Abstract
HBO1 acetylates lysine residues of histones and is involved in DNA replication and gene transcription. Two isoforms of JADE1, JADE1S and JADE1L, bind HBO1 and promote acetylation of histones in chromatin context. We characterized the role of JADE1-HBO1 complexes in vitro and in vivo during epithelial cell replication. Down-regulation of JADE1 by siRNA diminished the rate of DNA synthesis in cultured cells, decreased endogenous HBO1 protein expression, and prevented chromatin recruitment of replication factor Mcm7, demonstrating that JADE1 is required for cell proliferation. We used a murine model of acute kidney injury to examine expression of HBO1-JADE1S/L in injured and regenerating epithelial tissue. In control kidneys, JADE1S, JADE1L, and HBO1 were expressed in nuclei of proximal and distal tubular epithelial cells. Ischemia and reperfusion injury resulted in an initial decrease in JADE1S, JADE1L, and HBO1 protein levels, which returned to baseline during renal recovery. HBO1 and JADE1S recovered as cell proliferation reached its maximum, whereas JADE1L recovered after bulk proliferation had ceased. The temporal expression of JADE1S correlated with the acetylation of histone H4 on lysines 5 and 12, but not with acetylation of histone H3 on lysine 14, demonstrating that the JADE1S-HBO1 complex specifically marks H4 during epithelial cell proliferation. These data implicate JADE1-HBO1 complex in acute kidney injury and suggest distinct roles for JADE1 isoforms during epithelial cell recovery.
Cellular proliferation and differentiation are regulated in part by post-translational modification of histones. Acetylation of histones has been functionally linked to DNA replication, repair, and gene transcription.1–9 In general, it is believed that during DNA replication and transcription, coordinated bulk histone acetylation and deacetylation are required for proper remodeling of chromatin. In addition, acetylation of histones at specific lysine residues creates binding sites for recruitment of activators or inhibitors of gene transcription.6,9,10 Acetylation of specific lysine residues is executed by several families of histone acetyl transferase complexes.11–13 Most histone acetyl transferases (HATs) require protein partners and function within protein complexes to perform their specific actions. The cooperative interactions between proteins in HAT complexes provide functional regulation and substrate specificity.14–17
The protein, Gene for Apoptosis and Differentiation-1 (JADE1, also known as PHF17), has been originally identified by the yeast two-hybrid system as a protein partner of von Hippel-Lindau tumor suppressor (pVHL),18 which is the key regulator of cellular oxygen sensing pathway. JADE1 contains one canonical Cys4HisCys3 plant homeo domain (PHD) followed by a noncanonical extended PHD domain, which are zinc-binding motifs. JADE1 mRNA gives rise to two protein products: a full-length JADE1L consisting of 842 amino acids, and its truncated splice variant, JADE1S, that is missing a large C-terminal fragment of 333 amino acids. The short isoform of JADE1 is the most described JADE family protein so far.
We previously reported that endogenous JADE1S is localized to the cell nucleus and that ectopically expressed JADE1S possesses intrinsic transcriptional activity.15 We demonstrated that JADE1 promotes endogenous histone H4 acetylation by associating with a histone H4–specific endogenous HAT.15
Histone acetyltransferase HBO1 (MYST2, KAT711), was originally identified using a yeast two-hybrid screen as a HAT binding origin recognition complex-1.2,7 The 611-amino acid HBO1 polypeptide contains a serine-rich zinc finger followed by a 270-amino acid C-terminal MYST homology domain, which is also present in several other known members of this family. Information regarding the biological role of HBO1 is still limited. HBO1 has been implicated in regulation of DNA replication licensing, transcriptional regulation by the androgen receptor, progesterone receptor, lymphomagenesis, adipogenesis, and embryonic development.7,19–24 HBO1 also plays an important role in the cellular stress response.7,24,25
We recently reported the cooperative interactions between HBO1, JADE1 and inhibitor of growth 4/5 (ING4/5) in the formation of a HAT complex. We proposed that JADE1S targets a HAT to histone substrate in chromatin context15 via its PHD zinc fingers and demonstrated that JADE1 is crucial for HBO1 to acetylate histone H4 in a chromatin context. According to the recent report, HBO1 also interacts in vivo with another PHD zinc finger protein bromodomain-containing protein 1, BRD1 (BRPF2).17 These findings suggest that cellular activities of the HBO1 complex might be controlled by the presence of PHD zinc finger targeting proteins, such as ING4/5, JADE1/2/3, or BRD1.14,15,17,26
The in vivo roles of HBO1 and JADE1 have not been defined. Very few studies investigating the role of HBO1 and JADE1 were done using animal models or human subjects. HBO1 knockout mice were not viable.24 It has been suggested that HBO1 is required for transcriptional regulation of key developmental genes responsible for embryonic patterning, and for bulk histone H3K14 acetylation. In contrast to studies that reported cell cycle arrest in HBO1-depleted cultured cells, no defects were found in DNA replication or cell proliferation in HBO1 mutant primary embryonic fibroblasts or immortalized fibroblasts.24 In a study done with JADE1 hypomorphic mice generated by the gene trap method,27 the resultant JADE1 promoter-driven fusion protein was highly expressed in a regulated manner during embryogenesis, suggesting a developmental role for JADE1.
According to in vitro and cell culture studies, HBO1 and JADE1 have similar chromatin substrate specificities: both prefer either directly acetylating (HBO1) or potentiating (JADE1) acetylation of histone H4 in chromatin context.14–16 Surprisingly, HBO1 knockout mice embryos had unchanged levels of histone H4 acetylation, whereas histone H3K14 acetylation was significantly diminished.24 The substrate specificity of JADE1 in vivo has not been elucidated.
In this study, we further investigated the interactions between JADE1 and HBO1 in cultured cells, and present data revealing a role for JADE1-HBO complexes in DNA synthesis. Histone modifications have been implicated in tissue and organ regeneration.28 Thus, HAT complexes may control regenerative processes in various organs. To explore the interplay of JADE1 and HBO1 in a live animal for the first time, we examined the temporal and spatial expression of JADE1S, JADE1L, and HBO1 during organ injury and regeneration. We chose an established model of reversible acute kidney injury caused by ischemia-reperfusion (IR). The detailed analysis of JADE1S, JADE1L, HBO1, and potential substrates indicates in vivo interactions and substrate specificities for JADE1 and HBO1.
Materials and Methods
Cell Lines
H1299, HeLa, and 293T/17 were from ATCC (Manassas, VA). Conditionally immortalized mouse proximal tubule epithelial cell line (BUMPT) expressing SV40 T antigen was described previously.29 Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin-streptomycin (Invitrogen/Life Technologies, Carlsbad, CA). Subconfluent cells grown in 35-, 60-, or 100-mm dishes were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Antibodies and Chemicals
Rabbit polyclonal sera and affinity-purified antibodies specific for JADE1S, JADE1L, and HBO1 were custom generated (Proteintech Group, Chicago, IL; details are available on request). FLAG M5 and α-tubulin monoclonal antibodies, as well as polyclonal antibodies for FLAG, were from Sigma-Aldrich (St. Louis, MO). HA monoclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibodies for histones—acetyl-H4, acetyl-H3, H4acK5, and H4acK12—were from AbD Serotec (Kidlington, UK). Rabbit polyclonal antibodies specific for total H3 and total histone H4 were from Abcam (Cambridge, UK). Polyclonal antibodies for histone H3acK14 were from Millipore (Billerica, MA). Polyclonal rabbit Ki-67, monoclonal mouse cell division control protein 6 (cdc6) antibodies were from Bethyl Laboratories (Montgomery, TX), monoclonal mouse Cdc45, cyclin A, cyclin B1, and mini-chromosome maintenance protein 7 (Mcm7) were from Santa Cruz Biotechnology, monoclonal HIF2a was from Novus Biologicals (Littleton, CO), and monoclonal β-actin was from Sigma-Aldrich. Goat anti-mouse and anti-rabbit IgG-horseradish peroxidase conjugates were from Bio-Rad Laboratories (Hercules, CA). Protein A/G agarose was from Santa Cruz Biotechnology. Protease inhibitor mixture was from Roche Diagnostics (Indianapolis, IN). Alexa Fluor dye-labeled secondary antibodies were from Life Technologies.
