Amon and colleagues show that aneuploidy—the gain or loss of entire chromosomes—causes proteotoxic stress and makes cells prone to protein aggregation. The Hsp90 chaperone system is impaired in such cells, explaining the phenotype. Aggregate formation occurs soon after chromosome missegregation, indicating that gene copy number changes immediately impact protein homeostasis. Importantly, the study shows that aneuploidy predisposes prion formation and the aggregation of proteins implicated in neurodegenerative diseases.
Keywords: aneuploidy, proteotoxicity, folding, neurodegenerative disease
Abstract
Gains or losses of entire chromosomes lead to aneuploidy, a condition tolerated poorly in all eukaryotes analyzed to date. How aneuploidy affects organismal and cellular physiology is poorly understood. We found that aneuploid budding yeast cells are under proteotoxic stress. Aneuploid strains are prone to aggregation of endogenous proteins as well as of ectopically expressed hard-to-fold proteins such as those containing polyglutamine (polyQ) stretches. Protein aggregate formation in aneuploid yeast strains is likely due to limiting protein quality-control systems, since the proteasome and at least one chaperone family, Hsp90, are compromised in many aneuploid strains. The link between aneuploidy and the formation and persistence of protein aggregates could have important implications for diseases such as cancer and neurodegeneration.
Aneuploidy, defined as a karyotype that is not a multiple of the haploid complement, results in an unbalanced genome. This imbalance is generally not well tolerated in nature, as evidenced by the impaired fitness of aneuploid cells and organisms (for review, see Torres et al. 2008; Williams and Amon 2009). The condition also has a profound impact on human health. Aneuploidy is the leading cause of mental retardation and spontaneous abortions and a key characteristic of cancer, as >90% of all solid human tumors harbor aneuploid genomes (Weaver and Cleveland 2006).
Systematic analyses of aneuploid yeast and mouse cells suggest that aneuploidy causes chromosome-specific effects that are elicited by the duplication/deletion of individual genes or combinations of a small number of genes present on the aneuploid chromosome (Torres et al. 2007, 2010; Pavelka et al. 2010; Tang et al. 2011). Aneuploid yeast and mammalian cells also share a set of phenotypes, collectively called the aneuploidy-associated stresses (Torres et al. 2007; Williams et al. 2008; Tang et al. 2011), indicating that the aneuploid state per se impacts cell physiology. Aneuploidy impairs proliferation of budding and fission yeast cells as well as of mammalian cells under standard growth conditions (Baker et al. 2004; Niwa et al. 2006; Torres et al. 2007; Thompson and Compton 2008; Williams et al. 2008; Li et al. 2009; Pavelka et al. 2010), with a delay at the G1–S-phase transition being especially prominent (Niwa et al. 2006; Torres et al. 2007; Stingele et al. 2012). Whole chromosomal aneuploidies also lead to a transcriptional response. A gene expression signature similar to the environmental stress response (ESR) (Gasch et al. 2000) in budding yeast has been observed in aneuploid budding and fission yeast strains and Arabidopsis, mouse, and human cells (Sheltzer et al. 2012). Last, aneuploid cells exhibit phenotypes characteristic of disruption of protein homeostasis. Aneuploid budding yeast strains and trisomic mouse embryonic fibroblasts show increased sensitivity to compounds that interfere with protein folding and turnover (Torres et al. 2007, 2010; Pavelka et al. 2010; Tang et al. 2011). Understanding the phenotypes shared by many different types of aneuploidies is of particular importance, as this could provide insights into how an unbalanced karyotype impacts normal cellular physiology and disease states such as cancer. Here we investigate the consequences of one of the general effects of aneuploidy: disruption of cellular protein homeostasis.
Maintaining the proteome is essential for cell survival. Nascent peptides must be folded, proteins that are unfolded have to be refolded, and terminally damaged proteins must be degraded. Additionally, multiprotein complexes must be properly assembled. The cell relies on molecular chaperones to aid in the folding and refolding of proteins and the assembly of multiprotein complexes as well as on the 26S proteasome and vacuolar proteases to degrade proteins that are terminally misfolded (Tyedmers et al. 2010; Houck et al. 2012). The chaperone and ubiquitin–proteasome systems function in concert to ensure protein homeostasis. When cells experience proteotoxic stress—that is, when protein quality-control pathways such as the chaperone systems and the proteasomal degradation machinery are compromised or overwhelmed—misfolded proteins are not eliminated and aggregates form (Houck et al. 2012). Misfolded proteins not only inflict a fitness cost (Geiler-Samerotte et al. 2011), they are also associated with human disease. Highly structured aggregates have been linked to neurodegenerative pathologies, including Huntington's, Alzheimer's, and Parkinson's diseases, as well as prion diseases such as Kuru and Creutzfeld-Jacob syndrome (for review, see Goedert et al. 2010).
We previously generated 13 budding yeast strains harboring an additional copy of a single yeast chromosome, called disomes. These strains exhibit, among other deleterious phenotypes, increased sensitivity to high temperature and to inhibitors of protein synthesis and folding as well as chemical and genetic perturbation of proteasomal degradation (Torres et al. 2007, 2010). Furthermore, we found that increasing proteasomal degradation by deleting the gene encoding the deubiquitinating enzyme Ubp6 improves the proliferative abilities of a subset of disomic yeast strains. Together with the observation that the additional chromosomes are actively transcribed and translated (Torres et al. 2007, 2010; Pavelka et al. 2010), these studies suggest that aneuploidy alters the cell's proteome, resulting in proteotoxic stress, and implicate the ubiquitin–proteasome pathway in the survival of aneuploid cells. However, direct evidence of proteotoxicity in aneuploid cells and the role of chaperones in the generation of proteotoxic stress in aneuploid cells has thus far been lacking. Here we show that aneuploid yeast cells are prone to protein aggregate formation. Aneuploid yeast strains generated by a variety of different methods have defects in aggregate clearance and exhibit increased sensitivity to aggregate-prone proteins. The association between aneuploidy and protein aggregation uncovered in this study could have important implications for the pathology and treatment of diseases such as cancer and neurodegeneration, both of which have been associated with aneuploidy.
Results
Disomic yeast strains harbor a higher load of endogenous protein aggregates
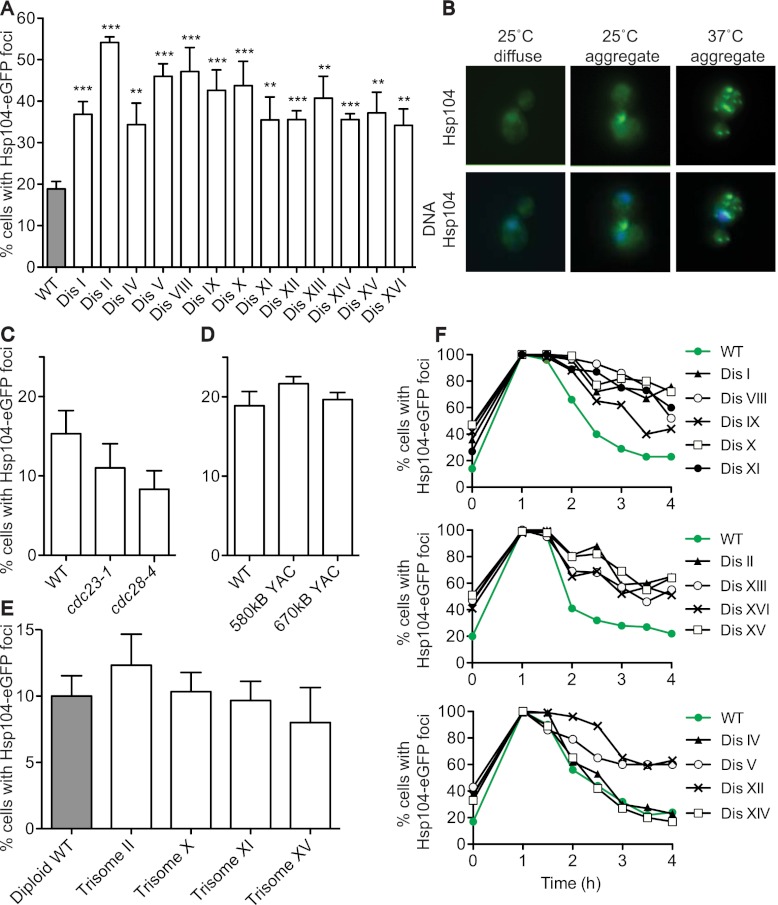
Introduction of whole chromosomes substantially alters the cell's proteome because most genes present on the additional chromosome are expressed according to gene copy number (Torres et al. 2007, 2010). This may impact protein homeostasis mechanisms. To test this possibility, we analyzed the subcellular localization of the disaggregase Hsp104. Under standard growth conditions, the Hsp104 chaperone fused to eGFP is diffusely localized throughout the cell, but the protein also colocalizes with protein aggregates, manifesting as Hsp104-eGFP foci (Fig. 1A,B; Liu et al. 2010). All 13 disomic strains analyzed showed a significant increase in the percentage of cells harboring Hsp104-eGFP foci compared with the euploid control (Fig. 1A,B). Increased aggregate formation was not due to slowed proliferation caused by aneuploidy (Torres et al. 2007) because temperature-sensitive cdc23-1 and cdc28-4 strains grow slowly at the permissive temperature but do not harbor additional aggregates (Fig. 1C). Our data further suggest that it is the increased protein load generated from the additional chromosome that leads to increased protein aggregation. We did not observe an increase in the percentage of cells with Hsp104-eGFP foci in strains that contain yeast artificial chromosomes (YACs) (Fig. 1D) that carry human DNA but generate no yeast proteins and very few if any other peptides and proteins (Foote et al. 1989; Torres et al. 2007). As protein aggregates are the consequence of misfolded proteins, our data suggest that aneuploid cells are challenged to fold proteins efficiently and/or to process protein aggregates appropriately.
