Abstract
Objective
The use of electronic medical record (EMR) data is necessary to improve clinical research efficiency. However, it is not easy to identify patients who meet research eligibility criteria and collect the necessary information from EMRs because the data collection process must integrate various techniques, including the development of a data warehouse and translation of eligibility criteria into computable criteria. This research aimed to demonstrate an electronic medical records retrieval system (ERS) and an example of a hospital-based cohort study that identified both patients and exposure with an ERS. We also evaluated the feasibility and usefulness of the method.
Design
The system was developed and evaluated.
Participants
In total, 800 000 cases of clinical information stored in EMRs at our hospital were used.
Primary and secondary outcome measures
The feasibility and usefulness of the ERS, the method to convert text from eligible criteria to computable criteria, and a confirmation method to increase research data accuracy.
Results
To comprehensively and efficiently collect information from patients participating in clinical research, we developed an ERS. To create the ERS database, we designed a multidimensional data model optimised for patient identification. We also devised practical methods to translate narrative eligibility criteria into computable parameters. We applied the system to an actual hospital-based cohort study performed at our hospital and converted the test results into computable criteria. Based on this information, we identified eligible patients and extracted data necessary for confirmation by our investigators and for statistical analyses with our ERS.
Conclusions
We propose a pragmatic methodology to identify patients from EMRs who meet clinical research eligibility criteria. Our ERS allowed for the efficient collection of information on the eligibility of a given patient, reduced the labour required from the investigators and improved the reliability of the results.
Keywords: Oral & Maxillofacial Surgery, Public Health, Statistics & Research Methods
Article summary.
Article focus
The focus of this work was to establish a pragmatic methodology to efficiently collect information from electronic medical records (EMRs) about patients who meet clinical research eligibility criteria.
Key messages
The use of EMR data is necessary to improve clinical research efficiency. However, it is not easy to identify patients who meet research eligibility criteria and collect necessary data from EMRs because the data collection process must integrate various techniques, including the development of a data warehouse and the translation of eligibility criteria into computable criteria. An efficient ERS and a standardised data processing model that integrates these techniques are essential to facilitate clinical research that utilises EMRs.
Strengths and limitations of this study
Our method uses a specialised data model for patient identification in clinical research and efficient data conversion that does not depend on the EMR database structure when converting narrative criteria to computable criteria.
We propose that computable criteria should not be a result of the automated conversion of narrative criteria but rather a result of research preparation involving medical concepts that are not expressed logically or explicitly in the narrative criteria. Therefore a large amount of the conversion of the eligibility criteria to computable criteria should be executed at the protocol development stage.
It is important to further discuss protocol standardisation, including eligibility criteria representation for computable use.
Enabling medical records retrieval system use in and across multiple institutions is an important future task.
Background
Medical information technology has recently advanced in many countries, and enormous amounts of clinical data are already stored as electronic medical records (EMRs). Utilising the data collected in EMRs is necessary to improve clinical research efficiency.1–3 An EMR is a large database of patient data and is used in observational research to investigate the relationships among diseases, treatments and outcomes,4–7 to conduct surveillance for rare drug reactions,4 8 and to recruit patients for clinical trials.9–13 However, it is not easy to identify patients who meet research eligibility criteria and collect necessary information from EMRs.2 3 Herein, we describe three major issues concerning EMR-based observational studies: EMR patient data retrieval function, eligibility criteria protocol representation and EMR data accuracy.
To identify patients who meet research eligibility criteria, it is necessary to obtain various types of information stored in EMRs by subject, for example, diagnosis and prescribed medications. However, the EMR database is designed to facilitate online transaction processing for rapid and detail-oriented clinical information searches on individual patients, and the current EMR system does not facilitate this retrieval function.2 3 14 Data warehouses are essential components of data-driven decision support. To allow for efficient research analyses, EMR data must first be warehoused to enable data analyses across patient populations.15–21 However, healthcare data modelling is difficult and time consuming because of the complexity of the medical knowledge involved. Thus, the most common approaches to clinical data warehouse modelling are variations on the entity-attribute-value (EAV) model,22–28 where data are stored in a single table with three columns: entity identification, attribute and attribute value. The EAV design has advantages, including flexibility and ease of storage; however, it requires transforming EAV data into another analytical format before analysis.25 28 Online analytical processing (OLAP) is most frequently used for searching data stored in the data warehouse.29–31 OLAP systems in relational databases are typically designed based on Kimball's star schema.32 However, the star schema was devised to facilitate online measurement analyses. In healthcare, this method can be used to dynamically gather online analyses of numeric data (eg, a specific dose of a drug for a specific disease) in clinical practice. Therefore, this method is not suitable for identifying patients who meet the complicated eligibility criteria for a given clinical research study. Data-modelling methods that facilitate the identification of patients and enable the collection of necessary information from EMRs remain to be established.28
Current eligibility criteria are written in a text format that cannot be computationally processed. Additionally, to be applied in actual EMR, eligible criteria need to be integrated with the data model of EMRs.33 Several investigations have sought to establish computable eligibility criteria.34–41 However, there is no consensus regarding a standard patient information model,33 and the eligibility criteria are not yet completely standardised. Using natural language processing (NLP) technologies to convert the text format of eligibility criteria to a computer or to extract patient identifications from EMRs is far from perfect without human intervention.3 42 43
Current EMRs have been used to support claims for medical service fees and the treatments administered to each patient; therefore, data gathered specifically for research purposes may be incomplete and unreliable.2 3 44
Although various investigations on each technique are executed individually, standardised methods must still be established that integrate these techniques, facilitate the identification of patients who are eligible for clinical research, and collect necessary information from EMRs.
