Abstract
Background
Drug interactions in oncology are common place and largely ignored as we tolerate high thresholds of ‘toxic’ drug responses in these patients. However, in the era of ‘targeted’ or seemingly ‘less toxic’ therapy, these interactions are more commonly flagged and contribute significantly towards poor ‘quality of life’ and medical fatalities.
Objective
This review and opinion article focuses on alteration of chemotherapeutic pharmacokinetic profiles by drug interactions in the setting of polypharmacy. The assumption is that the drugs, with changes in their pharmacokinetics, will contribute towards changes in their pharmacodynamics.
Methods
The examples cited for such drug–drug interactions are culled from published literature with an emphasis on those interactions that have been well characterized at the molecular level.
Results
Although very few drug interaction studies have been performed on approved oncology based drugs, it is clear that drugs whose pharmacokinetics profiles are closely related to their pharmacodynamics will indeed result in clinically important drug interactions. Some newer mechanisms are described that involve interactions at the level of gene transcription, whereby, drug metabolism is significantly altered. However, for any given drug interaction, there does not seem to be a comprehensive model describing interactions.
Conclusions
Mechanisms based drug interactions are plentiful in oncology; however, there is an absolute lack of a comprehensive model that would predict drug–drug interactions.
Keywords: drug metabolism, orphan nuclear receptors, pharmacokinetic–pharmacodynamic relationships, pregnane X receptor, transcriptional regulation
1. Introduction
The purpose of a critical review of the topic of pharmacokinetic cancer drug interactions stems from a vast majority (> 90%) of patients with cancer who consume several medications (i.e., intake of more than two different drug types). Furthermore, for a number of cancer drugs, there is a good correlation between pharmacokinetics (PK) (what the body does to the drug) and pharmacodynamics (PD) (what the drug does to the body) (Table 1). This implies that perturbations in PK may impact PD (e.g., toxicity and efficacy) [1–5]. Moreover, in a questionnaire based review of cancer patients (n = 405) ~9 and 77% of potential drug interactions were considered of major and moderate severity, respectively. In the same study, >55% of potential drug interactions were classified as pharmacokinetic [6,7]. Overall (all disciplines), drug–drug interactions cause roughly 2.8% of all hospitalizations, and using a cost-of-illness model, represent 245,280 hospital admissions/year, costing the health care system ~ $1.3 billion [8,9]. Hence, the study of mechanisms that relate to alterations in chemotherapeutic PK profiles by drug–drug interactions is significant and clinically relevant.
Table 1.
| Drug | PK variable | PD effect | Ref. |
|---|---|---|---|
| 5-fluorouracil | AUC | Neutropenia and thrombopenia | [1,2] |
| CPT-11 | Biliary index | Diarrhea | [3] |
| Carboplatin | AUC | Thrombocytopenia | [4] |
| Cyclophosphamide | AUC | Toxicity and survival | [5] |
PD: Pharmacodynamics; PK: Pharmacokinetics.
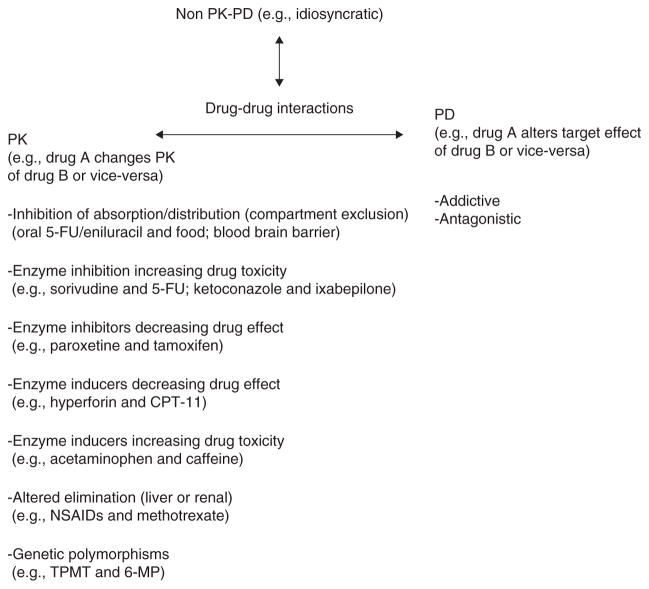
Drug–drug interactions in oncology may be viewed broadly in three categories: i) those that alter PK, ii) those that alter PD, and iii) those that apparently do not have observable effects on PK or PD but are known to give erratic, perhaps rare, side effects that may often be termed ‘idiosyncratic’ (Figure 1). Within these categories, this opinion paper focuses on the first category, namely, drug interactions that alter chemotherapy PK. In this category, there are at least seven broad categories that summarize the mechanistic basis for drug interactions. These include changes in absorption/transport, enzyme inhibition increasing drug toxicity, enzyme inhibitors decreasing drug effect, enzyme inducers decreasing drug effect, enzyme inducers increasing drug toxicity, altered elimination and genetic polymorphisms in all metabolic processes. For each of these categories, I will select a relevant example that has a clearly defined mechanistic basis.
