This review surveys the extensive literature on muscle development and regeneration. It looks at the chromatin-level molecular basis for lineage determination and the choice between myogenic versus nonmyogenic fates.
Keywords: cardiomyocyte, satellite cell, Smarcd3, fibrosis, TGFβ
Abstract
Developmental biologists have defined many of the diffusible and transcription factors that control muscle differentiation, yet we still have only rudimentary knowledge of the mechanisms that dictate whether a myogenic progenitor cell forms muscle versus alternate lineages, including those that can be pathological in a state of disease or degeneration. Clues about the molecular basis for lineage determination in muscle progenitors are only now emerging from studies of chromatin modifications that avail myogenic genes for transcription, together with analysis of the composition and activities of the chromatin-modifying complexes themselves. Here we review recent progress on muscle determination and explore a unifying theme that environmental cues from the stem or progenitor niche control the selection of specific subunit variants of the switch/sucrose nonfermentable (SWI/SNF) chromatin-modifying complex, creating a combinatorial code that dictates whether cells adopt myogenic versus nonmyogenic cell fates. A key component of the code appears to be the mutually exclusive usage of the a, b, and c variants of the 60-kD structural subunit BAF60 (BRG1/BRM-associated factor 60), of which BAF60c is essential to activate both skeletal and cardiac muscle programs. Since chromatin remodeling governs myogenic fate, the combinatorial assembly of the SWI/SNF complex might be targeted to develop drugs aimed at the therapeutic reduction of compensatory fibrosis and fatty deposition in chronic muscular disorders.
TWO ROADS DIVERGED IN A WOOD, AND I—I TOOK THE ONE LESS TRAVELED BY, AND THAT HAS MADE ALL THE DIFFERENCE.
FROM “THE ROAD NOT TAKEN” BY ROBERT FROST
Fortunately for most people, building new muscle is taken for granted, yet replacing damaged or degenerating muscle is one of the most important challenges faced by regenerative medicine. Acquired or congenital disease distorts or undermines the impressive homeostatic mechanisms that have evolved to maintain muscle mass and strength in proportion to workload in healthy individuals, leading to a significant incidence of patient morbidity and mortality (for reviews on muscle development, degeneration, and regeneration, see Charge and Rudnicki 2004; Tedesco et al. 2010; Laflamme and Murry 2011; Mercola et al. 2011; Fan et al. 2012). The debilitating clinical presentations reflect the essential roles played by muscle—the heart as a mechanical pump, and skeletal muscle as the engine of movement and locomotion as well as a vast reservoir of protein and carbohydrate and as a generator of heat.
For more than a century, it has been known that adult human skeletal muscle is capable of regeneration (Brack and Rando 2012; Wang and Rudnicki 2012). Mononuclear cells located sublaminally within skeletal myofibers, first described by Mauro (1961) as satellite cells, are considered to be the principal contributor to muscle growth in the adult, although additional stem and progenitor cells outside the basal lamina with myogenic potential might also contribute to muscle regeneration (Peault et al. 2007). Adult cardiac muscle, once thought to be nonregenerative, is now recognized to be capable of limited self-renewal that has also been attributed to stem cells, although their nature and origin remain less well characterized than those in skeletal muscle (Laflamme and Murry 2011; Mercola et al. 2011). However, even the robust regenerative response of skeletal muscle is insufficient to sustain repair and regeneration over time, such as in chronic diseases and during aging.
Despite their different embryological origins, physiological functions, and anatomical structures, cardiac and skeletal muscles share certain mechanisms of cell fate specification that are informative to consider together in the context of stem cell renewal. Analogous mechanisms include externally triggered signaling cascades that control chromatin-modifying complexes, which make key myogenic loci accessible to the transcriptional machinery. In particular, the heterogeneous and dynamic composition of the switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex appears to play an important role in committing multipotent progenitors to a myogenic fate. Thus, a major focus of this review is on the alternate usage of specific SWI/SNF subunit variants by both cardiac and skeletal muscle progenitor cells in response to signals in the damaged and regenerative environment. We examined the idea that assembly of SWI/SNF with certain alternate variants—in particular, the BAF60 (BRG1/BRM-associated factor 60) variants a, b, and c—directs a progenitor cell on a road to muscle differentiation versus one to alternate lineages and that this mechanism might be responsible for maladaptive responses, such as fibro-adipogenic degeneration of cardiac and skeletal muscles. Furthermore, we argue that the epigenetics of muscle cell commitment might govern a tradeoff between regeneration and pathological remodeling of cardiac and skeletal muscle and that a deeper understanding of the underlying molecular mechanisms might illuminate points for therapeutic intervention.
The hard road to regenerate injured muscle
Skeletal muscle fibers are syncytial cells consisting of several hundred nuclei within a continuous cytoplasm. Myofibers arise during development by fusion of mononuclear myoblasts, and once mature, they are renewed during homeostasis by spontaneous accretion of new nuclei (physiological myonuclear turnover) or by episodes of postnatal growth and regeneration through the fusion of myoblasts derived from satellite cells. Within days after injury, activated myoblasts proliferate and rapidly generate new myofibers of small caliber that progressively mature into larger fibers, which are morphologically and physiologically identical to undamaged muscle. Homeostatic replacement in healthy adults is robust, with reported evidence that ∼1%–2% of myonuclei are replaced each week in undamaged rat soleus and extensor digitorum longus muscle (Schmalbruch and Lewis 2000). Such a myogenic reserve necessitates a large pool of stem and/or progenitor cells. Satellite cells are thought to comprise the major stem cell pool for regeneration, but their involvement in homeostatic replacement remains less certain in mammals (Lepper et al. 2009, 2011). Satellite cells are estimated to comprise between 1% and 11% of nuclei in a given muscle fiber (Charge and Rudnicki 2004). Satellite cells arise developmentally from somites (Montarras et al. 2005; Relaix et al. 2005) and reside beneath the basal lamina tightly apposed to the myofiber plasma membrane in a quiescent state awaiting activation in response to injury or homeostatic turnover. In adults, the satellite cell pool can be replenished through asymmetric division of replicating satellite cells, leading to differentiating and “stem” daughter cells (Shinin et al. 2006; Kuang et al. 2007; Rocheteau et al. 2012). Multiple reports suggest that other cell types in the adult might participate in homeostatic or regenerative muscle renewal, including muscle resident stem cells (MRSCs), endothelial progenitor cells (EPCs), and bone marrow-derived cells (BMCs); however, the actual contribution of these cell populations to regeneration and myonuclear turnover under physiological conditions is still unclear (for review, see Charge and Rudnicki 2004; Tedesco et al. 2010).
