Abstract
The mercury-resistance transposon Tn5053 inhibits restriction activity of the type I restriction-modification endonuclease EcoKI in Escherichia coli K12 cells. This is the first report of antirestriction activity of a non-conjugative transposon. The gene (ardD) coding for the antirestriction protein has been cloned. The ardD gene is located within the tniA gene, coding for transposase, on the complementary strand. The direction of transcription is opposite to transcription of the tniA gene.
Keywords: restriction-modification system type I, antirestriction protein, mercury-resistance transposon, overlapping genes
Introduction
Conjugative plasmids and conjugative transposons contain the ardA, ardB and ardC genes, coding for antirestriction proteins. The ArdA, ArdB and ArdC proteins specifically inhibit type I restriction-modification enzymes (Delver et al., 1991; Belogurov et al., 1993, 2000; Serfiotis-Mitsa et al., 2010). The ArdA proteins simultaneously inhibit restriction (endonuclease) and modification (methylase) acitivity of these enzymes (Delver et al., 1991; McMaahon et al., 2009), while the ArdB proteins inhibit only restriction activity of the enzymes (Belogurov et al., 1993; Serfiotis-Mitsa et al., 2010). These proteins differ considerably in both primary and tertiary structure. The ArdA proteins (165–170 amino acids) carry a considerable negative charge (−25: −30) and belong to the family of DNA mimic proteins, because their spatial structure is similar to the double-helical DNA in B form (McMaahon et al., 2009). The ArdB proteins (145–153 amino acids) usually carry a small negative charge (−1: −6) and form a structure of a compact tetraeder (Serfiotis-Mitsa et al., 2010). The presence of the ardA and ardB genes helps mobile elements to overcome the restriction barriers, providing efficient ‘horizontal’ gene transfer between bacteria of various species and genera.
We have previously shown that the merR gene Tn5053, cloned in the vector pUC19 and introduced in Escherichia coli K12 strain JM83 shows an antirestriction effect against a type I restriction enzyme EcoKI. The presence of the merR gene in the cell increased the plating efficiency of the bacteriophage λ.0 with non-modified DNA about five- to seven-fold (Rastorguev et al., 1999). MerR is a transcriptional regulator of the mer operon. Here we demonstrate that the full-length mercury-resistance transposon Tn5053, when introduced in a bacterial cell within the vector pUC19, inhibits restriction activity of the EcoKI enzyme, decreasing it about 100-fold. We showed that a new gene, designated ardD, codes for a protein that shows antirestriction activity against EcoKI. This gene is located within the tniA gene (encoding transposase) on the complementary strand.
Materials and methods
Bacterial strains, bacteriophage, and plasmids
Relevant characteristics of the bacterial strains, bacteriophage and plasmids used in this study are described in Table 1. Routine cell growth was carried out at 37 °C in Luria–Bertani (LB) medium supplemented with antibiotics as appropriate.
Table 1.
Escherichia coli strains and plasmids used in this study
| Name | Genotype or description | Source or reference |
|---|---|---|
| Strain | ||
| AB1157 | F- thr-1, leu-6, proA2, his-4, thi-1, argE3, lacY1, galK2, ara14, xyl-5, mtl-1, tsx-33, rpsL31, supE44, r+m+ | N.E. Murray, UK |
| NK114 | ΔclpX::kan, derivative of AB1157 | N.E. Murray, UK |
| TG-1 | thi relA supE44 hsdR17 hsdM Δ (lac-proAB) [F′traD36 proAB lacIqZΔM15] | VKPM ‘GosNIIgenetika’ |
| MC1061 | araD139 Δ(araA-leu)7697 Δ lacX74 galK16 galE15 mcrA0 relA1 rpsL150 spoT1 mcrB1 hsdR2 | VKPM ‘GosNIIgenetika’ |
| Plasmid | ||
| pUC19 | ColE1 origin, Ampr | Fermentas, Lithuania |
| pTZ57R | ColE1 origin, Ampr | Fermentas, Lithuania |
| pKLH53.1 | Ampr, Hgr, pUC19 with Tn5053 (8500 bp) of chromosome Xanthomonas sp. W17 inserted between PvuII/DraI and NdeI sites. | Kholodii et al. (1995) |
| pKLH53.1tniQ1 | Deletion between the Acc651 and HpaI sites of pKLH53.2 inactivating tniB and tniQ | Kholodii et al. (1995) |
| pKLH53.1tniQ2 | 730-bp deletion between the ClaI and Acc651 sites within tniQ in pKLH53.1 | Kholodii et al. (1995) |
