Abstract
PCR- Restriction fragment length polymorphism (RFLP) is a time saving and accurate technique to differentiate closely related organisms. In the regions endemic for visceral leishmaniasis (VL) in India, various species of morphological similar sand fly exists but only female Phlebotomus argentipes is the vector for VL. In the present study primers were designed targeting the 18S rRNA encoding gene that showed amplification in all the major sand fly species found in India. The amplified fragments were further digested using the Hinf I or Hpa II restriction enzymes. Each of the restriction enzyme produced species specific restriction patterns, which can easily be used to identify specific sand fly species. This technique can be employed in the identification of the species of the sand flies.
Keywords: 18S rRNA, Leishmaniasis, PCR, Phlebotomine, RFLP, Sand fly
Leishmaniasis is one of the most severe forms of vector borne infectious diseases. It is caused by about 21 Leishmania species and transmitted by the bite of female sand flies belonging to the genus Phlebotomus in the Old World and Lutzomyia in the New World. There are about 500 known species of Phlebotomine sand flies but only about 30 of these are considered as vector (Desjeux 1996; Herwaldt 1999). The Bihar state of India is endemic for visceral leishmaniasis, the most severe form of leishmaniasis caused by L. donovani. Three species of Phlebotomine sand flies, Phlebotomus argentipes Annandale and Brunetti, Phlebotomus papatasi (Scopoli) and Sergentomyia babu Annandale have been reported from India, however, only female P. argentipes is considered as a vector for L. donovani (Swaminath et al. 2006).
To understand the transmission dynamics of vector borne diseases, accurate identification of vector species is important. Conventional methods of sand fly species identification requires expert entomologist to differentiate sand fly species, using morphological characteristics such as structure of mouth parts and terminal regions under the stereomicroscope. Morphological identification is labor intensive especially when applied for identification of large number of sand flies. However, molecular methods such as polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) appear to be more convenient and easier approach (Aransay 1999). Normal PCR-RFLP or multiplex PCR targeting polymorphism in the conserved genes of 18S rRNA, ITS or cytochrome b has been earlier reported for species identification (Khalid et al. 2010; Manonmani 2010; Surendran et al. 2005).
The present study aimed to develop a unique molecular tool combining single step PCR and RFLP method using polymorphism of 18S rRNA gene to identify P. argentipes, P. papatasi and S. babu commonly found in India.
Materials and Methods
Sand fly collection
The households of villages of Muzaffarpur district, Bihar which is endemic for VL, were selected for the collection of sand fly. The study was conducted during March-April 2011. CDC light traps (John W. Hock Co., USA) were installed inside house during twilight and taken out in the early morning of the next day. Trapped insects were collected in Petri dishes and kept at −20° C for 20 minutes for killing.
Morphological identification
All collected sand flies were morphologically identified using stereomicroscope (Carl Zeiss-Stemi 2000C, Germany) using Lewis keys (Lewis 1987) such as morphology of thorax and hairs of abdominal tergites and categorized into P. argentipes, P. papatasi and S. babu. All the identified sand flies were stored individually in 70 % ethanol and kept at −20°C until DNA was extracted.
DNA extraction
Sand fly DNA was extracted individually using High Pure PCR Template Preparation Kit (Roche, Germany) as per the manufacturer’s instructions after homogenizing sand flies using sterile polypropylene pestle. Quantification was done using NanoDrop 2000 (Thermo Scientific, USA) and stored at 4°C for PCR.
Design of the species identification PCR primers
Forward and reverse primers were designed within the conserved area of the 18S rRNA encoding sequence. Sequences of P. argentipes [GenBank: AJ244360], P. papatasi [GenBank: AJ244414] and S. minuta [GenBank: AJ244421 as sequence of S. babu is not available] were used (Fig. 1). Alignment of sequences was done in ClustalW2 multiple sequence alignment programme (Thompson et al. 2002). The forward primer was selected from the initial 100 bp region while reverse primer was selected from the next 500 bp region as 5’-TAGTGAAACCGCAAAAGGCTCAG-3’ (Forward primer) and 5’-CTCGGATGTGAGTCCTGTATTGT-3’ (Reverse primer).
