Abstract
The RNA-induced silencing complex (RISC) is a programmable gene-silencing machine involved in many aspects of eukaryotic biology. In humans, RISC is programmed or “loaded” with a small guide RNA by the action of a tri-molecular assembly termed the RISC-loading complex (RLC). The human RLC is composed of the proteins Dicer, TRBP and Argonaute2 (Ago2). To facilitate structural and biochemical dissection of the RISC-loading process we have developed a system for the in vitro reconstitution of the human RLC. Here, we describe in detail methods for the expression and purification of recombinant Dicer, TRBP and Ago2 and protocols for the assembly of RLCs and RLC-subcomplexes. We also describe several simple assays to observe the biochemical activities of the assembled protein complexes.
Keywords: RLC, RISC, Dicer, Argonaute, microRNA, RNAi
1. Introduction
RNA interference (RNAi) is a broad-spread eukaryotic mechanism of gene silencing that plays a fundamental role in many aspects of animal biology, including developmental timing, stem cell division, memory and learning. On the molecular level RNAi is mediated by a family of ribonucleo-protein complexes called RNA-Induced Silencing Complexes (RISC), which silence genes by mediating translational repression and degradation of targeted message RNAs (mRNA) (1). The versatility and power of RNAi arises from the fact that RISC can be programmed to target any nucleic acid sequence for silencing. RISC programming is therefore a critical cellular function, requiring the action of a specialized macromolecular assembly called the RISC-loading complex (RLC) (2,3,4).
On the molecular level, the RLC programs RISC with target sequence information by mediating the non-covalent binding, or “loading”, of a ~22 nucleotide RNA onto Argonaute proteins which are the core subunits of RISC. The small RNA functions as a guide for gene silencing through base pairing recognition of target mRNAs (5,6,7,8).
The mammalian RLC is a trimeric complex (350 kDa) composed of the proteins Dicer, TRBP and Argonaute-2 (Ago2) (3, 9). The main function of the human RLC is to load Ago2 with microRNAs (miRNA), an abundant class of 22-nucleotide regulatory RNAs that arise from endogenous pre-miRNA hairpin structures. The RLC first recognizes pre-miRNA and cleaves, or “dices”, it into a 22-nucleotide RNA duplex. Based on the stability properties of the duplex, one strand of RNA is selected to be the guide RNA for subsequent gene silencing and loaded into Ago2. The overall reaction is spontaneous and does not require any factors beyond the three proteins and a premiRNA (9).
The loading of Argonaute with a miRNA is perhaps the most important step in the mammalian RNAi pathway because this is the point at which RISC is programmed with its target sequence information. To insure fidelity in the process, Ago2 loading is coupled to the pre-miRNA recognition and dicing steps. The RLC also has the ability to distinguish which strand in a miRNA duplex is to be loaded into Ago2 as the silencing guide and which RNA strand is to be discarded as the “passenger” (10,11). This is an essential function because loading the incorrect RNA strand could lead to targeted silencing of an entirely different and unintended set of genes.
Here we describe detailed methods for expression and purification of each of the three protein components in the human RLC (9). We also describe methods for assembling the purified components into RLCs and assaying RLC activity. Methods described here should facilitate detailed biochemical and structural characterization of the RISC-loading mechanism and might be used to characterize various Ago-associated proteins.
2. Materials
2.1. RNA oligonucleotides
The following RNAs (synthesized by Dharmacon) are used: Drosophila pre-let-7 (pre-let-7), 5'-AAUGAGGUAGUAGGUUGUAUAGUAGUAAUUACACAUCAUACUAUACAAUGUG CUAGCUUUCU-3'; 37-nt A, 5’-UGAGGUAGUAGGUUGUAUAGUUUGAAAGUUCACGAUU-3’ and its complementary partner 37-nt B, 5’-UCGUGAACUUUCAAACUAUACAACCUACUACCUCAUU-3’; 21-nt guide RNA, 5’-UGAGGUAGUAGGUUGUAUAGU-3’; and 21-nt passenger RNA, 5’-UAUACAAUGUGCUAGCUUUCU-3’.
For radiolabelling, adenosine 5’-triphosphate, [γ-32P] (3000 Ci/mMol) (Perkin Elmer) and T4 Polynucleotide kinase (New England Biolabs) were used.
