Abstract
To investigate the role of genital shedding of herpesviruses in human immunodeficiency virus type 1 (HIV) transmission, we compared 20 HIV-infected men who did and 26 who did not transmit HIV to their sex partners. As described previously, HIV transmission was associated with the potential source partner having higher levels of HIV RNA in blood and semen, having lower CD4+ T cell counts, having bacterial coinfections in the genital tract, and not using antiretroviral therapy. This study extended these findings by observing significant associations between HIV transmission and the following characteristics, especially among therapy-naive potential source partners: seminal cytomegalovirus (CMV) shedding, seminal Epstein-Barr virus shedding, and levels of anti CMV immunoglobulin in blood plasma.
Keywords: Herpes viruses, HIV-1 transmission, genital coinfections, men who have sex with men, antiretroviral therapy
The risk of sexual human immunodeficiency virus type 1 (HIV) transmission correlates with the levels of HIV RNA in blood [1, 2] and semen [3, 4]. Although HIV levels mostly correlate between these 2 compartments [3], local genital factors, like bacterial [5] or viral coinfections [6–8], can increase HIV shedding in semen. Specifically, highly prevalent herpesviruses, including herpes simplex virus (HSV) [3, 7], Epstein-Barr virus (EBV) [9, 10], human herpes virus 8 (HHV-8) [10], and, perhaps most profoundly, cytomegalovirus (CMV) [8, 10], can have an important impact on HIV shedding. However, suppression of HSV replication with acyclovir among HIV-infected potential source partners was not effective in reducing HIV transmission [11], perhaps because the other herpesviruses, which are more prevalent, especially CMV and EBV, are not suppressed by acyclovir.
METHODS
Study Participants and Clinical Data
Individuals being evaluated for possible HIV infection were recruited for participation in the San Diego HIV Transmission Study [3, 12]. Participants recruited their HIV-positive sex partners, who were sorted as HIV transmitters or nontransmitters. In partnerships in which both men were infected with HIV, transmitters were confirmed by phylogenetic linkage (Viroseq 2.0; Applied Biosystems) of pol sequences of HIV from each partner (genetic distance, ≤1%) [3, 12], and the source partner was inferred on the basis of the estimated duration of infection of both partners [13]. For each source partner, semen was collected by masturbation [6, 12]. Screening for sexually transmitted infections (STIs; gonorrhea, chlamydial infection, and syphilis) was performed. In blood, CD4+ T-lymphocyte subsets were measured by flow cytometry (LabCorp), and the HIV RNA level was quantified (Amplicor HIV Monitor Test). HIV subtype was determined from HIV pol sequence data, using SCUEAL [14]. The studies were conducted with appropriate written consent and approved by the University of California–San Diego Human Research Protections program.
Viral and Antibody Quantification
As described previously, viral RNA and DNA were extracted from seminal plasma, and levels of HIV RNA and the DNA of 7 different herpesviruses were measured by real-time polymerase chain reaction [6, 10]. Anti CMV Immunoglobulin G (IgG) antibody levels were measured in blood plasma as described previously [15]
Statistical Analysis
Statistical analyses were performed using SAS (version 9.2). Viral load variables were transformed to base 10 logarithm values. Nonnormally distributed data were either dichotomized (as undetectable or detectable) or ordinalized (as undetectable, low viral level [<4 log10 copies/mL], or high viral level [≥4 log10 copies/mL]). Comparison of seminal viral shedding between groups was performed using a χ2 or Fisher exact test (for sparse data). Continuous variables were compared by a t test if their values were normally distributed; otherwise a Mann–Whitney U test was used. Relative risks (with 95% confidence intervals) of belonging to the transmitter group were determined by univariate analysis for each individual virus. A Cochran-Armitage test for trend was performed to determine the association between HIV transmission and seminal levels of HIV, CMV, and EBV, by ordinal categories. A multivariate logistic regression model was performed for factors that, in univariate analysis, were associated with being a transmitter. To avoid overfitting, the logistic regression model was repeated to evaluate whether CMV replication was associated with HIV transmission, with adjustment for 1 confounder at a time (HIV seminal shedding, HIV RNA level in blood, CD4+ T cell count, EBV shedding, estimated infection duration of < 90 days, and CMV IgG level). All analyses were also repeated after exclusion of patients with an STI.