Objective
We designed a pragmatic data processing model optimised for patient identification and for the collection of necessary information from EMRs for clinical research. These tools are implemented as an electronic medical records retrieval system (ERS).44
This research aimed to demonstrate an ERS and an example of a hospital-based cohort study that used the ERS to identify both patients and exposure. Another aim was to evaluate the feasibility and usefulness of the ERS, the method to convert text form eligible criteria to computable criteria, and a confirmation method to increase research data accuracy.
Materials and methods
Outline of our procedure for patient identification and data collection from the EMR
To identify patients who met the eligibility criteria for the clinical research in question, data were collected in the following ways:
The text form of the narrative criteria was converted into computable criteria.
A targeted patient list was created.
A flag was added for investigators to confirm the targeted patient list.
Reports were created for the investigators to confirm.
After confirmation by the investigator, the statistical analyses were executed.
EMR retrieval system
In our hospital, EMR use was introduced in 2005; approximately 800 000 cases of clinical information have already been stored. To comprehensively and efficiently collect information about patients participating in clinical research, we developed an ERS.44
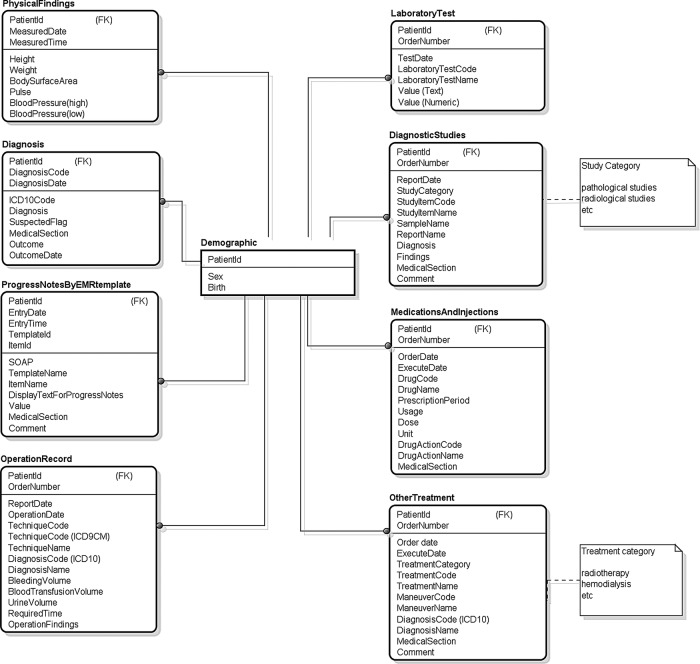
EMRs store various types of information, integrating billing, pharmacy, radiology, laboratory information and others.4 In creating the ERS database, we designed a new data model based on the star schema that was optimised for patient identification in clinical research. We identified nine data categories from EMRs that are useful for clinical research: demographic characteristics, physical findings, diagnostic studies, laboratory tests, diagnoses, progress reports on an EMR template,44 45 medications and injections, operation records and other treatments. We then designated these categories to ‘entities’. In our hospital, the diagnosis is managed by codes that were originally defined by our hospital and mapped with International Statistical Classification of Diseases (ICD) 10 codes46 for medical insurance purposes. Operations codes were also managed by codes that originally were defined by our hospital and mapped with ICD-9 Clinical Modification codes. We identified available columns (eg, ICD code, diagnosis date) from the EMR data model and designated these columns as ‘attributes’ of the entities.
Figure 1 presents our data model. In our model, all entities in a given schema are independent and complete; this allows for logical operations and for the creation of eligible patient lists for each respective parameter in a study. The target patient list is generated by combining these patient lists. The data model also supports the inference of medical concepts expressed in the eligibility criteria in reference to corresponding patient data accumulated in EMRs.33 34
Figure 1.
Data model for our electronic medical record retrieval system.
In our hospital, a replicate of the EMR database known as ‘Open DB’ was established for the secondary use of accumulated EMR data.7 A data mart for our ERS was created to ensure that the data retrieval process was practical and independent of the EMR system structure; the data mart was created on the relational database management system by extracting, transforming and loading (ETL) information from the Open DB.7 44 The ETL process is performed automatically once nightly except for the ‘Progress notes by EMR template’ entity, which is referred directly from the Open DB to ensure real-time visibility for the eClinical trial.44
An OLAP tool was installed to efficiently search through data from multiple patients.44 The OLAP tool runs in an Internet browser and can generate structured query language (SQL) based on predefined metadata (ie, a data model) by defining logical queries (ie, programmes) using a graphical user interface (GUI). Moreover, this tool allows reports on information retrieved from the browser to be transcribed using hypertext markup language (HTML). The reports are created in various formats, including portable document format (PDF), comma-separated values (CSV) and extensible markup language (XML).44
To protect personal information in medical records at our hospital, the EMR network is separated physically from other networks. Our data mart and OLAP servers are deployed in the same EMR network and managed using the same EMR security policies. Additionally, the use of our ERS is limited to clinical research approved by the ethics committee at our hospital, and only designated staff members at our centre are allowed to retrieve data. Our centre creates and manages ERS user identification separate from the EMRs. For the external output of CSV and other data, permission must be obtained from our department of medical informatics, and data extraction must be executed in the presence of supervisors who are responsible for protecting personal information at our hospital.
