, TLR-5, and TLR-9 relative to known controls in pooled plasma from CL mice. D: TLR-2 activation on HEK293 cells by no ligand (dashed line), portal blood (solid line) and heat-killed L. monocytogenes (dotted line) at 106, 107, and 108. Blood pooled from 10 mice. Samples were run in triplicate. E: TLR-5 Activation on HEK293 cells by no ligand (dashed line), portal blood (solid line), and flagellin (dotted line) at 100, 10, and 1 ng/mL. F: TLR-9 activation on HEK293 cells by no ligand (dashed line), portal blood (solid line), and CpG ODN 1826 (dotted line) at 1, 0.1, and 0.01 μg/mL.
Gut microbial density increases temporally and spatially, with the greatest bacterial loads found after weaning during adulthood in the distal gut2 (unpublished data). Bacterial density, however, can vary in relationship to the environment, mouse strain, sex, and age.11 Because >50% of gut bacteria cannot be cultivated,11 we assessed cecal bacterial density via PCR amplification of the bacterial 16S ribosomal gene and via flow cytometry. Cecal bacterial density measurements revealed that adult CL mice average >12 million bacteria per gram of stool, but CL siblings housed in the same room, cage, and specific pathogen free conditions yielded a wide range of bacterial levels (Figure 1B), likely attributable to bacterial population dynamics. As expected, cecal bacteria were undetectable in GF mice (Figure 1B), whereas AVMN mice had an 8.5-fold reduction in bacterial DNA relative to CL mice (P = 0.005) (Figure 1B).
Live, whole bacterial translocation to the liver rarely occurs under nonpathogenic homeostatic conditions12 (data not shown), but it is widely accepted that gut-derived MAMPs shed by commensals are able to penetrate the gut barrier. Yet, the amalgam of functionally active MAMPs able to translocate into portal and systemic circulation has not been fully characterized. It is also unclear whether physiologic levels of individual or combinations of MAMPs exert downstream effects on the liver. Therefore, we identified gut-derived PRR ligands by screening portal and systemic blood for MAMPs using an NF-κB activation bioassay (Figure 1C). PRR ligation by MAMPs from portal and systemic blood was observed for TLR-2, TLR-5, and TLR-9 but not for TLR-3, TLR-4, NOD1, or NOD2 (Figure 1C). Although TLR-4 and NOD ligands (LPS and PDG, respectively) are known to be detectable in the portal circulation, their physiologic concentrations may not be detectable in this bioassay or they might be in a form that does not trigger PRR signaling. Alternatively, LPS and PDG might be responsible for the observed TLR-2 activation because both are known to also bind to this receptor.
We next examined NF-κB activation in response to TLR-2 (Figure 1D), TLR-5 (Figure 1E), and TLR-9 (Figure 1F) ligation by circulating MAMPs in the portal blood compared with concentrations of known TLR ligands (heat-killed L. monocytogenes, flagellin, and CpG, respectively). Figure 1, E and F, indicate TLR-5 ligand concentration (presumably flagellin) ranges of 1 to 10 ng/mL and TLR-9 ligand concentration (presumably bacterial DNA) ranges of 0.01 to 0.1 μg/mL in portal circulation. The relative concentration of TLR-2 ligand is indeterminate because of infidelity of TLR-2 to a single ligand and recognition of multiple fungal and bacterial MAMPs.
KC Frequency and MHCII Expression Increase with Bacterial Density
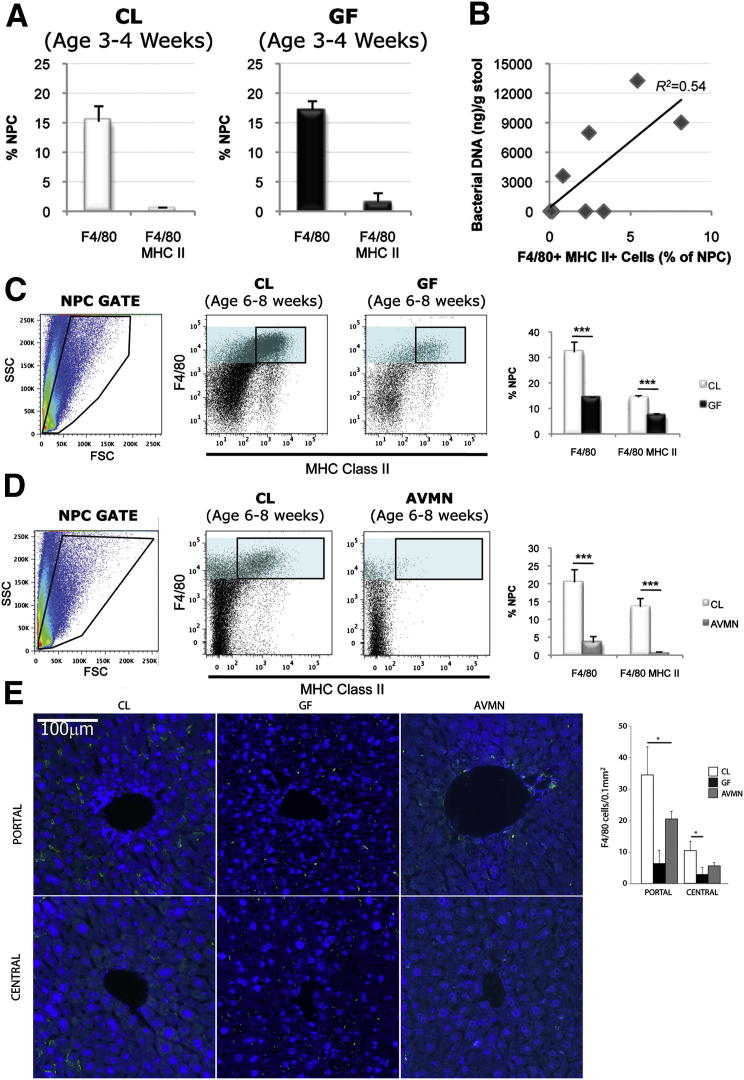
Ligation of a PRR (NOD1) by a gut-derived MAMP (PDG) results in expansion of specific mucosal immune cells (ie, regulatory T cells).13 Because liver NPCs, including KCs and LSECs, express many PRRs for MAMP detection,5 we determined whether gut bacteria stimulate expansion and/or recruitment of liver immune cell populations. Results indicate that nucleated sinusoidal cell number is reduced in GF livers (P = 0.009) (Supplemental Figure S1). However, the absolute numbers of NPCs isolated from CL and GF livers did not differ significantly (6.34 ± 1.2 million and 4.17 ± 1.2 million, respectively; P = 0.28).