Fig. 1.
ClustalW2 - Multiple Sequence Alignment of the nucleotide sequences of P. argentipes (PA), P. papatasi (PP) and S. babu (SB) for design of species specific primers and selection of restriction enzymes. Dark highlighted sequence selected for primer designing.
PCR amplification
PCR was performed in Veriti Thermal Cycler (Applied Applied Biosystems, Foster City, CA, USA). The reaction was carried out in a volume of 25µl using the pair of primers (10 pmol each), 1 U normal Taq DNA polymerase (New England Biolabs, UK) supplemented with MgCl2, 10X buffer, 100X BSA, 0.2 mM of dNTPs. For template 50 ng of extracted DNA was used while nuclease-free water (QIAGEN, Hilden, Germany) was used as negative control. PCR conditions were optimized as, initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 40 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min.
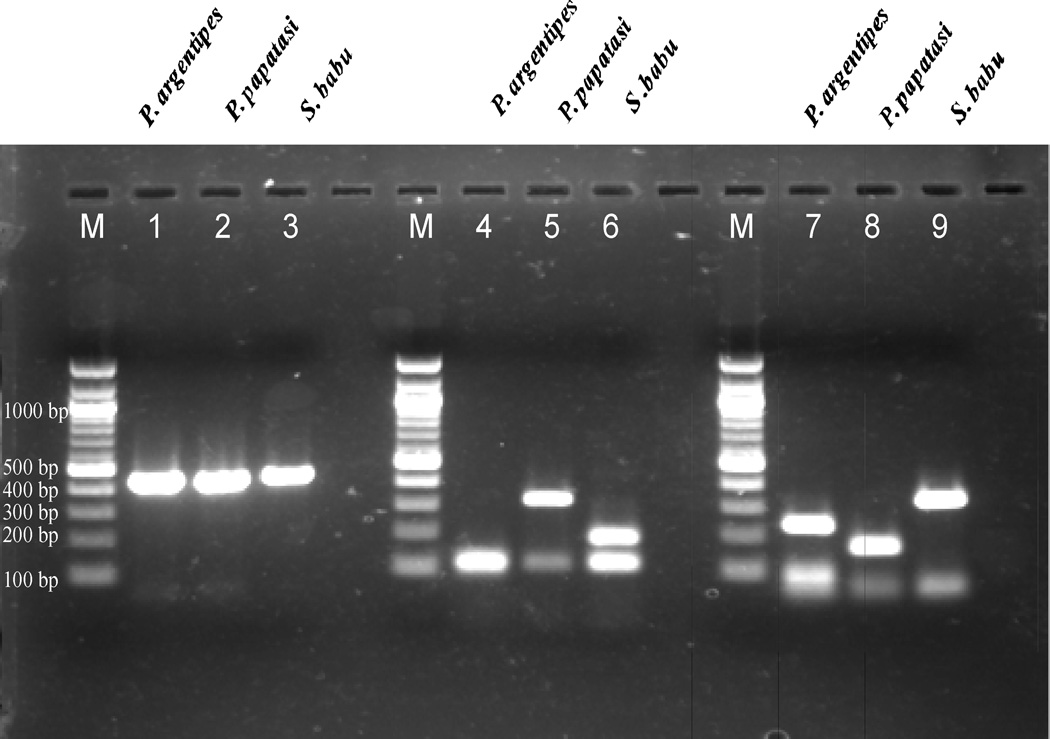
The amplified PCR products were run by electrophoresis on ethidium bromide (Merck, Darmastaft, Germany) stained 1.5% (w/v) agarose gel (Sigma-Aldrich, USA) in Tris-acetate ethylene di-amine tetra acetic acid (TAE) buffer. A DNA marker of 100-bp (New England Biolabs, UK) was loaded with the amplified product to determine the size of the product (Fig. 2).
Fig. 2.