RESULTS
Study Participants
Potential source partners were sorted into “transmitters” (n = 20) or “nontransmitters” (n = 26) according to the serostatus of their potential recipient partner and phylogenetic linkage of viruses from each partner. All source partners were HIV-infected MSM. Semen was collected within a mean of 73.9 days from the recipients' estimated infection date (for transmitters) and within a mean of 15.5 days from the date when the potential source partner was reported to our clinic (for nontransmitters). Detailed demographic characteristics are summarized in Table 1.
Table 1.
Demographic Characteristics and Predictors of Human Immunodeficiency Virus Type 1 (HIV) Seminal Shedding
| Factor | Transmitter (n = 20) | All Nontransmitters (n = 26) | Relative Riska (95% CI) | Pb | ART-Naive Nontransmitters (n = 21) | Relative Riska (95% CI) | Pb |
|---|---|---|---|---|---|---|---|
| White, non-Hispanic | 11 (55.0) | 9 (45.0) | … | .55 | 9 (47.6) | … | .64 |
| Age, y, mean | 33.3 | 33.5 | … | .92 | 33.7 | … | .87 |
| Estimated infection duration ≤90 d at baseline | 2 (10.0) | 8 (30.8) | … | .15 | 8 (30.8) | … | .07 |
| CD4+ T-cell count, cells/microL, mean | 438 | 595 | … | .03 | 559 | … | .09 |
| CD8+ T-cell count, cells/microL, mean | 810 | 1027 | … | .11 | 1059 | … | .12 |
| Blood plasma CMV IgG level, UI/mL, mean | 21.8 | 15.0 | … | .05 | 14.9 | … | .07 |
| Blood HIV RNA level, log10 copies/mL, mean | 4.6 | 3.9 | … | .05 | 4.4 | … | .44 |
| Any detectable HIV in semen | 16 (80.0) | 12 (46.2) | 2.6 (1.0–6.5) | .03 | 12 (57.1) | 1.4 (.9–2.2) | .18 |
| Semen HIV RNA level, log10 copies/mL | |||||||
| Not detected | 4 (20.0) | 14 (53.9) | Reference | 9 (42.9) | Reference | ||
| >0 and <4 | 11 (55.0) | 11 (42.3) | 2.3 (.9–5.9) | .1 | 11 (52.4) | 1.7 (.6–4.4) | .31 |
| ≥4 | 5 (25.0) | 1 (3.9) | 3.8 (1.5–9.6) | .01 | 1 (4.8) | 2.0 (.9–4.3) | .06 |
| Any detectable CMV DNA in semen | 16 (80.0) | 14 (53.9) | 2.1 (.9–5.3) | .12 | 10 (47.6) | 1.7 (1.0–2.8) | .05 |
| Semen CMV DNA level, log10 copies/mL | |||||||
| Not detected | 4 (20.0) | 12 (46.2) | Reference | 11 (52.4) | Reference | ||
| >0 and <4 | 4 (20.0) | 5 (19.2) | 1.8 (.6–5.4) | .39 | 3 (14.3) | 2.3 (.7–7.9) | .34 |
| ≥4 | 12 (60.0) | 9 (34.6) | 2.3 (.9–5.8) | .09 | 7 (33.3) | 1.7 (.9–3.2) | .14 |
| Any detectable EBV DNA in semen | 10 (50.0) | 5 (19.2) | 2.1 (1.1–3.9) | .03 | 5 (23.8) | 2.1 (.9–5.1) | .08 |
| Semen EBV DNA level, log10 copies/mL | |||||||
| Not detected | 10 (50.0) | 21 (80.8) | Reference | 16 (76.2) | Reference | ||
| >0 and <4 | 6 (30.0) | 3 (11.5) | 2.1 (1.0–4.1) | .12 | 3 (14.3) | 2.4 (.7–8.0) | .25 |