Figure 1. Mechanistic categories of drug interactions that affect drug PK.
PK: Pharmacokinetics; TPMT: Thiopurine methyltransferase; 6-MP: 6-Mercaptopurine.
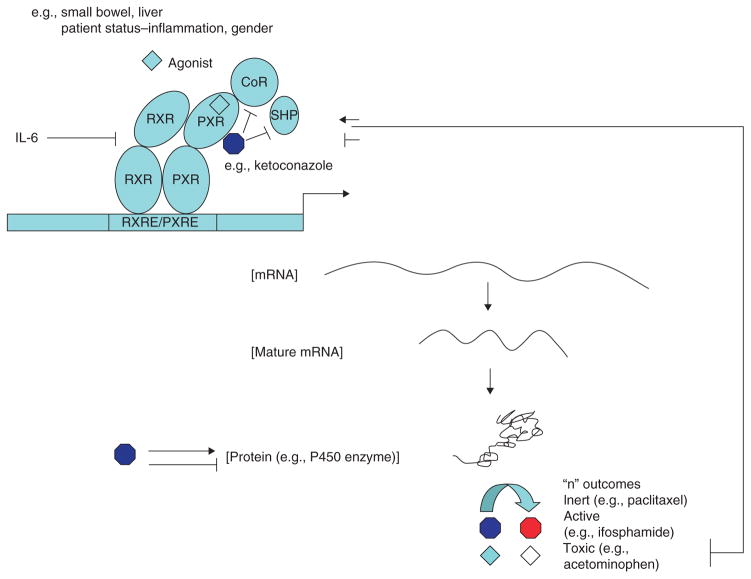
In addition to the mechanisms defined in these seven broad categories, most of which deal with enzymes and transporter activity, there is an emerging field of study in the transcriptional control of enzyme/transporter action. It is well established that most enzymes and transporters involved in xenobiotic metabolism are under transcriptional rather than translational control. This implies that enzyme/transporter induction or repression is tissue-specific and under control of master regulators (e.g., nuclear receptors). This has opened a new field of study whereby drug interactions may occur from alterations of transcription factor assembly through receptor competition resulting in altered drug clearance. There are numerous examples of how such interactions can alter chemotherapeutic efficacy and toxicity (e.g., hyperforin and irinotecan) (Figure 2).
Figure 2. Mechanisms involving ligand activation or inhibition (e.g., ketoconazole) of PXR that can alter expression of target genes (enzymes/transporters) affecting drug pharmacokinetics.
The same ligands that activate or inhibit PXR may also directly affect the downstream enzymes adding complexity to drug interactions.
PXR: Pregnane X receptor.
2. Mechanistic basis for drug–drug interactions
2.1 Changes in absorption/transport
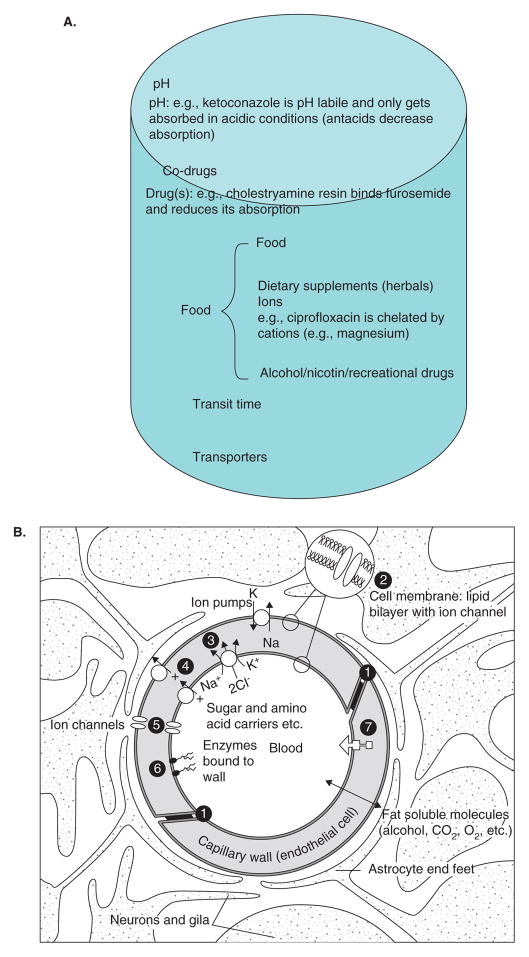
Oral drugs have to be absorbed and intravenous/cutaneous application of drugs has to be distributed (Figure 3A). These involve transporter based or facilitated movement of drugs from one compartment to the other. For example, ketoconazole (dibasic, pKa values ~ 6.51 and 2.94) when orally delivered, absolutely requires an acidic medium to maintain it in a form that could cross the gastric mucosa [10]. Therefore, consumption of antacids or drugs that block acid formation in the stomach (e.g., prevacid) could significantly inhibit ketoconazole absorption [6,7]. Another common basis for drug interaction occurs based on food consumption (mainly fatty meals) and drugs. For example, eniluracil is a potent inhibitor of the first enzyme in the catabolism of 5-flourouracil (5-FU) [11–16]. Hence, very small doses of 5-FU may be administered orally in conjunction with eniluracil. In investigations conducted at the University of Chicago Medical Center, it