We, therefore, hypothesized that gut bacteria contribute to quantitative and/or qualitative changes in the KC population. First, we evaluated KC frequency during adolescence when the gut microbiome is not fully established (data not shown). During adolescence, CL mice have 15.2% F4/80+ NPCs, and GF mice have 17.11% F4/80+ NPCs, although both CL and GF KCs are relatively immature/inactivated as reflected by the insignificant major histocompatibility complex class II (MHCII) expression (0.5% and 1.5% F4/80+MHCII+ NPCs, respectively) (Figure 2A). However, the KC population size doubles during the transition from adolescence to young adulthood in CL mice, whereas KC frequency in GF age-matched counterparts remains unchanged (32% and 14% F4/80+ NPCs in CL and GF adults, respectively) (Figure 2C). Moreover, adult CL mice have a significant population of MHCII+ KCs (14.4% NPCs), whereas adult GF mice (7.4% F4/80+MHCII+ NPCs) resemble their juvenile counterparts (Figure 2C and Supplemental Figure S2).
Figure 2.
The gut microbiota increase mature KC frequency and direct KC distribution. A: Percentage of NPCs isolated from livers of adolescent CL and GF mice expressing F4/80 (macrophage) or F4/80 and MHCII (mature macrophage) were similar. Representative of three independent experiments (n = 6 in the CL group; n = 6 in the GF group) (F4/80+P = 0.40; F4/80+ MHCII+P = 0.06). B: Correlative analysis revealed that mature KC frequency (F4/80+MHCII+ NPCs) was directly proportional to bacterial load (nanograms of bacterial DNA per gram of cecal stool); Pearson’s correlation coefficient = 0.73. C: Percentage of F4/80+ NPCs (cyan filled gate) or F4/80+MHCII+ NPCs (black lined gate) isolated from livers of adult CL mice was significantly greater than that of GF mice (n = 5 in the CL group; n = 5 in the GF group). Dot plots representative of 4 independent experiments. D: Percentage of F4/80+ NPCs (cyan filled gate) or F4/80+MHCII+ NPCs (black lined gate) isolated from livers of adult CL mice was significantly greater than AVMN mice (n = 5 in the CL group; n = 5 in the GF). Representative of two independent experiments. E: KC distribution in livers of CL, GF, and AVMN mice assessed by immunofluorescence staining of liver sections with F4/80 (green) and DAPI nuclear counterstain (blue). Analysis was performed by tissue cytometry (NearCYTE) (Supplemental Figure S3). Three lobular portal areas and central areas were each randomly selected from a total of three animals per group and F4/80+ cell numbers/0.1 mm2 counted. Representative images was shown in the figure. Scale bar = 100 μm. Adult CL, GF, and AVMN mice were used. *P < 0.05, **P < 0.025, ***P < 0.01.
Perplexed by the variability in CL KC frequency compared with GF KC frequency, we hypothesized that the former might be related to the aforementioned differences in gut bacterial loads among CL mice (Figure 1B). Results indicated that the number of mature KCs (F4/80+MHCII+ NPCs) is directly related to gut bacterial load (P = 0.024) (Figure 2B). If more gut bacteria are associated with higher KC numbers, then less gut bacteria should result in lower KC numbers. As expected, AVMN mice had a significant reduction in KCs compared with CL mice (3.5% and 20.4% NPCs in AVMN and CL mice, respectively) and lose/fail to develop the population of MHCII+ KCs (0.6% and 13.5% NPCs in AVMN and CL mice, respectively) (Figure 2D).
Twice as many KCs are located in the periportal compared with the centrilobular sinusoids.14 We therefore determined whether the KC deficiency observed in GF and AVMN mice was global or regional. CL mice exhibited three times as many KCs in the periportal regions (34.4 ± 8.9 cells/0.1 mm2) compared with the centrilobular regions (10.3 ± 3.0 cells/0.1 mm2), but GF and AVMN mice exhibited both reduced periportal (6.3 ± 4.2 cells/0.1 mm2 and 20 ± 2.5 cells/0.1 mm2) and centrilobular (2.8 ± 2.2 cells/0.1 mm2 and 5.5 ± 1.0 cells/0.1 mm2) numbers, respectively (Figure 2E and Supplemental Figure S3), although the greatest differences were seen in the periportal areas. The change in KC frequency could not be attributed to differences in liver size (P = 0.14, analysis of variance) (Supplemental Figure S4), lobular size (data not shown), or macrophage proliferation alone as assessed by Ki-67 expression (Supplemental Figure S5).
Gut Bacteria Affect KC Phenotype and Function
KCs link innate and adaptive immunity by interacting with MAMPs and circulating lymphocytes in the hepatic sinusoid via MHCII presentation of antigenic epitopes after endocytosis of circulating antigens.15 Loading of MHC molecules with antigenic peptide is often followed by up-regulation of co-stimulatory molecules CD80 and CD86.16 Because MHCII expression increased with higher bacterial loads (Figure 2, C and D), we next assessed expression of co-stimulatory molecules CD80 and CD86 on KC and phagocytic function (part of exogenous pathway of antigen presentation). Results revealed no significant differences in CD80/CD86 expression when comparing KCs from CL, GF, and AVMN mice, although KCs in GF and AVMN mice consistently expressed modestly lower levels (Supplemental Figure S6).
Because KCs characteristically exhibit low levels of co-stimulatory molecules compared with other professional antigen presentation cells,17 we assessed co-stimulatory molecule expression after flagellin stimulation, which is the most highly detectable MAMP found in both portal and systemic blood (Figure 1, C and E). Flagellin stimulation did not significantly increase or decrease co-stimulatory molecule expression by CL or GF KCs (Supplemental Figure S7).