Application to clinical research
We applied the system to a hospital-based cohort study performed at our hospital titled ‘Risk of osteomyelitis of the jaw induced by oral bisphosphonates (BP) in patients taking medications for osteoporosis: a hospital-based cohort study in Japan’,47 in which we identified eligible patients, extracted research data and evaluated the feasibility of our system. The ethics committee at Kyoto University Hospital approved this research. A different paper details the purpose, methods, results and discussion of this research.47
This research aimed to estimate the risks for osteomyelitis of the jaw in osteoporosis patients at our hospital who had been exposed to oral BP compared with those who had not.48 49
The eligibility criteria were as follows:
Inclusion criteria:
Patients diagnosed with osteoporosis and treated with osteoporosis medications at Kyoto University Hospital between November 2000 and October 2010.
Patients aged 20 years or older.
Exclusion criteria:
Patients with a history of treatment with radiation therapy to the maxillofacial region.
Patients with primary or metastatic tumours in the maxillofacial region.
Patients treated with intravenous BP.
The data collected were diagnosis, date of diagnosis, sex, birthdate and the doses and dates when osteoporosis medications, steroids, anticancer drugs, diabetes drugs and HbA1c tests were administered.
Conversion of the text form of the narrative criteria to computable criteria
To identify eligible patients and collect the necessary data from the EMRs, narrative criteria and data must be converted to computable criteria. Such computable criteria include entities, attributes, logical operators (ie, ‘and’ and ‘or’), codes and parameters.33–37 The clinical research purpose and clinical practice demands made it necessary to perform this task.
We manually executed the conversion from text eligibility criteria to computable criteria. As an example of the conversion from narrative criteria to computable criteria, we present the following two-step conversion procedure:
Step 1: Convert the narrative criteria into entity-level criteria
Medical concepts expressed as narrative criteria are mapped onto entities in the data model and converted into entity-level criteria. This task is manually performed at the protocol development stage of the study by the investigators. For each entity, a criterion is created to extract patients who meet each condition. If exclusive conditions for the same entity must be defined, a different criterion is created. Additionally, the list of codes for drugs and diagnoses (ie, ICD-10) is created, and the period of treatments and others are defined by investigators. In this study, we mapped ‘osteoporotic patients’ onto two entities (ie, ‘diagnosis’ and ‘medications and injections’) and converted it to a combination of two criteria (ie, ‘diagnosis of osteoporosis’ and ‘osteoporosis drug administration’). In the test research, we defined the entity-level criteria according to the entered diagnosis and ordered treatments rather than the diagnostic criteria of the disease. This process reflects that the test research aimed to estimate some risks of osteomyelitis of the jaw with BP administration instead of diagnosing osteoporosis patients accurately. The recorded diagnosis in the EMR was typically designed to ensure payment for medical claims. We thus sought to reduce the number of false-positives by extracting patients with a given treatment type.
Step 2: Convert entity-level criteria into attribute-level criteria (ie, computable criteria)
The abovementioned corresponding codes, date and parameters are mapped onto attributes of the entity-level criteria, and these factors become computable criteria.
Creating a targeted patient list
A targeted patient list is created from the entire set of patients for whom EMRs have been obtained by defining logical queries (ie, programmes defined by the GUI) based on the computable criteria included in the ERS.
Logical queries are first defined in the ERS to identify patients who meet the conditions for each criterion. The ERS automatically generates the SQL necessary for data extraction according to the logical queries. Logical queries are then defined to include or exclude eligible patients who meet each criterion for the demographic entity. The targeted patient list is created by executing the logical query. Figure 2 presents an example of an SQL automatically generated by the ERS.
Figure 2.
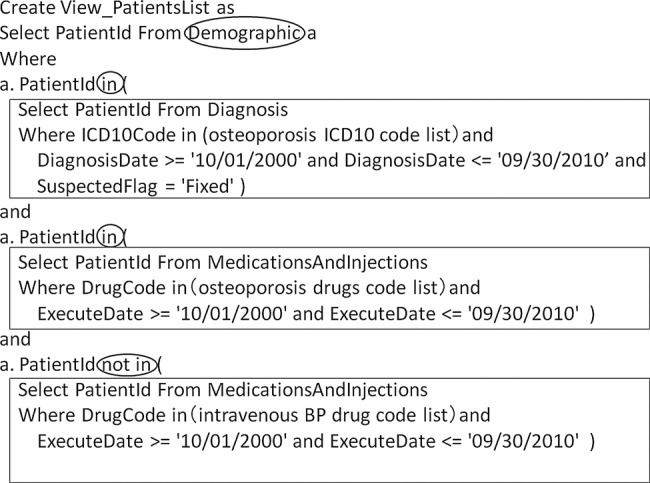
Example structured query language (SQL) to create the target patient list.
We thus designed our data model to enable the creation of a targeted patient list by defining the patients extracted from each criterion (ie, ‘in’ or ‘not in’) as conditions for the demographic entity that was the unique patient list for the entire hospital. If logical queries are defined using our method, even if the eligibility criteria are complicated, it is not necessary to dramatically change the SQL structure generated in the ERS.
Flagging entries for investigators to confirm
To improve research data accuracy, confirmation by the investigators is necessary. When confirmation is required, additional information is linked.
For the targeted patient list, logical queries are defined to flag certain items according to the investigators’ interest. Necessary logical queries are first defined for each criterion. Logical queries are then defined for addition to the patient list as ‘1’ if the data correspond or ‘0’ if they do not. Data sets created by these operations are joined by ‘union’ and pivoted on a cross-tabulation list using statistical analysis software. We show an example of an SQL generated by the ERS in figure 3.
Figure 3.