If skeletal muscle is abundantly endowed with stem or progenitor cells, why does regeneration not keep up with muscle loss during chronic disease? One roadblock to efficient regeneration might be that the continuous reduction in satellite cell number throughout a lifetime—or more rapidly in the face of chronic activation of muscle repair, such as in dystrophic muscles—exhausts the mitotic potential of the satellite cells and their progenitors. In support of this view, Duchenne's muscular dystrophy (DMD) patients exhibit a strikingly premature shortening of telomere lengths relative to healthy individuals (Decary et al. 2000). Consistent with this view, telomerase deficiency exacerbates the dystrophic phenotype of mdx mice (Sacco et al. 2010). The telomerase-null/mdx double-mutant mice showed signs of muscle aging that are reminiscent of clinical features of DMD patients and could be rescued by infusion of fresh satellite cells. Together, these findings suggest that telomere shortening depletes the pool of satellite cells. In addition, degradation of the cellular environment, or niche, might contribute to the decline in regenerative potential with age or chronic disease (Conboy and Rando 2005; Brack and Rando 2007; Gopinath and Rando 2008). Environmental changes that alter extrinsic cues controlling satellite cell function might reflect an adaptive mechanism that becomes maladaptive when chronically activated. For instance, transforming growth factor-β (TGFβ) family members control diverse processes in the myoblast and satellite cells ranging from proliferation and fusion to differentiation, and the patterns of expression change dynamically in regenerating muscles to control sequential stages of muscle repair (Kollias and McDermott 2008). Illustrating a switch from an adaptive to a maladaptive mechanism, functional levels of the TGFβ family member myostatin, a negative regulator of muscle mass, decline during active regeneration to permit satellite cell activation (Kirk et al. 2000; Kocamis et al. 2001). However, in chronic myopathic states, myostatin as well as TGFβ itself are elevated coincident with fibrosis, and recent interventions to antagonize signaling have been noted to restore muscle function in mouse models of muscular dystrophies (Minetti et al. 2006; Cohn et al. 2007). Collectively, these data support the idea that local environmental signaling can become maladaptive and undermine native regeneration. Moreover, certain of these maladaptive cues might actively hinder the replacement of new contractile tissue and favor the formation of fibrotic scar and fat deposition, as observed in late stages of many chronic muscular disorders.
Unlike skeletal muscle, the adult mammalian heart retains only a modest ability to regenerate, contrasting the efficient and scarless healing seen in lower vertebrates, such as newts, axolotls, and zebrafish (Oberpriller and Oberpriller 1974). Perhaps the most compelling evidence of myocardial regeneration in humans comes from 14C labeling that occurred during the era of aboveground atomic weapon testing (Bergmann et al. 2009). The production of 14C in the atmosphere followed by termination of aboveground testing in 1963 constitutes a global pulse-chase experiment from which the frequency of cardiomyocyte replacement was calculated to be 1% per year in young adults to 0.45% in the elderly. This estimate is 20-fold to 40-fold less than that derived from other criteria (e.g., Kajstura et al. 2010) but, over time, is numerically substantial, although the relevance to heart function is still controversial. Analogous 14C results have not yet been reported for human cardiac injury; however, a genetic pulse-chase study in mice revealed that up to 18% of cardiomyocytes bordering the myocardial infarction had regenerated 3 mo after injury, whereas homeostatic regeneration was below the threshold of detection, suggesting that injury boosts regenerative potential (Hsieh et al. 2007).
What is the source of new heart muscle cells? The murine pulse-chase study (Hsieh et al. 2007) incorporated a muscle-specific Cre to label pre-existing myocytes, revealing that new muscle cells arose de novo from formerly undifferentiated cells rather than by cell cycle re-entry of pre-existing cardiomyocytes. Although regeneration by replication of pre-existing cardiomyocytes occurs in neonatal mice, this capacity is greatly diminished by 1 wk after birth (Porrello et al. 2011). The postnatal decline reflects the maturational increase in multinucleated myocytes by nuclear division (endoreduplication) and the observation that mononuclear myocytes are more capable of undergoing cytokinesis in response to periostin and neuregulin (Kuhn et al. 2007; Bersell et al. 2009), perhaps indicating that replicative competence wanes with multinuclearity. Nonetheless, molecules like periostin, neuregulin, and fibroblast growth factor (FGF) with concomitant inhibition of p38 MAPK are capable of stimulating ∼0.2%–0.3% of cardiomyocytes to show evidence of cytokinesis (e.g., midbody localization of auroraB kinase) and preservation of myocardial structure and function after ischemic injury (Engel et al. 2006; Kuhn et al. 2007; Bersell et al. 2009). It will be important to determine how effectively reactivation of cell cycle as a regenerative strategy will translate to humans, since few mononuclear cardiomyocytes remain in the aged human heart, and infarction exacerbates this decline (Olivetti et al. 1995; Herget et al. 1997), suggesting a deleterious loss of cycling-competent cells when they are needed the most.
Partly as a consequence of their low abundance and uncertain molecular markers, it has been hard to generate a consensus on the nature and location of stem or progenitor cells in the adult heart. The problem is exacerbated by a low propensity to differentiate in vitro, historically poor retention, and the challenge of detecting the cells upon transplantation in vivo. Although key issues such as capacity for self-renewal and potential for forming other lineages in addition to cardiomyocytes (e.g., endothelial, smooth muscle cells, or fibroblast) remain a matter of some debate, multiple reports suggest that adult cardiac stem/progenitor cells express a key set of genes in common with embryonic cardiac progenitor cells, including GATA4, NKX2.5, TBX5, and MEF2c, suggesting a commonality of developmental and regenerative mechanisms. This panel has been consistently found in various cell populations regardless of the markers used for their enrichment, including CD117/KIT/c-kit, stem cell antigen-1 (Sca-1), an unknown human epitope that cross-reacts with anti-Sca1, expression of the transcription factor gene Isl-1, the side population (SP) dye efflux phenotype, epicardial-derived progenitor cells, or growth in tissue culture as “cardiospheres” analogous to the neurospheres produced by neural stem cells (for review, see Mercola et al. 2011; Sturzu and Wu 2011). By examining localized expression of developmental or stem cell markers (e.g., Nkx2.5 and c-kit), putative endogenous cardiac stem or progenitor cells have been reported to be induced by myocardial infarction in the subepicardial region of the ventricular wall and also between cardiomyocytes in the region bordering the infarction (Beltrami et al. 2003; Smart et al. 2011). Characterization with the available biomarkers remains imprecise, however, and important questions about the overlap between the various populations as well as their origin, heterogeneity, and developmental potential remain unresolved.