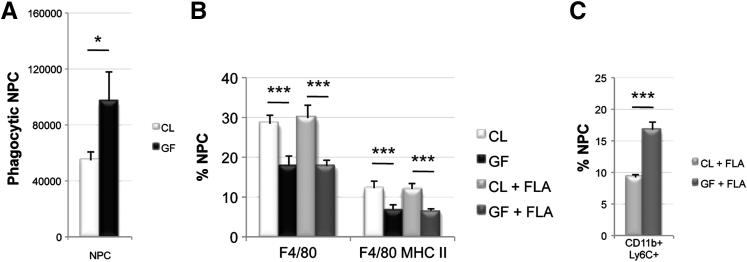
We next evaluated in vivo phagocytic activity by determining the number of active nonendothelial phagocytes after magnetic isolation of cells containing micromagnets (1.22 μm). The percentage of active phagocytes was 5.4% (54,716 phagocytes per million NPCs) in CL livers versus 9.7% (97,000 phagocytes per million NPCs) in GF livers (Figure 3A). The significantly increased phagocytic activity in GF mice suggests that MAMP exposure reduces environmental sampling, although overall collective environmental sampling might be maintained in CL mice due to increased KC numbers.
Figure 3.
Phagocytic function and in vivo response to antigenic stimulation. A: Phagocytosis (1.22 μm magnet uptake) assay reveals that GF mice have significantly more phagocytic activity than CL mice (P = 0.031). Representative of two independent experiments (n = 3 per group). B: KC frequency in CL and GF mice before and 24 hours after treatment with 100 ng of flagellin (FLA) reveals that FLA treatment does not cause significant differences in KC frequency. Representative of two independent experiments (n = 3 to 4 per group). C: Monocyte frequency (CD11b+Ly6C+ NPCs) 24 hours after FLA challenge results in greater liver monocyte influx in GF mice compared with CL mice. Representative of two independent experiments (n = 3 to 4 per group). Adult CL, GF, and AVMN mice were used. *P < 0.05, ***P < 0.01.
Because LPS can stimulate KC expansion,18 we determined whether flagellin (the most highly detectable MAMP in portal circulation) generated a CL phenotype in GF livers (Figure 3B). Neither total KC (F4/80+ NPCs) nor mature KC frequency (F4/80+MHCII+ NPCs) were altered by flagellin stimulation at physiologic concentration within the limitations of a 24-hour time course (ensuring no outside influence from environmental exposures to the GF mice). KC progenitor (monocyte) frequency, however, in GF livers nearly doubled that of CL livers after flagellin challenge (Figure 3C).
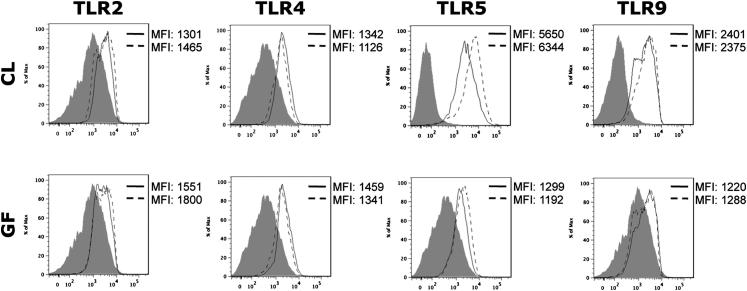
Constitutive MAMP Exposure Does Not Change PRR Expression by KCs
PRRs are conserved germline-encoded receptors,17 but expression levels of some PRRs (ie, TLR-4) can increase in response to stimulation.5 KCs express many PRRs constitutively (TLR-2, TLR-4, TLR-5, TLR-9, and NOD2),5 but we hypothesized that KC PRR expression might be dependent on constitutive MAMP exposure. Thus, TLR-2, TLR-4, TLR-5, and TLR-9 expression was evaluated before and after flagellin stimulation (Figure 4). Results reveal that CL KC and GF KC express surface TLR-2, TLR-4, TLR-5, and TLR-9 (Figure 4). TLR-5 expression was modestly increased in response to flagellin, but overall KC surface TLR expression did not appear to be dependent on exposure to gut bacteria/MAMPs (Figure 4).
Figure 4.
Germ status does not change PRR expression. TLR-2, -4, -5, and -9 expression on KCs from CL and GF mice before (solid line) or 24 hours after treatment (dashed line) with 100 ng of flagellin revealed no change in TLR expression. Isotype controls are shown in gray. Histograms are representative of two independent experiments (n = 3 to 4 per group). Adult CL and GF mice were used.