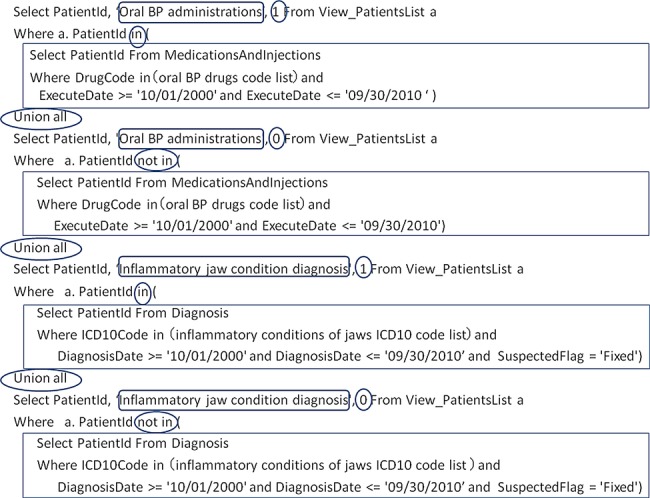
Example structured query language (SQL) to flag the target patient report for investigator confirmation.
Create reports for investigators to confirm
To help investigators confirm the targeted patient list, reports are created by linking the findings for diagnostic imaging, pathological diagnosis, operations and other findings. Investigators confirm these entries using the reports and EMR information, including progress notes and images. When the diagnosis history, medication, laboratory results, progress notes and other information are necessary, the same operation is executed for each instance. For example, the list of radiological findings involves ‘patient ID’, ‘study category’, ‘report name’, ‘diagnosis’, ‘findings’ and ‘comment’. The reports may improve the investigators’ confirmation efficiency because they prevent the need to refer to the medical records for each patient who needs confirmation.
Confirmation by the investigator and execution of the statistical analyses
The investigators confirm the accumulated data and execute the statistical analysis. In this test research, two oral and maxillofacial surgeons diagnosed cases by a chart review with an observation of imaging findings.47
Systemic evaluation
To evaluate our system, we collected information about the research period using the recall method. For the accuracy of the data collected by the ERS, we evaluated the results after they were confirmed by the investigator.
Results
Computable criteria, datasets and system evaluation
We present the computable criteria in table 1. To increase data accuracy, we collected all of the exclusion criteria for the investigators to confirm. As table 1 shows, we extracted information from EMRs. For investigator confirmation, we also reported all targeted patients using the following lists: osteoporosis drugs administered, oral BP administered, intravenous BP administered, diabetes drugs administered, anticancer drugs administered, steroid drugs administered, osteoporosis diagnoses, oral cancer diagnoses, patients diagnosed with inflammation of the jaw, patients diagnosed with other suspicious diseases, patients diagnosed with diabetes, HbA1c values, radiological findings, pathological findings and radioisotope findings. These data were extracted from the ERS for statistical analyses, presented in CSV format, and analysed using statistics software.
Table 1.
Computable criteria for our test research
| Criterion | Entity | Operator symbol | Attribute | Operator symbol | Parameter |
|---|---|---|---|---|---|
| Created a targeted patient list | |||||
| Inclusion criteria: osteoporosis diagnosis | Diagnosis | – | ICD10Code | In | (osteoporosis ICD10 code list) |
| and | DiagnosisDate | >= | ‘10/01/2000’ | ||
| and | DiagnosisDate | <= | ‘09/30/2010’ | ||
| and | SuspectedFlag | = | Fixed | ||
| Inclusion criteria: osteoporosis drug administrations | Medications and injections | – | DrugCode | in | (osteoporosis drugs code list) |
| and | ExecuteDate | >= | ‘10/01/2000’ | ||
| and | ExecuteDate | <= | ‘09/30/2010’ | ||
| Added a flag for investigators to confirm the targeted patient list | |||||
| Exclusion criteria: oral cancer diagnosis | Diagnosis | – | ICD10Code | in | (oral cancer ICD10 code list) |
| and | DiagnosisDate | >= | 10/01/2000’ | ||
| and | DiagnosisDate | <= | 09/30/2010’ | ||
| and | SuspectedFlag | = | Fixed | ||
| Exclusion criteria: intravenous BP administrations | Medications and injections | – | DrugCode | in | (intravenous BP drugs code list) |
| and | ExecuteDate | >= | ‘10/01/2000’ | ||
| and | ExecuteDate | <= | ‘09/30/2010’ | ||
| Oral BP administrations | Medications and injections | – | DrugCode | in | (oral BP drugs code list) |
| and | ExecuteDate | >= | ‘10/01/2000’ | ||
| and | ExecuteDate | <= | ‘09/30/2010’ | ||
| Inflammatory jaw condition diagnosis | Diagnosis | – | ICD10Code | in | (inflammatory conditions of jaws ICD10 code list) |
| and | DiagnosisDate | >= | ‘10/01/2000’ | ||
| and | DiagnosisDate | <= | ‘09/30/2010’ | ||
| and | SuspectedFlag | = | Fixed | ||
| Other suspicious disease diagnosis | Diagnosis | – | ICD10Code | in | (other suspicious disease ICD10 code list) |
| and | DiagnosisDate | >= | ‘10/01/2000’ | ||
| and | DiagnosisDate | <= | ‘09/30/2010’ | ||
| and | SuspectedFlag | = | Fixed | ||
| Diabetes diagnosis | Diagnosis | – | ICD10Code | in | (diabetes ICD10 code list) |
| and | DiagnosisDate | >= | ‘10/01/2000’ | ||
| and | DiagnosisDate | <= | ‘09/30/2010’ | ||
| and | SuspectedFlag | = | Fixed | ||
| Steroid drug administrations | Medications and injections | – | DrugCode | in | (steroid drugs code list) |
| and | ExecuteDate | >= | ‘10/01/2000’ | ||
| and | ExecuteDate | <= | ‘09/30/2010’ | ||
| Anticancer drug administrations | Medications and injections | – | DrugCode | in | (anticancer drugs code list) |
| and | ExecuteDate | >= | ‘10/01/2000’ | ||
| and | ExecuteDate | <= | ‘09/30/2010’ | ||
| Diabetes drug administrations | Medications and injections | – | DrugCode | in | (diabetes drugs code list) |
| And | ExecuteDate | >= | ‘10/01/2000’ | ||
| And | ExecuteDate | <= | ‘09/30/2010’ | ||
| HbA1c test execution | Laboratory test | – | Laboratory TestCode | in | (HbA1c test code) |
| and | TestDate | >= | ‘10/01/2000’ | ||
| and | TestDate | <= | ‘09/30/2010’ | ||
| Created reports for confirmation by the investigators | |||||
| Radiological finding reports | Diagnostic studies | – | ReportName | in | (report name list of oral region) |
| Pathological finding reports | Diagnostic studies | – | SampleName | contains | ‘bone’ |
| Or | SampleName | contains | ‘jaw’ | ||
| Radio isotope finding reports | Diagnostic studies | – | – | – | – |
BP, bisphosphonates; ID, identifications; ICD, International Classification of Diseases.