Like skeletal muscle, the damaged heart undergoes an initially adaptive healing process that can become debilitating under conditions of chronic heart disease following ischemic or other injury. In particular, the formation of a fibrotic scar and ventricular remodeling preserves the integrity of the ventricular wall after myocardial infarction yet can spread in the myocardium distal to the infarction, where it is a hallmark of hypertrophic cardiomyopathy and a likely contributor to arrhythmia and heart failure (Swynghedauw 1999). Whether specific signaling pathways that become up-regulated during ventricular remodeling or with cardiomyopathy also influence regeneration remains speculative; however, we can anticipate that some factors—TGFβ or Wnt, for instance—might adversely effect regeneration given that they act on cardiomyocyte development (see below). Thus, in both skeletal and cardiac muscle, it will be important to learn whether adaptive healing responses might adversely affect regenerative potential in chronic pathological states.
Strategies for therapeutic regeneration—lessons from development
Much can be learned about the basic concepts of commitment and renewal in the adult by analyzing muscle formation during early development. Skeletal and cardiac muscle, although derived from the mesoderm, arise from quite different sources and divergent mechanisms in the early vertebrate embryo. Most skeletal muscle forms from the somites, with craniofacial musculature arising from more anteriorly located mesoderm adjacent to the hindbrain. The diversity of skeletal muscle is controlled during development and postnatal remodeling by a set of basic helix–loop–helix (bHLH) myogenic regulatory factors (MRFs) together with pleiotropic transcription factors and epigenetic regulators (for review, see Tapscott 2005; Guasconi and Puri 2009; only briefly summarized here). A network of four MRFs governs commitment to the muscle lineage and terminal differentiation: myoblast determination protein (MyoD), myogenic factor 5 (Myf5), MRF4 (also known as Myf6), and myogenin (Puri and Sartorelli 2000). Although different muscle groups exhibit variations on the theme, Myf5 and MyoD generally specify proliferating myoblasts, whereas myogenin and MRF4 program these cells so that they exit cell cycle and adopt differentiated and specialized functions, such as contractile multinucleated myotubes. Satellite cells, set aside during development, express Paired box 7 (Pax7) (or Pax3 in a minority of muscle groups) but generally do not express detectable levels of MRFs until activation toward a myogenic lineage, when a majority of proliferating satellite cells induce MyoD and Myf5 (Zammit et al. 2004; Kuang et al. 2007). Asymmetric division of satellite cells is considered to give rise to both Myf5+ and Myf5− cells, potentially driven by Numb (which inhibits Notch signaling), Wnt7a, and low levels of MyoD, together balancing differentiation with maintenance of the satellite cell pool for future use (Conboy and Rando 2002; Shinin et al. 2006; Kuang et al. 2007; Le Grand et al. 2009; Rocheteau et al. 2012).
The MRF network during embryonic development of myoblasts is transcriptionally regulated by Pax3 and Pax7 as well as through the action of T-box and homeodomain transcription factors, including Sine oculis family members (six protein members), myogenic microRNAs (miRs), and other epigenetic modifiers (for review, see Braun and Gautel 2011). At a higher level, a tightly controlled, sequential activation of signaling pathways by secreted factors governs skeletal muscle differentiation during development and in the adult following episodes of exercise and injury. Key among these are members of the TGF-β, FGF, hepatocyte growth factor (HGF), insulin growth factor (IGF), Wnt bone morphogenetic protein (BMP) and Notch signaling, tumor necrosis factor-α (TNF-α), and other cytokine-activated pathways, which, in an orchestrated fashion during development or following injury, apportion renewal versus commitment to myogenic differentiation (for review, see Charge and Rudnicki 2004). This complex network of signals determines whether mesodermal-derived cells adopt the myogenic lineage or turn into nonmuscle fates, potentially contributing to fibrosis, heterotopic ossification, and adipogenesis.
Cardiac progenitors are not derived from the somatic mesoderm but instead arise from lateral mesoderm in response to a sequential action of diffusible proteins that constitute a stepwise inductive program. Multipotent progenitors in embryonic stem cells (ESCs) and embryos expressing Kdr and MesP1 become progressively committed with the expression of Nkx2.5, Isl-1, and Tbx5 (Yang et al. 2008; Blin et al. 2010). Unlike skeletal myogenesis, for which MyoD was first discovered based on its remarkable ability to convert fibroblasts into muscle (see above), cardiomyogenic commitment is not controlled by dominant bHLH factors. Instead, a complex transcriptional network consisting of GATA4, HAND2, MEF2C, MESP1, NKX2.5, and TBX5 directs cardiac muscle formation (Olson 2006). Despite this complex network, several recent reports show that a minimal set of transcription factors are capable of directing the formation of cardiomyogenic progenitors from dermal fibroblasts using MesP1 and Ets2 (Islas et al. 2012) and even immature cardiomyocytes from either embryonic mesoderm using Gata4, Tbx5, and BAF60c (Takeuchi and Bruneau 2009); fibroblasts with Gata4, Mef2c, and Tbx5 (Ieda et al. 2010; Inagawa et al. 2012; Qian et al. 2012); or the same with Hand2 (Song et al. 2012). The latter two instances reprogrammed cells directly in vivo following myocardial infarction and successfully added new cardiomyocytes to the myocardial wall, improved cardiac function, and reduced adverse remodeling, suggesting that this approach might be adapted therapeutically, although the efficiency of the approach has been questioned (Chen et al. 2012).
Exploiting the signal transduction, transcriptional, and epigenetic networks to boost regeneration is a logical concept, but promising drug targets have not yet emerged. One issue is selectivity: The signal transduction pathways that control stem or progenitor cell commitment and renewal largely overlap with other biological processes and/or have opposing functions at different stages of stem or progenitor cell renewal and differentiation. For instance, we can readily anticipate the complications involved with targeting signaling downstream from Wnt or BMP, which elicit both positive and negative effects depending on the stage of the stem or progenitor cell. The salient lesson from embryological development therefore might be that activating a minimal set of transcriptional and epigenetic machinery can direct cell type-specific differentiation and even reprogram a fibroblast into a cardiomyocyte, suggesting that therapeutic selectivity might be achieved by targeting specific nuclear events that set the epigenetic landscape to adopt a permissive state for a specific lineage commitment. In normal developmental or regenerative contexts, it appears that intranuclear signaling has the complexity and selectivity to broadcast external cues to the appropriate sites in chromatin to guide the transcriptional machinery toward selective activation of specific programs, leading to lineage determination and formation of terminally differentiated cells.
SWI/SNF and the regulation of cell commitment and differentiation
Ultimately, commitment of a stem cell to differentiate into a highly specialized cell type is dictated by the selective transcription of discrete constellations of genes, which together define a differentiation program and determine the specific lineage and phenotype adopted by multi- or pluripotent cells. Mammalian DNA is highly compacted into chromatin, which must be locally remodeled to permit transcription from specific loci (Cairns 2009). At its essence, cell type-specific transcription therefore is achieved through the selective remodeling of chromatin at discrete loci to allow the productive engagement of polymerase II (Pol II). Thus, the recent application of genome-wide techniques to human ESCs (hESCs) and adult stem cells have started to shed light on the epigenetic dynamics that regulate “localized” chromatin remodeling and accessibility to transcription factors in different cell types, including cardiomyocytes and skeletal muscle cells (Asp et al. 2011; Wamstad et al. 2012), and the relationship with the network of transcription factors (Paige et al. 2012) and external signals (Mullen et al. 2011).