Classical KC Progenitor Frequency Is Independent of Germ Status
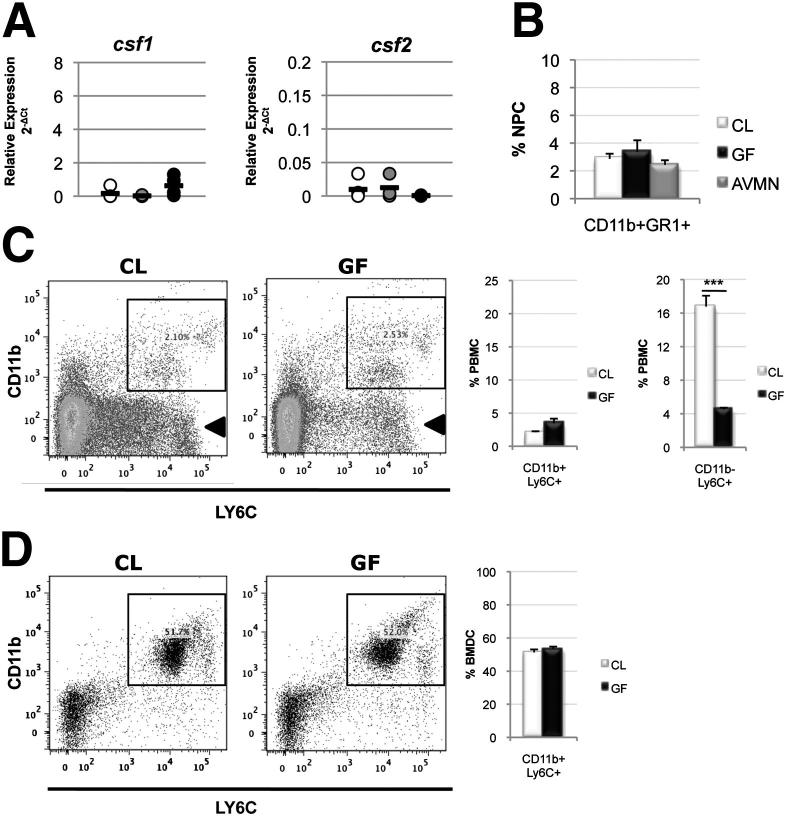
KCs are derived from bone marrow–derived monocytes under the influence of colony-stimulating factors as defined by the mononuclear phagocyte system.14 Therefore, the KC deficiency associated with low gut bacteria could be due to failure of KC progenitors to differentiate or deficiency of KC progenitors themselves. First, macrophage-colony stimulating factor (Csf1) and granulocyte-macrophage colony-stimulating factor (Csf2), which drive monocyte differentiation and promote KC survival, were not differentially expressed (cDNA assessed by RT-PCR) between CL (mean 2-ΔCt: csf1 0.16, csf2 0.009), AVMN (mean 2-ΔCt: csf1 0.02, csf2 0.01), and GF (mean 2-ΔCt: csf1 0.62, csf2 0.0006) livers relative to the Hprt1 internal control (Figure 5A). Second, monocyte frequency (percentage of CD11b+Ly6C+ or CD11b+GR1+ cells) in the liver (<3% of NPCs in CL, GF, and AVMN mice) (Figure 5B), circulation (CL mice, 2.1% PBMCs; GF mice, 3.4% PBMCs) (Figure 5C), and bone marrow (CL mice, 51.3% BMDCs; GF mice, 53.4% BMDCs) (Figure 5D) were unaffected by gut bacteria.
Figure 5.
KC progenitor development is independent of gut bacteria. A: Relative expression of monocyte colony-stimulating factor (csf1) (analysis of variance, P = 0.50) and granulocyte-monocyte colony-stimulating factor (csf2) (analysis of variance, P = 0.84) mRNA revealed no significant differences among the mouse groups. Black line represents mean expression, with Hrpt1 used as an internal control [n = 4 in the CL group (white circle), n = 4 in the AVMN group (gray circle), and n = 4 in the GF group (black circle)]. B: KC progenitors in the livers of CL, GF, and AVMN mice (analysis of variance, P = 0.663) revealed no difference among groups. Representative of three independent experiments (n = 3 to 6 in the CL group, n = 3 to 5 in the GF group, and n = 3 to 4 in the AVMN group). C: KC progenitors in systemic circulation (percentage of PBMCs) in CL and GF mice were not significantly different; CD11b+Ly6C+ PBMCs (lined gate) (P = 0.27). Circulating CD11b-Ly6C+ PBMC counts (arrow) were significantly lower in GF mice (n = 6 in the CL group; n = 6 in the GF group). D: KC progenitors in bone marrow (percentage of MDCs) in CL (n = 6) and GF (n = 6) mice were not significantly different (P = 0.37); CD11b+Ly6C+ BMDCs (lined gate). CL, GF, and AVMN mice were used. ***P < 0.01.
Notably, however, a population of circulating CD11b−Ly6C+ cells was found to be reduced in GF mice (Figure 5C). CD11b−Ly6C+ cells are not classically characterized as monocytes (myeloid lineage), but there is some plasticity within the mononuclear phagocyte system19; thus, this cell population cannot be entirely excluded as precursors to KCs or other cell types. Regardless, these findings indicate that the frequency of intrahepatic monocytes (classic KC progenitors) is independent of gut bacteria, and the mobilization of monocytes from the bone marrow to circulation is not defective. Therefore, another mechanism is responsible for the KC deficiency associated with gut bacteria deficits.
MAMP Exposure Induces ICAM-1 and LYVE-1 Expression by LSECs Increasing KC Retention
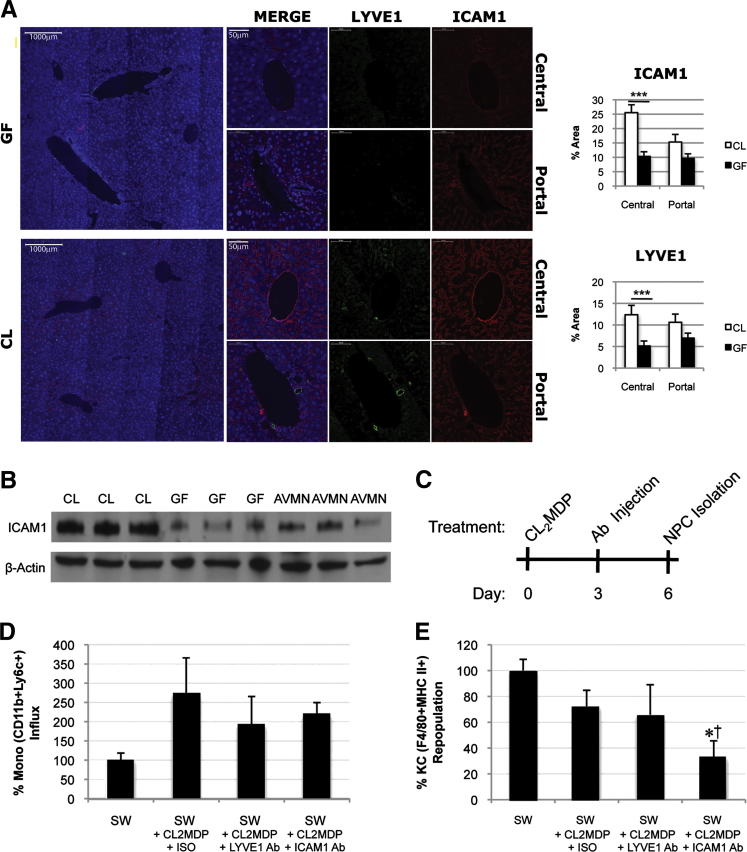
Monocyte recruitment to the liver in the infectious setting is stimulated by up-regulation of adhesion molecules on LSECs,20 although some adhesion molecules are thought to be constitutively expressed in the liver, including ICAM-1.21 Thus, we determined whether basal adhesion molecule expression is modulated by MAMPs and whether a change in adhesion molecule expression alters KC number. We first examined ICAM-1 expression on LSECs (LYVE1+ cells), but LYVE-1, an LSEC marker and adhesion molecule,22 was not consistently expressed between CL and GF mice (Figure 6A). Compared with expression in CL mice, quantitative morphometric examination of LYVE1 expression in GF livers was reduced (Figure 6A). ICAM-1 expression was similarly reduced in GF livers, and particularly so in the centrilobular regions (Figure 6A). We confirmed by Western blotting that whole liver ICAM-1 expression was significantly reduced in GF and AVMN mice (P < 0.001) (Figure 6B).