| ≥4 | 4 (20.0) | 2 (7.7) | 2.1 (1.0–4.4) | .17 | 2 (9.5) | 2.6 (.5–12.1) | .36 |
| Any detectable HSV-1 DNA | 0 | 0 | NA | 0 | NA | ||
| Any detectable HSV-2 DNAc | 0 | 0 | NA | 0 | NA | ||
| Positive HSV-2 serologyc | 8 (47.1) | 7 (28.0) | 1.6 (.8–3.3) | .21 | 6 (30.0) | 1.6 (.7–3.6) | .29 |
| Any detectable HHV-6 DNA | 1 (5.0) | 1 (3.9) | 1.2 (.3–4.8) | 1 | 1 (4.8) | 1.1 (.1–18.1) | 1 |
| Any detectable HHV-7 DNA | 3 (15.0) | 2 (7.7) | 1.4 (.6–3.2) | .64 | 2 (9.5) | 1.6 (.3–8.5) | .66 |
| Any detectable HHV-8 DNA | 2 (10.0) | 1 (3.9) | 1.6 (.7–3.8) | .57 | 1 (4.8) | 2.1 (.2–21.4) | .6 |
| Bacterial STId | 5 (26.3) | 0 | 2.7 (1.8–4.1) | .01 | 0 | NA | .05 |
Data are no. (%) of participants, unless otherwise indicated.
Abbreviations: ART, antiretroviral therapy; CI, confidence interval; CMV, cytomegalovirus; EBV, Epstein-Barr virus; HHV, human herpesvirus; HSV, herpes simplex virus; IgG, immunoglobulin G; NA, not available; STI, sexually transmitted infection.
a Calculated for being a transmitter in the presence of the coinfections.
b Calculated using χ2 or Fisher exact tests, for categorical variables, and the Student t test, for continuous variables.
c Data were only available for 42 subjects.
d Defined as chlamydial infection, gonorrhea, and/or syphilis.
Associations With HIV Transmission
To evaluate the impact of herpesvirus shedding on HIV transmission, we compared levels of HIV in blood, positivity for HIV in semen, counts of CD4+ T cells in blood, positivity for an STI, levels of CMV IgG in blood plasma, and positivity for DNA of 7 different herpesviruses in seminal plasma from 20 HIV-infected MSM who did and 26 HIV-infected MSM who did not transmit HIV to their sex partner (Table 1 and Figure 1). A subanalysis including only ART-naive subjects was also performed.
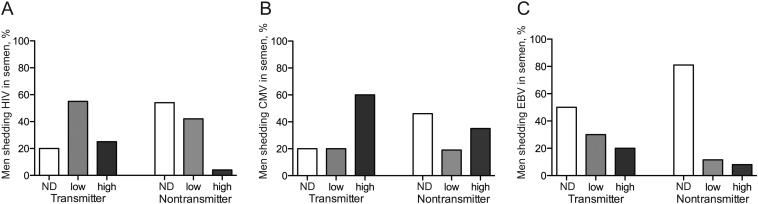
Figure 1.
Seminal human immunodeficiency virus type 1 (HIV) RNA levels and cytomegalovirus (CMV) and Epstein-Barr virus (EBV) DNA levels in samples from men who did and men who did not transmit HIV to their sex partners. A, Distribution of seminal HIV RNA shedding among transmitters and nontransmitters, by shedding level. B, Distribution of seminal CMV DNA shedding among transmitters and nontransmitters, by shedding level. C, Distribution of seminal EBV DNA shedding among transmitters and nontransmitters, by shedding level. A low level of shedding was defined as <4 log10 RNA or DNA copies/mL, and a high level of shedding was defined as >4 log10 RNA or DNA copies/mL. Abbreviation: ND, not detectable.