| pKLH53.1tniB2 | Insertion of the filled-in EcoRI fragment containing the Kmr cassette into the HpaI site within tniB of pKLH53.1 | Kholodii et al. (1995) |
| pKLH53.1tniA | Insertion of the SalI fragment with a Kmr cassette from pUC4K into the SalI site within tniA of pKLH53.1 | Kholodii et al. (1995) |
| pKLH53.2 | tni operon Tn5053 inserted in plasmid pACYC184 | Kholodii et al. (1995) |
| pTLHindIII-ClaI | HindIII-ClaI fragment from the mer operon of Tn5053 cloned in pUC19 | This study |
| pTL2.5 | HindIII fragment from the mer operon of Tn5053 cloned in pUC19 | This study |
| pTLΔHindIII | Obtained by treatment of pKLH53.1 with HindIII and subsequent ligation | This study |
| pTLORF5 | 2300-bp KpnI SalI fragment from pKLH53.1 cloned in pUC19 | This study |
| pSMΔORF5 | Obtained by treatment of pTLORF5 with Eco47III and subsequent ligation | This study |
| pORF5 | orf-5 cloned in pUC19 under the lac promoter | This study |
| Bacteriophage λvir | R. Devoret, France | |
Media and reagents
Luria–Bertani medium and LB agar (1.8% agar) were prepared according to Miller (1972). Antibiotics were added as required: ampicillin (100 μg mL−1), kanamycin (40 μg mL−1) and chloramphenicol (20 μg mL−1).
The enzymes for cloning were supplied by Fermentas.
DNA isolation, restriction, ligation and transformation
Hybrid plasmids and vectors were isolated using a kit from Qiagen. Chromosomal DNA was isolated from the cells at late exponential phase of growth; the cells were lysed with lysozym and sodium dodecyl sulphate and the lysate was then treated with phenol with subsequent DNA sedimentation in ethanol.
Restriction, ligation of DNA fragments, electrophoresis in agarose gel, isolation of DNA fragments from the gel by electroelusion and transformation of calcium cells were performed in E. coli as described (Sambrook et al., 1989).
Construction of recombinant plasmids
The plasmid pTLΔHindIII was obtained by treatment of pKLH53.1 with HindIII and subsequent ligation. The HindIII fragment of 2.5 kbp and HindIII-ClaI fragment from the mer operon of Tn5053 were cloned in pUC19 under the lac promoter: pTL2.5 (2.5-kbp HindIII fragment) and pTLHindIII-ClaI (HindIII-ClaI fragment). The fragment tniA,B,Q Tn5053 (2.3 kbp) was cloned in pUC19 under the lac promoter (pTLORF-5). Hybrid plasmid pSMΔORF-5 was obtained by eliminating the DNA between the Eco47III sites within the orf-5 gene in pTLORF-5 (see Fig.). In pORF-5, a 483-bp fragment from the tniA gene was cloned in pUC19 under the lac promoter (see Fig.). The DNA fragment containing the gene orf-5 was amplified by PCR using the following primers: Tn5053dir, 5′-GCAGAGGGTGACGGCCGGATGG-3′; Tn5053rev, 5′-CACGGCGATGCAGATGATCCACG-3′ and plasmid pKLH53.1 DNA as a template. Amplification was carried out at the conditions recommended by the manufacturer. The amplification product was purified by electrophoresis and cloned in T-vector pTZ57R. A 483-bp fragment was then recloned into pUC19 at XbaI and BamHI restriction sites to construct pORF-5. For the other plasmid constructs of the pKLH series see Kholodii et al. (1995).
Fig 2.
Nucleotide sequence of the gene orf-5 (ardD) and amino acid sequence of its product (147 amino acids), encoded by the complementary strand of the gene tniA in transposon Tn5053. RBS is shown in italics and underlined, a putative antirestriction motif is shown in italics and Eco47III sites are underlined.
Estimation of antirestriction activity
The antirestriction activity of plasmid was defined as the efficiency of plating (EOP) of unmodified phage λ.0 on the experimental (plasmid-bearing) strain divided by the EOP on the plasmidless restricting strain (Delver et al., 1991). The EOP (in Table 2 designated К) was calculated as: phage titre on the restricting strain (NK114)/phage titre on a nonrestricting strain (TG-1). Unmodified phages, denoted by λ.0, were grown on E. coli TG-1 r−m−, which lost restriction and modification functions. All assays were performed in triplicate and at least 50 phage plaques per plate per experiment were counted. Experiments were performed on numerous days with fresh samples and control experiments performed each day. Little variation was observed during the replicate experiments. The standard deviation for the antirestriction results is 25% or less.