was apparent that even small changes in oral 5-FU dosing in the context of eniluracil therapy would result in substantial changes in 5-FU PK and PD. Therefore, it became critical to determine the effects of food consumption on dosing of these two orally administered drugs. In a randomized, open-label, two-way crossover study, 12 patients received eniluracil (50 mg, orally) on days 1 and 2 and 5-FU (20 mg/m2, orally) on day 2 following either a 2-h fast or 20 min after a standard meal in 7-day treatment intervals. Timed blood samples were collected during the first two treatment periods and 5-FU concentrations determined by GC/MS. Data were analyzed and pharmacokinetic parameter estimates were obtained using a non-compartmental, two-stage and population analysis method. In fasted individuals, the clearance/bioavailability of 5-FU was estimated to be 5.6 l/h. The mean absorption lag-time was 0.24 h and was followed by rapid absorption of 5-FU. Administration of 5-FU and eniluracil with food resulted in a decrease in the 5-FU absorption rate constant by 90%. As a result, the Cmax of 5-FU was decreased by 21% and the time to Cmax was increased 2.9-fold. Clearance of 5-FU, relative bioavailability and AUC remained unchanged with co-administration of food. Similar results were obtained using all three data analysis methods. Administration of food with oral 5-FU and eniluracil slowed absorption of 5-FU and decreased 5-FU Cmax, but did not affect AUC [7]. The implications of these findings are significant, in that, it is known that 5-FU Cmax levels are associated with neuropathy (i.v. dosing) [17]. Therefore, controlling Cmax levels with this therapy would be important to avoid inadvertent side effects.
Figure 3. Mechanistic basis for drug interactions.
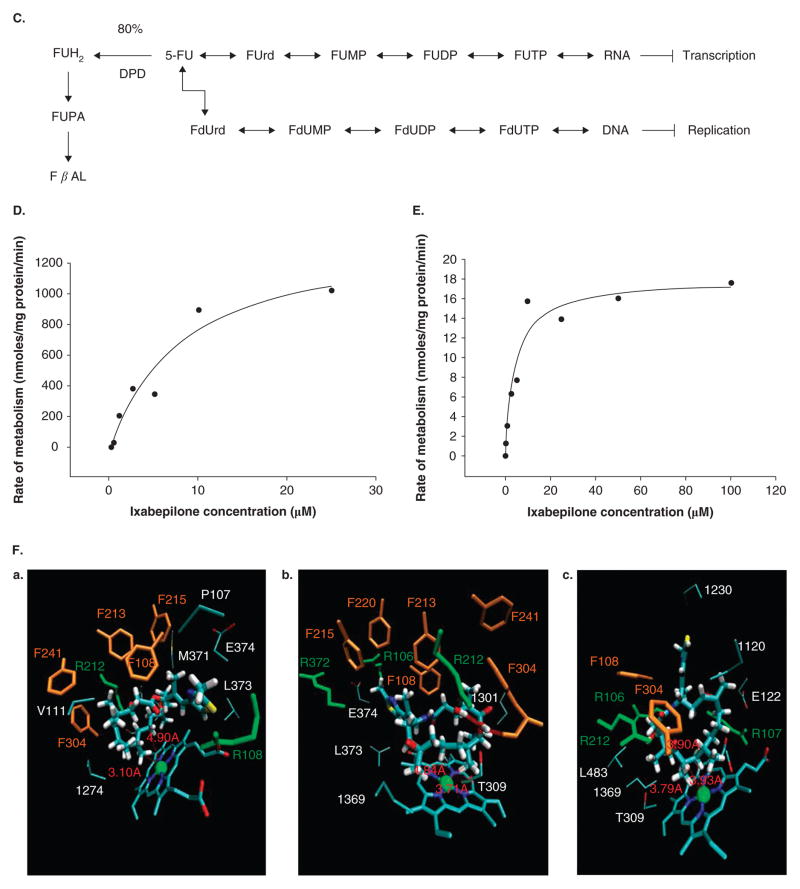
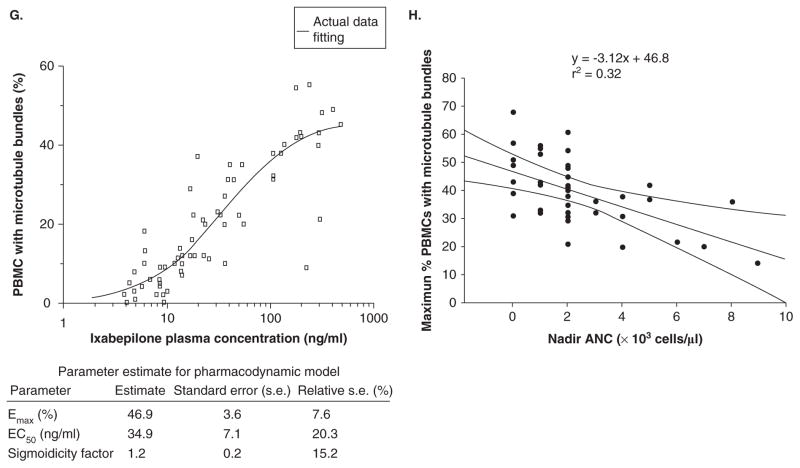
A. Factors affecting absorption and distribution of drugs. B. Components of the blood–brain barrier. Diagram of a cerebral capillary enclosed in astrocyte end-feet. Characteristics of the blood–brain barrier are indicated: (1) tight junctions that seal the pathway between the capillary (endothelial) cells; ii) the lipid nature of the cell membranes of the capillary wall that makes them a barrier to water-soluble molecules; (3i), (4) and (5) represent some of the carriers and ion channels; (6) the ‘enzymatic barrier’ that removes molecules from the blood; and vii) the efflux pumps that extrude fat-soluble molecules