Figure 1.
Disomic yeast strains harbor an increased protein aggregate load. (A) Wild-type (WT) and disomic yeast strains containing an HSP104-eGFP fusion were grown to exponential phase in YEPD, and the percentage of cells harboring Hsp104-eGFP foci was determined. n = 3; SEM, n = 100 cells per time point; (**) P < 0.005; (***) P < 0.0005; Student's t-test. Strains, in order, are A31392, A31393, A31394, A31395, A31396, A31397, A31398, A31399, A31400, A31401, A31402, A31403, A31404, and A31405. (B) Images of Hsp104-eGFP aggregates. Aggregates are in green, and DNA is in blue. (C,D) Wild-type (A25654), cdc23-1 (A29766), and cdc28-4 (A29765) strains (C) and strains harboring YACs containing 580 kb (A28922) or 670 kb (A28925) of human DNA (D) were grown as in A to determine the percentage of cells with Hsp104-eGFP aggregates. (E) Quantification of Hsp104-eGFP foci in trisomic yeast strains grown as in A. Strains, in order, are A31406, A31407, A31408, A31409, and A31410. Note that the number of cells with Hsp104-eGFP aggregates in diploid cultures is lower than in haploid cultures. The basis for this is unclear at present. (F) Strains grown at 25°C were shifted to 37°C. The percentage of cells with Hsp104-eGFP aggregates was determined at the indicated times after temperature shift. n = 100 cells per time point. Two replicas of this experiment are shown in Supplemental Figure S1. The strains are the same as in A.
We also found that, as with all other aneuploidy-associated phenotypes (Torres et al. 2007), increasing ploidy suppressed aggregate formation. The percentage of cells harboring Hsp104-eGFP foci in diploid strains carrying an additional chromosome (trisomic strains) is significantly lower than that of haploid strains with an extra chromosome (Fig. 1, cf. A and E). Many subunits of protein complexes require the assistance of protein chaperones to fold. These proteins then acquire a stable conformation by binding to the complex's other subunits. If one of the components is present in excess and cannot exist stably as an uncomplexed subunit, it requires the continuous assistance of chaperones to prevent aggregation (Tyedmers et al. 2010). As a result, chaperones cannot assist other folding reactions, and the general folding capacity of the cell is reduced. The observation that increasing ploidy reduces aggregate formation suggests that the proteotoxic stress in aneuploid cells could, in part, be a result of stoichiometric imbalances caused by the proteins encoded on the unbalanced chromosomes. Decreasing the ratio of uncomplexed proteins to complexed proteins reduces the protein aggregate load of aneuploid yeast. The observation that aggregate formation in many trisomic strains is not elevated compared with diploid controls further suggests either that cells have the ability to compensate for some genomic imbalances and/or that diploids have a higher folding capacity than haploids.
Adaptation to proteotoxic stress is delayed in disomic yeast strains
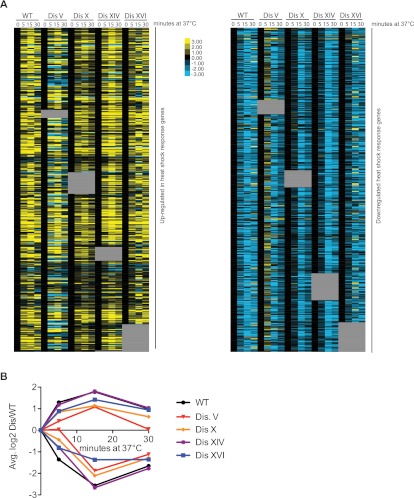
If disomic yeast strains experience increased proteotoxic stress, they may be delayed in responding or adapting to conditions that induce proteotoxicity. To test whether disomic yeast strains are delayed in adapting to proteotoxic stress-inducing growth conditions, we monitored Hsp104-eGFP foci after shift to high temperature (37°C). Virtually all wild-type and disomic cells contained Hsp104-eGFP foci within an hour of temperature shift (Fig. 1F). However, whereas wild-type cells cleared the aggregates by 4 h, all disomes except for disomes IV and XIV adapted to heat stress with slower kinetics (Fig. 1F; Supplemental Fig. S1). This delayed adaptation to high temperature was not due to an inability to mount a heat-shock response, as judged by microarray analysis of aneuploid cells adapting to thermal stress (Fig. 2). Activation of the unfolded protein response (UPR) in the endoplasmic reticulum was also unaffected in aneuploid strains; splicing of the UPR gene HAC1 in the disomes was similar to that in wild-type cells under both normal conditions and conditions in which the UPR is induced (Supplemental Fig. S2).
Figure 2.
The heat-shock response is intact in disomic yeast strains. Wild-type (WT) and disomic yeast strains were grown at 25°C and shifted to 37°C. RNA samples were taken 0, 5, 15, and 30 min after shift. RNA extracted from wild-type cells grown continuously at 25°C was used as reference for all samples. Data were mined for genes involved in the heat-shock response (Gasch et al. 2000), and those present in the extra chromosome were removed from the analysis (gray boxes). The data set was split into those genes that are up-regulated and those that are down-regulated in the heat-shock response. (A) The expression of genes involved in the heat-shock response in disomic yeast after shifting to 37°C. Yellow shows up-regulated genes, and blue shows down-regulated genes. Shown are the unclustered data after being zero-transformed. (B) The average expression changes of up-regulated (above the X-axis) and down-regulated (below the X-axis) genes displayed in A are shown.
Although aneuploid yeast strains can mount a heat-shock response, the proteotoxic stress that we observed in disomic cells under standard growth conditions (25°C, YEPD) is not sufficient to induce a canonical heat-shock response in most disomes (Supplemental Fig. S3; Torres et al. 2007). This lack of a heat-shock response is expected, as this response is tailored to an acute stressor and is transient in nature (Gasch et al. 2000). In contrast, aneuploidy is a chronic stress, and adaptation to the aneuploid state may have taken place. It is, however, noteworthy that disomes IV and XIV, which adapt to heat shock with the same kinetics as wild-type cells (Fig. 1F; Supplemental Fig. S1), up-regulate genes involved in the heat-shock response even under normal growth conditions (Supplemental Fig. S3). Analysis of the abundance of the chaperones Hsp104, Ssa1, Sse2, and Hsp42 further confirmed the absence of a canonical heat-shock response (Supplemental Fig. S4). Although aneuploidy is not sufficient to induce the canonical heat-shock response, most disomic yeast strains show a transcriptional response reminiscent of the ESR (Torres et al. 2007), which encompasses a subset of the heat-shock response (Gasch et al. 2000). HSP104 RNA levels, for example, increase with degree of aneuploidy (Sheltzer et al. 2012). Taken together, our results indicate that disomic yeast strains experience proteotoxic stress that is evidenced by increased aggregate burden under both normal growth conditions and conditions that induce proteotoxicity.