Table 2.
Comparison of antirestriction activity of cloned fragments and deletion and insertion mutants of the transposon Tn5053
| Plasmid | Coefficient of restriction (K)* | Restriction relief (R)† |
|---|---|---|
| pUC19 (control) | 1.0 × 10−5‡ | 1 |
| pKLH53.1 | 1.1 × 10−3 | 110 |
| pKLH53.1tniA | 1.0 × 10−3 | 100 |
| pKLH53.1tniB2 | 9.5 × 10−4 | 95 |
| pKLH53.1tniQ2 | 1.2 × 10−3 | 120 |
| pKLH53.1tniQ1 | 9.6 × 10−4 | 96 |
| pTLΔHindIII | 1.0 × 10−5 | 1 |
| pTLHindIII-ClaI | 1.0 × 10−5 | 1 |
| pTL2.5 | 1.0 × 10−5 | 1 |
| pKLH53.2 | 1.0 × 10−5 | 1 |
| pTLORF-5 | 1.1 × 10−3 | 110 |
| pSMΔORF-5 | 1.0 × 10−5 | 1 |
| pORF-5 | 5.3 × 10−3 | 530 |
The coefficient of restriction (K) was determined as the ratio of the titre of phage λ.0 on strain NK114 r+m+ to the titre of the same phage on strain TG-1 r−m−.
The restriction relief factor R = K+/K−, where K+ is K for NK114 with a plasmid, and K− is K for NK114 without a plasmid.
Mean of three independent experiments.
Results
Antirestriction activity of the transposon Tn5053, its deletion and insertion mutants
Plasmids with antirestriction activity
Data on antirestriction activity of the recombinant plasmid pKLH53.1, containing Tn5053, are given in Table. The factor of restriction relief (R) is about 100. We suspected that the nucleotide sequence of the mercury-resistance transposon Tn5053 contains a fragment encoding an antirestriction protein. We used both insertion and deletion mutants of Tn5053 for all transposition genes (tni) as well as plasmid constructs containing various fragments of the Tn5053 DNA, while searching for the locus responsible for the antirestriction activity (Fig. 1). The results of searches for the determinant of antirestriction activity within Tn5053 are shown in Table 2. It is evident that neither insertion (plasmids pKLH53.1tniA, pKLH53.1tniB2) or deletion (plasmids pKLH53.1tniQ2 and pKLH53.1tniQ1) mutations of the tni genes have any effect on antirestriction activity: about 100-fold decrease in EcoKI restriction level is preserved.
Fig 1.
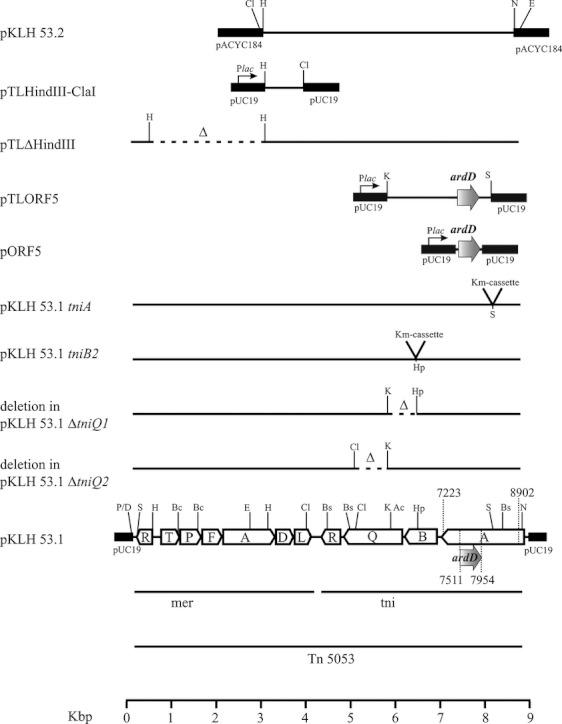
Structure of pKLH53.1, subcloned fragments, insertion and deletion mutants. EV, EcoRV; Bc, BclI; Bs, BssHII; Cl, ClaI; D, DraI; E, EcoRI; Ac, Acc65I; H, HindIII; Hp, HpaI; K, KpnI; N, NdeI; P, PvuII; S, SalI.