For preparing denaturing polyacrylamide gels, acrylamide 40% w/v solution (EMD); Urea (Fisher Chemicals); N, N, N', N'-Tetramethyl ethylenediamine TEMED (Fisher Bioreagents); ammonium persulfate (MP Biomedicals).
2X denaturing loading buffer: 95% formamide, 18 mM EDTA, 0.025% SDS, 0.1% xylene cyanol and 0.1% bromophenol blue.
RNA gel running buffer: 0.5 X Buffer TBE (1X TBE: 89 mM Tris base, 89 mM boric acid, 2.5 mM EDTA).
RNA gel apparatus (Dan-Kar Corp) and power supply (VWR).
For visualization, storage phosphor screen and Phosphorimager (Amersham, Healthcare Life Sciences).
For precipitating RNA, ethanol 200 proof (Sigma Aldrich) and 3 M sodium acetate, pH 5.2 (Fisher Bioreagents).
2.2. Baculovirus Production and Amplification
Human Dicer, Argonaute (Ago2) and TRBP cDNA clones (Open Biosystems).
pFastBac HTa vector (Invitrogen).
MAX Efficiency DH10BAC Competent cells (Invitrogen, Carlsbad, CA) and LB media.
LB agar plates containing 50 µg/ml kanamycin, 7 µg/ml gentamicin, 10 µg/ml tetracycline, 40 µg/ml Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 100 µg/ml bromo-chloro-indolyl-galactopyranoside (X-gal) (100 µg/ml).
DNA Miniprep solutions P1, P2 and N3 (Qiagen), isopropanol, 70% ethanol.
Fugene6 (Roche Applied Science
ESF-921, Sf-9 cell media (Expression Systems, Woodland, CA).
2.3. Protein expression and purification
ESF-921, Sf-9 cell media (Expression Systems, Woodland, CA).
Lysis buffer: 300 mM NaCl, 50 mM Sodium phosphate dibasic pH 8.0, 10 mM imidazole pH 8.0, 0.5% Triton X-100, 5% glycerol, 1mM tris(2-carboxyethyl)phosphine) (TCEP), 1 tablet of EDTA-free protease inhibitor cocktail (Roche) per 25 ml buffer.
Lysis: Dounce tissue grinder (Kimble Chase Life Science and Research Products LLC).
Ni-NTA resin (Qiagen).
Wash Buffer: 300 mM NaCl, 50 mM Sodium phosphate dibasic heptahydrate pH 8.0, 20 mM imidazole pH 8.0, 5% glycerol, 1mM TCEP (tris(2-carboxyethyl)phosphine).
Elution Buffer: 300 mM NaCl, 50 mM Sodium phosphate dibasic heptahydrate pH 8.0, 300 mM imidazole pH 8.0, 5% glycerol, 1mM TCEP (tris(2-carboxyethyl)phosphine).
TEV protease: purified in house by standard Nickel affinity purification, or alternatively may be purchased (Invitrogen).
Dialysis membrane (10,000 Da molecular weight cut-off) (Spectrum Labs).
HisTrap NiNTA column (Pharmacia) (GE Healthcare).
Superdex 200 16/60 column (Pharmacia) (GE Healthcare).
Gel filtration Buffer: 300 mM NaCl, 50 mM Hepes pH 8.0, 5% glycerol, 1mM TCEP (tris(2-carboxyethyl)phosphine).
Bradford dye reagent (Bio-rad).
2.4. RLC reconstitution
Gel filtration Buffer: 300 mM NaCl, 50 mM Hepes pH 8.0, 5% glycerol, 1mM TCEP (tris(2-carboxyethyl)phosphine).
Superose 6 10/30 column (GE Healthcare).
SDS cassette gel (Expedeon).
Tris-Tricine-SDS Running Buffer with Bisulfite (Expedeon).
2.5. RNA Filter-Binding Assay
BA-85 nitrocellulose filter (to retain protein-RNA complexes) (Whatman).
Hybond-N+ nylon membrane (to retain free RNA) (Amersham Biosciences).
MiniFold-1 Dot-Blot System (Whatman).
Membrane soaking buffer: 20 mM Hepes, pH 7.5.
RNA renaturing buffer: 10 mM Tris-Cl (pH 7.5), 1.5 mM Mg2+ and 50 mM NaCl.
Reaction buffer: 20 mM Hepes (pH 7.5), 60 mM KCl, 5 mM EDTA, 1 mM DTT, 0.01% Igepal-680 and 0.1 mg/ml tRNA.