Figure 6.
Constitutive ICAM-1 expression by LSECs is induced by the gut microbiota and permits KC repopulation. A: ICAM-1 (red) and LYVE-1 (green) expression on LSECs [DAPI (blue nuclear stain)]. Original magnification, ×10 (representative images of liver section); ×30 (representative images of portal/central regions). Quantification of immunofluorescence area per high-power field revealed significantly higher ICAM-1 and LYVE-1 expression in CL mice (n = 3 in the CL group; n = 3 in the GF group). Five fields per region per mouse for a total of 15 images per group were used for analysis. ***P < 0.01. B: Western blot reveals higher hepatic ICAM-1 expression in CL mice compared with GF and AVMN mice (n = 3). C: Injection scheme for repopulation model; CL2MDP was used to deplete KCs on day 0. Either ICAM-1 or LYVE-1 blockade was performed on day 3 after KC depletion. KC and monocyte frequency was assessed by flow cytometry on day 6 afrer KC depletion. D–E: Monocyte influx (CD11b+Ly6C+ NPCs) (D) and untreated CL Swiss Webster mice were used as negative controls (100% influx). KC repopulation (F4/80+MHCII+ NPCs) (E). Untreated CL SW mice were used as negative controls (100% repopulation). Representative of two independent experiments (n = 3 to 4 per group). CL, AVMN, and GF mice were used. *P < 0.05 experimental group versus negative control; †P < 0.05 experimental group versus positive control.
A repopulation model was next used to determine whether LYVE-1 and ICAM-1 participate in shaping the KC profile under noninfectious conditions using a liposome-entrapped clodronate (CL2MDP) KC depletion protocol (Figure 6C). Monocyte influx and KC repopulation were tracked after ICAM-1 or LYVE-1 blockade. Neither ICAM-1 nor LYVE-1 blockade completely limited liver monocyte trafficking after KC depletion (Figure 6D). However ICAM-1, but not LYVE-1, blockade impeded KC repopulation (Figure 6E). ICAM-1 blockade reduced monocyte influx by 53% (P = 0.32), and KC population was stunted by 68% (P = 0.04). LYVE-1 blockade reduced monocyte influx by 80% (P = 0.27), and KC population was stunted by 35% (P = 0.41). Results indicate that monocyte recruitment is not dependent on ICAM-1 alone, but full restoration of the mature KC population after depletion appears to be ICAM-1 dependent (Figure 6, D and E).
Reduced KC Frequency Limits Hepatic Ischemia/Reperfusion Injury after Orthotopic Liver Transplantation
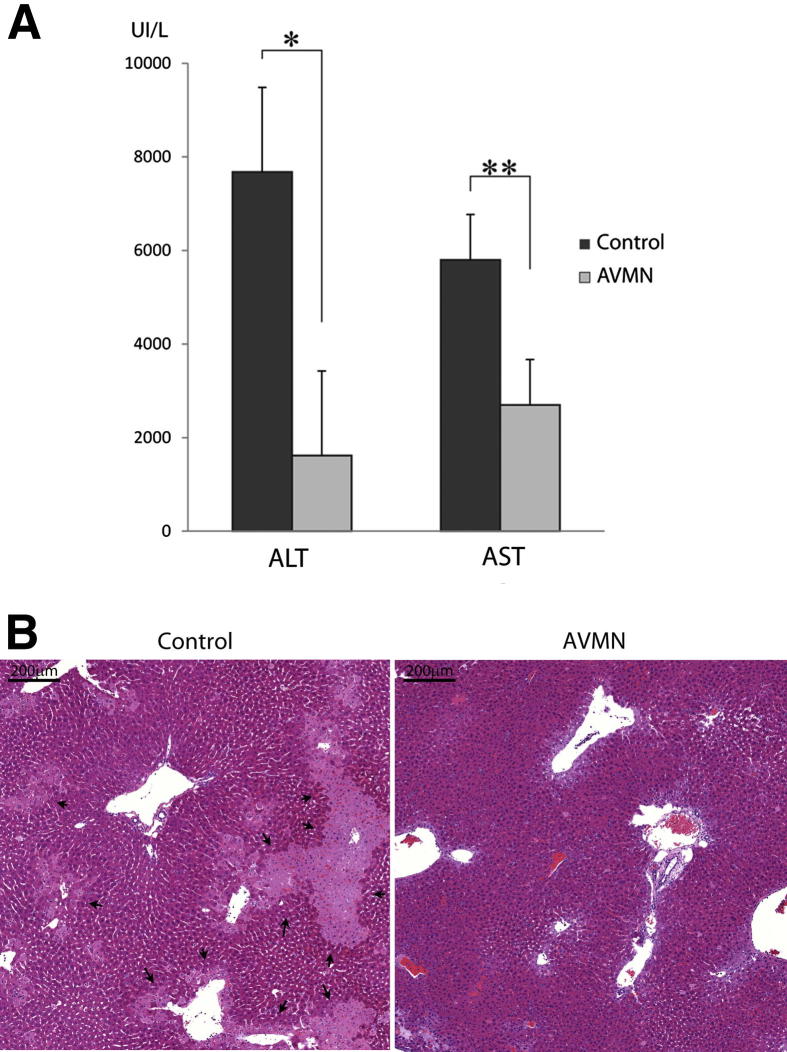
KCs can either promote or prevent liver injury, depending on the insult and context. In hepatic ischemia/reperfusion injury after orthotopic liver transplantation, KCs can release hepatotoxic mediators that promote the injury.23 For example, activated KCs increase liver injury, whereas inactivated KCs abrogate liver injury induced by ischemia/reperfusion (reviewed in Shiratori et al23). We tested, therefore, the hypothesis that reduced KC frequency and lower MHCII expression, as seen in GF and AVMN mice, might also lessen injury after cold preservation/reperfusion and transplantation. As expected, KC deficiency secondary to gut bacteria reduction profoundly reduces hepatocellular damage as evidenced by significantly lower alanine aminotransferase and aspartate aminotransferase levels and liver histopathologic findings (Figure 7). AVMN-treated mice had less coagulative-type hepatocyte necrosis and less sinusoidal neutrophil infiltration, particularly near the areas of necrosis, both of which were slightly accentuated in perivenular regions.