Meiotic and mitotic chromosome missegregation leads to protein aggregate formation
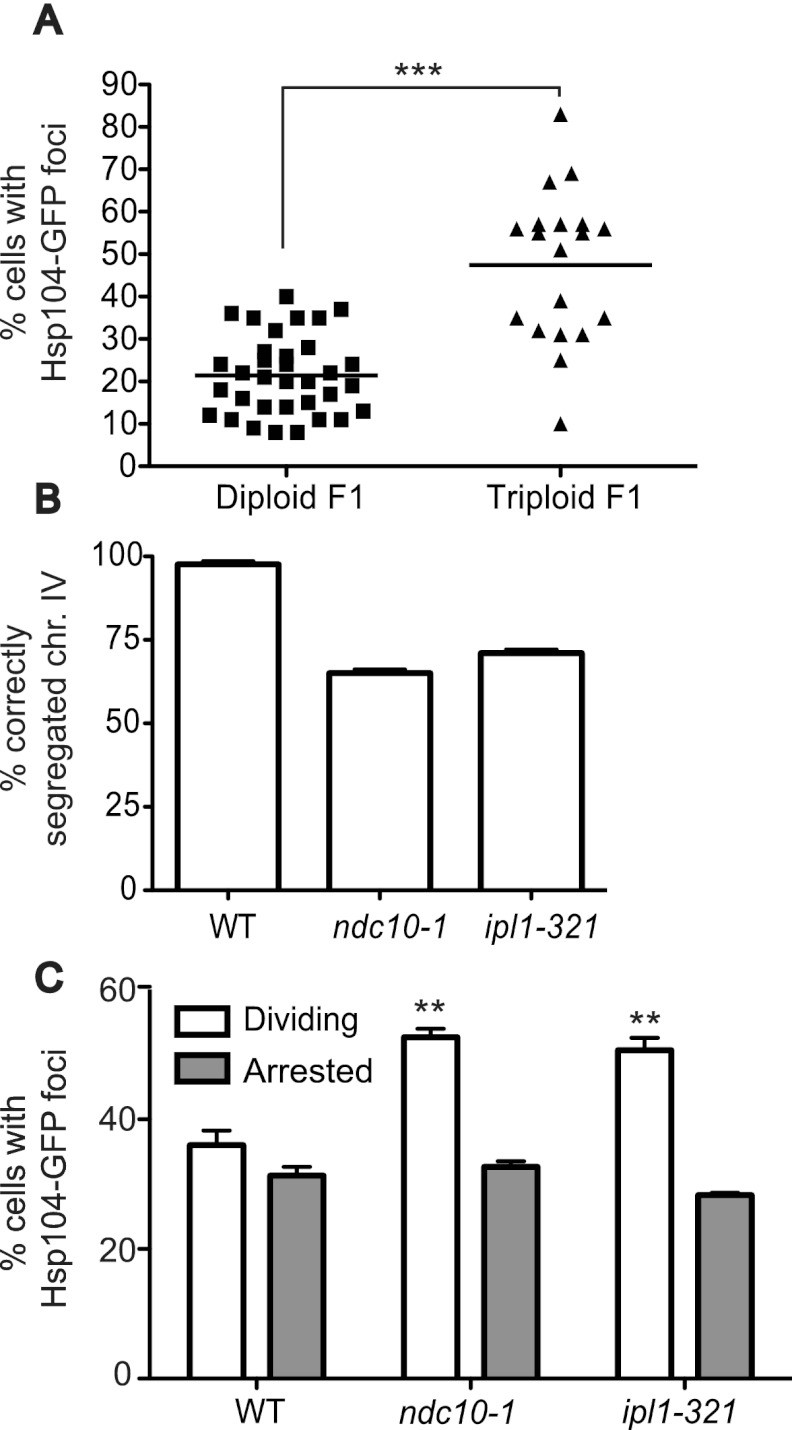
Increased protein aggregation was observed not only in strains harboring single chromosomal aneuploidies, but also in aneuploid cells that arose from meiotic and mitotic nondisjunction. Triploid cells induced to undergo meiosis produce highly aneuploid progeny with karyotypes ranging from diploid to highly aneuploid (St Charles et al. 2010). The majority of the aneuploid progeny is inviable (Parry and Cox 1970), but some genetically unstable aneuploid strains can be obtained (Sheltzer et al. 2011; Zhu et al. 2012). As colony formation is a prerequisite for the analysis, we were only able to analyze those aneuploids that were healthy enough to form colonies. Nevertheless, analysis of 19 products of triploid meioses showed that the percentage of cells harboring Hsp104-eGFP foci was increased in most strains (Fig. 3A).
Figure 3.
Meiotic and mitotic chromosome nondisjunction causes increased Hsp104-eGFP focus formation. (A) The percentage of cells with Hsp104-eGFP aggregates was analyzed in the progeny of diploid (A28220) or triploid (A28219) strains 3 d after germination. n = 100 cells per strain. Note that although many of the progeny from the triploid meioses will harbor multiple aneuploidies, some will also be euploid or will have become euploid as they proliferate. (B,C) Wild-type (WT) (A5244), ndc10-1 (A28204), and ipl1-321 (A16154) mutants harboring a GFP-marked chromosome IV, and wild-type (A25654), ndc10-1 (A27681), and ipl1-321 (A27682) mutants harboring Hsp104-eGFP were arrested in G1 with pheromone at 25°C, followed by release at 30°C in either the presence (10 μg/mL; Arrested) or absence (Dividing) of nocodazole. Samples were taken after 3 h to determine the percentage of cells that correctly segregated chromosome IV (B) and that harbored Hsp104-eGFP foci (C). We note that the temperature shift during this experiment may inflate the percentage of cells harboring aggregates in all strains, as they will be adapting to the temperature shift.
Chromosome missegregation during mitosis also resulted in aggregate formation. Strains harboring temperature-sensitive alleles of genes encoding the kinetochore component Ndc10 or the Aurora B kinase Ipl1 were arrested in G1 and released to progress through the cell cycle at the semipermissive temperature of 30°C. Under these conditions, 35% of ndc10-1 and 29% of ipl1-321 cells were unable to segregate a GFP-marked chromosome IV (Fig. 3B), indicating that dramatic chromosome missegregation occurs under these growth conditions. Aggregate formation was increased as early as 3 h after release from the pheromone-induced G1 arrest (Fig. 3C). Importantly, this increase in Hsp104-eGFP foci depended on cell division. When ndc10-1 or ipl1-321 cells were induced to undergo a synchronous cell cycle at 30°C but chromosome segregation was prevented by treating cells with the microtubule-depolymerizing drug nocodazole, Hsp104-eGFP focus number did not increase (Fig. 3C). We conclude that the percentage of cells harboring Hsp104-eGFP-decorated protein aggregates is increased in most, if not all, aneuploid strains and that aggregates form soon after chromosome nondisjunction.
Aneuploid strains fail to efficiently fold the protein quality-control sensor von Hippel-Lindau protein (VHL)
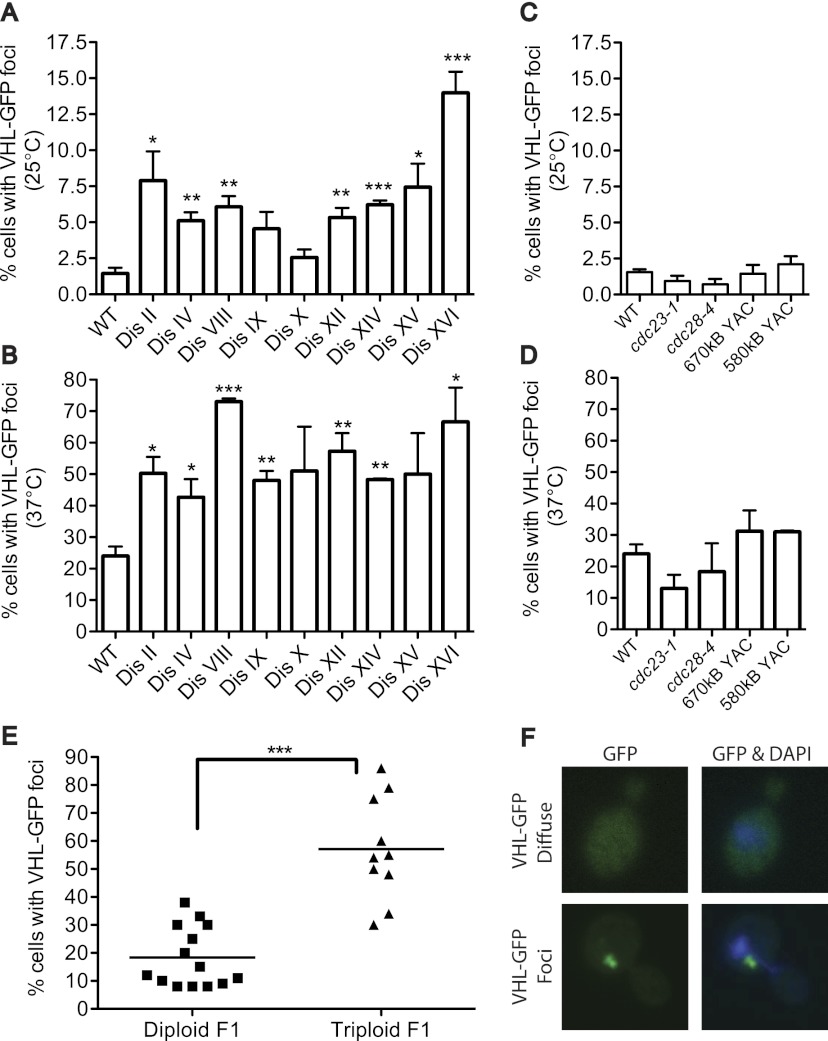
Protein aggregates in disomic yeast strains could be the result of proteins generated from the additional chromosomes overwhelming and/or impairing chaperones. To test this idea, we challenged the cell's protein quality-control pathways using the well-studied substrate, the human VHL. VHL is unable to fold without its binding partners, Elongin B and Elongin C. When human VHL is expressed in yeast in the absence of Elongin B and Elongin C, the protein is quickly ubiquitinated and degraded (Supplemental Fig. S5; McClellan et al. 2005; Kaganovich et al. 2008). The quality-control pathways involved in the elimination of misfolded VHL are known: Folding-defective VHL is shuttled from Hsp70 to an Hsp90 complex that enables degradation by the ubiquitin–proteasome system (McClellan et al. 2005). When any of these pathways are defective, misfolded VHL forms aggregates that are seen as foci in cells expressing VHL as a GFP fusion (Fig. 4F; Kaganovich et al. 2008). Disomic yeast strains grown under nonstress conditions (25°C, YEPRG) (Fig. 4A) and under conditions of heat stress (2 h at 37°C) (Fig. 4B) showed increased VHL focus formation; slow-growing mutants or strains carrying human DNA did not (Fig. 4C,D). The failure to process misfolded VHL-GFP was not specific to the disomic strains but was also observed in the progeny of triploid meioses (Fig. 4E). We note that haploid strains obtained from diploid meioses harbored a higher percentage of cells with VHL-GFP foci than the haploid control strain analyzed in the experiment shown in Figure 4A. We suspect that germination and colony growth on selective medium places an increased burden on the cell's protein quality-control systems compared with growth in liquid rich medium (YEPRG) at 25°C. We conclude that targeting of VHL-GFP for proteasomal degradation is compromised in aneuploid cells because the protein quality-control pathways (chaperones and/or the proteasome) of the cells either are defective or are functional but overwhelmed by changes in the cell's proteome caused by the aneuploid state.