As described previously [3, 4], there was a positive correlation between the levels of HIV in semen and in blood (P < .01), and significantly higher HIV levels in blood plasma were detected for the transmitter group (4.6 vs 3.9 log10 HIV RNA copies/mL; P = .05). Overall, 28 patients had detectable HIV in semen, all of whom had also detectable HIV in blood. Of the 18 patients with undetectable HIV in seminal plasma, 5 had HIV levels in blood of <400 copies/mL and were receiving ART at the time of semen collection. Having detectable seminal plasma HIV was associated with being in the transmitter group rather than in the nontransmitter group (80% vs 46%; P = .02), with a relative risk of 2.6, while an HIV seminal level of >4 log10 copies/mL had a relative risk of 3.8. A significantly lower mean CD4+ T-cell count was also found in the transmitter group (438 vs 595 cells/microL; P = .03). Transmitters presented significantly higher levels of CMV IgG in blood, compared with nontransmitters (21.8 vs 15 lU/mL; Supplementary Figure 1). In univariate analysis, being in the transmitter group was marginally associated with detectable seminal CMV (P = .12) and EBV (P = .03), compared with being in the nontransmitter group. When CMV was detectable (compared with undetectable), there was a 2.1 relative risk of being in the transmitter group; the relative risk was 2.3 when CMV seminal levels were >4 log10 copies/mL (compared with undetectable; P = .09). With EBV shedding, there was also a relative risk of 2.1 for belonging to the transmitter group; an effect of EBV level was not observed. The presence of any bacterial STI had a relative risk of 2.7 for belonging to the transmitter group. Overall, seminal shedding of HSV, HHV-6, HHV-7, and HHV-8 was low (0%–15%; Table 1) and was not significantly associated with belonging to either transmission group, by univariate analysis.
Inclusion of only ART-naive subjects in the analysis revealed that HIV levels in blood (P = .44) and HIV seminal shedding (P = .18) were not significantly different between transmission groups. Only seminal HIV levels of >4 log10 copies/mL remained associated with being in the transmitter group (P = .06). A higher frequency of CMV detection in semen (P = .05) and, marginally, of EBV detection in semen (P = .08) was found among ART-naive transmitters, compared with ART-naive nontransmitters. The presence of bacterial STI also remained statistically different (P = .05), and a trend was still observed for CMV IgG level (P = .07). Since bacterial STIs are a potential confounder, we repeated the univariate analysis, including only ART-naive subjects without documented STIs. No major change was observed in the effect sizes for HIV, CMV, and EBV shedding in semen, although decreased sample size yielded marginally increased P values.
A test for trend was performed between HIV transmission and seminal levels of HIV, CMV, and EBV, and there was a dose-response relationship between HIV levels and HIV transmission for all partner pairs (P < .01) and for partner pairs in which source partners had no STI (P = .05). The dose response for HIV was marginal in analysis involving only pairs in which the source partner was ART naive (P = .06). There was a trend of an association between CMV and the transmission group when the entire cohort was evaluated (P = .07), but this association became statistically significant in the cohort limited to ART-naive source partners (P = .04) and the cohort limited to source partners without STIs (P = .04). For EBV, only trends were observed in each data set (P = .06–.17).
A multivariable logistic regression analysis to evaluate CMV shedding and HIV transmission was performed. Most factors were highly associated. In particular, high CMV shedding was associated with detectable seminal HIV (P = .05), EBV shedding (P = .01), lower CD4+ T cell counts (P = .07), and higher blood HIV levels (P = .10) but not with bacterial STIs or CMV IgG levels. The multivariate analyses were performed with only CMV shedding (as a dichotomous outcome) and each of the potential confounders, entered 1 at a time, using the entire data set and the data sets restricted to ART-naive subjects and to ART-naive subjects without bacterial STIs (Supplementary Table 1). CMV shedding was consistently found to have a positive association with transmission, with odds ratios ranging from 2.49 (after adjustment for EBV in the entire data set) to 4.36 (after adjustment for HIV levels in blood in the ART-naive data set ).