Constructs
FLAG-tagged and HA-tagged HBO1, FLAG-JADE1S, and FLAG-JADE1L cDNAs were described previously.14
Transfection of Cultured Cells with cDNA and siRNA
The following siRNA oligonucleotides products (Dharmacon RNAi Technologies/Thermo Fisher Scientific, Lafayette, CO) were used for the experiments in this study: JADE1, On-Targetplus SMARTpool, human PHF17 (79960) (#L-017133-01-0010); HBO1/MYST2, (11143), On-Targetplus SMARTpool (L-017668-00-0010); and nontargeting control siRNA oligonucleotides (D-001210-01, Dharmacon RNAi Technologies). Lipofectamine 2000 (Invitrogen) was used for transfection of all of the siRNA oligonucleotides as well as cDNAs following the manufacturer’s protocol.
Analysis of Endogenous Histones in Nuclear Fraction
Histone extraction was done as described previously.15 Cultured H1299 cells were lysed with cold 10 mmol/L Tris buffer (pH 8), containing 0.6% NP-40, 0.15M NaCl, 1 mmol/L EDTA, protease inhibitors (Roche Pharmaceuticals, Basel, Switzerland), and 5 mmol/L sodium butyrate. After 5-minute incubation on ice, nuclei were pelleted at 1200 × g, at 4°C, and resuspended in 150 μL of 0.4 N H2SO4 on ice. After 20 minutes of incubation, nuclear debris was removed by centrifugation at 13,000 × g for 10 minutes at 4°C. The supernatant was treated with 1.5 mL of cold 20% trichloroacetic acid for 10 minutes at 4°C. Pellets were separated by centrifugation at 13,000 × g, 4°C, and rinsed with 0.1% HCl in acetone and, subsequently, with acetone alone. The final pellet was resuspended in reducing SDS sample buffer.
Fractionation of Cultured Cells
Cells were fractionated into soluble and chromatin-enriched fractions according to the protocol described previously with some changes.7 Cells were lysed for 15 minutes in situ with 10 mmol/L HEPES (pH 7.8) buffer containing 10 mmol/L KCL, 1.5 mmol/L MgCl2, 0.1% TritonX100, protein inhibitors (Roche Pharmaceuticals), and phenylmethylsulfonyl fluoride using 500 μL per 100-mm cell culture dish. Pellets were separated by low-speed centrifugation at 1300 × g for 5 minutes. Supernatants were further cleared by centrifuging at 13,000 × g for 10 minutes and frozen for further analysis (supernatants). The pellets obtained after low-speed centrifugation were resuspended in the same buffer except for omitting TritonX100 and centrifuged at 13,000 × g for 5 minutes. Supernatants were discarded, and the chromatin-enriched pellets frozen for analysis.
Synchronization of Cultured Cells with Nocodazole
H1299 cells were transfected with small-interfering RNA (siRNA) and allowed to grow for 48 hours. To arrest cells in the G2/M stage of the cell cycle, nocodazole (100 ng/mL) was added to cultures for 14 to 16 hours. Nocodazole was removed by centrifugation at 1300 × g for 4 minutes, and cells were re-suspended and plated onto fresh culture dishes for cell cycle recovery.
Synchronization of HDF Cells by Serum Depletion, and Transfection with siRNA
Human diploid fibroblast (HDF) cells were synchronized in quiescence according to the previously described protocol.30 Briefly, confluent HDF were serum deprived for 16 to 24 hours. Cells were trypsinized, plated at 40% to 50% confluence, and maintained in 0.1% fetal bovine serum for an additional 24 hours before siRNA transfection. To deplete JADE1, quiescent HDF cells were transfected with siRNA (Dharmacon Research) twice on consecutive days according to manufacturer protocols. Cells were maintained in 0.1% fetal bovine serum throughout the course of siRNA transfection. To stimulate cell cycle reentry, cells were treated with fresh medium containing 10% fetal bovine serum. At indicated times after cell cycle entry, cells were harvested for analysis of DNA synthesis by [3H]-thymidine incorporation.
Rates of [3H]-Thymidine Incorporation
The protocol was described elsewhere.30 HDF cells were replated into 12-well plates at 40% to 50% density (2.5 × 105 cells per well as determined by quantitation with a hemocytometer) and serum starved overnight. Each experiment was set up in triplicate. Additional plates with identical samples were set up and used for DNA extraction and quantitation. At various times after release from quiescence, cells were incubated for 2 hours with 2 μCi of [3H]-thymidine (1 μCi/mL; Perkin Elmer, Waltham, MA). The medium was aspirated, and monolayers were fixed and then washed once with 1 mL of 5% trichloroacetic acid. The fixed monolayers were solubilized in a mixture of 1% SDS and 0.3 N NaOH. Incorporated radioactivity was determined by scintillation counting. The same protocol was used to determine rates of thymidine incorporation into H1299 cells, except for omitting the serum starvation and synchronization steps. DNA concentrations were determined by isolating DNA from parallel wells using DNAsol. Isolated DNA was quantified by spectrophotometric absorbency at 260 nmol/L. Thymidine counts were standardized to total DNA contents.
Immunofluorescent Labeling of Whole Cells
H1299 cells grown on chamber slides were rinsed with PBS and then fixed with 4% paraformaldehyde for 20 minutes at room temperature. After incubation, cells were rinsed with PBS three times and then permeabilized with 0.1% Triton X-100 for 15 minutes at room temperature. Blocking was performed with 3% horse serum and 1% bovine serum albumin for 1 hour at room temperature. Primary antibodies in 1% bovine serum albumin, 0.05% Tween-TBS, were incubated overnight at 4°C, and secondary antibodies in 0.05% Tween-TBS were incubated for 1 hour at room temperature. Coverslips were mounted using Vectashield (Vector Laboratories, Burlingame, CA) mounting medium for fluorescence, with DAPI. Images were analyzed on an Olympus DSU spinning disk confocal microscope (Olympus, Tokyo, Japan) and edited in ImageJ 1.46 (NIH, Bethesda, MD) software.
All experiments were repeated at least two times and yielded essentially the same results.
Immunohistochemistry
Murine kidneys were fixed in 4% paraformaldehyde, embedded into paraffin blocks, and sectioned onto glass slides (5 μm thickness). Epitope retrieval was achieved by heating tissue sections in 0.1 mol/L sodium citrate buffer (pH 6.0), using a microwave. Immunohistochemistry of kidney sections was performed using Vectastain ABC kit (Vector Laboratories) following the manufacturer’s protocol and as previously described.31 Detection of the antibody was performed with a DAB (3, 3′-diaminobenzidine) kit (Vectastain; Vector Laboratories). Nuclei were visualized by staining sections with hematoxylin. Images of stained tissue sections were analyzed with an Olympus Bx51 Model UDO3 light microscope. Photos of stained kidney images were taken with an Olympus DP71 microscope camera using 20× or 40× magnification lens.