Figure 7.
Reduced KC frequency limits liver injury mediated by ischemia reperfusion after cold storage and transplantation. A: After 24 hours of cold preservation and 6 hours after reperfusion after orthotopic liver transplantation, AVMN mice (n = 5) had significantly lower alanine aminotransferase (P = < 0.001) and aspartate aminotransferase (P = 0.003) levels compared with CL mice (n = 5). B: Liver histologic analysis confirmed noticeably less injury in the AVMN mice that harbor significantly fewer KCs. The foci of coagulative-type necrosis were irregularly shaped and more often located in perivenular regions (arrows). *P < 0.001, **P < 0.01.
Discussion
Uniqueness of the liver as an immunologic organ resides, in large part, in the enrichment of specialized leukocyte subsets that are not found at the same concentrations in other organs,24 such as the high proportion of natural killer, natural killer T, and CD5+ B cells and the highest concentration of macrophages (KCs) of any organ. We report that gut bacteria and constitutive MAMP exposure via portal blood determine KC number, MHCII expression, and phagocytic/sinusoidal content monitoring, which in turn significantly influence how KCs and the liver respond to injury after cold preservation and orthotopic transplantation.
Germline KC PRR expression, regardless of gut bacteria, and MAMP exposure trigger KC maturation/activation characterized by increased MHCII+ KCs, decreased phagocytic activity, but persistently low co-stimulatory molecule expression. MAMPs, therefore, help shape a relatively tolerogenic KC phenotype under normal conditions, which in turn likely contributes to T-cell tolerance along with KC secretion of IL-10 and transforming growth factor-β.25
Previous studies report that monoassociation of GF mice with Lactobacillus delbrueckii,26 inoculation of GF rodents with Saccharomyces cerevisiae,27 and monoassociation of GF rats with E. coli18 all lead to increased KC numbers. Komatsu et al21 previously reported dynamic regulation of liver endothelial ICAM-1 expression by gut bacteria. Increased LSEC ICAM-1 expression has been associated with monocyte/macrophage recruitment during bacterial infection22 and KC expansion in old age.28
We have confirmed, amalgamated, and furthered previous observations by reporting that under physiologic conditions gut bacteria i) release a unique cocktail of MAMPs into circulation, which up-regulate LSEC ICAM-1 expression; ii) density directly increases KC numbers and activation/maturational status, including a tolerogenic profile; iii) density fluctuations occur naturally or can be induced via broad-spectrum antibiotic treatment, which in turn influence KC-dependent functions; and iv) markedly increase susceptibility to preservation/reperfusion liver injury after cold storage and orthotopic liver transplantation. Previous studies in humans and rats found a direct correlation between gut-derived bacterial products in the recipient and the severity of preservation/reperfusion injury.29–31
Other potential influences of the gut microbiome and MAMPs on KC function include clearance of bacteria, phagocytosis of malignant cells, platelet aggregates, activated complement components, antigen presentation to T cells/activation, and T-cell tolerance.32
In summary, we found a strong correlation between gut bacterial density, which can vary widely because of bacterial population dynamics or antibiotic treatment, and KC density and maturational status/functionality, which in turn can influence many aspects of hepatic pathophysiology, such as ischemia/reperfusion injury, tolerance induction, and bacteria/virus clearance.23,25 Continuing research, therefore, of the effect of the quantity, quality, and relative composition of gut bacteria on portal MAMPs and liver physiology and pathophysiology is warranted.
Footnotes
Supported by NIH/National Institute of Diabetes and Digestive and Kidney Diseases grants F31DK089902-01 (N.C.) and PO1 A1081678 (A.J.D.), the Thomas E. Starzl Transplant Endowment Fund (A.J.D.), and University of North Carolina grant P30 DK34987, Center for Gastrointestinal Biology and Disease (CGIBD) core support. Cl2MDP was a gift from Roche Diagnostics GmbH (Mannheim, Germany).
Supplemental Data
Nucleated hepatic sinusoidal cell quantification; 10 high-power fields per region per group (n = 3) of H&E sections were used to quantify sinusoidal cell count. GF and CL mice were used. Original magnification, ×40. *P < 0.05.
Absolute KC frequency (product of absolute NPC count and mean KC percentage, n = 6), representative of three independent experiments. *P < 0.05.
NearCYTE analysis of whole slide images for KC enumeration. Three steps were performed to confirm identification of cell subsets for enumeration. Step 1: Define threshold for positivity. Step 2: Nuclear segmentation according to the size of nuclei and DAPI staining intensity. Step 3: Classification according to immunofluorescent signal from KC marker F4/80. Right panel shows an example of an original image used for cell enumeration representing immunofluorescent staining of KCs (F4/80 green, DAPI blue). Middle panel represents the nuclear segmentation step in which all cell nuclei are identified and highlighted green. Each nucleus contains central red dots, which represent the nuclear geographic center. Left panel indicates the classification step. Briefly, each nucleus is segmented into eight sections. If more than three segments are F4/80+, the cell is classified as a KC.