Although chromatin remodeling and nucleosome disruption frequently result from the concerted and regulated activity of a multitude of enzymes and multiprotein complexes, the essential enzymatic activity is typically provided by the SWI/SNF chromatin remodeling complexes that use ATP hydrolysis to disrupt the interaction between DNA and histone octamers (de la Serna et al. 2006; Cairns 2009). Other nonredundant chromatin remodeling complexes endowed with ATPase activity include the ISWI, CHD, and INO80 families of remodelers (Hargreaves and Crabtree 2011). In addition to the ATPase subunit (either BRM1 or BRM), SWI/SNF complexes are composed of 10 other subunits assembled in different combinations (Clapier and Cairns 2009; Wu et al. 2009) that are involved in virtually all cell processes, including maintenance of pluripotency and lineage determination in hESCs (Lessard and Crabtree 2010; Singhal et al. 2010). Such versatile activity appears to be conferred on SWI/SNF by a combinatorial assembly of the 11 subunits, encoded by 20 genes, theoretically capable of yielding hundreds of complexes (Fig. 1; discussed in detail in Wu 2012). Overall, the composition of SWI/SNF complexes, which varies by cell type and differentiation stage, might alter the functional interactions with other chromatin modifiers and transcription factors, dictating the specificity for target genes. For instance, a switch from BAF53a to BAF53b is an essential determinant for multipotent neural stem and progenitor cells to become post-mitotic neurons (Lessard et al. 2007). Thus, the combinatorial association of SWI/SNF subunits appears to generate a functional diversity that might be critical for controlling exquisitely selective and cell type-specific developmental and regeneration decisions.
Figure 1.
Combinatorial logic of SWI/SNF assembly and lineage diversification. The chromatin remodeling catalytic core of SWI/SNF consists of one of the two ATPase subunits, BRG1 or BRM, with BAF47, BAF155, and BAF170. Emerging evidence suggests that selective activation of lineage-specific constellations of genes might be achieved through the combinatorial assembly of variants of the other BAF structural subunits, which are bridged to sites in chromatin by their interactions with specific transcription factors; for example, MyoD and GATA4 for skeletal and cardiac myogenesis, respectively. Hundreds of combinations are possible, yielding a potentially rich lineage determining “code” (discussed in detail in Wu 2012) that might drive the selection of myogenic versus nonmyogenic cell fates.
Recent evidence suggests that the functional diversity of SWI/SNF complexes confers the potential to respond to a variety of extracellular cues and reciprocally influence the ability of extrinsic signals to activate gene expression, suggesting a means to link environmental cues to the execution of cell-specific programs (Fig. 2). Ho et al. (2011) recently provided genome-wide evidence that interaction with BRG1 determines which loci in chromatin become accessible to STAT3 that enters the nucleus in response to LIF signaling. Interestingly, based on the observation that continued occupancy of STAT3 sites by esBAF (the SWI/SNF complex in ESCs) depends partially on STAT3, the investigators speculated that mutual reinforcement by the two factors for occupying critical sites in chromatin is essential to maintain pluripotency. Relevant to muscle determination, p38 directly phosphorylates BAF60c (Simone et al. 2004), leading to preferential incorporation of preassembled BAF60/MyoD into Brg1-based SWI/SNF and thereby enabling MyoD to activate muscle-specific genes during myoblast differentiation (Forcales et al. 2011). The conclusion from these and similar studies in other systems indicates that different combinations of nonenzymatic, structural subunits, referred to as BRG1/BRM-associated factors (BAFs), appear to generate a variety of subcomplexes possibly involved in dynamic and tissue-specific interactions with transcription factors and other chromatin-modifying enzymes. Although the precise mechanisms by which SWI/SNF complexes receive and interpret external signals is not well understood in any context, it is attractive to speculate that the heterogeneous assembly of cell fate-selective SWI/SNF complexes comprises a combinatorial “code” for converting extracellular inductive signals into discrete developmental or regenerative programs of gene expression.
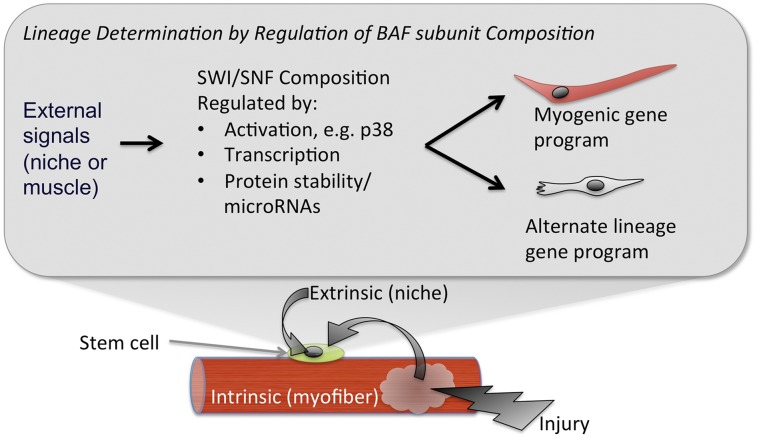
Figure 2.
A model for environmental control over stem cell fate. Regulation of SWI/SNF subunit composition might connect environmental cues to the differentiation response of a stem cell. Signals that increase the expression and usage of BAF subunits that favor activation of myogenic genes would be regenerative. In contrast, maladaptive signals in chronic myopathy could be hostile for regeneration by biasing the SWI/SNF combinatorial “code” against myogenesis and in favor of pathological fibrosis or adipocyte deposition.
Combinatorial SWI/SNF code for muscle determination
The assembly of the SWI/SNF subunits, which are encoded by many distinct genes, can potentially give rise to hundreds of SWI/SNF subcomplexes with different functions in distinct cell types (Wu et al. 2009; Wu 2012). An initial level of complexity is provided by the mutually exclusive presence of one of the two ATPase subunits: either BRG1 or BRM (Hargreaves and Crabtree 2011). Moreover, the finding that addition of the SWI/SNF components BAF47, BAF155, and BAF170 to BRG1 or BRM was sufficient to generate chromatin remodeling activity to a level comparable with that of the whole complex suggested that these subunits compose the “core” enzymatic machinery to remodel chromatin (Phelan et al. 1999). This evidence also raised the question of the other subunit's functions. One answer is that at least certain BAF components direct the complex to particular genetic loci through interactions with cell type-specific transcriptional activators (see Fig. 1), histones, or other chromatin-modifying complexes.
