Green and colleagues perform a global analysis of circadian-controlled poly(A) tails and identify hundreds of mRNAs that display dynamic rhythmic polyadenylation states. They identify three distinct classes of mRNAs with rhythmic poly(A) tails. Interestingly, class III mRNAs are controlled not by transcription, but by rhythmic cytoplasmic polyadenylation, and are regulated by the components of the cytoplasmic polyadenylation machinery, CPEB2 in particular, which are themselves rhythmically expressed during the day.
Keywords: circadian, polyadenylation, deadenylation, post-transcriptional mechanism
Abstract
Poly(A) tails are 3′ modifications of eukaryotic mRNAs that are important in the control of translation and mRNA stability. We identified hundreds of mouse liver mRNAs that exhibit robust circadian rhythms in the length of their poly(A) tails. Approximately 80% of these are primarily the result of nuclear adenylation coupled with rhythmic transcription. However, unique decay kinetics distinguish these mRNAs from other mRNAs that are transcribed rhythmically but do not exhibit poly(A) tail rhythms. The remaining 20% are uncoupled from transcription and exhibit poly(A) tail rhythms even though the steady-state mRNA levels are not rhythmic. These are under the control of rhythmic cytoplasmic polyadenylation, regulated at least in some cases by cytoplasmic polyadenylation element-binding proteins (CPEBs). Importantly, we found that the rhythmicity in poly(A) tail length is closely correlated with rhythmic protein expression, with a several-hour delay between the time of longest tail and the time of highest protein level. Our study demonstrates that the circadian clock regulates the dynamic polyadenylation status of mRNAs, which can result in rhythmic protein expression independent of the steady-state levels of the message.
Dynamic control of gene expression is a hallmark of the circadian system, in which cell-autonomous circadian clocks drive thousands of rhythmic output genes that ultimately produce daily rhythms in many types of physiology and behavior (Akhtar et al. 2002; Panda et al. 2002; Storch et al. 2002; Ueda et al. 2002; Duffield 2003; Welsh et al. 2004; Reddy et al. 2006; Koike et al. 2012). Circadian gene expression has been studied largely at the level of transcriptional control, and steady-state mRNA levels have been the primary readouts, due in part to the ease of making these measurements, particularly at the genome-wide level. However, recent findings have challenged the current transcription-centric model. For example, the clock is quite insensitive to large fluctuations of transcription rate (Dibner et al. 2009), and in mouse livers, almost 50% of the rhythmic proteins do not have rhythmic steady-state mRNA levels (Reddy et al. 2006). Moreover, circadian rhythms can exist in red blood cells devoid of nuclei (O'Neill and Reddy 2011; O'Neill et al. 2011). Finally, recent RNA sequencing (RNA-seq) experiments have demonstrated that 78% of mouse liver mRNAs that have rhythms in their steady-state levels are not transcribed rhythmically (Koike et al. 2012). These all suggest that regulatory mechanisms beyond transcription can also drive rhythmic physiology.
Although transcription drives mRNA synthesis, protein expression patterns also reflect regulation at the level of RNA splicing, 3′ end cleavage and polyadenylation, nuclear export, RNA stability, and translation. Post-transcriptional processes begin even while the RNA is still being transcribed and occur in a complex and highly coordinated manner, involving large complexes of proteins (Pawlicki and Steitz 2010). Over the last decade, the importance of post-transcriptional regulation has become clear with the discovery of sophisticated mechanisms, including many RNA-binding proteins, ribonucleases with specific functions, and the extensive machinery that conducts microRNA-mediated control of mRNA stability and translation to ultimately control gene expression. Although significant progress has been made in this area, understanding of post-transcriptional mechanisms still lags behind that of transcriptional processes.
One important post-transcriptional regulatory mechanism is control of the poly(A) tail length of mRNAs. These tracts of adenosines at the 3′ end of most eukaryotic mRNAs are critical for controlling translatability and stability. Poly(A) tail length regulation has been shown to play a critical role in some biological processes, such as oocyte maturation, mitotic cell cycle progression, cellular senescence, and synaptic plasticity (Gebauer et al. 1994; Groisman et al. 2002, 2006; Huang et al. 2002, 2006; Novoa et al. 2010). Mutations in the canonical polyadenylation signal hexanucleotide and misregulation of proteins involved in mRNA 3′ end processing lead to numerous diseases in humans (Danckwardt et al. 2008). This type of regulation allows cells to alter protein levels rapidly without requiring de novo transcript synthesis or processing.
Changes in poly(A) tail length can occur throughout the lifetime of an mRNA. In the nucleus, processing of 3′ ends of newly synthesized mRNAs is accompanied by the addition of a long poly(A) tail of ∼250 nucleotides (nt) (Kuhn and Wahle 2004). Upon export out of the nucleus, cytoplasmic poly(A)-binding protein (PABPC) binds to the tail and stabilizes the mRNA. PABPC also binds to the translation initiation factor eIF4G, which in turn binds to the cap-binding protein eIF4E to form a “closed-loop” circular structure, thereby promoting translation initiation (Tarun and Sachs 1996). Cytoplasmic deadenylation can result in poly(A) tail shortening to promote translational silencing and mRNA degradation. The deadenylation process is normally biphasic (Brown and Sachs 1998; Yamashita et al. 2005), in which long poly(A) tails are first gradually shortened by the PAN2–PAN3 complex to ∼100 nt, followed by additional shortening by the CCR4–CAF1 complex to 8–12 nt in mammals. Since deadenylation is a rate-limiting step for mRNA degradation and the rate of deadenylation is mRNA-specific, the regulation of poly(A) tail length is considered to be mRNA-specific and is an important determinant of mRNA half-life. Alternatively, poly(A) tails can be elongated in the cytoplasm by a process called cytoplasmic polyadenylation (Richter 2007). Much of the biochemistry of this process has been elucidated in Xenopus early development, although it is also conserved in other species, including Drosophila and mice (Gebauer et al. 1994; Wu et al. 1998; Groisman et al. 2002; Huang et al. 2002, 2006; Alarcon et al. 2004; Coll et al. 2010; Novoa et al. 2010). In the best-studied examples, the cytoplasmic polyadenylation element-binding protein (CPEB or CPEB1) has been shown to bind to cytoplasmic polyadenylation elements (CPEs) in the 3′ untranslated region (UTR) of the target mRNA, resulting in the recruitment of a cytoplasmic polyadenylation complex consisting of the cytoplasmic poly(A) polymerase GLD2 and other proteins to elongate the poly(A) tail and activate translation (Richter 2007). Phosphorylation of CPEB1 regulates the relative activity of the deadenylase and poly(A) polymerase, thereby controlling the tail length (Richter 2007). Much less is known about the function of three other CPEBs (CPEB2, CPEB3, and CPEB4), which are more closely related to each other than to CPEB1 (Huang et al. 2006). CPEB2, CPEB3, and CPEB4 have been reported to bind with higher affinity to non-CPE sequences (although they may also bind CPEs with lower affinity) and have N-terminal domains that are not conserved with CPEB1, including the lack of the regulatory phosphorylation site that is present on CPEB1 (Huang et al. 2006; Hagele et al. 2009; Novoa et al. 2010; Hosoda et al. 2011). While mechanisms of transcriptionally coupled poly(A) tail length control in the nucleus have been extensively studied and are considered to be default modifications, the mechanisms in the cytoplasm are much less well understood, especially in mammalian somatic cells, despite the fact that this mechanism can add flexibility and variety to overall gene expression independent from nuclear transcription/polyadenylation.