Plasmids without antirestriction activity
Deletion of the major part of the mer operon (plasmid pTLΔHindIII) completely removed the effect of antirestriction (Table). We assumed that the location of the gene coding for an antirestriction protein is within the mer operon. However, the recombinant plasmids pTLHindIII-ClaI and pTL2.5 with fragments HindIII-ClaI and HindIII from the mer operon (without the merR gene) in vector pUC19 show no antirestriction effect (Table 2). No antirestriction effect was also observed for the hybrid plasmid pKLH53.2, containing all the genes tni Tn5053 under its own promoter (in vector pACYC184; Fig., Table 2). A paradox appeared: the mer operon together with the transposition genes (tni) of Tn5053 produce an antirestriction effect, while the plasmids with separately cloned mer operon or tni genes show no antirestriction effect.
Construction of recombinant plasmids containing orf-5 and evaluation of their antirestriction activity
We considered that the nucleotide sequence coding for the ORF with antirestriction activity is located within the region of the tni genes, but orientated in reverse to the direction of transcription of the tni genes. Consequently, the coding strand for this ORF is the same as for the mer operon. If so, transcription of this DNA fragment passes from the side of the mer operon. We analysed the DNA sequence from the region of the tni genes of Tn5053 in reverse direction, and found several orfs. Of main interest was orf-5, encoding a negatively charged protein with a motif close to the antirestriction motif of the proteins Ard (Fig. 2). The protein ORF-5 contains 147 amino acid residues of summary charge −1. It is encoded by orf-5 at positions 7511–7954 on the complementary strand of the tniA gene (positions numbered according to the nucleotide sequence of Tn5053, deposited in DBJ/EMBL/GenBank under accession number L40585). The nucleotide sequence located upstream of the initiation codon (AGAGGGT) is virtually identical to the canonical ribosome binding site (RBS) sequence (AGGAGGT). Note that other ORFs found along the complementary strand in the region of the genes tni Tn5053 do not contain RBS sequence upstream of the initiation codon.
To test the hypothesis of antirestriction activity of orf-5, we constructed a hybrid plasmid using the 2300-bp KpnI-SalI DNA fragment from orf-5 containing region tniA,B,Q. This fragment was cloned under the lac promoter in vector pUC18 (pTLORF-5, Fig. 2). Introduction of this plasmid into cells of strain NK114 produced an antirestriction effect similar to that observed for the wild-type Tn5053, about 100-fold (Table 2). Internal deletion in the orf-5 gene was produced by Eco47III restriction endonuclease treatment of pTLORF-5. In the resulting plasmid pSMΔORF-5, a major part of orf-5 (245 bp; nucleotides 7621–7866 in the L40585 sequence) was deleted, including the putative antirestriction motif VVDVVDDKA (Fig. 2). The antirestriction effect in E. coli NK114 cells, containing pSMΔORF-5, disappeared completely (Table 2). For further evaluation of the role of orf-5 in this antirestriction effect, we amplified orf-5 together with the RBS and cloned them in pUC19 under the lac promoter (for details see Materials and methods). After the plasmid obtained (pORF-5) was introduced into NK114 cells, the antirectriction factor R was estimated. Plasmid pORF-5 showed a considerable antirestriction effect: efficiency of the λ.0 phage plating was about 500-fold higher than the control level (cells with pUC19) (Table 2).
Discussion
It has been shown that the genes encoding the antirestriction proteins (ArdA, ArdB, ArdC) may be located within conjugative plasmids and conjugative transposons (Delver et al., 1991; Belogurov et al., 1993, 2000; McMaahon et al., 2009; Serfiotis-Mitsa et al., 2010). Here we show for the first time that a similar gene is also present within a non-conjugative transposon (Tn5053). Analysis of the deduced amino acid sequence of ORF-5 revealed that this protein has no similarities to the known Ard proteins (ArdA, ArdB and ArdC types) except the ‘antirestriction’ motif conserved for all known Ard proteins. This suggests that ORF-5 may be classified as a new type of Ard protein, which we designate ArdD. The N-terminal region of ArdD has a high degree of similarity (about 39% identity and 53% similarity) to the region of the MerR protein (312–367 amino acids) of Desulfovibrio vulgaris strain ‘Miyazaki F’ (NCBI reference sequence YP_002436545.1; Fig.). Interestingly, the total negative charge of homologous sequences ArdD and MerR is virtually the same, −5 and −7, respectively. The location of the ardD gene appears to be unusual: inside a transposition gene (tniA) with transcription at the complementary strand (Fig. 1). Overlapping genes in bacterial genomes are rare. For example, most strains of Shigella flexneri 2a and enteroaggregative E. coli carry a highly conserved chromosomal locus which encodes a 109-kDa secreted mucinase Pic and, on the opposite strand in overlapping fashion, an oligomeric enterotoxin ShET1, encoded by the setA and setB genes. The setB gene is transcribed from a promoter which lies more than 1.5 kb upstream of the setB gene (Behrens et al., 2002). According to our data, the ardD gene promoter is also located distantly from the ardD gene in the region of the mer operon, at a distance of more than 3 kbp. We suggest that other non-conjugative transposons may also contain genes that encode products that can inhibit the restriction endonucleases, thereby efficient overcoming restriction barriers. Note that the tniA gene is usually present in integrons and composite transposons conferring antibiotic resistance and is widely distributed among environmental and clinical bacteria. As an example, the transposon Tn6006 contains a nucleotide sequence identical to ardD in the tniA gene. The Tn6006 transposon belongs to the group of recombinant transposons containing integrons (Fluit & Schmitz, 1999; Labbate et al., 2008).