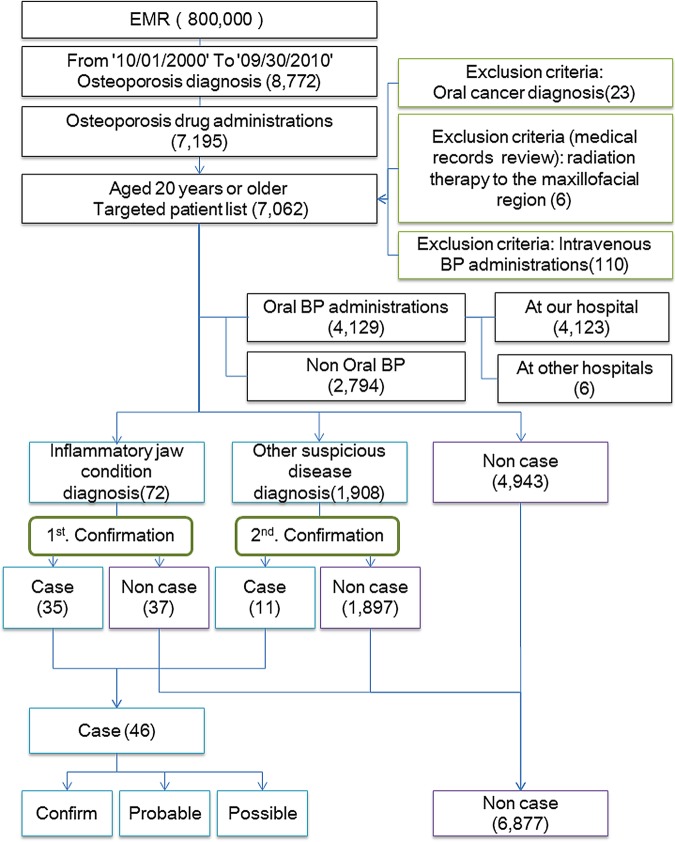
Among the approximately 800 000 cases at our hospital, 8772 were categorised using the terms ‘Inclusion criteria: Osteoporosis diagnosis’; among this group, 7195 were further categorised using ‘Inclusion criteria: Osteoporosis drug administration’. We then calculated the time that had elapsed since the osteoporosis diagnosis, determined that 7062 patients were aged 20 years or older, and created a targeted patient list. Among those on the targeted patient list, 23 patients were placed under the heading ‘Exclusion criteria: Oral cancer diagnosis’, 110 under ‘Exclusion criteria: Intravenous BP administration’, 4200 under ‘Oral BP administration’, 84 under ‘Inflammatory jaw condition diagnosis’, 2064 as ‘Other suspicious disease diagnosis’, 1700 as ‘Diabetes diagnosis’, 4551 as ‘Steroid drug administration’, 904 as ‘Anticancer drug administrations’, 1055 as ‘Diabetes drug administrations’ and 3641 as ‘HbA1c test execution’. Because of the end point considered, patients who were classified under ‘Inflammatory jaw condition diagnosis’ or ‘Other suspicious disease diagnosis’ were confirmed using predefined hierarchical diagnostic criteria by investigators who performed the statistical analyses and arranged the research results. We show the schema of data collection and confirmation as figure 4.47
Figure 4.
Schema of data collection and confirmation.
The accuracy of the data extracted by the ERS was then characterised. Reviewing the medical records revealed that 2817 patients were not labelled as ‘Oral BP administration’, including seven (one who received intravenous BP) treated at other hospitals. Six patients had been treated with radiation therapy to the oral and maxillofacial regions. Among the 72 patients classified under ‘Inflammatory jaw condition diagnosis’, 35 cases and 37 non-cases were identified.
The data extraction period lasted approximately 3 months. Ten meetings were held during the protocol development stage to create and validate the computable criteria and the list of codes for various drugs and diagnoses (ie, ICD-10). The time required for logical query definition when using the ERS was approximately 20 h. The investigator confirmations and statistical analyses took approximately 4 months.
Discussion
We identified eligible patients for this research and extracted the data necessary for confirmation by investigators and for statistical analyses.