The functional relevance of SWI/SNF heterogeneity for cell fate selection was suggested by genetic deletion of individual subunit variants that, despite seemingly redundant structure and activity, resulted in lineage-specific effects. Repeated in multiple contexts and cell types, these findings suggest that variants of the individual subunits play discrete roles. For instance, knocking out either of the two highly homologous ATPases BRG1 or BRM differentially impairs the enzymatic activity of the SWI/SNF complex across lineages. Whereas BRM-null mice are viable and do not show any apparent developmental defects (Reyes et al. 1998), the BRG1-null phenotype reveals a requirement during the earliest stages of embryo formation (Bultman et al. 2000). While the specific roles of BRG1 and BRM specialization remain unknown in most cell types, conditional deletion of BRG1 early in the myocardial lineage revealed that it is uniquely important for maintaining the expression of fetal myosin heavy chain (MHC) isoforms and morphology in embryonic cardiomyocytes, since BRG1-deficient cardiomyocytes undergo premature differentiation, resulting in trabeculation defects (Hang et al. 2010; Takeuchi et al. 2011). Conversely, BRG1 is down-regulated coincident with the β-MHC (fetal isoform)-to-α-MHC (adult isoform) transition and is re-expressed after pathological stimuli, leading to reactivation of fetal genes and hypertrophy (Hang et al. 2010). Interestingly, BRG1 shows dosage-sensitive interdependence with cardiac transcription factors Nkx2.5 and Tbx5, indicating that haploinsufficiency of cardiac transcription factors in congenital heart disease (CHD) might arise from an imbalance between transcription factors and the SWI/SNF complexes (Takeuchi et al. 2011).
Different combinations of the structural subunits assembled into the SWI/SNF complex have also been shown to elicit distinct myogenic cell fate effects. RNAi of BAF60c in mice leads to severe cardiac defects, including single ventricle, shortened outflow tract, and hypotrabeculation (Lickert et al. 2004), and deletion of BAF60c in zebrafish affects cardiac progenitor formation and migration to the developing heart tube (Lou et al. 2011). Similarly, BAF250a deletion suggested involvement in the formation of cardiac progenitor cells (Lei et al. 2012). Deletion of BAF180 causes hypoplastic ventricle development reminiscent of defects in retinoic acid signaling, consistent with its role in the transcriptional response to ligands and nuclear receptors (Wang et al. 2004).
Less is known about how subunit composition programs skeletal myogenesis. Early work indicated an essential role for both BRG1 and BRM in MyoD-activated transcription of muscle genes during fibroblast conversion into muscle cells (de la Serna et al. 2001). Subsequently, however, BRG1 became appreciated as the SWI/SNF ATPase that is recruited to regulatory elements of myogenic genes in skeletal myoblasts (Simone et al. 2004; Ohkawa et al. 2007) necessary for the activation of muscle coding and noncoding (myomiRs) transcripts (Mallappa et al. 2010; Forcales et al. 2011). In contrast, the relative contribution of BRM to the activation and maintenance of muscle gene expression has not been systematically explored in skeletal myoblasts.
The view that BAF variant usage controls muscle differentiation mirrors the conclusions drawn from phenotypes observed in other systems following experimental ablation of individual variants of SWI/SNF structural subunits (for review, see Wu et al. 2009; Wu 2012). Together, the biochemical and genetic knockout results support the paradigm that the particular BAF variant composition of the SWI/SNF complex configures it to regulate lineage-specific gene expression (Fig. 1).
BAF60 variants and the molecular regulation of mesodermal lineage commitment
The three BAF60 variants comprise perhaps the best example of how variant exchange can confer cell specificity on the SWI/SNF complex. During evolution, vertebrates acquired three variants of the BAF60 subunits—BAF60a (SMARCD1), BAF60b (SMARCD2), and BAF60c (SMARCD3)—that are encoded by separate genes and show specific functions. Among them, BAF60c is preferentially expressed in developing heart and somites during embryogenesis (Lickert et al. 2004). Of note, BAF60c was one of the first SWI/SNF subunits described to directly interact with transcriptional activators (Debril et al. 2004). Genetic and functional studies have established the essential role of BAF60c in cardiac and skeletal myogenesis. Bruneau and colleagues (Lickert et al. 2004) made the seminal discovery that RNAi-mediated BAF60c silencing in the embryos leads to defects in heart morphogenesis and results in abnormal cardiac and skeletal muscle differentiation. The cardiogenic role of BAF60c depends on an interaction with cardiac transcription factor GATA4 and a supporting role for Tbx5 (Lickert et al. 2004). Together, these factors promote differentiation of early mouse mesoderm into heart (Takeuchi and Bruneau 2009), suggesting the ability to direct uncommitted mesoderm to the cardiac fate.
Further work from Forcales et al. (2011) demonstrated an equivalent activity of BAF60c during skeletal muscle differentiation and elucidated the molecular mechanism by which BAF60c promotes tissue-specific activation of gene expression. Association of BAF60c with MyoD occurs in undifferentiated myoblasts prior to the activation of muscle gene expression and leads to the formation of the “pioneer” complex, which is devoid of the core SWI/SNF components Brg1 and BAF47, on the chromatin of MyoD target genes (Forcales et al. 2011). In addition, chromatin immunoprecipitation (ChIP)-on-chip analysis demonstrated that BAF60c knockdown impairs MyoD binding to target genes, indicating that it is required to provide MyoD access to target sites in repressive chromatin of undifferentiated myoblasts. MyoD typically binds the regulatory elements of target genes containing the CANNTG sequence, termed an E-box (for review, see Puri and Sartorelli 2000), and genome-wide studies using ChIP coupled with high-throughput sequencing (ChIP-seq) have revealed a pervasive binding of MyoD across the genome in myoblasts prior to differentiation (Cao et al. 2010). These and previous studies (Blais et al. 2005; Cao et al. 2006) also indicated that the promoter context is important for activation by MyoD during the myoblast-to-myotube transition. In particular, surrounding recognition sites in compacted chromatin are needed to bind “pioneer” factors such as PBX1, which facilitate access of tissue-specific transcription factors (e.g., MyoD) to target genes in compacted chromatin (Berkes et al. 2004). In undifferentiated myoblasts, MyoD binding to chromatin correlates with local histone hyperacetylation (Cao et al. 2010), possibly presetting muscle loci for nucleosome remodeling. Since BAF60c is detected together with MyoD but without the catalytic SWI/SNF subunits, a subsequent step appears necessary to assemble SWI/SNF and activate chromatin, possibly in response to extracellular differentiation stimuli. This model is in accordance with a general idea that transcription factors cooperate to predetermine chromatin accessibility (Biddie et al. 2011; John et al. 2011) and that BAF60c or other SWI/SNF subunits may “prime” adjacent nucleosomes for disruption upon the engagement of the SWI/SNF chromatin remodeling complex. Indeed, upon exposure to differentiation cues, such as the promyogenic signals that activate p38 (Forcales and Puri 2005), the BAF60c–MyoD complex is incorporated into a Brg1-based SWI/SNF complex that