Mean liver (LM) and body mass (BM). Adult GF (n = 12), CL (n = 12), and AVMN (n = 8) mice were used; BM (analysis of variance, P = 0.029), LM (analysis of variance, P = 0.31), and LM/BM ratio (analysis of variance, P = 0.14).
Liver cell proliferation. Ki-67 (green) indicates cell that has entered cell cycle. Representative images shown from CL, GF, and AVMN mice; 15 to 20 images per region per group (n = 3) used for quantification. Both hepatocytes and sinusoidal Ki-67+ cells were included in the quantification. Original magnification, ×40.
Co-stimulatory molecule expression (CD80 and CD86) by KC from CL, GF, and AVMN mice revealed no significant difference in the percentage of F4/80+ NPCs expressing CD80 (>90% for each group, P = 0.50) or in the percentage of F4/80+ NPCs expressing CD86 (>20% for each group, P = 0.17). Isotype controls are shown in gray. Histograms are representative of two independent experiments (n = 5 to 6 per group).
Co-stimulatory molecule expression on KC from CL and GF mice before (black solid line) and 24 hours after treatment (black dashed line) with 100 ng of flagellin (FLA) revealed no significant difference. Isotype controls are shown in gray. Histograms are representative of two independent experiments (n = 3 to 4 per group).
Supplemental Data
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2012.09.010.
References
- 1.Ley R.E., Peterson D.A., Gordon J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Bouskra D., Brezillon C., Berard M., Werts C., Varona R., Boneca I.G., Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456(7221):507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 3.Smith K., McCoy K.D., Macpherson A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Clarke T.B., Davis K.M., Lysenko E.S., Zhou A.Y., Yu Y., Weiser J.N. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seki E., Brenner D.A. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48(1):322–335. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- 6.Steib C.J., Gerbes A.L. Signaling pathways in liver diseases Kupffer cells. Signaling Pathways in Liver Disease. In: Dufour J.-F., Clavien P.-A., editors. Springer; Berlin: 2010. pp. 69–78. [Google Scholar]
- 7.Nadkarni M.A., Martin F.E., Jacques N.A., Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(Pt 1):257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 8.Qian S., Demetris A., Murase N., Rao A., Fung J., Starzl T. Murine liver allograft transplantation: tolerance and donor cell chimerism. Hepatology. 1994;19(4):916–924. doi: 10.1002/hep.1840190418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueki S., Castellaneta A., Yoshida O., Ozaki K., Zhang M., Kimura S., Isse K., Ross M., Shao L., Stolz D., Thomson A., Demetris A., Geller D., Murase N. Hepatic B7 homolog 1 expression is essential for controlling cold ischemia/reperfusion injury after mouse liver transplantation. Hepatology. 2011;54(1):216–228. doi: 10.1002/hep.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueki S., Dhupar R., Cardinal J., Tsung A., Yoshida J., Ozaki K., Klune J., Murase N., Geller D. Critical role of interferon regulatory factor-1 in murine liver transplant ischemia reperfusion injury. Hepatology. 2010;51(5):1692–1701. doi: 10.1002/hep.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fushuku S., Fukuda K. Inhomogeneity of fecal flora in separately reared laboratory mice, as detected by denaturing gradient gel electrophoresis (DGGE) Exp Anim. 2008;57(2):95–99. doi: 10.1538/expanim.57.95. [DOI] [PubMed] [Google Scholar]
- 12.Menard S., Cerf-Bensussan N., Heyman M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol. 2010;3(3):247–259. doi: 10.1038/mi.2010.5. [DOI] [PubMed] [Google Scholar]
- 13.Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., Taniguchi T., Takeda K., Hori S., Ivanov I.I., Umesaki Y., Itoh K., Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naito M., Hasegawa G., Ebe Y., Yamamoto T. Differentiation and function of Kupffer cells. Med Electron Microsc. 2004;37(1):16–28. doi: 10.1007/s00795-003-0228-x. [DOI] [PubMed] [Google Scholar]
- 15.Roberts R.A., Ganey P.E., Ju C., Kamendulis L.M., Rusyn I., Klaunig J.E. Role of the Kupffer cell in mediating hepatic toxicity and carcinogenesis. Toxicol Sci. 2007;96(1):2–15. doi: 10.1093/toxsci/kfl173. [DOI] [PubMed] [Google Scholar]
- 16.Murphy K.P. 7th ed. Garland Science; New York: 2008. Janeway’s Immunobiology. [Google Scholar]
- 17.Wu J., Meng Z., Jiang M., Zhang E., Trippler M., Broering R., Bucchi A., Krux F., Dittmer U., Yang D., Roggendorf M., Gerken G., Lu M., Schlaak J.F. Toll-like receptor-induced innate immune responses in non-parenchymal liver cells are cell type-specific. Immunology. 2010;129(3):363–374. doi: 10.1111/j.1365-2567.2009.03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Billiar T.R., Maddaus M.A., West M.A., Dunn D.L., Simmons R.L. The role of intestinal flora on the interactions between nonparenchymal cells and hepatocytes in coculture. J Surg Res. 1988;44(4):397–403. doi: 10.1016/0022-4804(88)90182-5. [DOI] [PubMed] [Google Scholar]
- 19.Hume D.A. The mononuclear phagocyte system. Curr Opin Immunol. 2006;18(1):49–53. doi: 10.1016/j.coi.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Body-Malapel M., Dharancy S., Berrebi D., Louvet A., Hugot J.P., Philpott D.J., Giovannini M., Chareyre F., Pages G., Gantier E., Girardin S.E., Garcia I., Hudault S., Conti F., Sansonetti P.J., Chamaillard M., Desreumaux P., Dubuquoy L., Mathurin P. NOD2: a potential target for regulating liver injury. Lab Invest. 2008;88(3):318–327. doi: 10.1038/labinvest.3700716. [DOI] [PubMed] [Google Scholar]
- 21.Komatsu S., Berg R.D., Russell J.M., Nimura Y., Granger D.N. Enteric microflora contribute to constitutive ICAM-1 expression on vascular endothelial cells. Am J Physiol Gastrointest Liver Physiol. 2000;279(1):G186–G191. doi: 10.1152/ajpgi.2000.279.1.G186. [DOI] [PubMed] [Google Scholar]