Some evidence suggests that poly(A) tail length regulation may take part in controlling circadian-regulated rhythmic gene expression. For example, the expression of a deadenylase, Nocturnin (gene name Ccnr4l), exhibits robustly rhythmic expression patterns (Wang et al. 2001; Garbarino-Pico et al. 2007; Kojima et al. 2010), and daily variations in poly(A) tail length have been previously reported for two mRNAs (Robinson et al. 1988; Gerstner et al. 2012). The extent of such regulation or the mechanism by which the circadian clock exerts such control is not known. To address this question, we performed a genome-wide screen to identify mRNAs that have diurnal rhythmicity in their poly(A) tail length. Our results demonstrate that rhythmic changes in poly(A) tail lengths are under the control of the circadian clock in both the nucleus and cytoplasm, including a novel role for CPEB2. Most importantly, the rhythmic poly(A) tail lengths correlate strongly with rhythmic protein expression, regardless of the steady-state mRNA expression levels or patterns.
Results
Identification of mRNAs with rhythmic poly(A) tail lengths
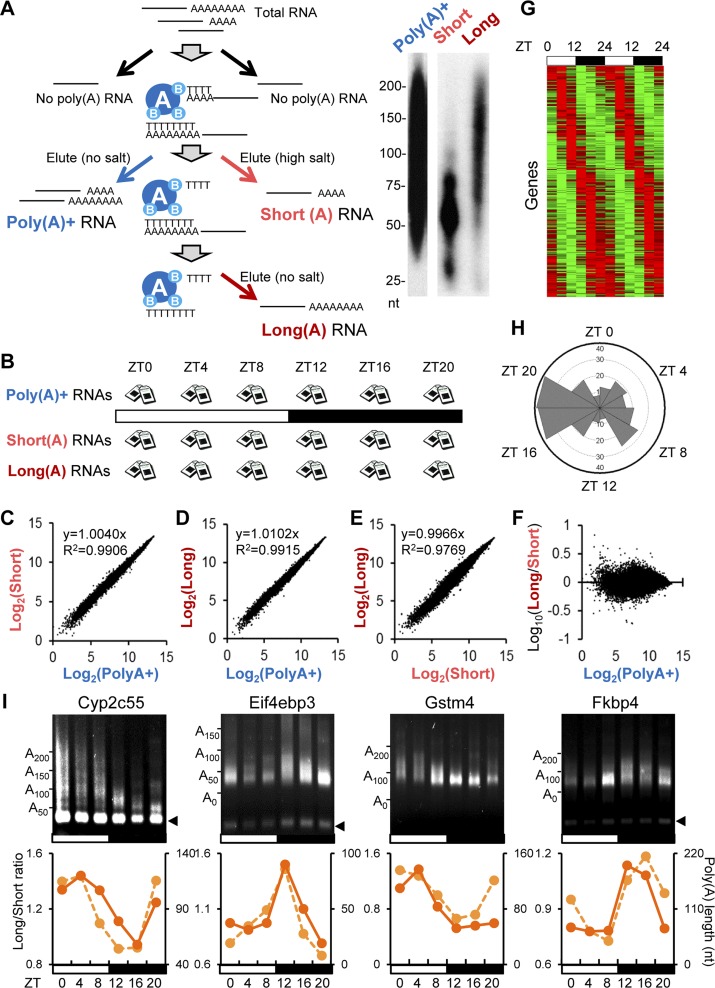
In order to determine the dynamics of mRNA poly(A) tail length throughout the circadian cycle, we isolated mRNAs from mouse livers collected every 4 h over a 24-h period in 12-h light:12-h dark (12L:12D) cycles, a condition that represents normal life. Peripheral tissues such as the liver do not receive light input directly, and these rhythms should reflect circadian control mechanisms from both the endogenous liver clocks and any rhythmic systemic signals that are entrained to the environment (Kornmann et al. 2007). These mRNAs were fractionated by oligo(dT) chromatography (Meijer et al. 2007) into populations that contained either short (<60-nt) or long (60- to 250-nt) poly(A) tails by varying the salt concentrations in the elution step (Fig. 1A). We also isolated nonfractionated mRNAs [the total poly(A)+ pool] for each time point as a reference. These RNA samples were subjected to microarray analysis using the Affymetrix Gene ST 1.0 chips (Fig. 1B) that do not require oligo(dT)-based sample preparation to avoid bias from the different poly(A) lengths in the different fractionated samples.
Figure 1.
A genome-wide screen identified mRNAs with rhythms in poly(A) tail length in mouse livers. (A) Oligo(dT) chromatography was used to separate total RNA into fractions with either short or long poly(A) tails by varying salt concentrations in the elution. Total polyA+ (nonfractionated) RNAs were also isolated as a reference. The poly(A) tail length (nucleotides) of each fraction was tested by 3′ end label assay. Representative gel images are shown on the right. (B) Shown is the tissue sampling scheme in which mouse livers (n = 3 per time point) were harvested every 4 h under 12L:12D conditions, and total RNA was extracted from each sample followed by RNA fractionation as depicted in A. Following the fractionation, each RNA sample was subjected to microarray analysis. (C–F) Comparison of the signal intensity at ZT0 of each mRNA in the polyA+ samples versus the short-tailed samples (C), in the polyA+ samples versus the long-tailed samples (D), in the short-tailed samples versus the long-tailed samples (E), and in the polyA+ samples versus the log10(long/short ratio) (F). The degree of correlation is shown in the top left corner of each graph. The data for other time points are shown in Supplemental Figure S1. (G) Phase-sorted heat map of the long/short ratio of PAR mRNAs. Red or green indicates higher or lower values in the long/short ratio (longer vs. shorter tails), respectively. Each line represents one mRNA. The heat map is double-plotted for easier visualization of rhythmicity. (H) Peak time distribution of PAR mRNAs. Peak time of poly(A) tail rhythms was calculated based on the long/short ratio of PAR mRNAs. Each wedge represents a 2-h bin. (I) Representative gel images of LM-PAT assay for PAR mRNAs. Each lane consists of pooled samples (n = 3 for each time point). Arrowheads on the right indicate the fragment derived from restriction enzyme treatment for verification of PCR specificity (see the Materials and Methods for details). Graphs are the comparison between the long/short ratio from the microarray (left Y-axis, dotted line) and the poly(A) tail length calculated from the LM-PAT assay (right X-axis, solid line).
Analysis of the nonfractionated [total poly(A)+] mRNAs identified 699 mRNAs (6.9% of all expressed genes) with statistically rhythmic steady-state mRNA expression levels (Supplemental Fig. S1A; Supplemental Table S1). These will be referred to as rhythmically expressed at the steady-state level (RESS) mRNAs throughout. This group includes mRNAs from clock genes such as Bmal1, Clock, Period 1, Period 2, and Period 3 as well as mRNAs from many other clock-controlled genes (ccgs) that have previously been reported to be rhythmic in mouse livers (Akhtar et al. 2002; Panda et al. 2002; Storch et al. 2002; Ueda et al. 2002; Hughes et al. 2009). Furthermore, the distribution of phases corresponds well with those previously described (Supplemental Fig. S1B; Duffield 2003; Koike et al. 2012). Therefore, our total steady-state mRNA profiles from the Affymetrix Gene ST 1.0 chips are consistent with previously published analyses.
Since microarray data only provide information on the expression level of each mRNA within each sample, this does not directly reveal the changes in poly(A) tail length. In order to determine differences in poly(A) tail length, it was necessary to normalize for these rhythmic changes in expression level, as seen in the total poly(A)+ mRNA samples. The correlation between poly(A)+ samples with either short or long tail samples at each time point was quite strong, reflecting the overall expression levels of these mRNAs within each fraction (Fig. 1C,D; Supplemental Fig. S2A,B); however, the relationship between short and long tail samples, although still strongly correlated, was less strong (Fig. 1E; Supplemental Fig. S2C), revealing the differences in poly(A) tail length of each gene. We thus defined the status of the poly(A) tail for each mRNA as the ratio of expression level from the “long tail” and “short tail” fractions (the “long/short” ratio), and indeed, this normalization largely eliminated the correlation of the expression levels in each fraction (Fig. 1F; Supplemental Fig. S2D). Statistical analysis identified 237 mRNAs (2.3% of expressed mRNAs) that exhibited statistically significant rhythmicity in the long/short ratio (Fig. 1G; Supplemental Table S2). These “poly(A) rhythmic” (PAR) mRNAs include mRNAs with peak tail lengths at all phases of the daily cycle but with disproportionally higher numbers of mRNAs with peak long/short ratios during the night at ZT16–ZT20 (Zeitgeber time, where ZT0 is defined as time [hours] of lights on and ZT12 is defined as time of lights off) (Fig. 1H).