Quantitation of Immunohistochemistry Staining
Images were analyzed by two blinded investigators. Four representative fields (40× lens) per sample were analyzed by counting stained and total nuclei. The number of positive-stained nuclei was normalized to the number of total nuclei (ImageJ; NIH). Average of four values and SD was calculated in Excel software (Microsoft Office Professional Plus 2010, Redmond, WA).
Acute Kidney Injury after Ischemia and Reperfusion
IR injury in mice was performed according to established protocols as previously described.32 Briefly, male C57Bl/6 mice underwent bilateral renal artery clamping for 28 minutes under anesthesia with tribromoethanol. Blood samples were collected from the tail vein for blood urea nitrogen (BUN) measurements daily or via intracardiac puncture after the animals were euthanized for tissue collection.
Unilateral ischemic rat kidneys were generated as described.31 Both post-ischemic and contralateral control kidneys were either frozen in liquid nitrogen or fixed for immunohistochemistry. Forty minutes of renal vascular occlusion in rats results in a marked reduction in glomerular filtration rate as measured 1 to 3 hours after reperfusion.
Results
JADE1 in DNA Replication
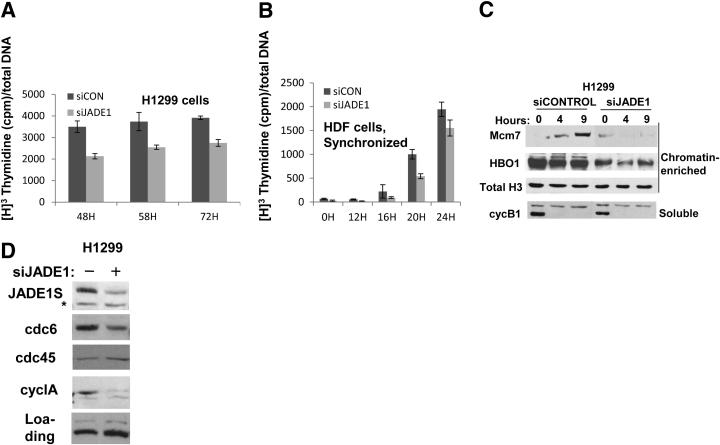
The histone acetyl transferase HBO1 has been implicated in DNA replication licensing and cell cycle progression.7,26,33,34 We previously reported that JADE1 is required for HBO1 to acetylate histone H4 in chromatin context.14,15 Thus, JADE1 might be required for cell proliferation. We examined the effects of JADE1 down-regulation on rates of DNA synthesis in cultured cells. Transient transfection of cells with JADE1-specific siRNA resulting in down-regulation of JADE1 protein lead to reduction of DNA synthesis rates in two different cell types (Figure 1, A and B). Importantly, the effect was reproduced in wild-type human dermal fibroblasts (Figure 1B), which are primary untransformed cells.
Figure 1.
JADE1 down-regulation inhibits DNA replication, HBO1 abundance, and Mcm7 chromatin recruitment. A and B: JADE1 down-regulation inhibits DNA replication in H1299 and HDF cells. A: Cells were transiently transfected with siRNA specific for JADE1 (siJADE1) or control siRNA (siCON). B: HDF cells were synchronized in G0 by serum starvation. Thymidine incorporation into DNA was determined as described in Materials and Methods. C: H1299 cells were transfected with siJADE1 or siCON, and cells synchronized in G2/M. Cell cycle was resumed after nocodazole removal (0 hours). At times, indicated chromatin-enriched and soluble fractions were collected. Fractions were analyzed by Western blots for HBO1, Mcm7, histone H3 (loading control), and cyclin B1 protein expressions. D: Asynchronous H1299 cells were lysed, and whole-cell protein extracts were generated 48 hours post-transfection with siJADE1 or siCON. Lysates were analyzed for JADE1S protein, replication factors cdc6 and cdc45, and cyclin A expression by Western blot. Asterisk indicates a non-specific band.
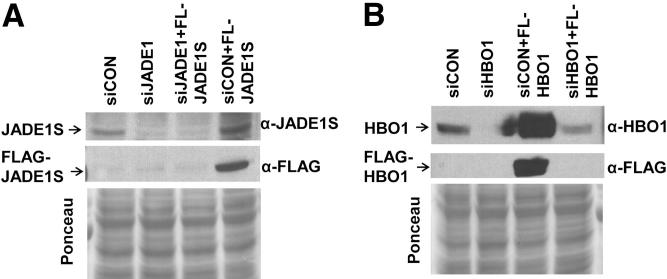
It has been shown that HBO1 is required for recruitment of key replication licensing factors Mcm2 and Mcm6.7 We predicted that like HBO1, down-regulation of JADE1 might affect Mcm2–7 recruitment to chromatin. Indeed, in cells synchronized by nocodazole, JADE1 down-regulation prevented Mcm7 recruitment to chromatin (Figure 1C). In addition, JADE1 down-regulation caused at least a twofold decrease in total levels of replication factor cdc6 and cell cycle marker cyclin A (Figure 1D). The specificities of new antibodies were verified by down-regulation of endogenous proteins with corresponding siRNA and following detection of protein bands on Western blots (Figure 2). So far, these data demonstrate that JADE1 is required for DNA synthesis, and suggest an HBO1-dependant mechanism of action.
Figure 2.
JADE1S and HBO1 antibody verification. Western blots. A: JADE1S antibody (α-JADE1S) verification. H1299 cells were transfected with nontargeting control siRNA (siCON), siRNA for JADE1 (siJADE1), FLAG-JADE1S cDNA expression vector, or combinations. B: HBO1 antibody (α-HBO1) verification. Experimental design was same as A except siRNA specific for HBO1 and FLAG-HBO1 cDNA expression vectors were used. Two days post-transfection, cells were lysed, and protein lysates were analyzed by Western blot.
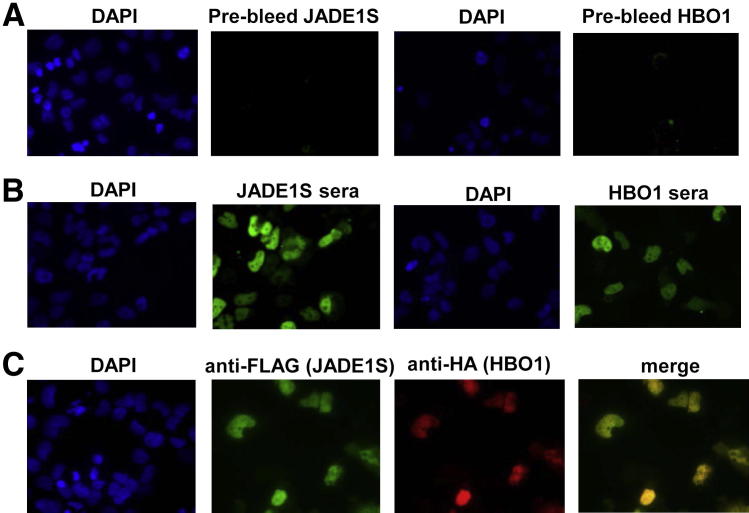
Our previous work has demonstrated that transiently transfected and overexpressed JADE1 and HBO1 proteins dramatically increase the abundance of each other,14 suggesting mutual stabilization. Here, we examined the effects of JADE1 siRNA on HBO1 expression. JADE1 down-regulation dramatically decreased levels of endogenous HBO1, supporting our previous observations, and further proving their mutual interaction in live cells (Figure 1C). Cell fractionation and immunoprecipitation studies suggest that JADE1S and HBO1 proteins are localized predominantly to the cell nuclei.8,15 We assessed JADE1S and HBO1 subcellular localization in whole cells. FLAG-JADE1S and HA-HBO1 cDNAs were cotransfected into the cells, and protein expression was visualized by immunofluorescence. Parallel samples of cells were stained with the indicated specific rabbit anti-sera or monoclonal tag-specific antibodies. JADE1S and HBO1 both colocalize to the cell nuclei, further proving their interaction in live cells (Figure 3).