We asked the chart reviewers to evaluate the system in a questionnaire about ‘the effect of computer programming support for data retrieval from the EMR’, ‘the result of the data retrieval’, ‘the positive and negative aspects of our ERS use’ and ‘the aspects of our method that should be improved’. The investigators evaluating the system mentioned that the following points: (1) the method enabled them to extract the necessary data for diagnosis and drug administration without exception; (2) by screening the entire patient population at the hospital using the ERS, they could identify not just eligible patients in the department of oral and maxillofacial surgery but all eligible patients, which reduced the study bias and (3) by creating reports for confirmation, it enabled investigators to devote their time to reading images, thus effectively reducing the time required for reviewing medical records. The aspects of our method that should be improved are the ‘lack of claim data’ and the ‘administrative complexity of EMR data use’. No negative aspects of our ERS use were noted.
The ERS allowed for the collection of information on patient eligibility by efficiently combining clinical information. Although we did not compare our method with other methods, our proposed method reduced the labour normally required from investigators and improved the reliability of test research results, which indicated that it was useful.
To design the ERS database, we designed a new data model optimised for patient identification. The main differences between our data model and the star schema were as follows: (1) demographic data, which were presented in list form in our EMR system, were presented as a fact-less fact table and (2) date, time, measurements and text information were presented in dimension tables.32 The most significant characteristic of our method for patient identification is the use of a specialised data model in clinical research and the ability to execute a large number of conversion tasks at the protocol development stage. Data can be converted efficiently in a way that does not depend on the EMR database structure when converting narrative criteria to computable criteria. In this research, we considered whether data were extracted directly from EMRs at the protocol development stage. However, EMR data were recorded in a sequential format for every medical practice, and the database structure was complicated. Comprehending the location and meaning of the necessary data thus required tremendous effort. It was difficult to make precise logical queries for patient identification. However, because our ERS data model was arranged by subjects (eg, tests, diagnosis), it was easy to interpret the available information. Due to the standardisation of computable criteria and SQL possible with the ERS, it was also possible to create computable criteria in little time. Additionally, verifying the patient identification accuracy was easy because it was possible to test each individual criterion.
The SQL generated by our ERS does not reduce the time required for data retrieval. Our ERS also cannot retrieve information that is not in the data model. Current EMRs do not store all necessary data for clinical research, including information related to pregnancy, performance status, cancer stage, availability of transportation to the hospital, specific tests that are not typically performed, drug regimen, outcomes (including death) and adverse events. Additionally, all tests are not administered to all patients, and necessary information may have been recorded in medical records at another hospital.44 To facilitate EMR use in clinical research, it is necessary to accumulate as much of this information as possible. In the hospital, much of this information does not integrate well with EMRs, including test reports stored only in the departmental system.50 However, it is important to utilise this information. Additionally, enabling ERS use in and across multiple institutions is also an important future task.
Currently, most clinical research studies that use data from EMRs are planned according to the concept that the primary use of EMRs is for clinical practice and a secondary use is for clinical research.44 Therefore, most investigators attempt to convert the text form eligibility criteria that already have been defined on a protocol to computable criteria at the data collecting stage.35 36 However, we propose that computable criteria should not be a result of the automated conversion of narrative criteria but rather a result of research preparation involving medical concepts that are not expressed logically or explicitly in the narrative criteria. Some medical concepts may be interpreted differently depending on the research and the investigator caring for the patients. Additionally, current eligibility criteria are vague or complex, and they do not consider the use of the actual EMR. To convert computable criteria appropriately, high-level medical decisions to answer the research question are required. Therefore, we thought that a large amount of the conversion of the eligibility criteria to computable criteria should be executed at the protocol development stage. In addition, the conversion process should be divided into entity-level conversions that require higher medical decisions and attribute-level conversions. To reduce the burden of conversion, it may be useful to apply NLP technology for the conversion from entity-level criteria to attribute-level criteria. Moreover, it is important to further discuss protocol standardisation, including eligibility criteria representation for computable use. For instance, the attribute-level criteria that describe the search conditions in detail may be useful in global studies to address diseases that vary according to the diagnostic criteria used in each country.
Concerning EMR data accuracy, the ICD10 code (osteomyelitis of the jaw) sensitivity was 48.6% (35/72). The investigators reported six simple diagnosis errors, seven oral BP administrations at other hospitals, and six patients who were treated with radiation therapy in the oral and maxillofacial region.47 For the accuracy of current EMRs, the investigators had to confirm the information. However, the EMRs provided rich confirmation data and were useful in improving research data accuracy. In this study, we checked the data from actual EMRs manually and identified patients precisely and extensively using coded information, narrative information, and images. However, only information from existing EMRs was available. Current EMRs have a high degree of flexibility in data entry and are not currently managed for research purposes, which decreases their reliability. It is necessary to improve data quality through quality control without placing too much of a burden on clinical practice. Alternatively, it may be possible to organise data sufficiently before research use.51–53 Standardising the terminology and exchange formats used in the healthcare setting has facilitated international discourse.46 54–58 It is necessary to further discuss not only clinical practice but also research purposes, particularly how to utilise various standards when using EMRs beyond the hospital setting.
Conclusion
We propose a pragmatic method for EMR-based observational studies. Our ERS is already used to support hospital-based cohort studies, clinical trial recruitment and the eClinical trial infrastructure44 at our centre. We believe an efficient ERS and standardised data processing model are essential to facilitate clinical research that utilises EMRs.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the staff of the department of medical informatics of Kyoto University Hospital for their generous support.
Footnotes
Contributors: KY designed the study, developed the ERS system, identified the computable eligibility criteria, wrote logical queries, collected data and wrote the manuscript. ES is grant holder who designed the study, developed the ERS system and wrote and edited the manuscript. TY designed and conducted the ‘Risk of osteomyelitis of the jaw induced by oral bisphosphonates in patients taking medications for osteoporosis: a hospital-based cohort study in Japan’ (BRONJ study) study and the current study, identified the computable eligibility criteria and wrote and edited the manuscript. KA and MY designed and conducted the BRONJ study. ST designed the study and provided comments and feedback. KB is the principal investigator of the BRONJ study. MY owns the ERS system and supervised the study. MF supervised the study and provided comments and feedback. All of the authors read and approved the final manuscript.