is competent to remodel the chromatin and activate transcription from MyoD target loci (Forcales et al. 2011). Phosphorylation of Thr 229 in human BAF60c2 (which corresponds to the sole BAF60c isoform in mice) by differentiation-activated p38α kinase is the signal that enables the recruitment of the Brg1-based SWI/SNF complex to sequences previously bound by the BAF60c–MyoD complex (Simone et al. 2004; Forcales et al. 2011). This two-step model of SWI/SNF recruitment to target genes via their prior interactions with tissue-specific transcription factors can explain why BAF60c was detected on the chromatin of skeletal muscle (with MyoD) and cardiac genes (with GATA4 and Tbx5) in the absence of essential SWI/SNF components, including Brg1 and Brm (Takeuchi and Bruneau 2009; Forcales et al. 2011). When translated to cardiac myogenesis, the signal-dependent interactions between the BAF60c and the Brg1-based SWI/SNF complex could be mediated by phosphorylation events, analogous to the p38-mediated phosphorylation of BAF60c in skeletal muscle. Whether these regulatory signals rely on p38 or other kinases is currently unknown. The external signals that control p38 kinase α and β activation in muscle progenitor cells include inflammatory cytokines released within the regenerative environment, such as TNFα and high-mobility group box 1 (HMGB1), and cell-to-cell interactions (Guasconi and Puri 2009; Krauss 2010). However, other upstream regulators of signaling that program SWI/SNF selectivity are currently unknown; thus, a major task ahead is to link external cues to the control of chromatin remodeling. Potential pathways are suggested by functional interactions of BAF60c with Notch (Takeuchi et al. 2007), Wnt/β-catenin (Klaus et al. 2012) pathways in cardiac cells, and Nodal/TGFβ signaling in ESC-derived cardiac progenitors (Cai et al. 2012; M Mercola, unpubl.), all of which might be consistent with the presence of multiple phosphorylation sites in BAF60c detected by mass spectrometry studies (PL Puri, unpubl.).
Future prospects
Overall, the finding that ectopic expression of BAF60c enables tissue-specific transcriptional activators to drive cell fate selection during development—or even convert one cell type into another—supports the concept that altering the composition of SWI/SNF components might be sufficient to direct remodeling of discrete loci and promote cell type-specific gene expression. Furthermore, the emerging biochemical data suggest that the combinatorial composition of mutually exclusive BAF variants in each SWI/SNF complex dictates the affinity for transcription factors that recognize lineage-specific constellations of genes. Thus, during cardiac and skeletal myogenesis, environmental signals stimulate the expression of BAF60c and the formation of BAF60c-based SWI/SNF complexes in progenitor cells, thereby activating skeletal and cardiac muscle-specific gene activation. The net effect is to broadcast extrinsic cues to the genome in order to instruct mesoderm-derived progenies to adopt the cardiac and skeletal muscle lineages. A corollary of this thesis is that elevated expression of BAF60a (and, to a lesser extent, BAF60b) biases the SWI/SNF complex to activate alternative cellular programs, such as fibroblasts. Extended to other structural subunits, this concept emphasizes the importance of combinatorial assembly for the dynamic formation of cell-specific SWI/SNF complexes that underlie normal development and, potentially, both adaptive and maladaptive responses to pathological cues.
At this point, the notion of subunit heterogeneity as a code for lineage specification seems well established, even though we lack a complete description of the rules that associate a particular subunit composition with a cognate lineage selection. A major gap in our understanding is the signaling that controls subunit selection. It is critical to address this issue, since doing so will address how extrinsic cues, such as from the stem cell niche, drive myogenesis forward. For skeletal myogenesis, our fragmentary understanding positions p38α as regulating BAF60c, as discussed above. A largely unexplored layer of regulation to fine-tune BAF subunit composition appears to be imposed by miRs. It is known, for instance, that replacement of BAF53a and BAF45a in neural stem cell progenitors with BAF53b and BAF45b generates the SWI/SNF identity of post-mitotic neurons and that the BAF53 subunit switch is achieved by the selective expression of specific miRs, miR-9 and miR-124, which down-regulate BAF53a in favor of BAF53b (Yoo et al. 2011). Thus, a miR-mediated exchange of SWI/SNF components controls the optimal composition of SWI/SNF complexes to respond to environmental changes that accompany neurogenesis. Although not yet demonstrated, we can anticipate that an analogous miR-based mechanism governs myogenic BAF variant expression during development and postnatal life, perhaps acting as a fulcrum that tips the balance toward myogenic versus alternative fates such as fibroblasts and adipocytes. Although miR-based therapeutics such as anti-miRs or miR mimics have not yet been approved for degenerative muscle diseases, targets exist (e.g., Condorelli et al. 2010; Patrick et al. 2010), and >20 candidate siRNA/shRNA and miR therapeutics have been evaluated in clinical trials (Burnett and Rossi 2012). Moreover, the first anti-miR candidate therapeutic—miravirsen, a chemically modified oligonucleotide designed to inhibit miR-122—has advanced to a phase II trial (Santaris Pharma, NCT01200420) to treat chronic hepatitis C virus infection.
From a translational perspective, the regulation of SWI/SNF subunit composition is likely to have important implications for understanding aspects of muscle pathology. Given that the formation of a SWI/SNF complex containing the p38α substrate BAF60c is instrumental to perpetuating muscle gene expression in myogenic progenitors, it is tempting to speculate that inclusion of BAF60a or BAF60b might render the complex refractory to promyogenic cues (such as p38 signaling) and even make progenitors responsive to external signals directing alternative cell fates. In skeletal muscle, the divergent cell fibroblast and adipocyte lineages share a common multipotent progenitor with skeletal myoblasts (Asakura et al. 2001; Wada et al. 2002). Of note, the recent identification of muscle-derived interstitial cells that can adopt mesodermal-derived lineages and contribute, either directly or indirectly, to muscle regeneration (Mitchell et al. 2010; Uezumi et al. 2010) indicates that multiple cell types regulate muscle homeostasis and regeneration depending on their ability to adopt specific lineages. For instance, fibro-adipogenic progenitors (FAPs) have been implicated in the formation of fibrotic scar and fatty deposition in dystrophic muscles (Uezumi et al. 2011). Thus, the control of linage determination of adult mesodermal derivatives, such as FAPs, by the alternative usage of SWI/SNF subunit variants might ultimately influence whether a damaged muscle fiber is repaired or undergoes fibro-adipogenic degeneration. Despite the paucity of examples of environmental signals that control alternative BAF60 variant usage, we can anticipate that extracellular signals that control developmental and pathological processes may direct BAF variant selection. For instance, it will be interesting to identify the upstream signaling network that controls selection of BAF60c for cardiac and skeletal myogenesis versus BAF60a (SMARCD1), which has been implicated in transcriptional control of lipid metabolism (Li et al. 2008).