- 22.Shi C., Velazquez P., Hohl T.M., Leiner I., Dustin M.L., Pamer E.G. Monocyte trafficking to hepatic sites of bacterial infection is chemokine independent and directed by focal intercellular adhesion molecule-1 expression. J Immunol. 2010;184(11):6266–6274. doi: 10.4049/jimmunol.0904160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolios G., Valatas V., Kouroumalis E. Role of Kupffer cells in the pathogenesis of liver disease. World J Gastroenterol. 2006;12(46):7413–7420. doi: 10.3748/wjg.v12.i46.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Racanelli V., Rehermann B. The liver as an immunological organ. Hepatology. 2006;43(2 Suppl 1):S54–S62. doi: 10.1002/hep.21060. [DOI] [PubMed] [Google Scholar]
- 25.You Q., Cheng L., Kedl R.M., Ju C. Mechanism of T cell tolerance induction by murine hepatic Kupffer cells. Hepatology. 2008;48(3):978–990. doi: 10.1002/hep.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.dos Santos L.M., Santos M.M., de Souza Silva H.P., Arantes R.M., Nicoli J.R., Vieira L.Q. Monoassociation with probiotic Lactobacillus delbrueckii UFV-H2b20 stimulates the immune system and protects germfree mice against Listeria monocytogenes infection. Med Microbiol Immunol. 2011;200(1):29–38. doi: 10.1007/s00430-010-0170-1. [DOI] [PubMed] [Google Scholar]
- 27.Martins F.S., Rodrigues A.C., Tiago F.C., Penna F.J., Rosa C.A., Arantes R.M., Nardi R.M., Neves M.J., Nicoli J.R. Saccharomyces cerevisiae strain 905 reduces the translocation of Salmonella enterica serotype Typhimurium and stimulates the immune system in gnotobiotic and conventional mice. J Med Microbiol. 2007;56(Pt 3):352–359. doi: 10.1099/jmm.0.46525-0. [DOI] [PubMed] [Google Scholar]
- 28.Le Couteur D.G., Warren A., Cogger V.C., Smedsrod B., Sorensen K.K., De Cabo R., Fraser R., McCuskey R.S. Old age and the hepatic sinusoid. Anat Rec (Hoboken) 2008;291(6):672–683. doi: 10.1002/ar.20661. [DOI] [PubMed] [Google Scholar]
- 29.Arai M., Mochida S., Ohno A., Arai S., Fujiwara K. Selective bowel decontamination of recipients for prevention against liver injury following orthotopic liver transplantation: evaluation with rat models. Hepatology. 1998;27(1):123–127. doi: 10.1002/hep.510270120. [DOI] [PubMed] [Google Scholar]
- 30.Mochida S., Arai M., Ohno A., Fujiwara K. Bacterial translocation from gut to portal blood in the recipient as a factor of hypercoagulopathy in hepatic sinusoids after orthotopic liver transplantation in rats. Transplant Proc. 1997;29(1-2):874–875. doi: 10.1016/s0041-1345(96)00183-2. [DOI] [PubMed] [Google Scholar]
- 31.Fiorini R.N., Shafizadeh S.F., Polito C., Rodwell D.W., Cheng G., Evans Z., Wan C., Belden S., Haines J.K., Birsner J., Lewin D., Wasiluk K.R., Dunn D.L., Schmidt M.G., Chavin K.D. Anti-endotoxin monoclonal antibodies are protective against hepatic ischemia/reperfusion injury in steatotic mice. Am J Transplant. 2004;4(10):1567–1573. doi: 10.1111/j.1600-6143.2004.00549.x. [DOI] [PubMed] [Google Scholar]
- 32.Bilzer M., Roggel F., Gerbes A.L. Role of Kupffer cells in host defense and liver disease. Liver Int. 2006;26(10):1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleated hepatic sinusoidal cell quantification; 10 high-power fields per region per group (n = 3) of H&E sections were used to quantify sinusoidal cell count. GF and CL mice were used. Original magnification, ×40. *P < 0.05.
Absolute KC frequency (product of absolute NPC count and mean KC percentage, n = 6), representative of three independent experiments. *P < 0.05.
NearCYTE analysis of whole slide images for KC enumeration. Three steps were performed to confirm identification of cell subsets for enumeration. Step 1: Define threshold for positivity. Step 2: Nuclear segmentation according to the size of nuclei and DAPI staining intensity. Step 3: Classification according to immunofluorescent signal from KC marker F4/80. Right panel shows an example of an original image used for cell enumeration representing immunofluorescent staining of KCs (F4/80 green, DAPI blue). Middle panel represents the nuclear segmentation step in which all cell nuclei are identified and highlighted green. Each nucleus contains central red dots, which represent the nuclear geographic center. Left panel indicates the classification step. Briefly, each nucleus is segmented into eight sections. If more than three segments are F4/80+, the cell is classified as a KC.
Mean liver (LM) and body mass (BM). Adult GF (n = 12), CL (n = 12), and AVMN (n = 8) mice were used; BM (analysis of variance, P = 0.029), LM (analysis of variance, P = 0.31), and LM/BM ratio (analysis of variance, P = 0.14).
Liver cell proliferation. Ki-67 (green) indicates cell that has entered cell cycle. Representative images shown from CL, GF, and AVMN mice; 15 to 20 images per region per group (n = 3) used for quantification. Both hepatocytes and sinusoidal Ki-67+ cells were included in the quantification. Original magnification, ×40.
Co-stimulatory molecule expression (CD80 and CD86) by KC from CL, GF, and AVMN mice revealed no significant difference in the percentage of F4/80+ NPCs expressing CD80 (>90% for each group, P = 0.50) or in the percentage of F4/80+ NPCs expressing CD86 (>20% for each group, P = 0.17). Isotype controls are shown in gray. Histograms are representative of two independent experiments (n = 5 to 6 per group).
Co-stimulatory molecule expression on KC from CL and GF mice before (black solid line) and 24 hours after treatment (black dashed line) with 100 ng of flagellin (FLA) revealed no significant difference. Isotype controls are shown in gray. Histograms are representative of two independent experiments (n = 3 to 4 per group).