2.6. Dicing Assay
Reaction buffer: 100 mM NaCl, 40 mM Hepes, pH 7.5, 1 mM DTT, 3 mM MgCl2.
Pre-let-7 hairpin RNA, 32P 5’ end labeled 37 nt-A and cold 37 nt-B RNA.
RNA gel: 14% acrylamide gel (23 g Urea, 2.5 ml 10X TBE buffer, 17.5 ml 40% polyacrylamide, water to 50 ml, 71 µl TEMED, and 350 µl 10% Ammonium persulfate).
2x denaturing loading buffer: 95% formamide, 18 mM EDTA, 0.025% SDS, 0.1% xylene cyanol and 0.1% bromophenol blue.
2.7. Slicing Assay
Reaction buffer: 0.1 mg/ml yeast tRNA, 20 mM Tris-HCl (pH 7), 50 mM KCl, 5% glycerol, 1.5 mM MgCl2.
21 nt guide RNA, 37 nt-A sense target RNA.
2.8. RISC-loading activity Assay
Reaction buffer: 0.1 mg/ml yeast tRNA, 20 mM Tris-HCl (pH 7), 50 mM KCl, 5% glycerol, 1.5 mM MgCl2.
Pre-let-7 hairpin RNA, 37 nt-A and -B RNAs, 21 nt guide and passenger RNAs.
3. Methods
3.1. Radiolabeling RNA oligos
7 µl synthetic RNA substrates (5 µg/µl) are mixed with 1 µl adenosine 5’- triphosphate, [γ-32P] (3000 Ci/mMol), 1 µl T4 Polynucleotide kinase and 1 µl T4 Polynucleotide kinase buffer and incubated for 1 hour at 37°C.
Unreacted ATP is removed by passing the reaction mixture through an Illustra MicroSpin G-25 column. 2x denaturing loading buffer is then added to each sample. The volume of the flow through is estimated and an equal volume of 2X denaturing loading buffer is added prior to gel purification.
RNA samples are loaded into a denaturing 14% polyacrylamide gel poured by mixing 23 g urea, 2.5 ml 10X TBE buffer, 20 ml 40% polyacrylamide, plus water to a final volume of 50 ml. Once the urea is dissolved 71 µl TEMED and 350 µl 10% ammonium persulfate are added to induce polymerization. The gel is run with 0.5X TBE buffer at a constant power of 20 watts.
The RNA gel is wrapped in a layer of plastic wrap and exposed to a storage phosphor screen for 1 – 3 minutes and visualized by Phosphorimaging. The imaged gel is then printed at full size (100%) so that the printed image has dimensions identical to those of the gel. The printed image is placed under the gel to help identify the physical position of the desired RNA. In order to align the printed image of the gel with the actual gel “hot dots”, or spots of 32P placed on small pieces of filter paper which are then placed on top of the gel prior to exposure and imaging, can be employed. The desired band is cut out of the gel with a clean razorblade, crushed with a sterile needle, resuspended in approximately twice the volume of water and incubated on a rocker overnight at 4°C.
The following day the aqueous solution is moved to a fresh eppendorf tube and centrifuged briefly to pellet any residual gel fragments. The volume of the liquid is then estimated and added to a new eppendorf tube containing 2.5 times (v/v) of 100% ethanol and 0.1 times (v/v) of 3 M sodium acetate, pH 5.2. The mixture is incubated at −80°C for 30 minutes to precipitate the RNA.
The precipitated RNA is centrifuged at maximum speed in a table-top centrifuge for 10 minutes and the supernatant is carefully removed and discarded as radioactive waste. The pellet is subsequently washed with 1 ml of 70% ethanol, and centrifuged briefly again. The supernatant solution is removed and the pellet resuspended in 20 µl double-distilled water.
3.2. Baculovirus Production and Amplification
Baculoviruses separately expressing His6-tagged Dicer, TRBP and Ago2 are generated using a modified version of the Bac2Bac system (Invitrogen). This protocol begins after insertion of cDNA clones of human Dicer, TRBP and Ago2 individually into the plasmid pFastBac HTA, which appends a His6-tag and recognition sequence for the Tobacco Etch Virus (TEV) protease appended to the Nterminus of each protein. Plasmid DNAs are transformed into DH10BAC cells by mixing 250 ng of DNA with 10 µl of competent cells and incubating on ice for 20 minutes.