Figure 3.
JADE1S and HBO1 nuclear colocalization. Whole-cell immunofluorescence. A and B: JADE1S and HBO1 antibody verification. H1299 cells grown on chamber slides were cotransfected with FLAG-JADE1S and HA-HBO1 cDNA plasmids. Two days post-transfection, cells were processed for immunofluorescent staining with antibodies as indicated. A: Pre-bleed control sera. Pre-bleed sera used to generate negative control; JADE1S and HBO1 nuclear localization. JADE1 and HBO1 sera. JADE1S or HBO1 sera stains overexpressed as well as endogenous proteins (B). C: HA-HBO1 and FLAG-JADE1S nuclear co-localization. HA- and FLAG- antibody. HA-tag and FLAG-tag antibodies stain only overexpressed proteins and show high levels of colocalization.
JADE1 and HBO1 Complex Regulation during in Vivo Recovery following Ischemia-Reperfusion Injury
JADE1S has been identified by the two-hybrid system as a partner of pVHL,18 a tumor suppressor protein involved in oxygen sensing,35 suggesting a potential role for JADE1S in the hypoxic stress response. To investigate the role of JADE1 during tissue injury and regeneration, a murine model of renal ischemia-reperfusion was used.32 The pathogenesis of murine ischemic acute kidney injury has been well characterized and is similar to human ischemic kidney injury. The initial injury is followed by regeneration of kidney tubules that is required for recovery of kidney function. On the cellular level, the initial injury leads to loss of epithelial cells and cell growth arrest. Thereafter, surviving tubular epithelial cells reconstitute the tubule in a process requiring activation, dedifferentiation, proliferation, and redifferentiation.36 We set out to examine HAT HBO1 complex expression profiles in this model.
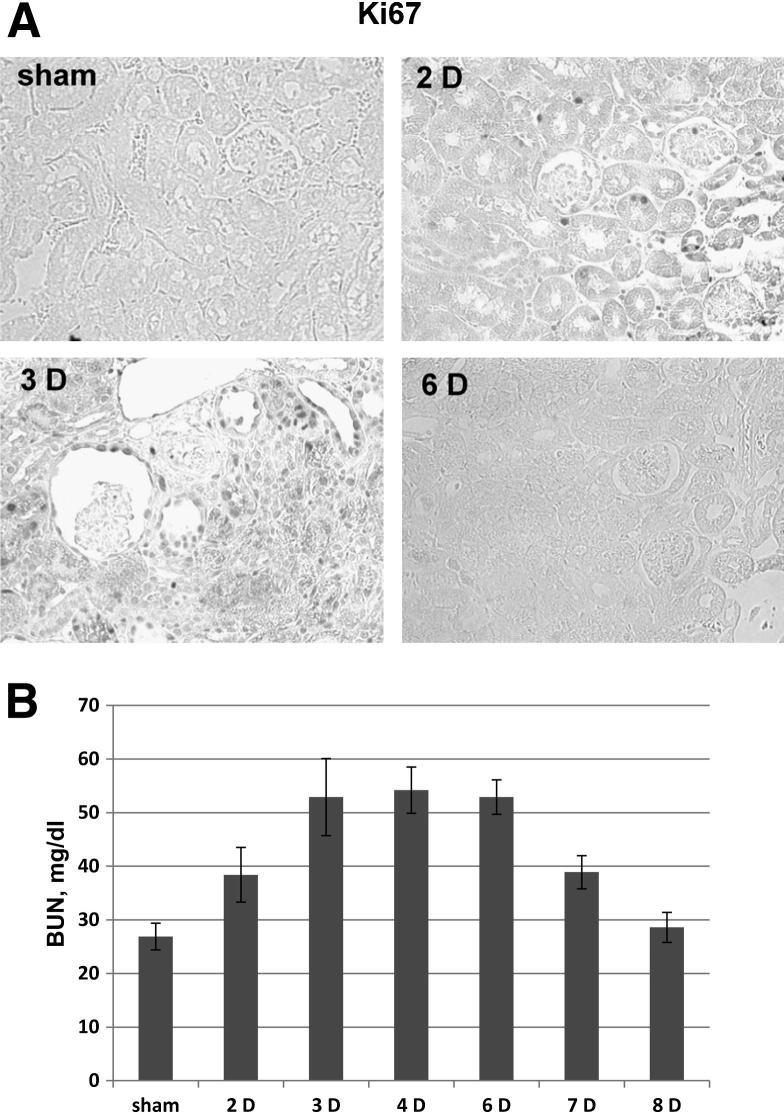
Mice kidneys were subjected to 28 minutes of ischemia by bilateral renal artery clamping and were followed up to 8 days post injury. Kidneys were removed, and the percentage of positive cells was determined by immunohistochemistry with antibodies specific for the two proteins of the minimal functional HBO1 complex, JADE1S and HBO1, as well as for JADE1L protein partner of HBO1 (Figures 4, 5, and 6). Renal function was monitored by serial BUN measurements, and cell proliferation was assessed by Ki-67 staining (Figure 5, A and B).
Figure 4.
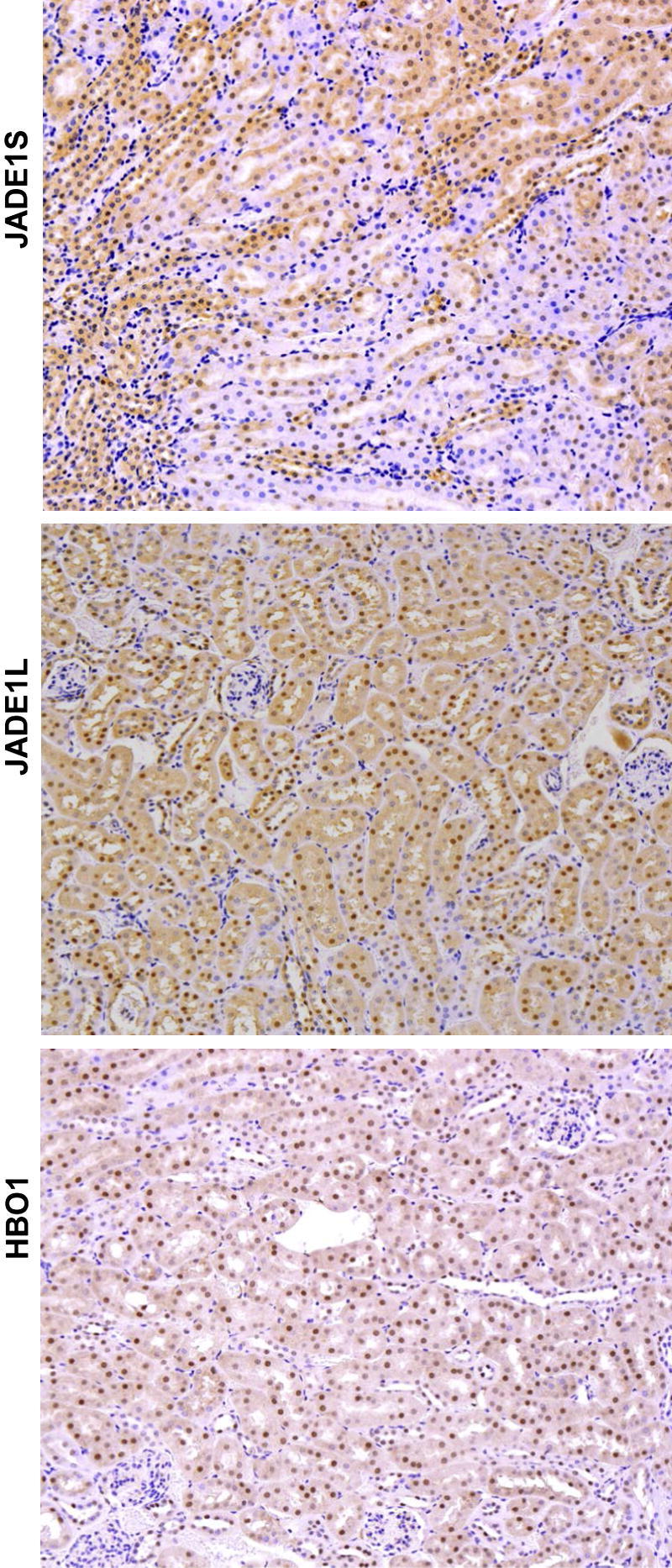
JADE1S, JADE1L, and HBO1 proteins are highly expressed in mice kidney tubular epithelial cells and are localized predominantly to nuclei. Whole-tissue immunohistochemistry. Normal mice kidney sections were processed for immunohistochemistry and stained with antibodies (dilution 1:1000) as indicated. Nuclei were visualized with hematoxylin counterstain.