In order to independently validate our microarray results, we measured the poly(A) tail lengths of PAR mRNAs using the ligation-mediated poly(A) tail length (LM-PAT) assay (Salles and Strickland 1999). Due to the heterogeneous nature of poly(A) tail lengths, signals from LM-PAT assays are often detected as smears. Therefore, the average of the range of sizes detected by densitometry analysis was used to assign poly(A) tail length from this assay. The LM-PAT assays revealed that the poly(A) tail length of the 38 tested mRNAs, including the four shown in Figure 1I (other examples are shown in Fig. 5, below; Supplemental Figs. S3, S4, S7), exhibited clear diurnal changes in their poly(A) tail length, with the phases of the poly(A) length fluctuations well-matched to the long/short ratios obtained from microarray study (Figs. 1I, 5 [below]). We therefore concluded that the long/short ratio provided reliable quantitative information on the relative poly(A) tail length.
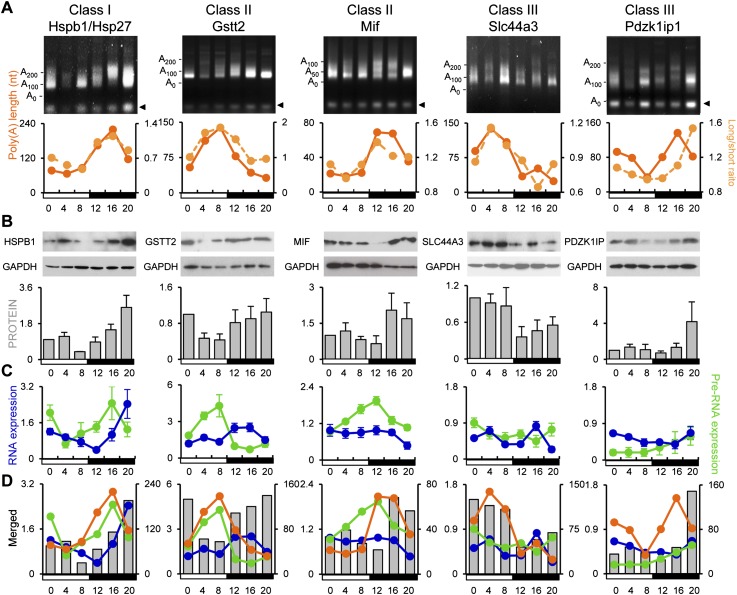
Figure 5.
Length of the poly(A) tail correlates with protein expression. The patterns of poly(A) tail length (orange solid line) and long/short ratio obtained from microarray data (orange dotted line) (A); protein expression (B); and pre-mRNA (green line) and steady-state mRNA levels (blue line) of Hspb1 (class I), Gstt2 (class II), Mif (class II), Slc44a3 (class III), and Pdzk1ip1 (class III) (C) are depicted. The error bars are omitted on the composite graphs in D for simplicity. Relative protein expression level was calculated by ImageJ software, and the expression at ZT0 was set as 1. The arrowhead in A indicates the fragment derived from restriction enzyme treatment for verification of PCR specificity (see the Materials and Methods for details).
LM-PAT assays also displayed multiple differently sized PCR products for some mRNAs (for example, Hsd17b6 in Supplemental Fig. S4), and the phase of poly(A) fluctuation appeared synchronized. These are probably derived from alternative polyadenylation, which occur in almost 30% of all genes (Di Giammartino et al. 2011), although our microarray analysis would not discriminate these mRNAs.
PAR mRNAs can be categorized into three groups
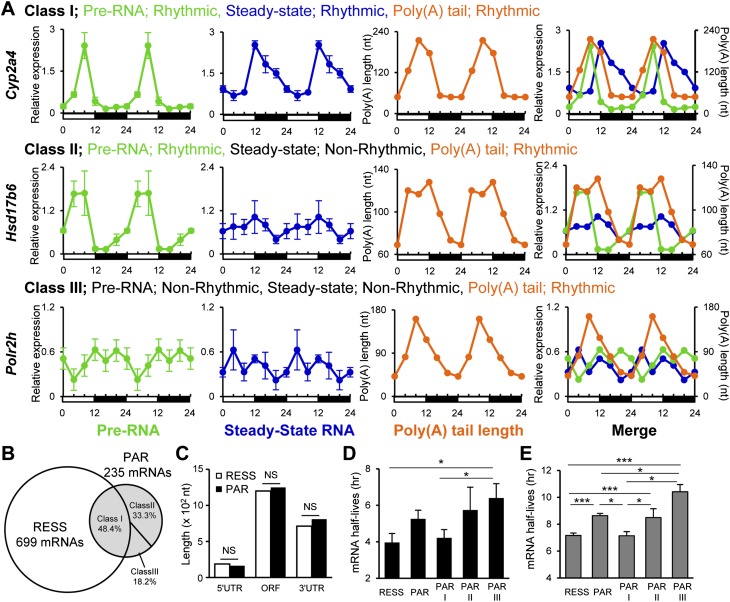
Since polyadenylation is often coupled with transcription, we analyzed whether these rhythmic poly(A) tails were the result of rhythmic transcription. Of PAR mRNAs, 43.4% had rhythmic steady-state mRNA expression levels, while the remaining 57.6% did not, based on the rhythmicity calculation by Circwave (available at http://hutlab.nl). Since the steady-state mRNA level reflects both RNA synthesis and degradation rates, we randomly picked 65 PAR mRNAs and measured their pre-RNA expression levels using intron-specific primers throughout the day as an indicator of active transcription. Rhythmic expression at the pre-RNA levels was observed in 81.5% of the tested PAR mRNAs, including all mRNAs that showed rhythmicity at the steady-state level. Based on the pre-mRNA and steady-state mRNA profiles, the PAR mRNAs were categorized into three classes: Class I PAR mRNAs (49.2%) were rhythmic in their poly(A) tail length and pre-RNA and steady-state mRNA levels, class II PAR mRNAs (32.3%) were rhythmic in their poly(A) tail length and pre-RNA expression but not in steady-state mRNA levels, and class III PAR mRNAs (18.5%) were rhythmic in their poly(A) tail length rhythms but not in pre-RNA or steady-state mRNA levels (Fig. 2A,B; Supplemental Fig. S3).
Figure 2.
PAR mRNAs fall into three classes. (A) Examples of the expression pattern of pre-RNA, steady-state RNA, and poly(A) tail length of an mRNA belonging to each class:Cyp2a4 (class I), Hsd17b6 (class II), and Polr2h (class III). The pre-mRNA (green) and steady-state mRNA levels (blue) (mean ± SEM, n = 3 for each time point) were measured by quantitative PCR (qPCR). Poly(A) tail length (orange) was calculated from LM-PAT assays of pooled samples (n = 3 for each time point). All of the graphs are double-plotted for easier visualization of rhythmicity, and error bars are not depicted on merged graphs for simplicity. Rhythmicity was determined by Circwave version 1.4. (B) Venn diagram showing the relationship of RESS and the three classes of PAR mRNAs. (C) The comparison of mean UTR and ORF lengths between RESS and PAR mRNAs. (NS) Not significant (Student's t-test). (D) The mRNA half-lives of PAR mRNAs (mean ± SEM) measured in AML12 cells. mRNAs were randomly picked from each category (RESS, n = 26; PAR, n = 24; PAR class I, n = 11; PAR class II, n = 5; and PAR class III, n = 8). (E) In silico analysis of mRNA half-lives of PAR mRNAs (mean ± SEM) using genome-wide mRNA half-life data from Sharova et al. (2009). All of the mRNAs available from database were included (RESS, n = 500; PAR, n = 137; PAR class I, n = 16; PAR class II, n = 13; and PAR class III, n = 8). (***) P < 0.0005, (*) P < 0.05 (Student's t-test).