Figure 4.
Aneuploid yeast display hallmarks of impaired protein quality control. (A) Wild-type (WT) and disomic yeast strains deleted for the multidrug transporter PDR5 and expressing a VHL-GFP fusion were grown in YEP 2% raffinose and 2% galactose at 25°C, and the percentage of cells with VHL-GFP foci was determined. n = 3; SEM; n = 300 cells per time point; (*) P < 0.05; (**) P < 0.005; (***) P < 0.0005; Student's t-test. Strains, in order, are A32076, A32077, A32078, A32079, A32080, A32081, A32082, A32083, A32084, A32085, A32086, A32087, A32088, and A32089. (B) Strains described in A were analyzed after a 2-h incubation at 37°C. n = 100. (C,D) cdc23-1 (A30461), cdc28-4 (A30462), and wild-type strains harboring YACs carrying human DNA (A29969 and A29971) were grown at 25°C (C) or shifted for 2 h to 37°C (D) to analyze VHL-GFP focus formation. (E) The percentage of cells with VHL-GFP foci was determined in the progeny of diploid (A28388) or triploid (A28389) strains 4 d after germination. n = 100 per colony. (F) Images of VHL-GFP aggregates. Aggregates are in green, and DNA is in blue.
Loss of UBP6 reduces aggregate burden in disomic yeast strains
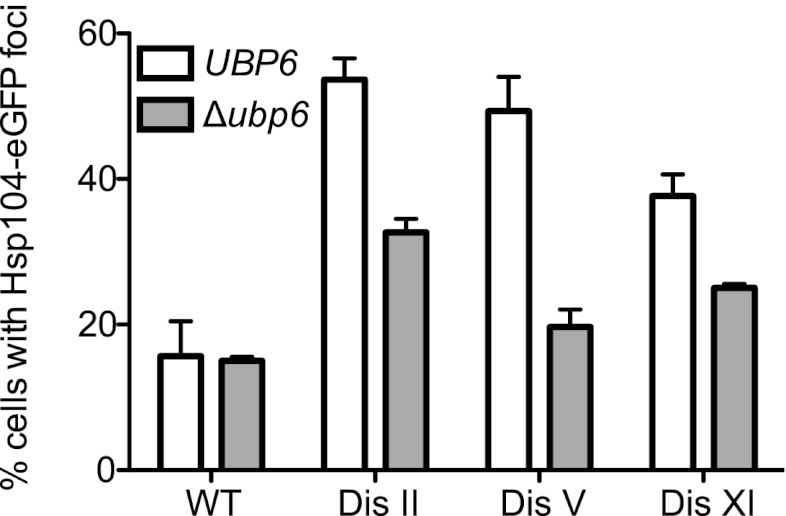
The high incidence of both endogenous and VHL protein aggregates in aneuploid strains suggests that aneuploidy negatively impacts protein folding and/or degradation of misfolded proteins. Previous studies showed that ubiquitin–proteasomal degradation is important for the survival of aneuploid yeast strains (Torres et al. 2007, 2010). Insufficient proteasome activity could also be responsible for increased aggregate formation in aneuploid cells. A prediction of this hypothesis is that increasing proteasome function decreases aggregate burden in aneuploid cells. To test this possibility, we examined the consequences of deleting UBP6 on Hsp104-eGFP focus formation in disomic yeast strains.
Ubp6 associates with the proteasome and removes ubiquitin chains from substrates. This not only allows for the recycling of ubiquitin, but also causes proteasome substrates to escape degradation. This is evident from the analysis of cells lacking UBP6. Degradation of all proteasome substrates analyzed to date is accelerated in such cells (Hanna et al. 2006; Peth et al. 2009). We deleted UBP6 in disome V and disome XI cells, whose proliferation improves when UBP6 is deleted, and in disome II cells, in which deleting UBP6 leads to decreased proliferation (Torres et al. 2010). Deletion of UBP6 reduced aggregate burden in all three disomic strains (Fig. 5). This finding is consistent with our previous observation that deletion of UBP6 causes attenuation of levels of proteins with high relative expression in all disomic strains analyzed, irrespective of whether deletion of UBP6 improves proliferation (Torres et al. 2010). Interestingly, in disome V strains, aggregate burden was reduced to almost wild-type levels when UBP6 was deleted (Fig. 5). This finding raises the interesting possibility that the increased proliferative abilities of disome V ubp6Δ cells are due to a reduction in protein aggregates. We conclude that enhanced proteasomal degradation reduces the aggregate burden in all disomic strains analyzed.
Figure 5.
Increased proteasome activity decreases aggregate burden in disomic strains. Wild-type (WT) (A3369), disome II (A33370), disome V (A33371), and disome IX (A33372) cells harboring a deletion of UBP6 and the HSP104-eGFP fusion were grown to exponential phase in YEPD, and the percentage of cells harboring Hsp104-eGFP foci was determined. n = 3; SEM, n = 100 cells per time point.
Hsp90 folding capacity is reduced in many disomic yeast strains
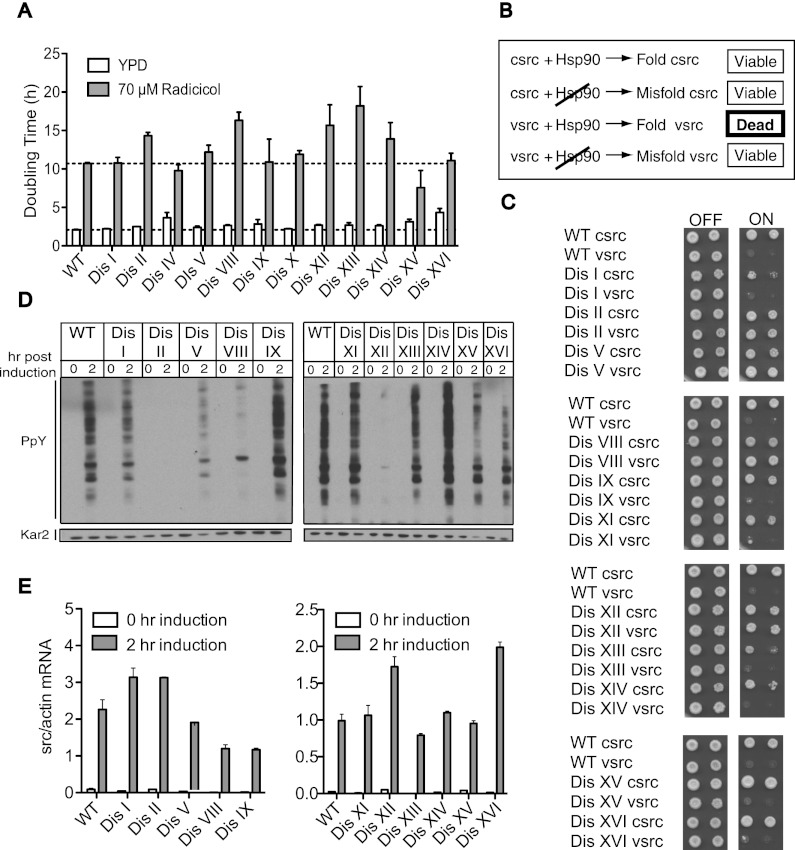
Are other protein quality-control systems also affected in aneuploid cells? Because VHL is an Hsp90 client, we explored the in vivo folding activity of Hsp90 and found it to be reduced in many disomic strains. Hsp90 is a highly abundant chaperone that, in concert with cochaperones, folds cytosolic proteins (McClellan et al. 2007; Franzosa et al. 2011). Consistent with previous results using the Hsp90 inhibitor geldanamycin (Torres et al. 2007), we found that several disomic yeast strains are more sensitive to the Hsp90 inhibitor radicicol than the euploid control strain (Fig. 6A).
Figure 6.