Figure 5.
Characterization of kidney injury and recovery during mice renal IR. A: Kidney tissue sections from sham, 2-day (2 D), 3-day (3 D), and 6-day (6 D) postoperative mice were processed for immunohistochemistry with Ki-67 antibody as described in Figure 4 and in Materials and Methods. Representative fields are shown. Full quantification of Ki-67 protein expression during the course of IR is presented in Figure 6. B: Assessment of BUN levels during mice renal IR. BUN was measured using a QuantiChrom BUN assay kit (Bioassay Systems, Hayward, CA) following the manufacturer’s protocol.
Figure 6.
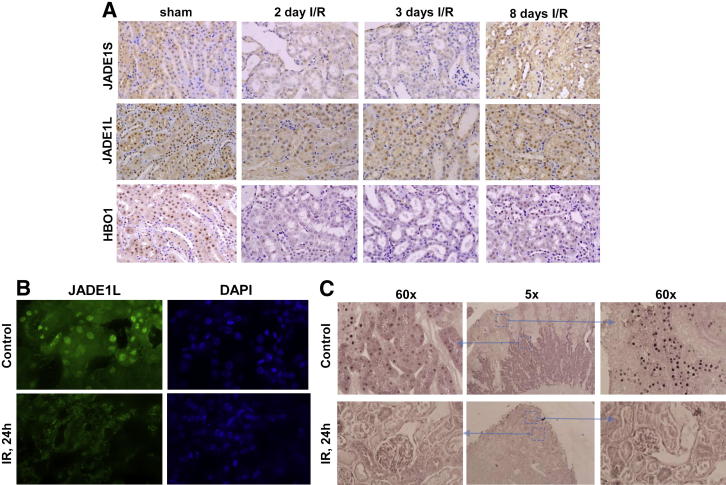
Expression of JADE1S, JADE1L, and HBO1 in tubular epithelium during kidney injury and regeneration. A: Immunohistochemistry on mice kidney sections was performed as described in Figure 4 and in Materials and Methods. Representative fields are shown. Quantifications of these protein expressions during the entire course of IR are presented in Figure 7. B: The effect of renal IR on JADE1L expression in mice tubular epithelium by immunofluorescence assay. Kidney sections were processed for immunofluorescence assay as described in Materials and Methods with JADE1L antibody (dilution 1:300). Alexa Fluor 488–conjugated secondary antibody (Invitrogen) was used to visualize endogenous protein expression. The nonspecific faint signal in the IR sample is due to autofluorescence. C: The effect of renal IR on JADE1S protein expression in tubular epithelium of rat kidneys. Unilateral renal IR in rats causes JADE1S protein down-regulation and recapitulates effects in mice. Injured and contralateral kidneys were processed for immunohistochemistry. The tissue sections were stained with JADE1S antibody (dilution 1:2000) as described in Figure 4. Areas in blue boxes are enlarged as indicated by arrows. Note that in rat kidneys, JADE1S protein is localized predominantly to the proximal tubular epithelium.
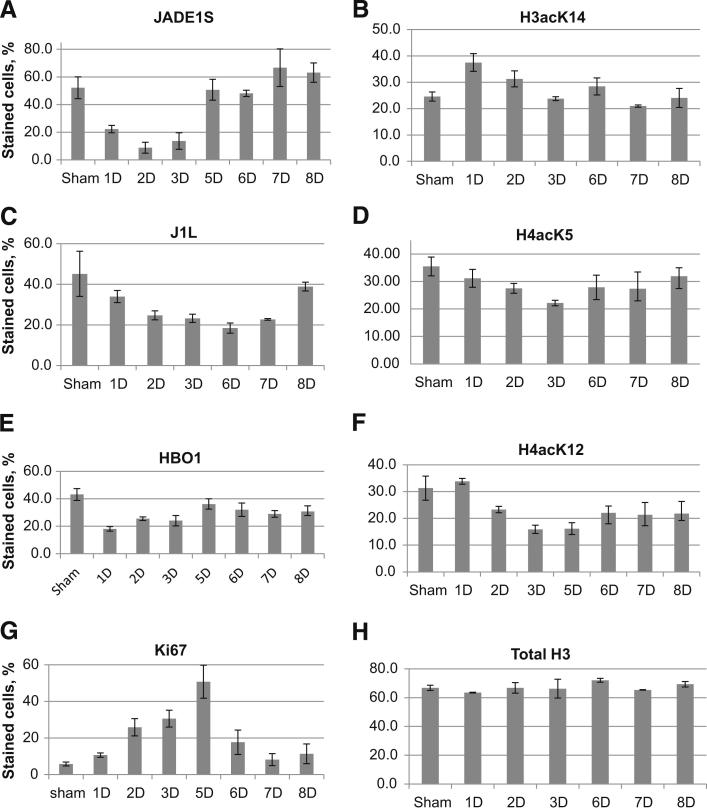
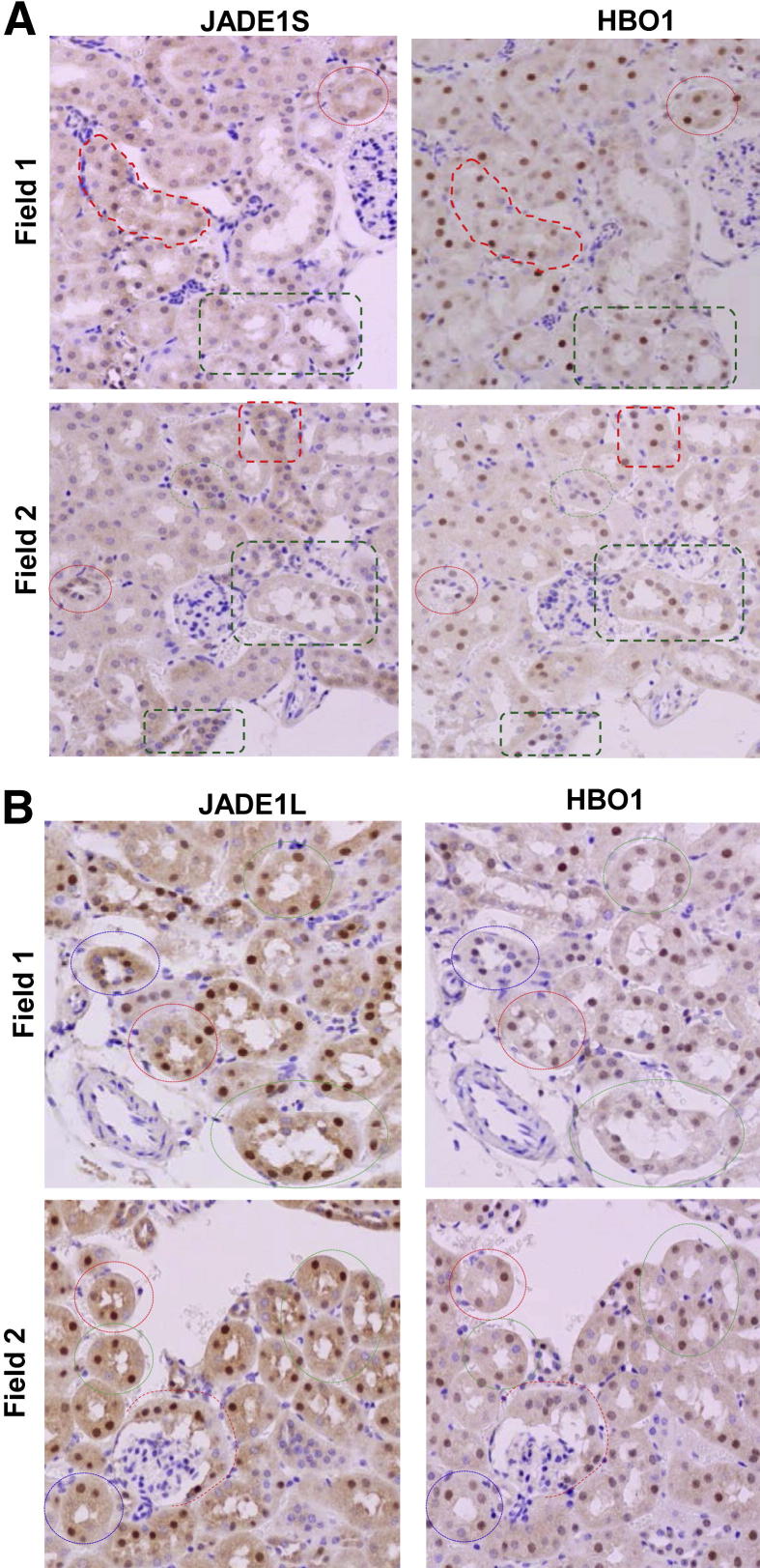
The components of the HAT HBO1 complex (ie, JADE1L, JADE1S, and HBO1) were highly expressed in the nuclei of epithelial cells in control kidneys (Figure 4). One day after IR injury, when the renal histology showed signs of significant damage, the fraction of cells positive for JADE1S and HBO1 proteins decreased by 56% and 58%, respectively (Figure 6 and Figure 7, A and E). Interestingly, the fraction of cells expressing the full-length isoform of JADE1 (JADE1L) decreased only by 28% (Figure 7C). The percentage of cells expressing all three proteins was restored during the course of recovery. Fractions of HBO1- and JADE1S-expressing cells were recovered by day 5 of reperfusion, which is the time when Ki-67 expression reached its maximum (Figure 7G). The recovery of the JADE1L-positive cell fraction was slower and only reached pre-ischemic levels 8 days after IR injury and after the fraction of Ki-67–positive cells almost returned to basal pre-ischemic levels (Figure 5A and Figure 7, C and G). Kidney IR injury performed in rats confirmed these observations. We observed similar changes in JADE1-positive tubular cell fractions during kidney injury and regeneration (Figure 6C). The synchronous temporal and spatial JADE1–HBO1 localization suggested in vivo interactions between JADE1 and HBO1 proteins. Indeed, comparison of consecutive kidney sections stained with JADE1 and HBO1 revealed identical areas of expression in the tubular epithelial cells (Figure 8).