Whether endogenous cardiac stem or progenitor cells give rise to both myogenic and scar tissue during adverse remodeling is less clear, but even if the progenitors differ, the principle that extrinsic cues might favor one lineage over the other is a potential substrate for pathogenesis. In both tissues, the reciprocal interplay between tissue-specific transcriptional activators and dynamic SWI/SNF composition might balance regeneration of contractile tissue versus formation of fibrotic scar and even fat deposition. Thus, pathological signals in chronic myopathies, by affecting BAF subunit composition, might dictate that the “road not taken” during fetal myogenesis might be selected and “make all the difference” as maladaptive remodeling. Therefore, understanding the combinatorial code for muscle regeneration and, in particular, how it is controlled by extrinsic cues might point to specific targets for interventions in regenerative medicine. Potential applications include redirecting cardiac fibroblasts to regenerate ischemic myocardium and promoting the sustained regeneration of skeletal muscles by adult stem cells in the treatment of neuromuscular dystrophies.
Acknowledgments
P.L.P. is an Associate Professor in the Sanford Children's Health Research Center at the Sanford-Burnham Medical Research Institute (SBMRI) and Senior Telethon Scientist of the Dulbecco Telethon Institute (DTI) and acknowledges support from the NIH (R01AR056712, R01AR052779, and P30AR061303), the Muscular Dystrophy Association (MDA), and the European Community's Seventh Framework Programme in the project FP7-Health-2009 ENDOSTEM 241440. M.M. is a Professor in the Sanford Children's Health Research Center at the SBMRI and a Professor of Bioengineering at the University of California at San Diego and acknowledges support from the NIH (R01HL083463, R01HL113601, and P30AR061303), the Mathers Charitable Foundation, and the Sanford Children's Health Center.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.207415.112.
References
- Asakura A, Komaki M, Rudnicki M 2001. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation 68: 245–253 [DOI] [PubMed] [Google Scholar]
- Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, Bowman C, Kluger Y, Dynlacht BD 2011. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci 108: E149–E158 doi: 10.1073/pnas.1102223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, et al. 2003. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114: 763–776 [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. 2009. Evidence for cardiomyocyte renewal in humans. Science 324: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14: 465–477 [DOI] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kuhn B 2009. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138: 257–270 [DOI] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. 2011. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell 43: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD 2005. An initial blueprint for myogenic differentiation. Genes Dev 19: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin G, Nury D, Stefanovic S, Neri T, Guillevic O, Brinon B, Bellamy V, Rucker-Martin C, Barbry P, Bel A, et al. 2010. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest 120: 1125–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Rando TA 2007. Intrinsic changes and extrinsic influences of myogenic stem cell function during aging. Stem Cell Rev 3: 226–237 [DOI] [PubMed] [Google Scholar]
- Brack AS, Rando TA 2012. Tissue-specific stem cells: Lessons from the skeletal muscle satellite cell. Cell Stem Cell 10: 504–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Gautel M 2011. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol 12: 349–361 [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 6: 1287–1295 [DOI] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ 2012. RNA-based therapeutics: Current progress and future prospects. Chem Biol 19: 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Guzzo RM, Wei K, Willems E, Davidovics H, Mercola M 2012. A nodal to TGFβ cascade exerts biphasic control over cardiopoiesis. Circ Res 111: 876–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ 2006. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J 25: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev Cell 18: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charge SB, Rudnicki MA 2004. Cellular and molecular regulation of muscle regeneration. Physiol Rev 84: 209–238 [DOI] [PubMed] [Google Scholar]
- Chen JX, Krane M, Deutsch MA, Wang L, Rav-Acha M, Gregoire S, Engels MC, Rajarajan K, Karra R, Abel ED, et al. 2012. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ Res 111: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Cohn RD, van Erp C, Habashi JP, Soleimani AA, Klein EC, Lisi MT, Gamradt M, ap Rhys CM, Holm TM, Loeys BL, et al. 2007. Angiotensin II type 1 receptor blockade attenuates TGF-β-induced failure of muscle regeneration in multiple myopathic states. Nat Med 13: 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Rando TA 2002. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell 3: 397–409 [DOI] [PubMed] [Google Scholar]
- Conboy IM, Rando TA 2005. Aging, stem cells and tissue regeneration: Lessons from muscle. Cell Cycle 4: 407–410 [DOI] [PubMed] [Google Scholar]
- Condorelli G, Latronico MV, Dorn GW II 2010. MicroRNAs in heart disease: Putative novel therapeutic targets? Eur Heart J 31: 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J 2004. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem 279: 16677–16686 [DOI] [PubMed] [Google Scholar]
- Decary S, Hamida CB, Mouly V, Barbet JP, Hentati F, Butler-Browne GS 2000. Shorter telomeres in dystrophic muscle consistent with extensive regeneration in young children. Neuromuscul Disord 10: 113–120 [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet 27: 187–190 [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN 2006. Chromatin remodelling in mammalian differentiation: Lessons from ATP-dependent remodellers. Nat Rev Genet 7: 461–473 [DOI] [PubMed] [Google Scholar]
- Engel FB, Hsieh PC, Lee RT, Keating MT 2006. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci 103: 15546–15551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CM, Li L, Rozo ME, Lepper C 2012. Making skeletal muscle from progenitor and stem cells: Development versus regeneration. Wiley Interdiscip Rev Dev Biol 1: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcales SV, Puri PL 2005. Signaling to the chromatin during skeletal myogenesis: Novel targets for pharmacological modulation of gene expression. Semin Cell Dev Biol 16: 596–611 [DOI] [PubMed] [Google Scholar]
- Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, Coutinho P, Saccone V, Consalvi S, Williams R, et al. 2011. Signal-dependent incorporation of MyoD–BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J 31: 301–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath SD, Rando TA 2008. Stem cell review series: Aging of the skeletal muscle stem cell niche. Aging Cell 7: 590–598 [DOI] [PubMed] [Google Scholar]
- Guasconi V, Puri PL 2009. Chromatin: The interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol 19: 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP 2010. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466: 62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR 2011. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herget GW, Neuburger M, Plagwitz R, Adler CP 1997. DNA content, ploidy level and number of nuclei in the human heart after myocardial infarction. Cardiovasc Res 36: 45–51 [DOI] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR 2011. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol 13: 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh PC, Segers VF, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT 2007. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 13: 970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagawa K, Miyamoto K, Yamakawa H, Muraoka N, Sadahiro T, Umei T, Wada R, Katsumata Y, Kaneda R, Nakade K, et al. 2012. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of gata4, mef2c, and tbx5. Circ Res 111: 1147–1156 [DOI] [PubMed] [Google Scholar]
- Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, et al. 2012. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci 109: 13016–13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogorek B, Ferreira-Martins J, Goichberg P, Rondon-Clavo C, Sanada F, et al. 2010. Cardiomyogenesis in the adult human heart. Circ Res 107: 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kirk S, Oldham J, Kambadur R, Sharma M, Dobbie P, Bass J 2000. Myostatin regulation during skeletal muscle regeneration. J Cell Physiol 184: 356–363 [DOI] [PubMed] [Google Scholar]