DISCUSSION
In this case-control study, we compared the shedding and viral levels of HIV and 7 different human herpesviruses in seminal plasma, CMV IgG levels, CD4+ T-cell counts, and bacterial coinfections between HIV-infected men who did (n = 20) or did not (n = 26) transmit HIV to their male sex partner. Similar to previous studies [1, 3–5], the transmitters had significantly higher levels of HIV in blood and semen, a decrease frequency of ART use, lower CD4+ T-cell counts, and a higher prevalence of bacterial STIs, compared with nontransmitters. Unique to this study, we observed a higher frequency of seminal CMV and EBV in the transmitter group, even after excluding the 5 potential source partners receiving ART in the nontransmitter group. In the subanalysis including only ART-naive individuals, HIV levels in blood and semen did not differ between transmission groups, suggesting that viral coinfections likely play a bigger role in HIV transmission when the potential source partner is not receiving ART. However, during ART, suppressed viral load becomes the main factor predicting lack of HIV transmission. After excluding the 5 source partners with STIs and after adjustment for other possible confounding factors, CMV shedding remained an independent predictor of HIV transmission.
As suggested in our previous study [10], a dose response was observed for CMV and HIV shedding, meaning that the relative risk of HIV transmission increased when viral levels for HIV and CMV were higher in semen. A similar effect was not seen for EBV.
In contrast to previous studies [3, 7, 10], we did not find differences in HHV-8 or HSV shedding between the groups, likely because of the small sample size.
HIV transmitters also presented higher levels of CMV IgG in blood as compared to nontransmitters, but CMV antibody levels were not associated with the frequency or level of CMV shedding in semen.
Only 10% of the source partners in the transmitter group had an estimated infection duration of <90 days, compared with 31% in the nontransmitter group (P = .15), suggesting that factors other than recent stage of infection and its associated high levels of HIV in blood contribute to sexual transmission. This study was limited in that samples were collected within at least 4 months of the putative transmission event (for transmitters) or report date (for nontransmitters), an interval during which the genital milieu might have changed.
Since many variables were highly correlated and because of the small sample size, we could not establish which factor was most important by means of a multivariate analysis that accounted for all the variables. We assessed whether CMV shedding was associated with HIV transmission, individually accounting for each potential confounder. Overall, seminal CMV replication was associated with HIV transmission independently of any other factor, and there was no confounder for the association of CMV shedding with HIV transmission.
In conclusion, this study provides new evidence that CMV and EBV replication in the male genital tract is associated not only with increased HIV seminal shedding, but also with HIV transmission, and this observation is especially true when the potential source partner is not receiving ART. Nevertheless, achievement of suppressed levels of HIV in blood and semen during ART remains the most effective strategy to reduce transmission of HIV among MSM. The results of this study are biologically interesting and provide important insights in the complex viral and immunologic dynamics in the male genital tract.
However, deferring ART in favor of therapy against herpesviruses is not justified, given the toxicity of currently available broad-spectrum herpesvirus therapy, and previous studies have demonstrated that HSV-specific therapy is also not warranted [11] for HIV prevention. Future studies should determine whether these observations hold true during ART and, if this is the case, whether adding CMV suppressive therapy to standard ART for high-risk patients or discordant couples may be clinically relevant if newer, less toxic therapies become available.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We are grateful to all the participants in the San Diego Primary Infection Cohort and the San Diego HIV-transmission Study.
S. G., S. R. M., D. D. R., J. A. Y., S. J. L., and D. M. S. designed the present study; S. R. M., S. G., J. A. Y., and B. C. performed the data analyses; S. G., S. R. M., and D. M. S. wrote the primary version of the manuscript; M. V. V. and S. G. performed the laboratory experiments; and all authors revised and approved the final manuscript.