In yeast, there is a correlation between poly(A) tail length and UTR length, mRNA length, and ORF size but not with mRNA stability (Beilharz and Preiss 2007). However, we did not observe any difference in the length of 5′ or 3′ UTRs or in ORF size between PAR and RESS mRNAs (Fig. 2C). We also did not find enrichment of any specific RNA sequence in the 5′ UTR, ORF, or 3′ UTR that could be responsible for poly(A) rhythmicity. However, we found significant differences in mRNA half-lives between the different PAR classes in the mouse hepatoma cell line AML12; class III mRNAs were the most stable, followed by class II mRNAs, with class I mRNAs the least stable and statistically indistinguishable from the RESS mRNAs (Fig. 2D). We also examined the half-lives of the RESS and PAR mRNAs using a genome-wide database of mRNA half-life measurements from mouse embryonic stem cells (Sharova et al. 2009). The difference in mRNA half-lives from this large-scale in silico analysis was consistent with our experimentally measured mRNA half-life differences, with class III mRNAs showing the longest half-lives, and class I mRNAs showing the shortest (Fig. 2E). These differences in mRNA stability correlate well with whether or not the steady-state mRNA levels are rhythmic but do not account for the presence or absence of poly(A) tail length rhythms.
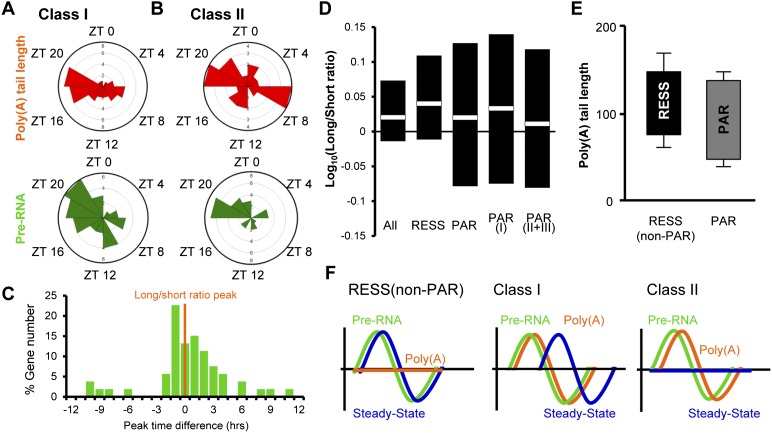
Class I and II PAR mRNAs have peak tail lengths in phase with their rhythmic pre-mRNA synthesis
The mechanisms to control the poly(A) rhythmicity of the class I/II PAR mRNAs most likely involve nuclear polyadenylation immediately following rhythmic transcription, since both classes exhibit rhythmic pre-RNA levels. In support of this idea, peak phase distributions of poly(A) and pre-RNA levels are similar (ZT18–ZT22) (Fig. 3A,B), although we also observed a second peak of poly(A) tail length during the day (ZT6–ZT8) for the class II mRNAs. Furthermore, when the peak pre-RNA level and the peak tail length were compared for each mRNA, we observed close correspondence of the phase of the longest poly(A) tails with the peak level of pre-RNA for most mRNAs tested (mean is +0.59 h) (Fig. 3C), suggesting that the rhythmic poly(A) lengths of both class I and II mRNAs reflect nuclear polyadenylation, coordinated with rhythmic transcription during classical 3′ end processing. The defining characteristics of these two classes are the differences in steady-state mRNA rhythmicity and mRNA stability (Fig. 2D,E), suggesting that the lack of rhythmicity in the class II PARs reflects the longer half-lives of these mRNAs. Indeed, theoretical analyses have shown that rhythmically expressed transcripts must have relatively short half-lives and that increased stability results in reduced rhythmicity (Wuarin et al. 1992).
Figure 3.
Rhythmic poly(A) tails of class I and II PAR mRNAs are the result of transcription-coupled poly(A) tail addition and slow deadenylation. (A,B) Peak phase distribution of the poly(A) tail length and pre-RNA expression of class I (A) or class II (B) mRNAs. (C) Distribution of peak time differences of pre-mRNA expression from the peak time of the long/short ratio of class I/II mRNAs from which the pre-mRNA level was measured. (D) The range and the average of the long/short ratio in different groups of mRNAs. The data from all time points were combined, regardless of the rhythmicity. Black bars represent the range (the top and the bottom of the black bars represent the average of the maximum and minimum of the long/short ratio of the day, respectively), while white boxes in the middle indicate the average of the ratio from all time points. (E) The range of poly(A) tails of 38 PAR and nine RESS (non-PAR) mRNAs. The top and bottom lines of each box represent the maximum and minimum poly(A) tail length (mean ± SEM). (F) Schematic representation of rhythmic characteristics of RESS (non-PAR) and class I and class II PAR mRNAs.
RESS mRNAs are rhythmic at the steady-state level and yet are distinct from class I/II PARs and do not exhibit poly(A) rhythmicity (Fig. 3F). We hypothesized that variations in the deadenylation and decay kinetics could also be different between RESS and PAR mRNAs. The average long/short ratios across all time points of RESS and class I PAR mRNAs (both of which have rhythmic steady-state mRNA levels) were indistinguishable (Fig. 3D). However, the ranges of the long/short ratios of all PAR classes were much broader than that of RESS mRNAs, especially the lower half (short tails) (Fig. 3D). This was supported by direct measurement of the tail lengths of the 38 PAR as well as nine RESS (non-PAR) mRNAs tested by the LM-PAT assays. The amplitude of poly(A) tail lengths [defined by (longest) − (shortest) poly(A) length], ranged from 122 nt ± 14 nt to 31 nt ± 7 nt in PAR mRNAs, compared with 151 nt ± 10 nt to 81 nt ± 9 nt in RESS (non-PAR) mRNAs (Fig. 3E). These data suggest that the PAR mRNAs can retain short poly(A) tails for a longer time before being degraded, whereas RESS mRNAs only exist in longer-tailed states. Therefore, we propose that the mechanism of poly(A) rhythmicity in both class I and II mRNAs results from the addition of long tails following rhythmic synthesis and subsequent deadenylation that does not cause immediate decay. This delay in decay is more pronounced in the class II mRNAs and results in the arrhythmic steady-state levels.
Even though class I PAR mRNAs make up 14.5% of the RESS mRNA population and have relatively short mRNA half-lives as well as rhythmic steady-state RNA expression (Fig. 2), the peak phase of the steady-state mRNA of class I was delayed by almost 5 h compared with that of pre-mRNA, whereas we did not observe this delay in non-PAR RESS mRNAs (Fig. 3F; Supplemental Fig. S5). It appears that rhythmic control of poly(A) tail length is somehow correlated with a delayed accumulation of steady-state mRNA and may be part of a regulatory mechanism to regulate the timing of mRNA/protein rhythmicity.