Figure 7.
Temporal expression profiles of HAT HBO1 complex proteins, acetylated histones, and Ki-67 in tubular epithelium during kidney injury and recovery time course. A–H: Immunohistochemistry was performed and protein expression was quantified as described in Figure 4 and Materials and Methods.
Figure 8.
Comparison of consecutive kidney sections revealed similarities in spatial expressions of JADE1 and HBO1 proteins in the tubular epithelial cells. A: Consecutive sections stained with JADE1S and HBO1 antibodies. B: Consecutive sections stained with JADE1L and HBO1 antibodies. Normal mice kidneys were processed for immunohistochemistry and consecutive tissue sections stained with indicated antibodies. Two identical fields within each tissue section are presented. Identical areas are enclosed in punctate shapes (compare same shapes/colors).
The apparent down-regulation of the fraction of tubular epithelial cells expressing HBO1 complex during injury and recovery suggested possible parallel changes of acetylated histones in these cells. Data on HBO1 specificities toward histone substrate are contradictory. In vitro and in cultured cells, the HBO1-JADE1 complex acetylates histone H4 most efficiently,14,33 specifically, HBO1 enzyme strongly prefers to acetylate lysine 5 and lysine 12 residues of histone H4 (H4K5 and H4K12).33 However, these results do not agree with the in vivo study in which knocking out HBO1 in mice embryo did not affect levels of histone H4 acetylation, but greatly diminished levels of lysine 14 residue of histone H3 (H3K14) acetylation.24 We assessed the fraction of H4acK12-, H4acK5-, and H3acK14-positive cells in kidney samples from uninjured mice and mice following IR injury. Down-regulation of the fraction of HBO1- and JADE1-positive cells was followed by a decrease in H4acK5- and H4acK12-positive cells (Figure 7, D and F). The fraction of acetyl-H4–positive cells reached its minimum 3 days after IR injury (Figure 7, D and F). The percentage of cells positive for both histone H4 marks recovered by day 8 of reperfusion. By contrast, the fraction of cells expressing H3acK14 histone mark followed the opposite dynamic: the initial increase was followed by a decrease to that of control levels (Figure 7B). Note that the expression of total histone H3 protein was unaffected during the entire time course (Figure 7H). Moreover, similar to the results previously reported for HBO1,33 transient transfection of cultured epithelial cells with siRNA specific for JADE1 resulted in down-regulation of H4acK12, but not H3acK14, histone acetylation mark (Figure 9). Taken together, these data suggest that the JADE1-HBO1 complex might be responsible for histone H4 acetylation in vivo, and implicate HBO1 complex activities in kidney recovery after ischemic injury.
Figure 9.