- Klaus A, Muller M, Schulz H, Saga Y, Martin JF, Birchmeier W 2012. Wnt/β-catenin and Bmp signals control distinct sets of transcription factors in cardiac progenitor cells. Proc Natl Acad Sci 109: 10921–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamis H, McFarland DC, Killefer J 2001. Temporal expression of growth factor genes during myogenesis of satellite cells derived from the biceps femoris and pectoralis major muscles of the chicken. J Cell Physiol 186: 146–152 [DOI] [PubMed] [Google Scholar]
- Kollias HD, McDermott JC 2008. Transforming growth factor-β and myostatin signaling in skeletal muscle. J Appl Physiol 104: 579–587 [DOI] [PubMed] [Google Scholar]
- Krauss RS 2010. Regulation of promyogenic signal transduction by cell–cell contact and adhesion. Exp Cell Res 316: 3042–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA 2007. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, Keating MT 2007. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med 13: 962–969 [DOI] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE 2011. Heart regeneration. Nature 473: 326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand F, Jones AE, Seale V, Scime A, Rudnicki MA 2009. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell 4: 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei I, Gao X, Sham MH, Wang Z 2012. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J Biol Chem 287: 24255–24262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan CM 2009. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, Fan CM 2011. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138: 3639–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard JA, Crabtree GR 2010. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol 26: 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR 2007. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD 2008. Genome-wide coactivation analysis of PGC-1α identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab 8: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG 2004. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432: 107–112 [DOI] [PubMed] [Google Scholar]
- Lou X, Deshwar AR, Crump JG, Scott IC 2011. Smarcd3b and Gata5 promote a cardiac progenitor fate in the zebrafish embryo. Development 138: 3113–3123 [DOI] [PubMed] [Google Scholar]
- Mallappa C, Nasipak BT, Etheridge L, Androphy EJ, Jones SN, Sagerstrom CG, Ohkawa Y, Imbalzano AN 2010. Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function. Mol Cell Biol 30: 3176–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A 1961. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol 9: 493–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercola M, Ruiz-Lozano P, Schneider MD 2011. Cardiac muscle regeneration: Lessons from development. Genes Dev 25: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti GC, Colussi C, Adami R, Serra C, Mozzetta C, Parente V, Fortuni S, Straino S, Sampaolesi M, Di Padova M, et al. 2006. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat Med 12: 1147–1150 [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Pannerec A, Cadot B, Parlakian A, Besson V, Gomes ER, Marazzi G, Sassoon DA 2010. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol 12: 257–266 [DOI] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M 2005. Direct isolation of satellite cells for skeletal muscle regeneration. Science 309: 2064–2067 [DOI] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147: 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberpriller JO, Oberpriller JC 1974. Response of the adult newt ventricle to injury. J Exp Zool 187: 249–253 [DOI] [PubMed] [Google Scholar]
- Ohkawa Y, Yoshimura S, Higashi C, Marfella CG, Dacwag CS, Tachibana T, Imbalzano AN 2007. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. J Biol Chem 282: 6564–6570 [DOI] [PubMed] [Google Scholar]
- Olivetti G, Giordano G, Corradi D, Melissari M, Lagrasta C, Gambert SR, Anversa P 1995. Gender differences and aging: Effects on the human heart. J Am Coll Cardiol 26: 1068–1079 [DOI] [PubMed] [Google Scholar]
- Olson EN 2006. Gene regulatory networks in the evolution and development of the heart. Science 313: 1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. 2012. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN 2010. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest 120: 3912–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peault B, Rudnicki M, Torrente Y, Cossu G, Tremblay JP, Partridge T, Gussoni E, Kunkel LM, Huard J 2007. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol Ther 15: 867–877 [DOI] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell 3: 247–253 [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA 2011. Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V 2000. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol 185: 155–173 [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D 2012. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M 2005. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435: 948–953 [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J 17: 6979–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S 2012. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148: 112–125 [DOI] [PubMed] [Google Scholar]
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, et al. 2010. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell 143: 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalbruch H, Lewis DM 2000. Dynamics of nuclei of muscle fibers and connective tissue cells in normal and denervated rat muscles. Muscle Nerve 23: 617–626 [DOI] [PubMed] [Google Scholar]
- Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S 2006. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol 8: 677–687 [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, P.L. P. 2004. p38 pathway targets SWI–SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 36: 738–743 [DOI] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR 2010. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141: 943–955 [DOI] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, et al. 2011. De novo cardiomyocytes from within the activated adult heart after injury. Nature 474: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. 2012. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzu AC, Wu SM 2011. Developmental and regenerative biology of multipotent cardiovascular progenitor cells. Circ Res 108: 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swynghedauw B 1999. Molecular mechanisms of myocardial remodeling. Physiol Rev 79: 215–262 [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG 2009. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459: 708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG 2007. Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci 104: 846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, et al. 2011. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun 2: 187 doi: 10.1038/ncomms1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132: 2685–2695 [DOI] [PubMed] [Google Scholar]
- Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G 2010. Repairing skeletal muscle: Regenerative potential of skeletal muscle stem cells. J Clin Invest 120: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K 2010. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol 12: 143–152 [DOI] [PubMed] [Google Scholar]
- Uezumi A, Ito T, Morikawa D, Shimizu N, Yoneda T, Segawa M, Yamaguchi M, Ogawa R, Matev MM, Miyagoe-Suzuki Y, et al. 2011. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci 124: 3654–3664 [DOI] [PubMed] [Google Scholar]
- Wada MR, Inagawa-Ogashiwa M, Shimizu S, Yasumoto S, Hashimoto N 2002. Generation of different fates from multipotent muscle stem cells. Development 129: 2987–2995 [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. 2012. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151: 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Rudnicki MA 2012. Satellite cells, the engines of muscle repair. Nat Rev Mol Cell Biol 13: 127–133 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang C, Skarnes WC, Tjian R 2004. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev 18: 3106–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI 2012. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim Biophys Sin (Shanghai) 44: 54–69 [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR 2009. Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. 2008. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453: 524–528 [DOI] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR 2011. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476: 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR 2004. Muscle satellite cells adopt divergent fates: A mechanism for self-renewal? J Cell Biol 166: 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]