Cells are then heat shocked at 42°C for 45 seconds and immediately moved back onto ice. After 2 more minutes, 800 µl of LB media is added to each transformation and cells are allowed to recover at 37°C for 5 hours with constant shaking.
After recovery, 10 µl of the transformation mixture is plated out on LB agar plates containing kanamycin, gentamicin, tetracycline, IPTG and X-gal. Plates are incubated at 37°C for 48 hours to allow large colonies to grow.
A large, well-isolated, white colony from each DH10BAC plate is identified and used to inoculate 2.5 ml of LB media containing 50 µg/ml kanamycin, 7 µg/ml gentamicin and 10 µg/ml tetracycline. Cultures are grown overnight (12–18 hours) at 37°C with vigorous shaking.
“Bacmid” DNA is isolated from each bacterial culture using the buffers from a QIAprep Spin Miniprep Kit, however the spin columns are not employed. Cells from 1.5 ml of each bacterial culture are pelleted by brief centrifugation in 1.5 ml eppendorf tubes using a tabletop microfuge. The supernatant solution is removed and the cell pellet resuspended in 250 µl buffer P1. Following resuspension of the cell pellet, 250 µl of buffer P2 is added to induce cell lysis. After a 3 minute incubation at room temperature 300 µl of buffer N3 is added and the tube is gently mixed and then centrifuged for 15 minutes at maximum speed on a tabletop microfuge. The supernatant liquid is then carefully transferred to a new microfuge tube containing 800 µl of 100% isopropanol. After mixing, the tube is centrifuged for 5 minutes at maximum speed to pellet the precipitated DNA. The supernatant solution is discarded and the DNA pellet is washed once with 800 µl of 70% ethanol. The DNA pellet is air dried for 5 minutes (or until all the ethanol is evaporated) before it is resuspended in 100 µl buffer EB. DNA concentration is determined by spectrometry. A typical yield is 100 µl of a 1 mg/ml DNA solution.
Sf-9 cells grown in Excel 420 media are added to a 6 well plate so that they are 70% confluent (~ 1 ×106 cells per well). Plated cells are incubated at 27°C for 10 minutes to allow cells to attach. The media is removed and the cells are covered with 3.5 ml of fresh Excel 420.
94 µl Excel 420 and 6 µl Fugene 6 are mixed in a tube by gentle tapping.
After 5 minutes 1 µg bacmid DNA is added and mixed by tapping.
After 15 minutes the transfection mixture is added to the cells in one well of a 6-well dish, swirling after every few drops. The plate is then incubated at 27 °C for 4–5 days for the initial generation of the virus.
After 4–5 days the liquid media, which contains the virus, is harvested and any cell debris are removed by centrifugation. The virus is then amplified by applying 200 µl of the virus-containing media to 1 ×106 of Sf-9 cells freshly plated in a 6-well dish. The remaining media, containing the initial virus is stored at 4°C.
After 2–4 days Step 11 is repeated two more times to obtain second and third amplifications of the virus. To obtain a large quantity of virus, the third amplification can be done in a sterile 500 ml flask containing 250 ×106 cells in 250 ml of Excel 420, infected with 5 ml of the second amplification of the virus and shaken at ~140 rpm.
3.3. Protein expression and purification
N-terminally His6-tagged Human Dicer, Ago2 and TRBP can be purified separately from Sf-9 cells infected with baculovirus bearing the cDNA copy of the desired protein. For each protein, 1.5×109 cells in 750 ml of Excel 420 media are infected with 25 ml of virus (third amplification) at 27 °C and then harvested 60–72 hours after infection.
Cells are pelleted by centrifugation at 2000×g for 10 minutes. Cell pellets are resuspended in 25 ml of ice-cold Lysis buffer. All subsequent steps are carried out on ice or at 4°C.
Cells are lysed by 7 strokes with a B pestle of a 40 ml Dounce tissue grinder.
Insoluble material is pelleted by centrifugation (15 min., 20,000xg) and the supernatant solution is applied to 2.5 ml (packed) of Ni-NTA resin in a 50 ml Falcon tube and gently rocked for 40 minutes (See Note 1).
The resin is pelleted by brief centrifugation and washed by resuspending in 45 ml of Wash buffer. The resuspended resin is pelleted again and subjected to 4 more rounds of washing.