Financial support. This work was supported by the Department of Veterans Affairs, the James Pendleton Charitable Trust, the US National Institutes of Health (awards AI69432, AI043638, MH62512, MH083552, AI077304, AI36214, AI047745, AI74621, GM093939, and AI080353, as well as award AI306214 [from the Centers for AIDS Research]), and the Swiss National Science Foundation (grant PASMP3_136983).
Potential conflicts of interest. D. D. R. has served as a consultant for Bristol-Myers Squibb, Gilead Sciences, Merck, Monogram Biosciences, Biota, Chimerix, Gen-Probe, Tobira, and Idenix Pharmaceuticals. D. M. S. has received grant support from ViiV Pharmaceuticals and consultant fees from Gen-Probe. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Quinn TC, Wawer MJ, Sewankambo N, et al. Viral load and heterosexual transmission of human immunodeficiency virus type 1. N Engl J Med. 2000;342:921. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 2.Gray RH, Wawer MJ, Brookmeyer R, et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357:1149–53. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 3.Butler DM, Smith DM, Cachay ER, et al. Herpes simplex virus 2 serostatus and viral loads of HIV-1 in blood and semen as risk factors for HIV transmission among men who have sex with men. AIDS. 2008;22:1667–71. doi: 10.1097/QAD.0b013e32830bfed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baeten JM, Kahle E, Lingappa JR, et al. Genital HIV-1 RNA predicts risk of heterosexual HIV-1 transmission. Sci Transl Med. 2011;3:77ra29. doi: 10.1126/scitranslmed.3001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson LF, Lewis DA. The effect of genital tract infections on HIV-1 shedding in the genital tract: a systematic review and meta-analysis. Sex Transm Dis. 2008;35:946–59. doi: 10.1097/OLQ.0b013e3181812d15. [DOI] [PubMed] [Google Scholar]
- 6.Gianella S, Strain MC, Rought SE, et al. Associations between the virologic and immunologic dynamics in blood and in the male genital. Tract J Virol. 2012;86:1307–15. doi: 10.1128/JVI.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS. 2006;20:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- 8.Sheth PM, Danesh A, Sheung A, et al. Disproportionately high semen shedding of HIV is associated with compartmentalized cytomegalovirus reactivation. J Infect Dis. 2006;193:45–8. doi: 10.1086/498576. [DOI] [PubMed] [Google Scholar]
- 9.Lisco A, Munawwar A, Introini A, et al. Semen of HIV-1-infected individuals: local shedding of herpesviruses and reprogrammed cytokine network. J Infect Dis. 2012;205:97–105. doi: 10.1093/infdis/jir700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gianella S, Morris SR, Anderson C, et al. Herpesviruses and HIV-1 drug resistance mutations influence the virologic and immunologic milieu of the male genital tract. AIDS. 2012 doi: 10.1097/QAD.0b013e3283573305. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celum C, Wald A, Lingappa JR, et al. Acyclovir and transmission of HIV-1 from persons infected with HIV-1 and HSV-2. N Engl J Med. 2010;362:427–39. doi: 10.1056/NEJMoa0904849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler DM, Delport W, Kosakovsky Pond SL, et al. The origins of sexually transmitted HIV among men who have sex with men. Sci Transl Med. 2010;2:18re1. doi: 10.1126/scitranslmed.3000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Little SJ, Frost SD, Wong JK, et al. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol. 2008;82:5510–8. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosakovsky Pond SL, Posada D, Stawiski E, et al. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol. 2009;5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parrinello CM, Sinclair E, Landay AL, et al. Cytomegalovirus immunoglobulin G antibody is associated with subclinical carotid artery disease among HIV-infected women. J Infect Dis. 2012;205:1788–96. doi: 10.1093/infdis/jis276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.