Class III mRNAs are under the control of cytoplasmic polyadenylation
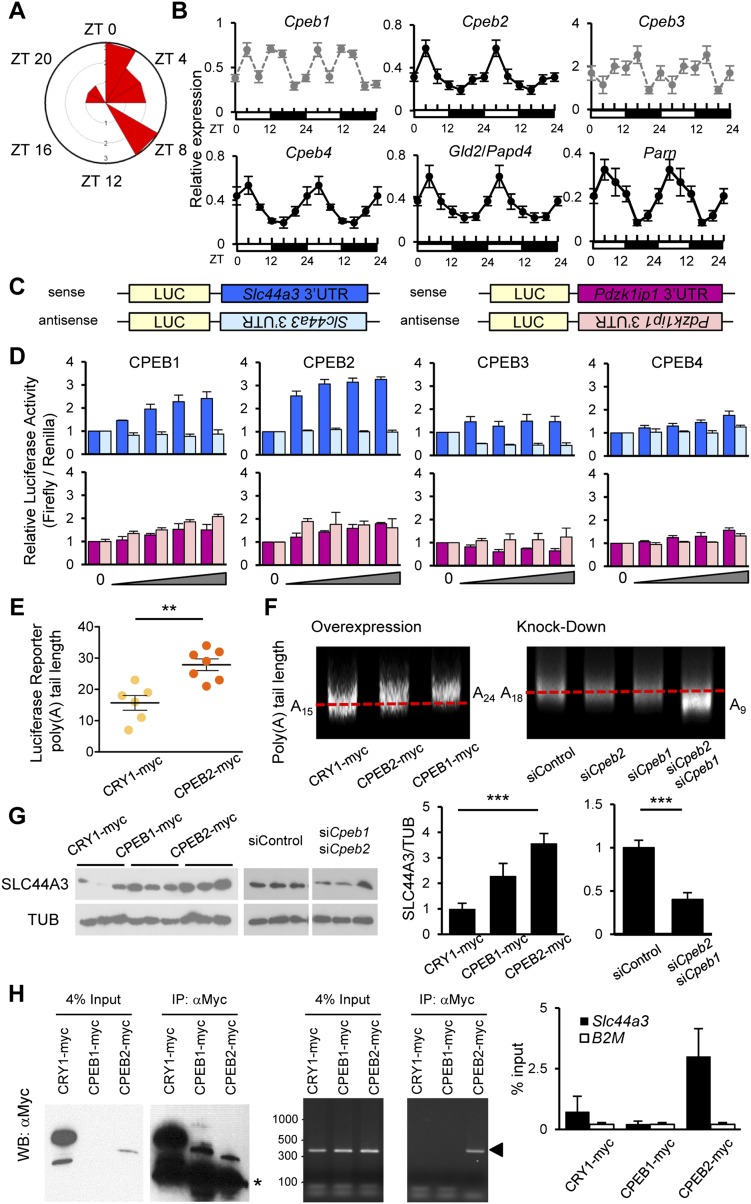
Contrary to class I/II mRNAs, class III mRNAs exhibit robust rhythmicity in their poly(A) tail length, yet are not rhythmically transcribed and have longer half-lives (Fig. 2). Therefore, class III PARs must employ transcription-independent mechanisms to control their rhythmic poly(A) tail lengths. Peak distribution analysis of class III PAR mRNAs revealed that >80% had their longest poly(A) tails during the day (Fig. 4A), which is distinct from the nighttime poly(A) rhythmic profile of class I and many of the class II mRNAs, further supporting our hypothesis. The steady-state mRNA level of several putative cytoplasmic polyadenylation machinery components in the liver, including Cpeb2, Cpeb4, Parn, and Gld2, were rhythmically expressed with phases similar to the majority of the class III PAR mRNAs, peaking in the early day (Fig. 4B). Therefore, we hypothesized that poly(A) rhythms of class III mRNAs are controlled by rhythmic cytoplasmic polyadenylation.
Figure 4.
Cytoplasmic polyadenylation regulates class III PAR mRNAs. (A) Peak phase distribution of the poly(A) tail length of PAR class III mRNAs. (B) Analysis of the steady-state mRNA expression level of components that are involved in cytoplasmic polyadenylation machinery shows that many are rhythmic. All of the graphs (mean ± SEM, n = 3 for each time point) are double-plotted for easier visualization of rhythms. Graphs with black circles and solid lines represent rhythmically expressed mRNAs, while graphs with gray circles and dotted lines indicate nonrhythmic mRNAs. Rhythmicity of each mRNA was calculated by Circwave version 1.4. (C) Schematic representation of luciferase reporter genes for Slc44a3 and Pdzk1ip1. (D) Relative luciferase activity of reporter genes harboring 3′ UTRs of Slc44a3 (top) or Pdzk1ip1 (bottom) when co-expressed with increasing doses of CPEB1–4. The expression of reporter genes without CPEB was set as 1. The color code for sense (dark bars) and antisense (light bars) is as shown in the schematic in C. (E) The poly(A) tail length of the mRNAs expressed from the luciferase reporter genes when co-expressed with Cryptochrome 1 (CRY1; negative control) or CPEB2 were measured by circularization RT–PCR followed by sequencing of several independent clones (n = 6 for CRY1; n = 7 for CPEB2). (F) LM-PAT assay for endogenous Slc44a3 when CPEB1 and/or CPEB2 were either overexpressed (left) or knocked down (right) in AML12 cells. (G) Representative images of endogenous SLC44A3 protein expression when CPEB1 and CPEB2 were overexpressed (left) or knocked down (right). Relative protein expression level shown on the right was calculated by ImageJ software from two (overexpression) and six (knockdown) independent experiments, and the expression of controls (CRY1-myc for overexpression or siControl for knockdown) was set as 1. (H) The CLIP assay demonstrates that CPEB2, but not CPEB1, interacts with Slc44a3 mRNA. (Left) Input and immunoprecipitated proteins used for CLIP assay. Proteins were immunoprecipitated and blotted with anti-Myc antibody. The asterisk represents the position of the IgG heavy chain used for immunoprecipitation. (Middle and Right) PCR products showing total and immunoprecipitated Slc44a3 mRNAs from the CLIP assay. RNAs precipitated with CPEB proteins were reverse-transcribed, and the Slc44a3 mRNA was detected by end-point PCR (middle) or quantified by qPCR (right) with primers specific to Slc44a3. The arrowhead in the middle panel represents the band with the expected size for Slc44a3 amplification. CRY1 and B2-microglobulin (B2M) were used as negative controls.
In order to test this possibility, we chose two class III mRNAs, Slc44a3 and Pdzk11ip, for further analysis because their poly(A) rhythms were anti-phase, peaking either in the daytime like the majority of the class III PAR mRNAs (Slc44a3) or in the nighttime (Pdzk1ip1) (Fig. 5A). CPEB regulation of the Slc44a3 and Pdzk1ip1 mRNAs was first examined by a luciferase reporter gene harboring the 3′ UTR of these mRNAs (Fig. 4C). Expression of either CPEB1 or CPEB2 caused a dose-dependent increase in luciferase activity of Slc44a3 reporter gene in a 3′ UTR sequence-specific manner, while expression of CPEB3 or CPEB4 had a minimal effect even at the highest dose (Fig. 4D). In contrast, the Pdzk1ip1 reporter genes did not respond to expression of any of the CPEB proteins (Fig. 4D), indicating that the effect of CPEB1 and CPEB2 was specific to the Slc44a3 3′ UTR. The increase in luciferase activity of the Slc44a3 reporter gene by CPEB2 correlates with the elongation of the poly(A) tail of the reporter-derived mRNA (Fig. 4E). Likewise, the poly(A) tail length of endogenous Slc44a3 was increased by CPEB1 or CPEB2 overexpression and decreased when both Cpeb1 and Cpeb2 were knocked down (Fig. 4F; Supplemental Fig. S6). These changes in poly(A) tail length of endogenous Slc44a3 were accompanied by changes in its protein expression level, in which overexpression of CPEB1 or CPEB2 increased SLC44A3 protein, while knockdown of both Cpeb1 and Cpeb2 decreased the SLC44A3 protein level (Fig. 4G). We found that the tail lengths of both the endogenous Slc44a3 mRNA and the reporter luciferase-Slc44a3 mRNA were generally very short in the AML12 cell line, resembling the short tails (20–30 nt) that we observed for endogenous Slc44a3 during the night in the liver (Fig. 5A). We also found that CPEB2, but not CPEB1, interacts with the Slc44a3 mRNA, as demonstrated by cross-linking immunoprecipitation (CLIP) followed by RT–PCR (Fig. 4H).