Protein is eluted from the washed resin with 7.5 ml of Elution buffer.
To remove the N-terminal His6-tag, 0.5 mg of TEV protease is added to the eluted protein. The protein solution is then dialyzed using a 10,000 Da molecular weight cut-off dialysis membrane against 1 L of wash buffer overnight.
Dialyzed protein is then passed through a 5 ml His-Trap column. The unbound material (protein without His6) is collected and concentrated to 1–2 ml using a 15 ml Amicon Ultra centrifugal filter.
The concentrated protein sample is applied to a Superdex 200 16/60 column equilibrated in gel filtration buffer (100 mM KCl, 5% glycerol, 1 mM DTT, 20 mM Hepes, pH 7.5). The column is run with gel filtration buffer at a flow rate of 1 ml/min, collecting 2.5 ml fractions. Fractions containing non-aggregated protein are pooled, concentrated to 5–10 mg/ml and used in subsequent reconstitution experiments.
Protein concentrations are determined by the Bradford Assay.
3.4. RLC reconstitution
Dicer (700 µg or 3 nmol), Ago2 (600 µg or 6 nmol) and TRBP (550 µg or 11 nmol, assuming a dimer) are mixed in 250 µl (final volume) of gel filtration buffer and incubated on ice for 10 min (See Notes 2 & 3).
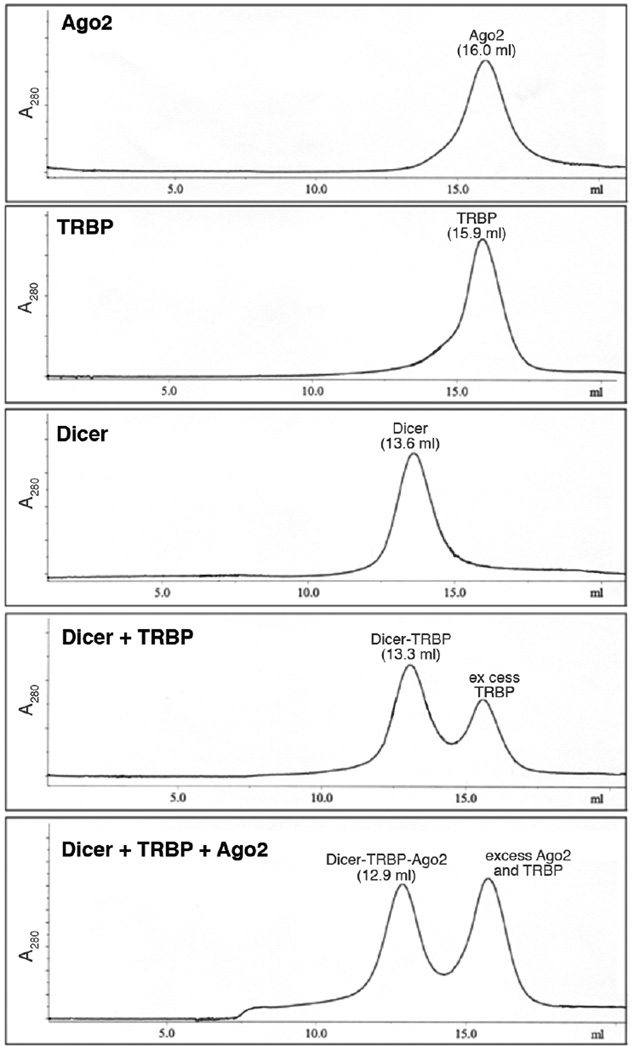
The protein solution is applied to a Superose 6 10/30 column equilibrated in gel filtration buffer. The column is run at a flow rate of 0.5 ml/min while collecting 0.5 ml fractions. Examples of the elution profiles of the various RLC components are shown in Fig. 1.
Fractions are analyzed by SDS-PAGE and those containing the RLC are pooled and concentrated to ~1.5 mg/ml (~4 µM). Aliquots are stored frozen at −80°C.
Figure 1. Superose 6 elution profile of RLC and RLC components.
The absorbance at 280 nm of the eluate is plotted against the elution volume for each protein sample. Protein components and elution volumes are indicated above each absorbance peak.
3.5. RNA Filter-Binding Assay
Prior to setting up the binding reaction, the RNA is annealed in a buffer containing 10 mM Tris-Cl (pH 7.5), 1.5 mM Mg2+ and 50 mM NaCl by heating to 65 °C for 7 minutes followed by snap cooling on ice.