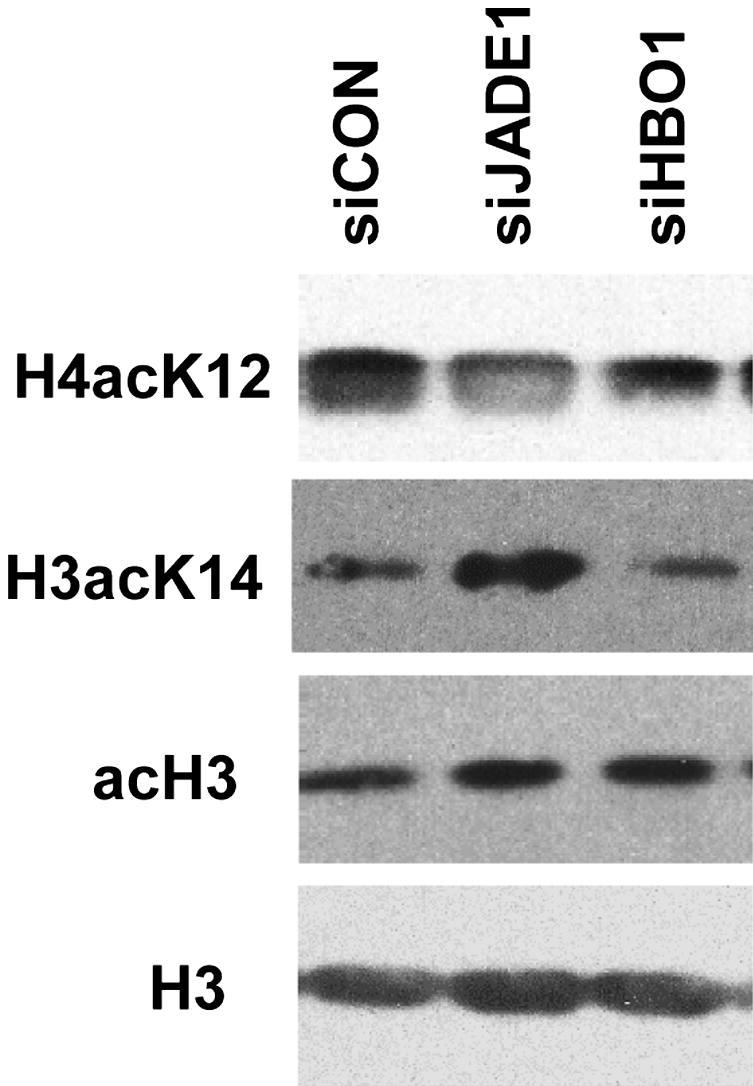
Down-regulation of JADE1 protein by siRNA attenuated levels of H4acK12, but not H3acK14, histone acetylation marks. H1299 cells were transiently transfected with siRNA specific for JADE1, HBO1, or control siRNA (siCON). Forty-eight hours post-transfection, cells were lysed, and total histones isolated from nuclear fractions as described in Materials and Methods. Acetylated histones were visualized by Western blots with indicated antibodies. Total histone H3 was used for loading control.
Discussion
JADE1S protein promotes bulk histone H4 acetylation in epithelial cell cultures and is required for HAT HBO1 acetyl transferase activities.14,15 In the current study, we extend our previous findings using siRNA to define the role of JADE1 during cell division. Furthermore, we characterized the temporal and spatial pattern of the JADE1-HBO1 complex expression in vivo during organ regeneration.
The role of JADE1 in cellular growth and proliferation has not been fully elucidated. Available information is based on cultured cell models overexpressing JADE1.37 According to a recent report, overexpression of JADE1S and JADE1L caused growth delay in cultured HeLa cells.38
In this study, we chose an siRNA-based protein down-regulation approach to avoid potential side effects of overexpression and cell toxicity caused by JADE1 protein excess (unpublished observations by M.V.P.). In addition, we used untransformed primary cell cultures (HDF cells) to better understand the role of JADE1. We have demonstrated that JADE1 protein knockdown diminishes the rate of DNA synthesis. JADE1 deficiency markedly decreases endogenous HBO1 protein levels, a finding that corroborates our previous report that JADE1 stabilizes the HBO1 protein.14 In addition, we have demonstrated that JADE1 is required for expression of replication factors cdc6 and G1/S-specific cyclin A during cell division (Figure 1). Moreover, similar to HBO1, JADE1 allows for the recruitment of the replication licensing factor Mcm7 to the chromatin-enriched fraction of cells that is required for DNA replication (Figure 1). HBO1 has been implicated in cellular stress response pathways, and it has been reported that HBO1 is regulated by nocodazole, hyperosmotic shock, and DNA replication fork arrest.7,25,39 p53 is thought to modulate enzyme activity of HBO1 during stress responses.25 It has been shown that HAT activity was required for HBO1 replication function and cell proliferation.38,33 Knocking down HBO1 in cultured cells inhibited bromodeoxyuridine incorporation, accumulation of cells in the S phase of the cell cycle, chromatin recruitment of DNA replication licensing factors Mcm2–6, which is similar to what we have seen with JADE1 knockdown (Figure 1). Taken together, these data demonstrate for the first time that JADE1 is a cofactor required for HBO1 replication function.
Renal tubule regeneration after acute ischemia-reperfusion injury has been well studied, both as a model of tissue regeneration and in the search for rational therapeutics for the treatment of acute kidney injury.40,41 The initial epithelial cell injury results in the loss of epithelial cells and cell growth arrest. This is followed by the activation, proliferation, and redifferentiation of surviving cells that repopulate the tubule to reestablish organ function. We show that all three HAT complex proteins—JADE1S, JADE1L, and HBO1—are highly expressed in murine kidneys, specifically in the nuclei of epithelial cells in the proximal and distal tubules; where, as was seen in cell culture studies, JADE1 and HBO1 proteins are colocalized (Figures 3, 4, 6, and 8). Immediately following ischemia-reperfusion injury, at a time when many cells are lethally injured and others are relatively dedifferentiated and undergoing reparative processes, the fraction of cells expressing JADE1S, JADE1L, and HBO1 decreased substantially. This was associated with a marked decrease in the fraction of cells expressing acetylated K5 and K12 histone H4 residues and a low level of epithelial cell proliferation. In stark contrast to this early post-ischemic period, during the regenerative phase, the JADE1S- and HBO1-positive fraction of cells recovered, and this recovery was temporally and spatially correlated with the acquisition of H4acK5 and H4acK12 histone H4 marks (Figure 7). Unexpectedly, during the regenerative phase, we observed a poor correlation between JADE1L, HBO1, and proliferation. The fraction of JADE1L protein–positive cells recovered only after proliferation had ceased, suggesting that JADE1L is not involved in cellular proliferation in vivo. Our findings point to an important role for JADE1S during cellular proliferation and organ regeneration, which is in keeping with our in vitro data (14,15 and above) but suggest that JADE1L may have unique HBO1-independent functions during redifferentiation.
In conclusion, we demonstrate that JADE1 is required in the HBO1-HAT complex for expression and chromatin recruitment of replication factors during cell cycling. JADE1 deficiency leads to degradation of HBO1 and prevents cell cycle progression in cultured epithelial cells. In vivo, the fraction of cells expressing JADE1 and HBO1 proteins decreases following ischemia-reperfusion injury. Cells positive for HBO1 and JADE1S, but not JADE1L, complexes accumulate during epithelial cell regeneration, indicating a key role for JADE1S during organ regeneration. Further studies will be required to define the apparently distinct functions of JADE1L and JADE1S during cell cycle progression and redifferentiation.
Acknowledgments
We thank members of Joe Bonventre’s lab for kindly sharing lab resources to perfect our immunohistochemistry technique. We thank Cyrus Vaziri for help with the thymidine incorporation assay, Herbert Cohen for helpful discussions in early studies, John Schwartz for continuous intellectual contributions, and Nirodhini Siriwardana for proofreading the manuscript.
Footnotes
Supported by NIH grant RO1 DK087910 and American Cancer Society grant IRG-72-001-32-IRG (M.V.P.), and in part by NIH grant 1KO8DK090143 (A.H.).