Agarose gel picture showing different digestion patterns for Hinf I and Hpa II. Lane 1–3: undigested PCR products for P. argentipes, P. papatasi and S. babu respectively. Lane 4–6: Hinf I digested products and lane 7–9: Hpa II digested products for the three sand fly species. (M: 100bp DNA ladder)
Sequencing of PCR products
Amplified PCR products were eluted from agarose gel using QIAquick Gel Extraction kit (Qiagen). Purified products were sequenced using the BigDye Terminatorv3.1 cycle sequencing kit (Applied Biosystems) following manufacturer’s instruction and further run through an ABI 3130 genetic analyser. The products were sequenced for both forward and reverse primer. The sequences were compared with those of P. argentipes, P. papatasi and S. minuta using NCBI-BLAST (http://www.ncbi.nlm.nih.gov/blast/tools) for alignment. Similarity between the sequences was found 100% for P. argentipes [GenBank: AJ244360] andP. papatasi [GenBank: AJ244414], and 98% for S. minuta [GenBank: AJ244421].
Restriction Fragment Length Polymorphism
For restriction digestion, nucleotide sequences of all the three sand fly species were aligned using CLUSTAL 2.1 multiple sequence alignment programme. Differences in between the sequences for all the three sand fly species were used to select specific restriction enzymes. Two restriction enzymes Hinf I and Hpa II were selected as they have different digestion sites (Fig. 1).To discriminate all the three species of sand fly, the amplified PCR products was subjected to restriction digestion with Hinf I (recognition site GANTC; time saver, New England Bio labs, UK) and Hpa II (recognition site GTTAAC; time saver, New England Bio labs, UK) restriction enzyme in supplied 10 X reaction buffer at 37°C for 10 min. The digested products were electrophoresed on a 1.5% (w/v) agarose gel and stained with ethidium-bromide.
Results
Total sand flies collected were 464 and grouped into S. babu (50%), P. argentipes (48%) and P. papatasi (1.5%) and rest were other species. For the present study P. argentipes (N=50), S. babu (N=50) and P. papatasi (N=7) were selected. The amplicon size of amplified PCR product of P. argentipes, P. papatasi and S. babu were found to be 454bp, 453 bp and 444 bp respectively. Sequence of S. babu was submitted in GenBank with accession number JN581685.
Restriction digestion of PCR products with Hinf I produced different digested fragments in all the three sand fly species (Table 1). On agarose gel, closely sized fragments appeared as single band while lower sized products could not be observed. Thus, the gel image showed single band of 121 bp for P. argentipes, two bands of 109 bp and 319 bp for P. papatasi and two bands of 120 bp and 191 bp for S. babu (Fig. 2).
Table 1.
Hinf I restriction digestion pattern
| Sand fly species |
Position of cutting sites | Length of restriction digested fragments (bp) |
Numbers of bands observed in 1.5 % (w/v) agarose gel |
|---|---|---|---|
| PA | 109, 198, 319, 331, 441 | 12, 12, 89, 109, 110, 121 | 1 band of 121 bp |
| PP | 319, 331, 440 | 12, 12, 109, 319 | 2 bands of 109 and 319 bp |
| SB | 191, 311, 323, 433 | 10, 12, 110, 120, 191 | 2 bands of 120 and 191 bp |
PA: Phlebotomus argentipes, PP: Phlebotomus papatasi, SB: Sergentomyia babu
While restriction enzyme Hpa II also had different cutting sites (Table 2). In gel image, there were three bands of 68 bp, 99 bp and 236 bp for P. argentipes, two bands of 68 bp and 168 bp for P. papatasi and two bands of 68 bp and 337 bp for S. babu. Thus, three different patterns were produced that were easily visible in agarose gel (Fig. 2).
Table 2.