5’ end-labeled pre-let-7 hairpin RNA (<2 pM) is incubated in a 50 µl reaction volume containing 20 mM Hepes (pH 7.5), 60 mM KCl, 5 mM EDTA, 1 mM DTT and 0.1 mg/ml tRNA.
The RNA is incubated with protein for 1 hour prior to application to filters.
For each experiment, two filters are used: a BA-85 nitrocellulose filter to retain protein-RNA complexes and a Hybond-N+ nylon membrane to retain free RNA (12). These filters are soaked in a buffer containing 20 mM Hepes, pH 7.5, for one hour prior to use in a 96-well dot blot apparatus.
A 40 µl aliquot from each reaction is applied to the top filter and then vacuum is applied to the apparatus to draw the samples through. The filters are not washed with additional buffer after the sample is drawn through.
After brief air-drying, the free and bound RNA is quantified by phosphorimaging of the filters.
3.6. Dicing Assay
Dicing substrates can be either an RNA hairpin (pre-let-7) or a dsRNA composed of two 37-nt RNA oligonucleotides (designated strand A and B) (See Note 4). For the hairpin RNA, 32P 5’ end labeled RNA (~10 nM) is incubated in reaction buffer (100 mM NaCl, 40 mM Hepes, pH 7.5, 1 mM DTT, 3 mM MgCl2) in a metal heat block at 65°C for 7 minutes, then snap-cooled by immediately placing the sample on ice prior to every assay. To form the 37 nt duplex substrate, 32P 5’ end labeled strand A (~10 nM) is mixed with an equal amount of unlabeled strand B RNA in reaction buffer. The RNA mixture is then denatured by placing in a 65°C metal heat block for 7 minutes and then slowly cooled to room temperature by moving the heat block (still containing the RNA sample) to the bench top.
Reactions are carried out in a total volume of 10 µl containing 100 mM NaCl, 40 mM Hepes, pH 7.5, 1 mM DTT and 3 mM MgCl2 with radiolabeled RNA at 37°C for 10–60 minutes in the presence of 100–200 nM RLC or Dicer alone.
RNA products are then resolved by denaturing PAGE (14% acrylamide-7M urea in 0.5xTBE).
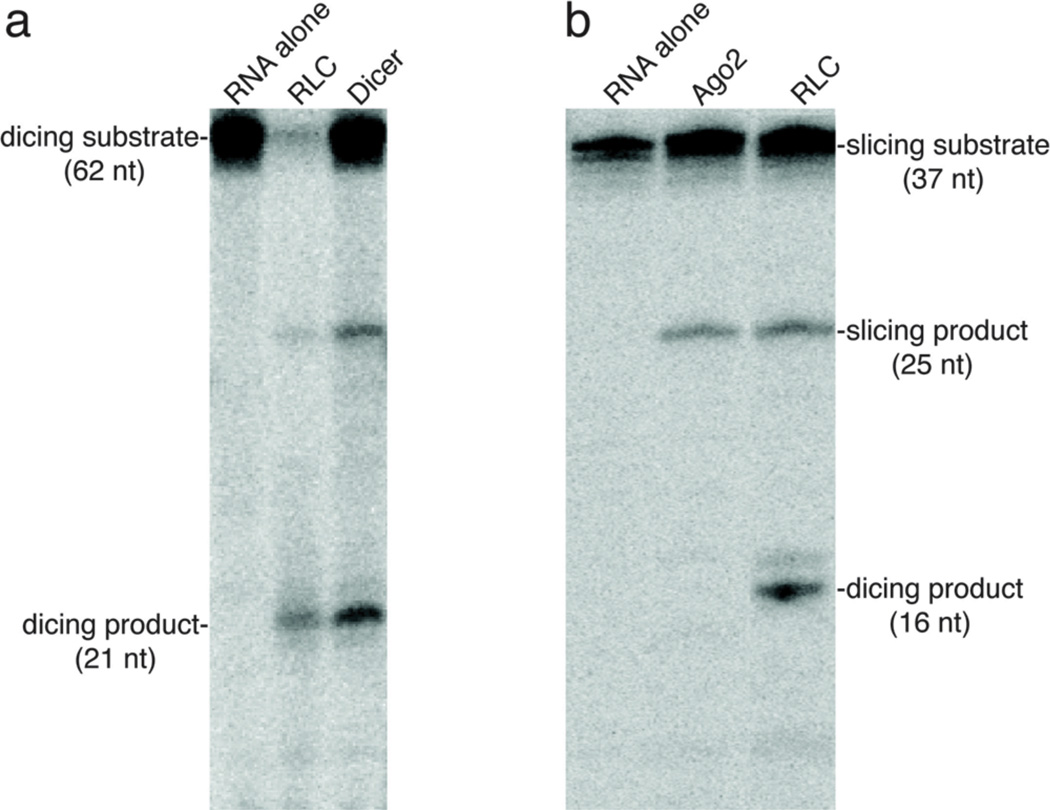
The gel is dried and visualized by phosphorimaging. An example of the results typically observed is shown in Fig. 2a.