Funding: This work was supported by the Coordination, Support and Training Program for Translational Research of the Ministry of Education, Culture, Sports, Science and Technology of Japan and by Grants-in-Aid for Scientific Research of Japan (23790566).
Competing interests: None.
Ethics approval: This research was approved by the ethics committee of Kyoto University Hospital.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No other data are available.
References
- 1.Embi PJ, Payne PR. Clinical research informatics: challenges, opportunities and definition for an emerging domain. J Am Med Inform Assoc 2009;16:316–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prokosch HU, Ganslandt T. Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods Inf Med 2009;48:38–44 [PubMed] [Google Scholar]
- 3.Wasserman RC. Electronic medical records (EMRs), epidemiology, and epistemology: reflections on EMRs and future pediatric clinical research. Acad Pediatr 2011;11:280–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean BB, Lam J, Natoli JL, et al. Review: use of electronic medical records for health outcomes research: a literature review. Med Care Res Rev 2009;66:611–38 [DOI] [PubMed] [Google Scholar]
- 5.Tannen RL, Weiner MG, Marcus SM. Simulation of the Syst-Eur randomized control trial using a primary care electronic medical record was feasible. J Clin Epidemiol 2006;59:25464. [DOI] [PubMed] [Google Scholar]
- 6.Williams JG, Cheung WY, Cohen DR. Can randomised trials rely on existing electronic data? A feasibility study to explore the value of routine data in health technology assessment. Health Technol Assess 2003;7: iii, v–x, 1–117 [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto K, Matsumoto S, Tada H, et al. A data capture system for outcomes studies that integrates with electronic health records: development and potential uses. J Med Syst 2008;32:423–7 [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto K, Matsumoto S, Yanagihara K, et al. A data-capture system for post-marketing surveillance of drugs that integrates with hospital electronic health records. Open Access J Clin Trials 2011;3:21–6 [Google Scholar]
- 9.Embi PJ, Jain A, Clark J, et al. Effect of a clinical trial alert system on physician participation in trial recruitment. Arch Intern Med 2005;165:2272–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell MK, Snowdon C, Francis D, et al. Recruitment to randomised trials: strategies for trial enrollment and participation study. The STEPS study. Health Technol Assess 2007;11:iii, ix-105 [DOI] [PubMed] [Google Scholar]
- 11.Dugas M, Lange M, Müller-Tidow C, et al. Routine data from hospital information systems can support patient recruitment for clinical studies. Clin Trials 2010;7:183–9 [DOI] [PubMed] [Google Scholar]
- 12.Thadani SR, Weng C, Bigger JT, et al. Electronic screening improves efficiency in clinical trial recruitment. J Am Med Inform Assoc 2009;16:869–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torgerson JS, Arlinger K, Käppi M, et al. Principles for enhanced recruitment of subjects in a large clinical trial: the XENDOS study experience. Control Clin Trials 2001;22:515–25 [DOI] [PubMed] [Google Scholar]
- 14.Kristianson KJ, Ljunggren H, Gustafsson LL. Data extraction from a semi-structured electronic medical record system for outpatients: a model to facilitate the access and use of data for quality control and research. Health Inform J 2009;15:305–19 [DOI] [PubMed] [Google Scholar]
- 15.Shim JP. Past, present, and future of decision support technology. Decis Support Syst 2002;33:111–26 [Google Scholar]
- 16.Prat N. A UML-based data warehouse design method. Decis Support Syst 2006;42:1449–73 [Google Scholar]
- 17.Park YT. An empirical investigation of the effects of data warehousing on decision performance. Inform Manag 2006;43:51 [Google Scholar]
- 18.Schlaps D, Schmid T. Data warehousing in clinical research and development—from clinical data to knowledge portals. Pharmind 2004;66:637–46 [Google Scholar]
- 19.Grant A, Moshyk A, Diab H, et al. Integrating feedback from a clinical data warehouse into practice organisation. Int J Med Inform 2006;75:232–9 [DOI] [PubMed] [Google Scholar]
- 20.Junttila K, Meretoja R, Seppälä A, et al. Data warehouse approach to nursing management. J Nurs Manag 2007;15:155–61 [DOI] [PubMed] [Google Scholar]
- 21.Rubin DL, Desser TS. A data warehouse for integrating radiologic and pathologic data. J Am Coll Radiol 2008;5:210–17 [DOI] [PubMed] [Google Scholar]
- 22.Johnson SB. Generic data modeling for clinical repositories. J Am Med Inform Assoc 1996;3:328–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadkarni PM, Brandt C. Data extraction and ad hoc query of an entity—attribute—value database. J Am Med Inform Assoc 1998;5:511–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anhøj J. Generic design of web-based clinical databases. J Med Internet Res 2003;5:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen RS, Nadkarni P, Marenco L, et al. Exploring performance issues for a clinical database organized using an entity-attribute-value representation. J Am Med Inform Assoc 2000;7:475–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinu V, Nadkarni P. Guidelines for the effective use of entity-attribute-value modeling for biomedical databases. Int J Med Inform 2007;76:769–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corwin J, Silberschatz A, Miller PL, et al. Dynamic tables: an architecture for managing evolving, heterogeneous biomedical data in relational database management systems. J Am Med Inform Assoc 2007;14:86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wade TD, Hum RC, Murphy JR. A dimensional bus model for integrating clinical and research data. J Am Med Inform Assoc 2011;1:96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardillo J, Mazón JN. Model-driven development of OLAP metadata for relational data warehouses. Comput Stand Interfac 2012;34:189–202 [Google Scholar]