Hsp90 folding capacity is limiting in many disomic strains. (A) Disomic yeast harboring a deletion of the multidrug transporter PDR5 were grown in YEPD or YEPD containing 70 μM radicicol at 30°C to determine their doubling time. The mean and SEM of three replicates are shown. Strains, in order, are A15549, A15551, A15553, A15555, A15557, A15559, A15561, A15563, A15566, A15567, A15569, A15571, and A15573. (B) Schematic of v-src/c-src Hsp90 assay. Hsp90 is required to fold c-src and toxic v-src. A reduction in Hsp90 activity results in misfolded v-src and restores cell viability. (C) Wild-type (WT) and disomic yeast strains carrying c-src or v-src under the galactose-inducible GAL1-10 promoter were grown under conditions in which expression is repressed (−URA 2% glucose) or induced (−URA 2% raffinose and galactose). Tenfold dilutions were plated. C-src strains, in order, are A32090, A32091, A32092, A32093, A32094, A32095, A32096, A32097, A32098, A32099, A32100, and A32101. V-src strains, in order, are A32102, A32103, A32104, A32105, A32106, A32107, A32108, A32109, A32110, A32111, A32112, and A32113. (D,E) Wild-type and disomic yeast strains harboring the GAL-v-src fusion were grown in YEP + 2% raffinose. Galactose was added, and the relative amount of v-src RNA (E) and total tyrosine phosphorylation (D) was determined before and after 2 h of v-src induction. V-src strains are in the same order as in C.
To examine Hsp90 activity, we analyzed the in vivo folding activity of the well-studied Hsp90 model substrate, the tyrosine kinase Src. Both c-src and the oncogenic form, v-src, depend on Hsp90 for folding (Kimura et al. 1995; Nathan et al. 1997). For unknown reasons, overexpression of v-src, but not c-src, is lethal in budding yeast (Xu and Lindquist 1993). Compromising Hsp90 activity suppresses v-src folding and activity and, consequently, this lethality (Fig. 6B; Nathan and Lindquist 1995; Nathan et al. 1997). We found that the toxicity of v-src was diminished in many aneuploid strains (Fig. 6C), suggesting a reduction in Hsp90 activity. To further explore the activity of v-src, we took advantage of the low levels of endogenous tyrosine phosphorylation in yeast that are dramatically increased when v-src is expressed from the galactose-inducible GAL1-10 promoter (Fig. 6D). Total tyrosine phosphorylation was reduced in disomes II, V, VIII, and XII (Fig. 6D), correlating well with these cells' ability to form colonies under v-src-inducing conditions (Fig. 6C). The inability to generate active v-src was not due to decreased mRNA expression, as v-src RNA levels in the disomes were as high as or higher than in wild-type cells (Fig. 6E). Reduced v-src activity was also observed in disomes I, XIII, XV, and XVI as judged by reduced tyrosine phosphorylation levels, but this decreased activity was not sufficient to allow growth on v-src-inducing medium (Fig. 6C,D). Our results show that eight out of 11 disomes exhibit reduced Hsp90 activity. Hsp90 may be overloaded by substrates that rely on this chaperone to be folded. It is also possible that the activity of the Hsp90 folding machinery is reduced. Given that many different disomic strains exhibit decreased Hsp90 activity, we favor the idea that the Hsp90 folding reservoir is depleted, rather than inactive, in aneuploid strains. Hsp90 is thought to serve a limited number of clients under normal growth conditions and to be present in excess (Borkovich et al. 1989; Neckers 2007). It is therefore surprising that Hsp90 activity appears limiting in many disomic strains. Perhaps under conditions of proteotoxic stress, Hsp90's folding repertoire is expanded. We conclude that many aneuploid cells experience saturation of the Hsp90 system.
Aneuploid strains are more susceptible to protein aggregates associated with human disease
Does aneuploidy also cause cells to be more susceptible to protein-folding defects associated with human disease? To address this question, we used an assay that measures the activity of the prion protein Sup35 and assessed toxicity associated with the glutamine-rich protein Huntingtin (Htt1) in disomic yeast strains.
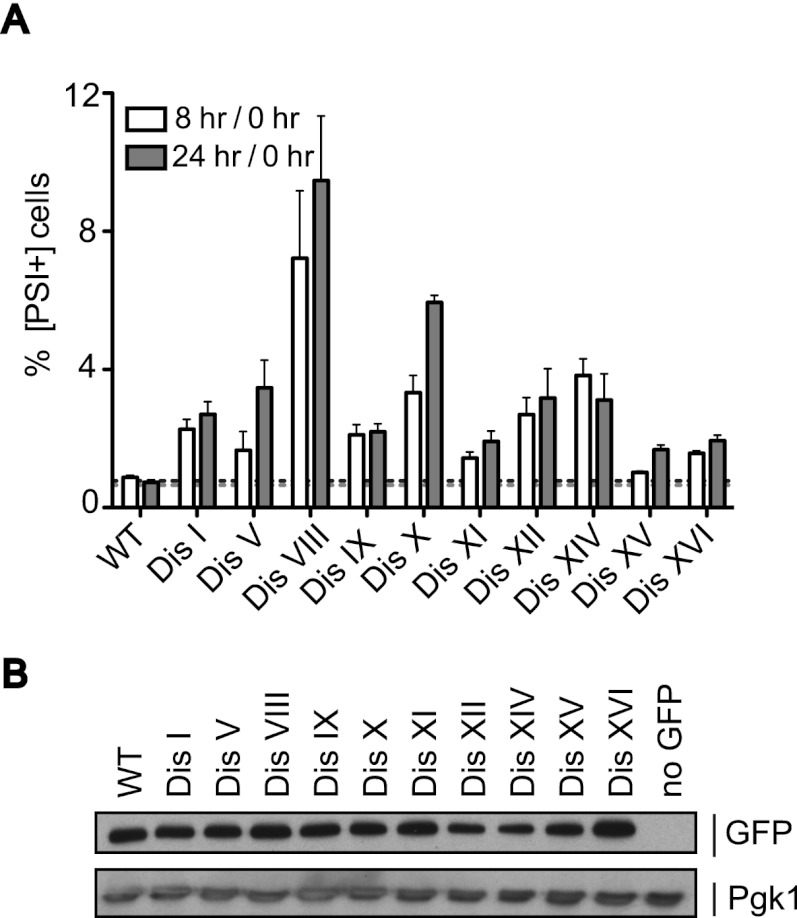
The prion [PSI+] is formed by Sup35, a subunit of the translation terminator complex. When Sup35 switches to the aggregated amyloid conformation [PSI+], much of the protein becomes unavailable to terminate translation, causing readthrough of stop codons (Liebman and Sherman 1979). Because the basal conversion frequency of Sup35 to its prion form is low (10−6–10−7), we used strains carrying a variant of the SUP35 gene (SUP35-R2E2) that increases the conversion frequency (Liu and Lindquist 1999; Cox et al. 2003) to study the effects of aneuploidy on Sup35 activity. We then used an assay where conversion from [psi−] to [PSI+] results in readthrough of three stop codons upstream of GFP, allowing expression of the fluorescent protein (Tyedmers et al. 2008). Single colonies obtained from a frozen stock were inoculated into rich medium, and the percentage of fluorescent cells was determined immediately after inoculation and after 8 and 24 h. All disomic strains tested showed an increase in the fraction of cells expressing GFP (Fig. 7A,B; Supplemental Fig. S6). Attempts to visualize the Sup35 aggregates by SDD-AGE (semidenaturing detergent-agarose gel electrophoresis) were unsuccessful. We therefore cannot exclude the possibility that mechanisms other than prion conversion lead to the observed increase in the percentage of GFP-positive cells in the disomic strains. We, however, favor the interpretation that Sup35 aggregates are in the detergent-soluble, small oligomer stage that precedes the large amyloid aggregates detectable by SDD-AGE (Halfmann et al. 2010) and/or that aggregates comprise a small fraction of total Sup35 protein and are thus undetectable by this technique.
Figure 7.
Disomic yeast strains show increased expression of the Sup35 prion reporter. (A) Single colonies of wild-type (WT) and disomic strains carrying the SUP35-R2E2 allele and a GFP construct preceded by three stop codons were inoculated into SC medium, and the percentage of fluorescent cells was determined after 0, 8, and 24 h by flow cytometry. Shown is the ratio of GFP+ cells after 8 and 24 h of growth to GFP+ cells immediately after inoculation (0 h). The mean and SEM of at least 12 single colonies are depicted. Strains, in order, are A31114, A29843, A29845, A29846, A29847, A29848, A31110, A31111, A31112, A29450, and A31113. (B) Strains in A and A22361 (no GFP control) were grown for 24 h, and total protein was extracted. GFP protein levels were analyzed by Western blot analysis. Pgk1 was used as a loading control.