Protein rhythms correlate with poly(A) tail length
In order to determine the biological relevance of poly(A) tail length fluctuation, we analyzed the protein expression profiles of PAR mRNAs representing all three classes: HSPB1/HSP27 (class I), GSTT2 and MIF (class II), and SLC44A3 and PDZK1IP1 (class III) (Fig. 5; Supplemental Fig. S7). As expected from its rhythmic steady-state mRNA profiles, the protein expression of HSPB1 was also rhythmic. However, GSTT2 and MIF, both of which lack rhythmic RNA expression at the steady-state level, and SLC44A3 and PDZK1IP1, which are not rhythmic at either the transcriptional or steady-state level, were also robustly rhythmic. We observed a similar phase relationship between the peak poly(A) tail length and the peak protein accumulation in all five cases, with the protein peaking ∼4–8 h after the time of the longest poly(A) tail (Fig. 5). These data suggest that poly(A) tail rhythms can generate rhythmic protein levels even when there is no rhythm in the steady-state mRNA levels.
Discussion
We found that the poly(A) tails of 2.3% of all expressed mRNAs in the mouse liver cycle in length daily and that these rhythmic poly(A) tail lengths correlate strongly with the ultimate circadian protein expression profiles of this tissue. It is likely that this number of PAR mRNAs underestimates the total number of mRNAs with cycling tail lengths due to the stringent criteria for assigning an mRNA as PAR as well as our methodology to use an oligo(dT) column that would not capture mRNAs with extremely short tails or lacking tails altogether.
Class I and II mRNAs, comprising 81.5% of the PAR mRNAs, use similar mechanisms for their poly(A) rhythmicity. Class I and II PARs are rhythmically transcribed, and the phase of poly(A) tail rhythms correlates well with pre-mRNA rhythms. It is most plausible that class I/II are rhythmically polyadenylated with long initial poly(A) tails as they are rhythmically transcribed in the nucleus, likely by the canonical poly(A) polymerase α (PAP α) that is also expressed rhythmically (Supplemental Fig. S8). However, it appears that class I/II PAR mRNAs can exist in short-tailed states (Fig. 3D,E), and this distinguishes these mRNAs from most RESS mRNAs whose steady-state levels are rhythmic but do not exhibit poly(A) tail rhythms. It has been long thought that mRNAs with short poly(A) tails are rapidly degraded, since deadenylation is a rate-limiting step that triggers mRNA decay (Cao and Parker 2001; Meyer et al. 2004). However, recent studies have shown that mRNAs can exist with a broad range of poly(A) tail lengths, and a considerable number of mRNAs that are well-expressed have poly(A) tails of >30 residues (Choi and Hagedorn 2003; Peng and Schoenberg 2005; Beilharz and Preiss 2007; Meijer et al. 2007; Wu et al. 2008). This was also observed in our analysis, in which a substantial number of short poly(A)-tailed mRNAs (<60 nt) were identified along with long poly(A)-tailed mRNAs (Fig. 1A). Since the mRNA half-lives of RESS and class I mRNAs are not different, the dynamics of deadenylation before decay must be different, by allowing the short poly(A)-tailed class I PAR mRNAs to accumulate without being degraded.
The class III mRNAs, comprising 18.5% of the PARs, are likely regulated by cytoplasmic polyadenylation mechanisms. The majority of class III PAR mRNAs had their longest poly(A) tail during the day, and these are most likely under the circadian control of canonical CPEB-dependent cytoplasmic polyadenylation, as many of the components of this machinery exhibit rhythmic expression patterns (Fig. 4B). However, this cannot be the only mechanism, as our analysis also identified mRNAs, such as Pdzk1ip1, that have poly(A) tail length rhythms in the opposite phase and are not affected by any of the four CPEBs (Fig. 4D). Other cytoplasmic polyadenylation mechanisms have been reported (Benoit et al. 2005; Coll et al. 2010), and noncanonical cytoplasmic poly(A) polymerases have been identified in a wide variety of organisms (Eckmann et al. 2011).
Our results demonstrate that Slc44a3, but not Pdzk1ip1, is under the control of CPEB-dependent cytoplasmic polyadenylation. CPEB1 and CPEB2 overexpression or knockdown resulted in the robust but modest changes in the poly(A) tail length of Slc44a3 as well as the Slc44a3 3′ UTR reporter gene, and changes in tail length correlated with changes in protein level and luciferase activity in AML12 (Fig. 4D–G). The magnitude of these changes was likely limited by the fact that we were only able to achieve a 30%–40% reduction in Cpeb1 and Cpeb2 levels in our knockdowns (Supplemental Fig. S6). Regardless, these data show that even small changes in the poly(A) length can have a significant impact on the protein expression level. Although our data suggest that both CPEB1 and CPEB2 regulate the poly(A) tail of Slc44a3 in AML12 cells, we think that CPEB2 likely plays a predominant role in rhythmic Slc44a3 poly(A) tail regulation in mouse livers. While Cpeb1 and Cpeb2 mRNAs are expressed at comparable levels in AML12 cells, Cpeb2 mRNA is expressed at ∼50-fold higher levels than Cpeb1 in mouse livers (data not shown), and Cpeb2 mRNA is robustly rhythmic in phase with the peak poly(A) tail length of the majority of class III mRNAs, while Cpeb1 mRNA levels are not. Furthermore, CPEB2 interacts with the Slc44a3 mRNA, while we could not detect any interaction by CPEB1 (Fig. 4H). However, we cannot rule out a role for CPEB1 in this process, since CPEB1 protein expression has been previously reported in the liver (Alexandrov et al. 2012). The RNA-binding sequence of CPEB2 has not been defined so far, and deletion mutants of the Slc44a3 3′ UTR reporter genes all interrupted the translational activation (data not shown). These suggest that CPEB2 recognizes RNA secondary structures or requires accessory proteins that also bind to Slc44a3 RNA to promote translational activation. In contrast to our observation that CPEB2 promotes translational activation of Slc44a3, previous studies have shown that CPEB2 can act as a translational repressor of the HIF-1α mRNA by slowing peptide elongation via interaction with the translation elongation factor eEF2 (Hagele et al. 2009; Chen and Huang 2011), suggesting that it has different mRNA-specific functions. How CPEB2 causes the translational activation of Slc44a3, whether it uses the previously described CPEB1-like strategy of regulating poly(A) tail length by balancing polyadenylation and deadenylation, and the identification of the PAPs and deadenylases that carry out these functions await further investigation.
Regulation of poly(A) tail length is traditionally considered to be unidirectional, going from long to short. However, much recent evidence has demonstrated that the ultimate poly(A) tail length is determined by a balance between concomitant deadenylation and polyadenylation, and this balance is controlled in a highly regulated and mRNA-specific manner. Our data indicate that the circadian system uses rhythmic polyadenylation and possibly deadenylation in mouse somatic cells to control rhythmic protein synthesis even in cases where the steady-state mRNA levels are not rhythmic. In fact, many components of the deadenylase and poly(A) polymerase machineries are under circadian control (Supplemental Fig. S8). This allows the circadian clock to temporally control protein expression independently from transcription. A previous study reported that nearly 50% of the rhythmic proteins that the investigators identified did not have rhythmic steady-state mRNA levels (Reddy et al. 2006), further supporting the idea that circadian protein expression can be regulated independently from rhythmic steady-state mRNA expression. We showed here that poly(A) tails are dynamic modulators of protein expression patterns in the liver and that the circadian clock regulates tail length by several different mechanisms. Therefore, the poly(A) tail length is carefully coordinated with time of day, and the impact on the circadian proteome is significant.