References
- 1.Agalioti T., Chen G., Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 2.Iizuka M., Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 3.Kuo M.H., Allis C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 4.Narlikar G.J., Fan H.Y., Kingston R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 5.Suganuma T., Workman J.L. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 6.Turner B.M. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 7.Iizuka M., Matsui T., Takisawa H., Smith M.M. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol. 2006;26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iizuka M., Takahashi Y., Mizzen C.A., Cook R.G., Fujita M., Allis C.D., Frierson H.F., Jr., Fukusato T., Smith M.M. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene. 2009;436:108–114. doi: 10.1016/j.gene.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson C.L., Laniel M.A. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Allis C.D., Berger S.L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., Shilatifard A., Workman J., Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 12.Roth S.Y., Denu J.M., Allis C.D. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 13.Sterner D.E., Berger S.L. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foy R.L., Song I.Y., Chitalia V.C., Cohen H.T., Saksouk N., Cayrou C., Vaziri C., Cote J., Panchenko M.V. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem. 2008;283:28817–28826. doi: 10.1074/jbc.M801407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panchenko M.V., Zhou M.I., Cohen H.T. von Hippel-Lindau partner Jade-1 is a transcriptional co-activator associated with histone acetyltransferase activity. J Biol Chem. 2004;279:56032–56041. doi: 10.1074/jbc.M410487200. [DOI] [PubMed] [Google Scholar]
- 16.Saksouk N., Avvakumov N., Champagne K.S., Hung T., Doyon Y., Cayrou C., Paquet E., Ullah M., Landry A.J., Cote V., Yang X.J., Gozani O., Kutateladze T.G., Cote J. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell. 2009;33:257–265. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishima Y., Miyagi S., Saraya A., Negishi M., Endoh M., Endo T.A., Toyoda T., Shinga J., Katsumoto T., Chiba T., Yamaguchi N., Kitabayashi I., Koseki H., Iwama A. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood. 2011;118:2443–2453. doi: 10.1182/blood-2011-01-331892. [DOI] [PubMed] [Google Scholar]
- 18.Zhou M.I., Wang H., Ross J.J., Kuzmin I., Xu C., Cohen H.T. The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J Biol Chem. 2002;277:39887–39898. doi: 10.1074/jbc.M205040200. [DOI] [PubMed] [Google Scholar]
- 19.Burke T.W., Cook J.G., Asano M., Nevins J.R. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem. 2001;276:15397–15408. doi: 10.1074/jbc.M011556200. [DOI] [PubMed] [Google Scholar]
- 20.Georgiakaki M., Chabbert-Buffet N., Dasen B., Meduri G., Wenk S., Rajhi L., Amazit L., Chauchereau A., Burger C.W., Blok L.J., Milgrom E., Lombes M., Guiochon-Mantel A., Loosfelt H. Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol Endocrinol. 2006;20:2122–2140. doi: 10.1210/me.2005-0149. [DOI] [PubMed] [Google Scholar]
- 21.Johmura Y., Osada S., Nishizuka M., Imagawa M. FAD24 acts in concert with histone acetyltransferase HBO1 to promote adipogenesis by controlling DNA replication. J Biol Chem. 2008;283:2265–2274. doi: 10.1074/jbc.M707880200. [DOI] [PubMed] [Google Scholar]
- 22.Sharma M., Zarnegar M., Li X., Lim B., Sun Z. Androgen receptor interacts with a novel MYST protein, HBO1. J Biol Chem. 2000;275:35200–35208. doi: 10.1074/jbc.M004838200. [DOI] [PubMed] [Google Scholar]
- 23.Stedman W., Deng Z., Lu F., Lieberman P.M. ORC, MCM, and histone hyperacetylation at the Kaposi’s sarcoma-associated herpesvirus latent replication origin. J Virol. 2004;78:12566–12575. doi: 10.1128/JVI.78.22.12566-12575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kueh A.J., Dixon M.P., Voss A.K., Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol. 2011;31:845–860. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iizuka M., Sarmento O.F., Sekiya T., Scrable H., Allis C.D., Smith M.M. Hbo1 Links p53-dependent stress signaling to DNA replication licensing. Mol Cell Biol. 2008;28:140–153. doi: 10.1128/MCB.00662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyon Y., Selleck W., Lane W.S., Tan S., Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzouanacou E., Tweedie S., Wilson V. Identification of Jade1, a gene encoding a PHD zinc finger protein, in a gene trap mutagenesis screen for genes involved in anteroposterior axis development. Mol Cell Biol. 2003;23:8553–8562. doi: 10.1128/MCB.23.23.8553-8562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maki N., Suetsugu-Maki R., Sano S., Nakamura K., Nishimura O., Tarui H., Del Rio-Tsonis K., Ohsumi K., Agata K., Tsonis P.A. Oocyte-type linker histone B4 is required for transdifferentiation of somatic cells in vivo. FASEB J. 2010;24:3462–3467. doi: 10.1096/fj.10-159285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinha D., Bannergee S., Schwartz J.H., Lieberthal W., Levine J.S. Inhibition of ligand-independent ERK1/2 activity in kidney proximal tubular cells deprived of soluble survival factors up-regulates Akt and prevents apoptosis. J Biol Chem. 2004;279:10962–10972. doi: 10.1074/jbc.M312048200. [DOI] [PubMed] [Google Scholar]
- 30.Song I.Y., Palle K., Gurkar A., Tateishi S., Kupfer G.M., Vaziri C. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J Biol Chem. 2010;285:31525–31536. doi: 10.1074/jbc.M110.138206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichimura T., Bonventre J.V., Bailly V., Wei H., Hession C.A., Cate R.L., Sanicola M. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem. 1998;273:4135–4142. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Gall J.M., Bonegio R.G., Havasi A., Hunt C.R., Sherman M.Y., Schwartz J.H., Borkan S.C. Induction of heat shock protein 70 inhibits ischemic renal injury. Kidney Int. 2011;79:861–870. doi: 10.1038/ki.2010.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyon Y., Cayrou C., Ullah M., Landry A.J., Cote V., Selleck W., Lane W.S., Tan S., Yang X.J., Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Miotto B, Struhl K: HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37:57–66 [DOI] [PMC free article] [PubMed]
- 35.Kaelin W.G., Jr. How oxygen makes its presence felt. Genes Dev. 2002;16:1441–1445. doi: 10.1101/gad.1003602. [DOI] [PubMed] [Google Scholar]
- 36.Bonventre J.V., Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest. 2011;121:4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou M.I., Foy R.L., Chitalia V.C., Zhao J., Panchenko M.V., Wang H., Cohen H.T. Jade-1, a candidate renal tumor suppressor that promotes apoptosis. Proc Natl Acad Sci U S A. 2005;102:11035–11040. doi: 10.1073/pnas.0500757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avvakumov N., Lalonde M.E., Saksouk N., Paquet E., Glass K.C., Landry A.J., Doyon Y., Cayrou C., Robitaille G.A., Richard D.E., Yang X.J., Kutateladze T.G., Cote J. Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol Cell Biol. 2012;32:689–703. doi: 10.1128/MCB.06455-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miotto B., Struhl K. JNK1 phosphorylation of Cdt1 inhibits recruitment of HBO1 histone acetylase and blocks replication licensing in response to stress. Mol Cell. 2011;44:62–71. doi: 10.1016/j.molcel.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphreys B.D., Valerius M.T., Kobayashi A., Mugford J.W., Soeung S., Duffield J.S., McMahon A.P., Bonventre J.V. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008;2:284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Yang L., Humphreys B.D., Bonventre J.V. Pathophysiology of acute kidney injury to chronic kidney disease: maladaptive repair. Contrib Nephrol. 2011;174:149–155. doi: 10.1159/000329385. [DOI] [PubMed] [Google Scholar]