Figure 2. Dicing and Slicing Assays.
a) Dicing assay—both the RLC or Dicer alone can process a pre-let-7 hairpin (63 nt) into a 21 nt guide RNA. b) Slicing assay—once loaded with a single stranded guide RNA, both Ago2 and the RLC will cleave the corresponding target RNA. The RLC can also produce a smaller 16 nt product. This is generated by the Dicer subunit of the RLC acting on target RNA that is annealed to excess guide RNA. Dicer removes 21 nt from the 37 nt target, which generates a 16 nt. product.
3.7. Slicing Assay
-
1.
RLC or Ago2 alone (100–200 nM) is loaded with a single stranded guide 21nt RNA by incubating with 100–200 nM guide RNA in a 10 µl reaction containing 50 mM KCl, 5% glycerol, 1.5 mM MgCl2 and 20 mM Tris-HCl pH 7 for 30 minutes at 30°C.
-
2.
The slicing reaction is started by the addition of 1–100 nM of 5’-end 32P labeled single stranded target 37 nt-A RNA and then carried out at 30°C for 10–60 minutes.
-
2.
The reaction is stopped by adding 10 µl of formamide denaturing loading buffer and heating the samples to 75°C for 5 min.
-
3.
Denatured samples are analyzed by electrophoresis on a 14% polyacrylamide-7M urea gel.
-
4.
The gel is dried and the labeled RNA substrates and products are detected by phosphorimager. An example of the results produced is shown in Fig. 2b.
3.8. RISC-loading activity Assay
The RISC-loading assay is very similar to the slicing assay except that dsRNA (Dicer substrate) is used as the source of the guide RNA. Reconstituted RLC (200 nM) is incubated with 200 nM pre-let-7 hairpin pre-miRNA in slicing assay buffer at 30°C for 30 min.
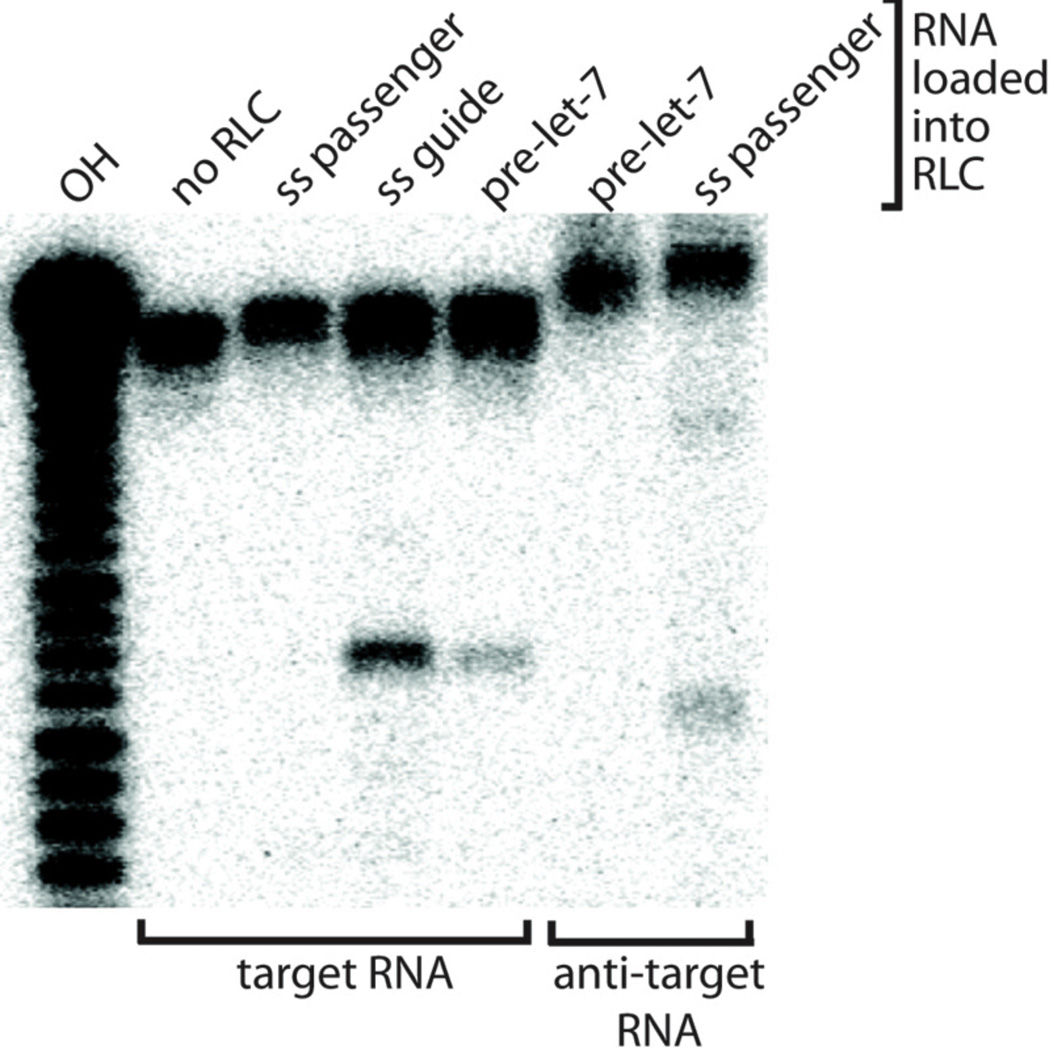
Radiolabeled target 37 nt-A or anti-target 37 nt-B RNA oligo (10 nM) is added and slicing activity is measured as described above. An example of the results produced from this assay is shown in Fig. 3.
Figure 3. RISC-loading Assay.
RLC can cleave target or anti-target (reverse complement of the target) RNA based on complementarity to the RNA loaded into the Ago2 subunit of the RLC. Lanes indicate addition of unlabeled single-stranded 21 nt guide RNA (ss guide), single-stranded 21 nt passenger RNA (ss passenger) or pre-let7 RNA to RLC.
Acknowledgements
We thank the members of the MacRae lab for helpful suggestions. Work on the Human RLC is funded by the NIH (R01 GM086701). IJM is a Pew Scholar in the Biomedical Sciences.