Materials and methods
RNA fractionation and 3′end-labeling assay
Male C57BL/6J mice were maintained on a 12L:12D cycle and fed ad libitum, and then livers were collected every 4 h. Animal experiments were conducted following the protocols approved by the Institutional Animal Care and Use Committees. RNAs were extracted from liver using Trizol (Invitrogen) according to the manufacturer's instructions. RNA fractionation was performed as described previously (Meijer et al. 2007) with slight modifications. In brief, a total of 80 μg of mouse liver total RNAs was isolated and resuspended in 400 μL of PolyATract GTC extraction buffer (Promega). RNAs were then mixed with 0.75 nmol of Biotinylated Oligo(dT) Probe (Promega) in dilution buffer (6× SSC, 10 mM Tris-HCl (pH 7.6), 1 mM EDTA, 0.25% SDS, 1% β-mercaptoethanol) and incubated for 10 min at 70°C. After centrifugation at 12,000g for 10 min at room temperature, supernatant was mixed with 600 μL of prewashed Streptavidin MagneSphere Paramagnetic Particles (Promega) and incubated for 15 min at room temperature while nutating. After three washes with 0.5× SSC at room temperature, short poly(A) RNAs were eluted by 0.075× SSC, and subsequently, long poly(A) RNAs were eluted by DEPC-treated water. Alternatively, polyA+ RNAs were eluted by DEPC-treated water immediately after the washing step. For bulk poly(A) tail length analysis, fractionated or polyA+ RNAs were 3′ end-labeled with [32P]-pCp with T4 RNA ligase (Epicentre) overnight at 4°C, followed by RNaseA/T1 (Fermentas) digestion to remove the bodies of the mRNAs. The nondigested poly(A) tails were then resolved by 7.5% denaturing PAGE and detected on X-ray film (Minvielle-Sebastia et al. 1991; Beilharz and Preiss 2009).
Microarray analysis
Fractionated or nonfractionated [total poly(A)+] RNAs were further purified using RNeasy MinElute Cleanup kit (Qiagen). For each time point, three independent samples were hybridized, each to an individual microarray chip (Affymetrix Mouse Gene ST 1.0). Raw data were analyzed and normalized by the quantile method through the dChip software (Li 2008). Rhythmic transcripts were identified using CircWave Batch version 3.3 (available at http://hutlab.nl), and cutoff thresholds were determined empirically and set as follows: PAR mRNAs (all time average expression > 200, amplitude {highest[long/short ratio] / lowest[long/short ratio]} > 1.4, one-way ANOVA < 0.05, sine curve fitting R2 > 0.5000) and RESS mRNAs (average expression over all time points > 200, amplitude [highest expression/lowest expression] > 1.4, one-way ANOVA < 0.05, sine curve fitting R2 > 0.7500). Probe sets were annotated using the Affymetrix annotation file as of February 2009. Heat maps were generated with the program Heatmap Builder version 1.0 (King et al. 2005).
LM-PAT assay
LM-PAT assay was performed as described previously (Salles and Strickland 1999) with a slight modification. In brief, nonfractionated RNAs (50 ng) were first incubated with 5′-phosphorylated oligo(dT)15 in the presence of T4 DNA ligase for 30 min at 42°C to anneal with poly(A) tails of RNAs, followed by an excess amount of anchor primer with oligo(dT)12 (5′-GCGAGCTCCGCGGCCGCGTTTTTTTTTTTT-3′) to anneal at the end of poly(A) tails, and were further incubated for 2 h at 12°C to complete ligation between oligo(dT)s. These oligo(dT)-annealed RNAs were then subjected to reverse transcription reaction using SuperScript II (Invitrogen) for cDNA synthesis. Aliquots of this cDNA were used as templates for PCR reactions with message-specific primers of our mRNAs of interest. Then, PCR products were digested by a restriction enzyme to confirm the specificity. Resulting DNA fragments were visualized by Alpha MultiphotoimagerII (AlphaInnotech), and poly(A) tail lengths were calculated by AlphaImager software (AlphaInnotech). Primer sequences and restriction enzymes used in this study can be found in Supplemental Table S3.
Quantitative PCR (qPCR)
Total RNAs were extracted as described above using Trizol reagent (Invitrogen), and then poly(A)-enriched RNAs were extracted by Poly(A)Tract system (Promega). Fifty nanograms of poly(A)-enriched RNAs (for steady-state mRNA measurement) or 4.5 μg of DNase I-treated total RNAs (for pre-mRNA measurement) was subjected to cDNA synthesis using SuperScript II (Invitrogen). qPCR was performed using ABI7900 (Applied Biosystems) with SYBR Power Green (Applied Biosystems). Primer sequences used in this study can be found in Supplemental Table S4. Rhythmicity analyses for individual genes were performed by CircWave version 1.4 (available at http://hutlab.nl), COSOPT (Panda et al. 2002), JTK_CYCLE (Hughes et al. 2010), and ARSER (Yang and Su 2010).
Western blot analysis
Mouse liver samples from each time point were homogenized in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 0.5% sodium deoxycholate, 1% Igepal CA-630, 5 mM DTT) containing protease inhibitor (Sigma). Equal amounts of each sample were separated by electrophoresis on an SDS–10% polyacrylamide gel before transfer to PVDF membrane (Bio-Rad). The membrane was then blocked with BLOTTO solution (0.1% Tween 20, 5% dry nonfat milk powder in Tris-buffered saline [TBS] at pH 7.4) for 1 h at room temperature. The membrane was treated with the primary antibody, anti-SLC44A3 (Abcam), anti-PDZK1IP (Abnova), anti-HSP27/HSPB1 (Santa Cruz Biotechnology), anti-MIF (Santa Cruz Biotechnology), anti-GSTT2 (a kind gift from Dr. John Hayes, University of Dundee), anti-Myc (Cell Signaling), anti-tubulin (Sigma), or anti-GAPDH HRP-conjugated (Sigma) overnight at 4°C. After washing, the blots were treated with secondary antibodies conjugated to horseradish peroxidase and were developed with a Chemiluminescence Western blotting kit (Roche).
Plasmid construction and luciferase assay
For luciferase reporter plasmids, luciferase gene from pGL3 (Promega) and 3′ UTR sequences of mouse Slc44a3 or Pdzk1ip1 amplified by PCR from mouse livers were cloned into pcDNA3.1 (Invitrogen). CPEB1 coding sequence was amplified from mCPEB1/pET30 (a kind gift from Dr. Joel Richter, University of Massachusetts Medical School) and CPEB2-coding sequence was amplified from MGC Clone (BC107349) and then cloned into pcDNA3.1/myc-His (Invitrogen). mCPEB3-HA and mCPEB4-myc were kind gifts from Dr. Shin-ichi Hoshino (Nagoya City University) and Dr. Joel Richter, respectively. The target sequences of siRNAs were as follows: Cpeb1 (sense, 5′-GUAUUAAGCAUGCUCCAAAtt-3′; antisense, 5′-UUUGGAGCAUGCUUAAUACtg-3′) and Cpeb2 (sense; 5′-CCUUAUUGAUAUCAUGAGAtt-3′; antisense, 5′-UCUCAUGAUAUCAAUAAGGga-3′). The control siRNAs were purchased from Ambion (Negative Control #1 siRNA).
The AML12 mouse hepatocyte cell line was maintained in DMEM/Ham F-12 medium (Invitrogen) supplemented with 10% FBS (Atlanta Biologica). For the measurement of mRNA half-lives, AML12 cells were treated with 1 μg/mL actinomycin D (Sigma) for 0, 1, 2, 3, 6, and 12 h, and the half-lives were calculated from the mRNA decay curve model with Prism (Graphpad). DNA transfection was performed by FuGENE6 (Roche) or Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions.
Luciferase assays were performed as previously reported (Kojima et al. 2010). A mixture of plasmid DNAs containing 100 ng of firefly luciferase reporter genes with the 3′ UTR of Slc44a3 or Pdzk1ip1, 10 ng Renilla luciferase reporter genes, and increasing amounts of CPEB-expressing plasmids (100, 200, 300, and 400 ng) were cotransfected to AML12 cells. Luciferase activities were measured ∼48 h after transfection.