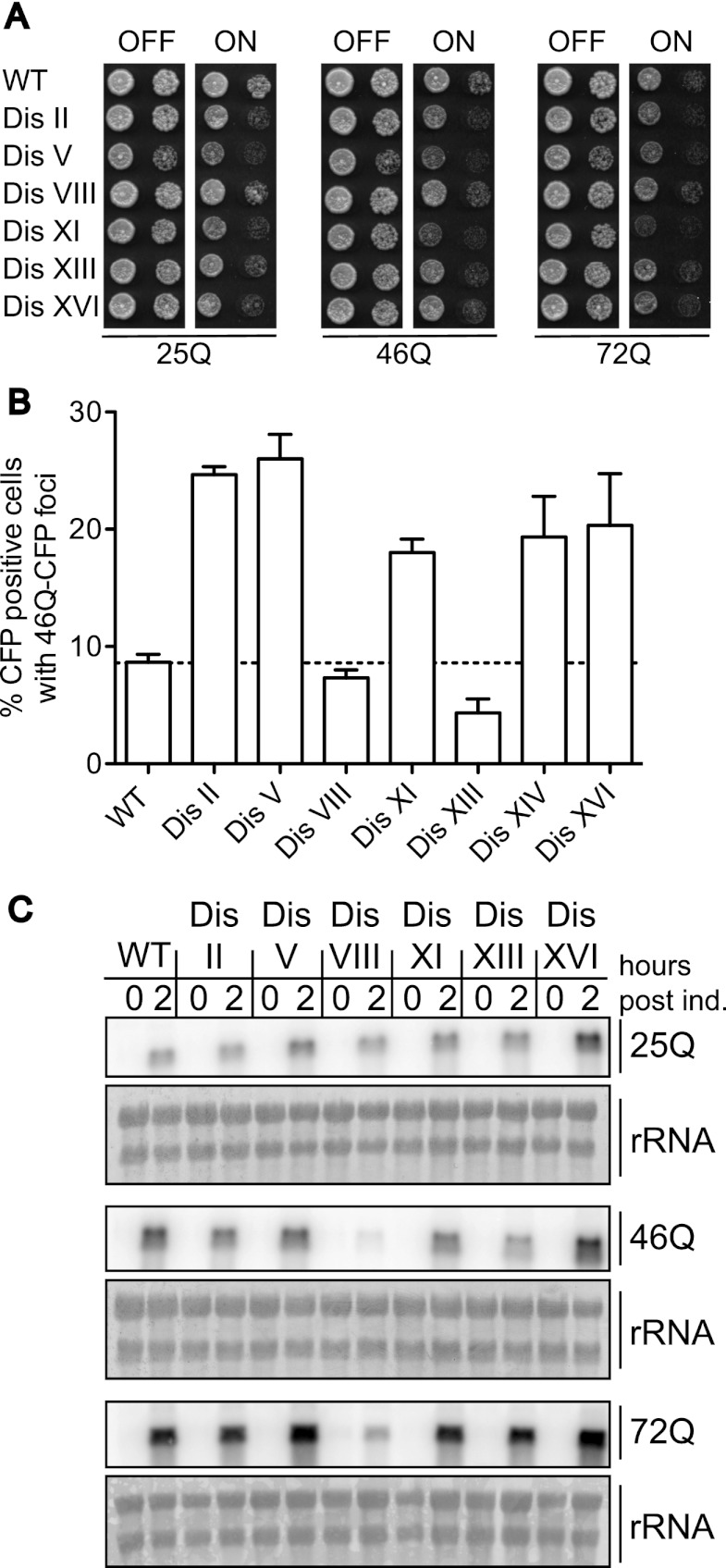
Huntington's disease is a neurodegenerative disease associated with β-sheet aggregates comprised mainly of the Htt1 protein (Goedert et al. 2010). Toxicity and disease phenotypes require that the polyglutamine (polyQ) stretch in its N terminus expand beyond 38 repeats (Duyao et al. 1993; The Huntington's Disease Collaborative Research Group 1993). We used yeast strains expressing a 17-amino-acid fragment of Htt1 exon 1 with polyQ tracts of varying length to determine the susceptibility of aneuploid yeast to expanded polyQ stretches (Duennwald et al. 2006). In euploid cells, Htt1 harboring 25 glutamine residues (25Q) is not toxic when expressed from the galactose-inducible GAL1-10 promoter, but Htt1 harboring 46 or 72 Qs causes toxicity (Duennwald et al. 2006). All disomes tested, except for disome VIII, exhibited increased sensitivity to Htt1-polyQ expression compared with the euploid control (Fig. 8A). Htt1-46Q-CFP aggregates are also accumulated more readily in many of the disomic strains analyzed (Fig. 8B). The lack of sensitivity of disome VIII is most likely due to reduced expression of the construct (Fig. 8C). We conclude that the proteotoxicity that afflicts aneuploid yeast cells can predispose them to the accumulation of protein aggregates associated with human diseases.
Figure 8.
Disomic yeast strains exhibit increased sensitivity to Huntingtin polyQ aggregates. (A) Wild-type (WT) and disomic strains harboring a GAL-Flag-HTT1(17AA)25QΔpro-CFP, GAL-Flag-HTT1(17AA)46QΔpro-CFP, or GAL-Flag-HTT1(17AA)72QΔpro-CFP construct were grown under conditions in which expression is repressed (YEPD) or induced (YPRG). Tenfold dilutions were plated. 25Q strains, in order, are A32114, A32115, A32116, A32117, A32118, A32119, and A32120. 46Q strains, in order, are A32121, A32122, A32123, A32124, A32125, A32126, A32127, and A32128. 72Q strains in order are A32129, A32130, A32131, A32132, A32133, A32134, and A32135. (B) Wild-type and disomic strains harboring the GAL-Flag-HTT1(17AA)46QΔpro-CFP construct were grown for 8 h in the presence of galactose to determine the percentage of cells with Htt1-46Q-CFP foci. n = 100. Shown are the mean and SEM of three independent experiments. (C) Expression of the GAL-HTT1(17aa)-Flag- nQ-CFP constructs. Strains were grown in YEP 2% raffinose to OD600 = 0.2 when 2% galactose was added. RNA was extracted from samples taken after 2 h, and the amount of HTT1-nQ-CFP RNA was determined via Northern blot analysis.
Discussion
Our studies of aneuploid yeast have revealed the dramatic effect of an unbalanced karyotype on cellular protein homeostasis. All aneuploid strains, irrespective of how they were generated or their karyotype, showed an increased protein aggregate burden. Aneuploid strains are prone to aggregation of endogenous proteins as well as of ectopically expressed hard-to-fold proteins such as polyQ stretch-containing proteins. We do not know which proteins comprise the aggregates observed in aneuploid cells. Obligate chaperone clients present in excess due to aneuploidy could accumulate and then either form aggregates themselves or interfere with the folding of other chaperone clients. Identifying aggregate constituents will distinguish between these two nonmutually exclusive possibilities.
Two protein quality-control systems, the proteasome and the Hsp90 chaperone, appear to be limiting in many aneuploid yeast strains. Increasing proteasome function by deleting UBP6 led to a decrease in aggregate burden in all aneuploid yeast strains analyzed. We also found that the Hsp90 substrate v-src was less active in many disomic strains, indicating that Hsp90 activity is limiting, which could contribute to aggregate formation in these strains. This latter result is surprising, as Hsp90 is highly abundant and its activity is thought to be in excess in cells (Borkovich et al. 1989; Neckers 2007). Perhaps this is not the case, especially under conditions of proteotoxic stress. Hsp90 may not be the only chaperone system limiting in aneuploid cells. We speculate that other protein-folding pathways are also saturated with different aneuploidies impacting different chaperone families to varying degrees depending on the identity of the proteins encoded on the extra chromosomes. Examining the activity of the different folding pathways in different disomic yeast strains will test this idea.
Why are aneuploid cells aggregate-prone?
Aggregates could be a result of overwhelmed folding pathways or could stem from reduced chaperone activity. Both alternatives are possible, but given that the aneuploid chromosomes are actively expressed, it is likely that excess proteins produced from the aneuploid chromosomes occupy chaperones and thereby reduce their availability to assist in the folding of their other clients. What determines the extent of aggregate formation is not yet known. It does not appear to correlate with either the degree of aneuploidy (by DNA content), total protein in excess, delay in G1, or doubling time. However, it is important to bear in mind that our aggregate analyses do not measure absolute amounts of aggregated proteins in cells and are not able to distinguish toxic from nontoxic aggregates. The ESR, which encompasses part of the heat-shock response, correlates with degree of aneuploidy (Torres et al. 2007; Sheltzer et al. 2012). We propose that protein aggregate burden correlates with the number of obligate chaperone clients and hence with the distribution of their encoding genes in the genome.
It may seem surprising that the cell's protein quality-control pathways cannot compensate for the presence of a single additional chromosome, which, depending on chromosome size, results in 2%–12% of the genome being imbalanced. Previous studies showed that even small amounts of misfolded proteins place a burden on the cell's protein quality-control systems and hence adversely affect cellular fitness. Expression of a single misfolded cytosolic protein at <0.1% of total protein leads to a significant decrease in proliferative abilities and the induction of a cytoplasmic unfolded protein response (Geiler-Samerotte et al. 2011).