Footnotes
Proteins are purified from cell lysates using Ni-NTA resin in batch instead of using a Ni-NTA column. This is very important because crude Sf-9 cell lysates are often very viscous and will often clog conventional chromatography columns. Once the bound resin has been washed a few times it can be moved to a column format, if desired.
A key step in reconstitution of Dicer containing complexes, such as the RLC, is ensuring that Dicer is the limiting reagent in the assembly reaction. This is because Dicer runs anomalously large through size exclusion columns and it is, therefore, essentially impossible to separate the assembled RLC from free Dicer by this method. Fortunately, keeping Dicer-associated proteins (TRBP and Ago2) in 2-fold excess of Dicer eliminates any detectable free Dicer from the reaction.
The reconstitution protocols described here are specific for the human RLC. However, the methods are quite general and therefore (with some optimization) may be extended to other Ago2-containing molecular assemblies. Likewise, omitting either Ago2 or TRBP from the reconstitution reaction, allows generation of Dicer- TRPB or Dicer-Ago2 complexes, respectively. We have not been successful in producing a stable Ago2-TRBP complex.
The Dicing assays described here use either an RNA hairpin or a 35-bp RNA duplex. However, Dicer enzymes display flexibility in substrate specificity and can act on basically any dsRNA that posses: 1) an open helical end, preferably with a 3’ overhang; 2) a region of duplex structure that extends at least 20 bp from the helical end. When presented with long dsRNA, Dicer will cleave the substrate multiple times, each time ~21 bp from the open helical end, until all products are equal to or less than 23 bp in length.
References
- 1.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 5.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 7.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 8.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 9.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 11.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 12.Batey RT, Sagar MB, Doudna JA. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]