Hpa II restriction digestion pattern
| Sand fly species |
Position of cutting sites |
Length of restriction digested fragments (bp) |
Numbers of bands observed in 1.5 % (w/v) agarose gel |
|---|---|---|---|
| PA | 236, 335, 403 | 51, 68, 99, 236 | 2 bands of 99 and 236 bp |
| PP | 167, 335, 403 | 53, 68, 167, 168 | 2 bands of 68 and 168 bp |
| SB | 327, 395 | 59, 68, 337 | 2 bands of 68 and 337 bp |
PA: Phlebotomus argentipes, PP: Phlebotomus papatasi, SB: Sergentomyia babu
Discussion
For the study of vector borne diseases, proper identification of the species of the vector is very important. Very little information is available regarding sand fly population in India in the VL endemic region. As female P. argentipes is considered as the vector for L. donovani, it is very important to differentiate it from other sand fly species. There are various species of sand flies found in the same habitat and they share several morphological features. Only expert entomologists can perform such differentiation as it requires consistent observation under stereomicroscope using various identification keys. Although morphological identification of different sand fly species is feasible when dealing with small samples size but it might be time consuming when large sample size is to be identified. Further, for better understanding of transmission dynamics of a vector borne infectious disease, a large sample size is needed which is collected at different time intervals. For such studies, a reliable molecular method of species identification is needed. With the technique described in the present study, species identification of sand flies in large numbers can be done in a short time with limited resources.
Other studies, using molecular methods have been done earlier to differentiate different species of sand flies. One such study has been reported to differentiate only P. argentipes from other sand flies (Surendran et al. 2005), while in another study, only two species P. argentipes and P. papatasi could be identified (Manonmani 2010). Similar studies from Greece, Cyprus (Aransay 1999) and Sudan (Khalid et al. 2010) have been reported but none of these species are found in India, and in most of these descriptions are morphological characterisation using microscopic method.
The present molecular assay for reliable identification of regional sand fly species may contribute to the knowledge of species distribution and better understanding of the entomologic aspects of the transmission of VL. This method combines a single step of PCR and RFPL thus it is less time consuming and at the same time multiple samples can be processed simultaneously. Each of the three sand fly species can be identified by visualizing the agarose gel in UV transilluminator as each species produces a unique pattern after restriction digestion that making it easier and convenient tool.
Acknowledgement
The study was funded by National Institute of Allergy and Infectious disease (NIAID), DMID funding mechanism: Tropical Medicine Research Center Grant number: P50AI074321. Puja Tiwary and Dinesh Kumar like to thanks CSIR for providing fellowships.
References
- Aransay AM, Scoulica E, Chaniotis B, Tselentis Y. Typing of sandflies from Greece and Cyprus by DNA polymorphism of 18S rRNA gene. Insect Mol Biol. 1999;8:179–184. doi: 10.1046/j.1365-2583.1999.820179.x. [DOI] [PubMed] [Google Scholar]
- Desjeux P. Leishmaniasis. Public health aspects and control. Clin Dermatol. 1996;14:417–423. doi: 10.1016/0738-081x(96)00057-0. [DOI] [PubMed] [Google Scholar]
- Herwaldt BL. Leishmaniasis. Lancet. 1999;354:1191–1199. doi: 10.1016/S0140-6736(98)10178-2. [DOI] [PubMed] [Google Scholar]
- Khalid N, Elnaiem D, Aboud M, Al Rabba F, Tripet F. Morphometric and molecular differentiation of Phlebotomus phlebotomus sandflies. Med Vet Entomol. 2010 doi: 10.1111/j.1365-2915.2010.00893.x. [DOI] [PubMed] [Google Scholar]
- Lewis D. Phlebotomine sandflies (Diptera:Psychodidae)from the Oriental region. Syst Entomol. 1987;12:163–180. [Google Scholar]
- Manonmani AM, Mathivanan A, Srinivasan R, Jambulingam P. Species-Diagnostic Polymerase Chain Reaction Assays for Phlebotomus argentipes and Phlebotomus papatasi Vectors of Leishmania. J Med Entomol. 2010;47:743–747. doi: 10.1603/me09195. [DOI] [PubMed] [Google Scholar]
- Surendran SN, Karunaratne SH, Adams Z, Hemingway J, Hawkes NJ. Molecular and biochemical characterization of a sand fly population from Sri Lanka: evidence for insecticide resistance due to altered esterases and insensitive acetylcholinesterase. Bull Entomol Res. 2005;95:371–380. doi: 10.1079/ber2005368. [DOI] [PubMed] [Google Scholar]
- Swaminath CS, Shortt HE, Anderson LA. Transmission of Indian kala-azar to man by the bites of Phlebotomus argentipes, ann and brun. 1942. Indian J Med Res. 2006;123:473–477. [PubMed] [Google Scholar]