- 30.Hettler M. Data mining goes multidimensional. Healthc Inform 1997;14:43.–, 48, 51–6 [PubMed] [Google Scholar]
- 31.Gordon BD, Asplin BR. Using online analytical processing to manage emergency department operations. Acad Emerg Med 2004;11:1206–12 [DOI] [PubMed] [Google Scholar]
- 32.Kimball R, Reeves L, Ross M, et al. The data warehouse lifecycle toolkit. New York: John Wiley, 1998 [Google Scholar]
- 33.Wang D, Peleg M, Tu SW, et al. Representation primitives, process models and patient data in computer-interpretable clinical practice guidelines: a literature review of guideline representation models. Int J Med Inform 2002;68:18. [DOI] [PubMed] [Google Scholar]
- 34.Weng C, Tu SW, Sim I, et al. Formal representation of eligibility criteria: a literature review. J Biomed Inform 2010;43:451–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lonsdale DW, Tustison C, Parker CG, et al. Assessing clinical trial eligibility with logic expression queries. Data Knowl Eng 2008;66:3–17 [Google Scholar]
- 36.Tu SW, Peleg M, Carini S, et al. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform 2011;44:239–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sordo M, Boxwala AA, Ogunyemi O, et al. Description and status update on GELLO: a proposed standardized object-oriented expression language for clinical decision support. Stud Health Technol Inform 2004;107:164–8 [PubMed] [Google Scholar]
- 38.Séroussi B, Bouaud J. Using OncoDoc as a computer-based eligibility screening system to improve accrual onto breast cancer clinical trials. Artif Intell Med 2003;29:153–67 [DOI] [PubMed] [Google Scholar]
- 39.CDISC ASPIRE: Integration of clinical research and EHR: Eligibility coding standards http://crisummit2010.amia.org/files/symposium2008/S14_Niland.pdf (accessed 26 Oct 2012).
- 40.CDISC Study Design Model: SDM-XML Version 1.0 http://www.cdisc.org/stuff/contentmgr/files/0/8c85b168e80d6834ded59339b55fdbc7/misc/cdisc_sdm_xml_1.0.pdf (accessed 26 Oct 2012).
- 41.US National Cancer Institute (NCI) caMATCH. https://cabig.nci.nih.gov/community/tools/caMATCH (accessed 31 Mar 2012).
- 42.Jagannathan V, Mullett CJ, Arbogast JG, et al. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. Int J Med Inform 2009;78:284–91 [DOI] [PubMed] [Google Scholar]
- 43.Pakhomov S, Weston SA, Jacobsen SJ, et al. Electronic medical records for clinical research: application to the identification of heart failure. Am J Manag Care 2007;13:281–8 [PubMed] [Google Scholar]
- 44.Yamamoto K, Yamanaka K, Hatano E, et al. An eClinical trial system for cancer that integrates with clinical pathways and electronic medical records. Clin Trials 2012;9:408–17 [DOI] [PubMed] [Google Scholar]
- 45.Matsumura Y, Kuwata S, Yamamoto Y, et al. Template-based data entry for general description in medical records and data transfer to data warehouse for analysis. Stud Health Technol Inform 2007;129(PT 1):412–16 [PubMed] [Google Scholar]
- 46.World Health Organization (WHO) International Classification of Diseases (ICD). http://www.who.int/classifications/icd/en/ (accessed 26 Oct 2012). [Google Scholar]
- 47.Yamazaki T, Yamori M, Yamamoto K, et al. Risk of osteomyelitis of the jaw induced by oral bisphosphonates in patients taking medications for osteoporosis: a hospital-based cohort study in Japan. Bone 2012;51:882–7 [DOI] [PubMed] [Google Scholar]
- 48.Fellows JL, Rindal DB, Barasch A, et al. ONJ in two dental practice-based research network regions. J Dent Res 2011;90:433–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vestergaard P, Schwartz K, Rejnmark L, et al. Oral bisphosphonate use increases the risk for inflammatory jaw disease: a cohort study. J Oral Maxillofac Surg 2012;70:821–9 [DOI] [PubMed] [Google Scholar]
- 50.Shortliffe EH, Cimino JJ. Biomedical informatics: computer applications in health care and biomedicine. Health Informatics series. New York: Springer, 2006 [Google Scholar]
- 51.McFadden E. Management of data in clinical trials. 2nd edn, Hoboken, NJ: Wiley-Interscience, 2007 [Google Scholar]
- 52.Zhengwu L, Jing S. Clinical data management: current status, challenges, and future directions from industry perspectives. Open Access J Clin Trials 2010;2:93–105 [Google Scholar]
- 53.Data Management Association (DAMA) International Data management body of knowledge. http://www.dama.org/i4a/pages/index.cfm?pageid=3364 (accessed 26 Oct 2012).
- 54.MedDRA MSSO MedDRA MSSO. http://www.meddramsso.com/ (accessed 31 Mar 2012).
- 55.US National Library of Medicine SNOMED clinical terms, http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html (accessed 31 Mar 2012).
- 56.Clinical Data Interchange Standards Consortium (CDISC) Study data tabulation model (SDTM). http://www.cdisc.org/sdtm (accessed 26 Oct 2012).
- 57.Huff S. Development of the logical observation identifier names and codes (LOINC) vocabulary. J Am Med Inform Assoc 1998;5:276–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Health level seven (HL7) http://www.hl7.com/ (accessed 26 Oct 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.