Fig 3.

Comparison of the amino acid sequences of ArdD (Tn5053) and MerR (Desulfovibrio vulgaris strain ‘Miyazaki F’). Aligment was done with the program NCBI blast. ArdD (2–58 amino acids) and MerR (312–367 amino acids) homologous regions: identity = 39%, similarity = 53%.
Acknowledgments
This study used equipment of centre of collective use of GosNIIgenetika. It was supported in part by the Russian Foundation for Basic Research (grant 10-04-00541), the Federal Program ‘Scientific and pedagogical innovation resources in Russia, 2009–2013’ (Contract P1070 from 4 June 2010) and The Ministry of Education and Science (Contract 16.522.11.7029).
References
- Behrens M, Sheikh J, Nataro JP. Regulation of the overlapping pic/set locus in Shigella flexneri and Enteroaggregative Escherichia coli. Infect Immun. 2002;70:2915–2925. doi: 10.1128/IAI.70.6.2915-2925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov AA, Delver EP, Rodzevich OV. Plasmid pKM101 encodes two nonhomologous antirestriction proteins (ArdA and ArdB) whose expression is controlled by homologous regulatory sequences. J Bacteriol. 1993;175:4843–4850. doi: 10.1128/jb.175.15.4843-4850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov AA, Delver EP, Agafonova OV, Belogurova NG, Lee LY, Ado CI. Antirestriction protein Ard (Type C) encoded by IncW plasmid pSa has a high similarity to the “protein transport” domain of TraC1 primase of promiscuous plasmid RP4. J Mol Biol. 2000;296:969–977. doi: 10.1006/jmbi.1999.3493. [DOI] [PubMed] [Google Scholar]
- Delver EP, Kotova VY, Zavilgelsky GB, Belogurov AA. Nucleotide sequence of the gene (ard) encoding the antirestriction protein of plasmid ColIb-P9. J Bacteriol. 1991;173:5887–5892. doi: 10.1128/jb.173.18.5887-5892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluit AC, Schmitz FJ. Class1 integrons, gene cassettes, mobility, and epidemiology. Eur J Clin Microbiol Infect Dis. 1999;18:761–770. doi: 10.1007/s100960050398. [DOI] [PubMed] [Google Scholar]
- Kholodii GY, Yurieva OV, Lomovskaya OL, Gorlenko Z, Mindlin SZ, Nikiforov VG. Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol Microbiol. 1995;17:1189–1200. doi: 10.1111/j.1365-2958.1995.mmi_17061189.x. [DOI] [PubMed] [Google Scholar]
- Labbate M, Chowdhury PR, Stokes HW. A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments. J Bacteriol. 2008;190:5318–5327. doi: 10.1128/JB.00199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaahon SA, Roberts GA, Jhonson KA, et al. Extensive DNA mimicry by the ArdA antirestriction protein and its role in the spread antibiotic resistance. Nucleic Acids Res. 2009;37:4887–4897. doi: 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Rastorguev SM, Letuchaia TA, Kholodi GY, Mindlin SZ, Nikiforov VG, Zavilgelsky GB. Antirestriction activity of metalloregulatory proteins ArsR and MerR. Mol Biol (Moscow) 1999;33:203–206. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Serfiotis-Mitsa D, Herbert AP, Roberts GA, Soares DC, White H, Blakely GW, Uhrin D, Dryden DT. The structure of the KlcA and ArdB proteins reveals a novel fold and antirestriction activity against type I DNA restriction systems in vivo but not in vitro. Nucleic Acids Res. 2010;38:1723–1737. doi: 10.1093/nar/gkp1144. [DOI] [PMC free article] [PubMed] [Google Scholar]