Importantly, the generation of misfolded proteins requiring the assistance of the cell's protein quality-control pathways is a common occurrence in aneuploid cells. It is well established that many subunits of protein complexes only acquire a stable conformation by binding to other subunits of the complex (Imai et al. 2003; Boulon et al. 2010). Thus, every single polypeptide produced by genes located on aneuploid chromosomes that normally has a binding partner is—in the disomes—in excess. For example, if in euploid cells, 1% of a subunit of a heterodimeric protein complex is present in excess due to variability in subunit expression and must be eliminated, the number of proteins that needs to be eliminated rises to 102% in cells that carry an additional copy of the gene encoding one of the two subunits. This scenario applies to all proteins encoded on the extra chromosome that require a binding partner to acquire a stable conformation. We propose that this dramatic change in protein stoichiometries leads to an increased burden on the protein quality-control pathways of the cell. Individual subunits present in excess require the continuous assistance of chaperones, preventing chaperones from assisting other folding reactions and reducing the general folding capacity of the cell, and thus interfere with their essential function of mediating folding of essential proteins (Hartl et al. 2011). The fact that the aggregate phenotype was ameliorated when the ratio of uncomplexed proteins to properly complexed proteins was decreased by increasing base ploidy (as in trisomic strains) suggests that the proteotoxicity observed in aneuploids is indeed in part the result of the protein stoichiometry imbalances caused by aneuploidy, although it is also possible that diploid cells are more efficient at clearing aggregates. We furthermore propose that an additional burden on the protein quality-control machinery is generated by the overproduction of proteins encoded on the extra chromosomes that require chaperones for their function, such as protein kinases and WD40 repeat proteins. In summary, aneuploidy impacts protein homeostasis in multiple ways so that even small unbalanced chromosomes have a significant impact on the cell's protein quality-control systems.
Aneuploidy in cancer and neurodegenerative diseases
Our results have important implications for how we think about the impact of aneuploidy on human disease. Solid tumors, which are highly aneuploid, have long been deemed chaperone addicted (Neckers 2007; Workman et al. 2007; Powers et al. 2008). Eliminating HSF1, the master regulator of the heat-shock response, results in a lower incidence of tumors in mice (Dai et al. 2007). This dependence on chaperones has been attributed to the importance of chaperones for the folding of oncogenes. Our studies suggest that the aneuploid nature of tumors contributes to their dependence on chaperones such as Hsp90 for survival. We further suggest that aneuploidy could contribute to neurodegenerative diseases such as Huntington's or Alzheimer's diseases. The human brain is a naturally aneuploid organ, with one-third of fetal neurons and 10% of adult neurons being aneuploid (Rehen et al. 2001, 2005; Yurov et al. 2007a,b). Our finding that aneuploidy causes proteotoxic stress, including polyQ aggregate formation, raises the interesting possibility that aneuploidy reduces the cell's capacity to eliminate protein aggregates and/or increases the propensity for aggregate formation. Thus, their aneuploid nature may predispose neurons to protein aggregation diseases. Interestingly, mice chimeric for trisomy 16 (one of the mouse models of Down's syndrome) have been previously associated with increased susceptibility and poor prognosis when injected with the Scrapie prion protein (Epstein et al. 1991). Further studies of the proteotoxicity associated with aneuploidy could therefore provide important insights into tumorigenesis and neurodegenerative diseases and may even pave the way for the development of novel treatments for these diseases.
Materials and methods
Strains and plasmids
The strains used in this study are described in Supplemental Table S1 and are derivatives of W303. Strains were constructed using PCR-based methods described by Longtine et al. (1998). The generation of disomic strains has been described previously (Torres et al. 2007). Karyotypes of all disomic and trisomic strains were confirmed by comparative genome hybridization (Torres et al. 2007). The YACs used in this study have been previously described (Foote et al. 1989). The pGAL-VHL-GFP fusion is described in Kaganovich et al. (2008). GAL-HTT1(17aa)-Flag-25Q-CFP, GAL-HTT1(17aa)-Flag-46Q-CFP, and GAL-HTT1(17aa)-Flag-72Q-CFP are described in Duennwald et al. (2006).
Analysis of endogenous protein aggregates in disomic yeast strains
For analysis of endogenous aggregates, strains carrying an Hsp104-eGFP fusion were grown in YEPD medium. Exponentially growing cells were fixed in 3.7% formaldehyde by adding 0.1 mL of 37% formaldehyde to 1 mL of cells. Cells were then permeabilized in 1% Triton/potassium phosphate, washed, and resuspended in KPi/sorbitol. The percentage of cells harboring Hsp104-eGFP foci was determined in at least 100 cells per sample. Foci were defined as GFP dots that were visible without the aid of a camera.
Analysis of endogenous aggregates in the progeny of diploid and triploid meioses
Diploid and triploid cells were sporulated. Tetrads were dissected on YEPD plates. Colonies that grew up were diluted in water, and Hsp104-eGFP foci were analyzed as described above in at least 100 cells per colony. We note that since this analysis relied on colony growth, the most severe aneuploids that could not form colonies could not be analyzed.
Analysis of endogenous aggregates upon chromosome missegregation
Haploid strains harboring the temperature-sensitive ndc10-1 or ipl1-321 alleles were arrested in YEPD + 10 μg/mL α-factor at room temperature. After 90 min, 5 μg/mL α-factor was added. One-hundred-eighty minutes after the initial α-factor addition, cells were washed and released into prewarmed (30°C) YEPD. To half of the culture, 15 μg/mL nocodazole was added. Hsp104-eGFP foci were analyzed 3 h after release from the G1 arrest as described above.
High-temperature adaptation time courses
Cells were grown to exponential phase in YEPD at 25°C. Cells were collected by filtration and resuspended in prewarmed (37°C) YEPD. Samples were taken at the indicated times (0 h is immediately before shifting temperature). Hsp104-eGFP foci were counted in at least 100 cells per time point as described above.
Analysis of cellular protein quality-control using the VHL-GFP reporter
Strains deleted for the multidrug transporter PDR5 harboring the GAL-VHL-GFP fusion were grown at 25°C in YEP 2% raffinose and 2% galactose to OD600 = 0.2 and split in two. Half of the culture was maintained at 25°C, and the other half was shifted to 37°C. Samples were taken 2 h later and fixed with 3.7% formaldehyde, and the percentage of cells harboring VHL-GFP foci was determined. GFP foci were counted without the aid of a camera, and any cell with a visible focus was counted as a cell harboring a focus. At least 300 cells were counted for cultures grown at 25°C, and at least 100 cells were counted for cultures grown at 37°C.
Analysis of VHL aggregates in the progeny of diploid and triploid meioses
Diploid and triploid cells were sporulated, and tetrads were dissected on plates lacking leucine and containing 2% raffinose and 2% galactose. Colonies were resuspended in water, and the percentage of cells with VHL-GFP foci was determined as described above. As this analysis relied on colony growth, the most severe aneuploids that could not form colonies could not be analyzed.
Effects of radicicol on the growth rate of disomic yeast strains
Disomes deleted for the multidrug transporter PDR5 were inoculated at OD600 = 0.1 in YEPD in 96-well plates either lacking or containing 70 μM radicicol in freshly made medium. OD600 was measured every 15 min on a plate reader (Synergy2, Biotek) for 24 h. Doubling times were calculated using the exponential growth phase of each culture.
Assessing v-src and Htt1-polyQ glutamine toxicity
Tenfold serial dilutions were prepared and spotted onto the appropriate medium: medium lacking uracil and containing either 2% glucose or 2% raffinose and 2% galactose for GAL-v-src, GAL-c-src-containing strains, and YEPD or YEP 2% raffinose and 2% galactose for GAL-HTT1(17aa)-Flag-nQ-CFP-harboring strains. Plates were imaged after 3 d of growth at 25°C.
Western blot analyses
Cells were harvested by adding an equal volume of 10% trichloroacetic acid to the cell culture and were incubated on ice for at least 20 min. Cells were then washed with 1.5 mL of acetone. The dried pellet was resuspended in 100 μL of protein breakage buffer (50 mM Tris at pH 7.5, 1 mM EDTA, 2.75 mM DTT, Roche Complete protease inhibitor, used per the manufacturer's instructions). One-hundred microliters of glass beads was added, and the cells were broken by beating for 2.5 min on a Biospec minibead beater. Fifty microliters of 3× SDS sample buffer was added, and the samples were boiled for 5 min, then centrifuged for 5 min. An equal volume of lysate was loaded onto a 10% SDS–polyacrylamide gel, electrophoresed, and transferred to a nitrocellulose membrane. Hsp104-eGFP was detected using a mouse anti-GFP antibody (JL-8, Clontech) at a 1:1000 dilution. Pgk1 was detected using a mouse anti-Pgk1 antibody (A-6457, Molecular Probes) at a 1:5000 dilution. Ssa1-3HA, Sse2-3HA, and Hsp42-3HA were detected using a mouse anti-HA antibody (HA.11, Covance) at a 1:1000 dilution. Total phosphotyrosine levels were detected using a mouse anti-phosphotyrosine antibody (4G10, Millipore) at a 1:1000 dilution. The secondary antibody was a sheep anti-mouse antibody coupled to horseradish peroxidase (NA931, GE Healthcare) and was used at a 1:2000 dilution. Kar2 was detected using a rabbit anti-Kar2 antibody at a 1:200,000 dilution, followed by donkey anti-rabbit antibody coupled to horseradish peroxidase (NA9340, GE Healthcare) used at a 1:2000 dilution. Bands were detected using Amersham ECL Plus substrate according to the manufacturer's instructions.