CLIP assay
CRY1-myc, CPEB2-myc, or CPEB1-myc plasmids (1 μg) were transfected into AML12 cells and then lysed in immunoprecipitation buffer (20 mM HEPES at pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.5 mM DTT, 0.1% Triton X-100) supplemented with protease inhibitor cocktail (Sigma). The cell lysates were UV-irradiated for 15 min on ice and then centrifuged at 12,000g for 10 min at 4°C. The supernatant was incubated with either myc or HA antibody in the presence of protein A/G agarose (Pierce) overnight and then immunoprecipitated. After several washes, the immunoprecipitates were divided into two parts: One was analyzed on Western blot, and another was used for RNA isolation and subsequent RT–PCR. The primer sequences used in this study can be found in Supplemental Table S4.
Circularizing RT–PCR (cRT–PCR)
cRT–PCR was performed as previously described (Mullen and Marzluff 2008) with slight modifications. In brief, after transfection, RNA was extracted and then treated with DNase I as well as tobacco acid pyrophosphatase (Epicentre) for DNA digestion and decapping, respectively. Subsequently, RNAs were treated with T4 RNA ligase (Epicentre) for circularization and then reverse-transcribed using gene-specific primer. PCR primers were designed to the 5′ and 3′ end of RNA sequence so that poly(A) tail length could be included in the amplified fragments. To ensure the amplification of target gene, nested PCR was performed. PCR products were then TA-cloned and sequenced to measure poly(A) tail length. Primer sequences used in this study can be found in Supplemental Table S4.
Acknowledgments
We thank Dr. Edward Fox at the Microarray Core Facility of Dana-Farber Cancer Institute for performing microarray analyses, and Dr. Cornelia de Moor at the University of Nottingham for technical assistance for oligo(dT) chromatography. We also thank Dr. John Hayes, Dr. Joel Richter, Dr. Shin-ichi Hoshino, and Dr. Tadashi Baba for providing antibodies or DNA plasmids. We also thank Hyun Jeung Sung and Laura Weseman for mouse colony management, and other Green laboratory members for useful discussions. This work was supported by National Institutes of Health Grants GM076626 and GM090247 (to C.B.G.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.208306.112.
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP 2002. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12: 540–550 [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER, Richter JD 2004. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem 11: 318–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov IM, Ivshina M, Jung DY, Friedline R, Ko HJ, Xu M, O'Sullivan-Murphy B, Bortell R, Huang YT, Urano F, et al. 2012. Cytoplasmic polyadenylation element binding protein deficiency stimulates PTEN and Stat3 mRNA translation and induces hepatic insulin resistance. PLoS Genet 8: e1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T 2007. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13: 982–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T 2009. Transcriptome-wide measurement of mRNA polyadenylation state. Methods 48: 294–300 [DOI] [PubMed] [Google Scholar]
- Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M 2005. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev Cell 9: 511–522 [DOI] [PubMed] [Google Scholar]
- Brown CE, Sachs AB 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol 18: 6548–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R 2001. Computational modeling of eukaryotic mRNA turnover. RNA 7: 1192–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PJ, Huang YS 2011. CPEB2–eEF2 interaction impedes HIF-1α RNA translation. EMBO J 31: 959–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH, Hagedorn CH 2003. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc Natl Acad Sci 100: 7033–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll O, Villalba A, Bussotti G, Notredame C, Gebauer F 2010. A novel, noncanonical mechanism of cytoplasmic polyadenylation operates in Drosophila embryogenesis. Genes Dev 24: 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE 2008. 3′ End mRNA processing: Molecular mechanisms and implications for health and disease. EMBO J 27: 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Sage D, Unser M, Bauer C, d'Eysmond T, Naef F, Schibler U 2009. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J 28: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield GE 2003. DNA microarray analyses of circadian timing: The genomic basis of biological time. J Neuroendocrinol 15: 991–1002 [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Rammelt C, Wahle E 2011. Control of poly(A) tail length. Wiley Interdiscip Rev RNA 2: 348–361 [DOI] [PubMed] [Google Scholar]
- Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, Green CB 2007. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA 13: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Xu W, Cooper GM, Richter JD 1994. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J 13: 5712–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner JR, Vanderheyden WM, Lavaute T, Westmark CJ, Rouhana L, Pack AI, Wickens M, Landry CF 2012. Time of day regulates subcellular trafficking, tripartite synaptic localization, and polyadenylation of the astrocytic fabp7 mRNA. J Neurosci 32: 1383–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD 2002. Translational control of the embryonic cell cycle. Cell 109: 473–483 [DOI] [PubMed] [Google Scholar]
- Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD 2006. Control of cellular senescence by CPEB. Genes Dev 20: 2701–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagele S, Kuhn U, Boning M, Katschinski DM 2009. Cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 bind to the HIF-1α mRNA 3′-UTR and modulate HIF-1α protein expression. Biochem J 417: 235–246 [DOI] [PubMed] [Google Scholar]
- Hosoda N, Funakoshi Y, Hirasawa M, Yamagishi R, Asano Y, Miyagawa R, Ogami K, Tsujimoto M, Hoshino S 2011. Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. EMBO J 30: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD 2002. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and αCaMKII mRNA polyadenylation at synapses. EMBO J 21: 2139–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, Richter JD 2006. CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J 25: 4865–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet 5: e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K 2010. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JY, Ferrara R, Tabibiazar R, Spin JM, Chen MM, Kuchinsky A, Vailaya A, Kincaid R, Tsalenko A, Deng DX, et al. 2005. Pathway analysis of coronary atherosclerosis. Physiol Genomics 23: 103–118 [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Gatfield D, Esau CC, Green CB 2010. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS ONE 5: e11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Wahle E 2004. Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678: 67–84 [DOI] [PubMed] [Google Scholar]
- Li C 2008. Automating dChip: Toward reproducible sharing of microarray data analysis. BMC Bioinformatics 9: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, de Moor CH 2007. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res 35: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E 2004. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit Rev Biochem Mol Biol 39: 197–216 [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F 1991. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol Cell Biol 11: 3075–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Gallego J, Ferreira PG, Mendez R 2010. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol 12: 447–456 [DOI] [PubMed] [Google Scholar]
- O'Neill JS, Reddy AB 2011. Circadian clocks in human red blood cells. Nature 469: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ 2011. Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320 [DOI] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA 2010. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol 20: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schoenberg DR 2005. mRNA with a <20-nt poly(A) tail imparted by the poly(A)-limiting element is translated as efficiently in vivo as long poly(A) mRNA. RNA 11: 1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. 2006. Circadian orchestration of the hepatic proteome. Curr Biol 16: 1107–1115 [DOI] [PubMed] [Google Scholar]
- Richter JD 2007. CPEB: A life in translation. Trends Biochem Sci 32: 279–285 [DOI] [PubMed] [Google Scholar]
- Robinson BG, Frim DM, Schwartz WJ, Majzoub JA 1988. Vasopressin mRNA in the suprachiasmatic nuclei: Daily regulation of polyadenylate tail length. Science 241: 342–344 [DOI] [PubMed] [Google Scholar]
- Salles FJ, Strickland S 1999. Analysis of poly(A) tail lengths by PCR: The PAT assay. Methods Mol Biol 118: 441–448 [DOI] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS 2009. Database for mRNA half-life of 19,977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res 16: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83 [DOI] [PubMed] [Google Scholar]
- Tarun SZ Jr, Sachs AB 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15: 7168–7177 [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. 2002. A transcription factor response element for gene expression during circadian night. Nature 418: 534–539 [DOI] [PubMed] [Google Scholar]
- Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC 2001. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol 1: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA 2004. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14: 2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD 1998. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron 21: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng Y, Zhou T, Zhang MQ, Wu CI, Wang SM 2008. Poly A- transcripts expressed in HeLa cells. PLoS ONE 3: e2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Falvey E, Lavery D, Talbot D, Schmidt E, Ossipow V, Fonjallaz P, Schibler U 1992. The role of the transcriptional activator protein DBP in circadian liver gene expression. J Cell Sci Suppl 16: 123–127 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Yang R, Su Z 2010. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 26: i168–i174 [DOI] [PMC free article] [PubMed] [Google Scholar]