Determination of Sup35 function
The GFP readthrough assay to assess Sup35 function was performed essentially as described previously (Tyedmers et al. 2008). Briefly, strains were streaked from frozen stocks on selective medium (−His, G418) and allowed to grow to single colonies for 3 d. Single colonies were then resuspended in 200 μL of SC, and 100 μL was analyzed immediately by flow cytometry to determine the percent of GFP-positive cells (n > 10,000). The remaining 100 μL was used to inoculate 3-mL SC cultures, which were grown at room temperature and maintained in exponential phase. Samples were taken for flow cytometry analysis after 8 and 24 h of growth, and the percentage of GFP-positive cells was determined.
Analysis of polyQ aggregates
Strains harboring the GAL-HTT1(17aa)-Flag-46Q-CFP construct were grown at 25°C in YEP 2% raffinose and 2% galactose to OD600 = 0.4. Samples were taken and fixed with 3.7% formaldehyde, and the percentage of cells harboring polyQ-CFP foci was determined. CFP foci were counted without the aid of a camera, and any cell with a visible focus was counted as a cell harboring a focus. At least 100 were counted for each replicate.
Cell imaging
For the analysis of Hsp104-eGFP and VHL-GFP foci, cells were fixed in 3.7% formaldehyde by adding 0.1 mL of 37% formaldehyde to 1 mL of cells. Cells were then permeabilized in 1% Triton/potassium phosphate, washed, and resuspended in DAPI /KPi/sorbitol. Microscopy was performed using a Zeiss Axioplan 2 microscope with a Hamamatsu OCRA-ER digital camera. Image analysis was performed with Openlab 4.0.2 software.
Northern blot analysis
Total RNA was purified by phenol extraction and isopropanol precipitation as described in Hochwagen et al. (2005). Ten micrograms of total RNA was separated on a 1.1% agarose gel containing 6% formaldehyde and 40 mM MOPS (pH 7.0). Gels were blotted in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate at pH 7.0) onto Hybond-XL membranes (Amersham Biosciences). Blots were probed overnight with radioactively labeled HAC1 or GAL-HTT1(17aa)-Flag-nQ-CFP-specific probes.
Heat-shock microarrays
Strains were grown in YEPD at 25°C to OD600 = 0.2, collected by filtration, and shifted to prewarmed (37°C) YEPD. Samples were collected 0, 5, 15, and 30 min after temperature shift. Wild type grown at 25°C was used as a reference for all samples. RNA preparation and microarrays were performed as described previously (Torres et al. 2007). Briefly, total RNA was ethanol-precipitated and further purified over RNeasy columns (Qiagen). RNA (325 ng) was labeled using the Agilent Low RNA Input fluorescent linear amplification kit. Reactions were performed as directed except half the recommended reaction volume and one-quarter the recommended Cy-CTP amount were used. Dye incorporation and yield were measured with a NanoDrop spectrophotometer. Equal amounts of differentially labeled control and sample cDNA were combined such that each sample contained at least 2.5 pmol of dye. Samples were fragmented, combined with hybridization buffer, boiled for 5 min, and applied to a microarray consisting of 60-mer probes for each yeast ORF (Agilent). Microarrays were rotated for 17 h at 60°C in a hybridization oven (Agilent). Arrays were then washed according to the Agilent SSPE wash protocol and scanned on an Agilent scanner. The image was processed using the default settings with Agilent Feature Extraction software. All data analysis was performed using the resulting log2 ratio data and filtered for spots called as significantly over background in at least one channel. Data were normalized to account for the extra chromosomes as previously described (Torres et al. 2007). Data were mined for genes that comprise the heat-shock response (Gasch et al. 2000). The full data set has been deposited in the Gene Expression Omnibus under the accession number GSE40073.
Acknowledgments
We thank B. Vincent, D. Jarosz, and M. Duennwald for reagents; J. Boulin for technical assistance; and S. Lindquist, F. Solomon, and members of the Amon laboratory for suggestions and critical reading of this manuscript. This work was supported by the National Institutes of Health (GM056800 to A.A) and a Ludwig Fund Graduate Fellowship (to A.O.). A.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.207407.112.
References
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36: 744–749 [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol 9: 3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. 2010. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 39: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B, Ness F, Tuite M 2003. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics 165: 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S 2006. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci 103: 11045–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M, et al. 1993. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet 4: 387–392 [DOI] [PubMed] [Google Scholar]
- Epstein CJ, Foster DB, DeArmond SJ, Prusiner SB 1991. Acceleration of scrapie in trisomy 16–diploid aggregation chimeras. Ann Neurol 29: 95–97 [DOI] [PubMed] [Google Scholar]
- Foote SJ, Thompson JK, Cowman AF, Kemp DJ 1989. Amplification of the multidrug resistance gene in some chloroquine-resistant isolates of P. falciparum. Cell 57: 921–930 [DOI] [PubMed] [Google Scholar]
- Franzosa EA, Albanese V, Frydman J, Xia Y, McClellan AJ 2011. Heterozygous yeast deletion collection screens reveal essential targets of Hsp90. PLoS ONE 6: e28211 doi: 10.1371/journal.pone.0028211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci 108: 680–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Clavaguera F, Tolnay M 2010. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci 33: 317–325 [DOI] [PubMed] [Google Scholar]
- Halfmann R, Alberti S, Lindquist S 2010. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol 20: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D 2006. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127: 99–111 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M 2011. Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332 [DOI] [PubMed] [Google Scholar]
- Hochwagen A, Wrobel G, Cartron M, Demougin P, Niederhauser-Wiederkehr C, Boselli MG, Primig M, Amon A 2005. Novel response to microtubule perturbation in meiosis. Mol Cell Biol 25: 4767–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck SA, Singh S, Cyr DM 2012. Cellular responses to misfolded proteins and protein aggregates. Methods Mol Biol 832: 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72: 971–983 [DOI] [PubMed] [Google Scholar]
- Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K 2003. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J 22: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Yahara I, Lindquist S 1995. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science 268: 1362–1365 [DOI] [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P 2009. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol 185: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Sherman F 1979. Extrachromosomal ψ+ determinant suppresses nonsense mutations in yeast. J Bacteriol 139: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Lindquist S 1999. Oligopeptide-repeat expansions modulate ‘protein-only' inheritance in yeast. Nature 400: 573–576 [DOI] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, Nystrom T 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140: 257–267 [DOI] [PubMed] [Google Scholar]
- Longtine MS, Fares H, Pringle JR 1998. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol 143: 719–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Scott MD, Frydman J 2005. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121: 739–748 [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J 2007. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131: 121–135 [DOI] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S 1995. Mutational analysis of Hsp90 function: Interactions with a steroid receptor and a protein kinase. Mol Cell Biol 15: 3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S 1997. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci 94: 12949–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L 2007. Heat shock protein 90: The cancer chaperone. J Biosci 32: 517–530 [DOI] [PubMed] [Google Scholar]
- Niwa O, Tange Y, Kurabayashi A 2006. Growth arrest and chromosome instability in aneuploid yeast. Yeast 23: 937–950 [DOI] [PubMed] [Google Scholar]
- Parry EM, Cox BS 1970. The tolerance of aneuploidy in yeast. Genet Res 16: 333–340 [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A, Besche HC, Goldberg AL 2009. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell 36: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MV, Clarke PA, Workman P 2008. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 14: 250–262 [DOI] [PubMed] [Google Scholar]
- Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, Chun J 2001. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci 98: 13361–13366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, et al. 2005. Constitutional aneuploidy in the normal human brain. J Neurosci 25: 2176–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A 2011. Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A 2012. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci 109: 12644–12649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Charles J, Hamilton ML, Petes TD 2010. Meiotic chromosome segregation in triploid strains of Saccharomyces cerevisiae. Genetics 186: 537–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 8: 608 doi: 10.1038/msb.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, Amon A 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 180: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924 [DOI] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A 2008. Aneuploidy: Cells losing their balance. Genetics 179: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S 2008. Prion switching in response to environmental stress. PLoS Biol 6: e294 doi: 10.1371/journal.pbio.0060294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B 2010. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 11: 777–788 [DOI] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW 2006. Does aneuploidy cause cancer? Curr Opin Cell Biol 18: 658–667 [DOI] [PubMed] [Google Scholar]
- Williams BR, Amon A 2009. Aneuploidy: Cancer's fatal flaw? Cancer Res 69: 5289–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P, Burrows F, Neckers L, Rosen N 2007. Drugging the cancer chaperone HSP90: Combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann N Y Acad Sci 1113: 202–216 [DOI] [PubMed] [Google Scholar]
- Xu Y, Lindquist S 1993. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci 90: 7074–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Kutsev SI, Pellestor F, Beresheva AK, Demidova IA, Kravets VS, et al. 2007a. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS ONE 2: e558 doi: 10.1371/journal.pone.0000558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurov YB, Vorsanova SG, Iourov IY, Demidova IA, Beresheva AK, Kravetz VS, Monakhov VV, Kolotii AD, Voinova-Ulas VY, Gorbachevskaya NL 2007b. Unexplained autism is frequently associated with low-level mosaic aneuploidy. J Med Genet 44: 521–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Pavelka N, Bradford WD, Rancati G, Li R 2012. Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet 8: e1002719 doi: 10.1371/journal.pgen.1002719 [DOI] [PMC free article] [PubMed] [Google Scholar]