that have crossed into the cells. From http://answers.com/topic/blood-brain-barrier; courtesy Malcolm Segal. C. Enzymatic biotransformation pathway of 5-flourouracil and DPD. D. Concentration-dependent metabolism of ixabepilone in human liver microsomes. E. Concentration-dependent metabolism of ixabepilone in human cDNA expressed CYP3A4 enzymes F. Results of dynamic docking simulations: 3D representations of the two ligands docked into the active site of CYP3A4 after equilibration and minimization. The closest side chains of the vicinal residues (< 3.0 Å) are represented. Phenylalanine side chains belonging to the CYP3A4 phenylalanine cluster are displayed in orange, basic side chains (Arg) in green. a. Epothilone B; b. one docking mode of ixabepilone. For sake of clarity, the orientation of the representation has been changed with respect to panels a and c; c. the other docking mode of ixabepilone; b and c have similar docking energies according molecular mechanics calculations (see [42]). G. Relationship of percent PBMCs with microtubule bundles and ixabepilone plasma concentration (see [42]). H. Relationship between percent PBMCs with microtubule bundles and nadir ANC. The line describes a negative linear fit equation with r2 = 0.32 and curved lines represent 95% CI.
ANC: Absolute neutrophil count; DPD: Dihydropyrimidine dehydrogenase; FUH2: Dihydrofluorouracil; PBMC: Peripheral blood mononuclear cell.
The blood–brain barrier (BBB) functions to protect the brain from changes in the levels in blood of ions, amino acids, peptides and other substances. The barrier is located at the brain blood capillaries that are unique. First, the cells that make up the walls of these vessels (the endothelium) are sealed together at their edges by tight junctions that form a key component of the barrier. These junctions prevent water-soluble substances in the blood from passing between the cells and, therefore, from freely entering the fluid environment of the brain cells. Second, these capillaries are enclosed by the flattened ‘end-feet’ of astrocytic cells (one type of glia), which also act as a partial, active, barrier. Thus, the only way for water-soluble substances to cross the BBB is by passing directly through the walls of the cerebral capillaries, and because their cell membranes are made up of a lipid/protein bilayer, they also act as a major part of the BBB [18]. Toxic fat-soluble xenobiotics require transporters to carry them through into the brain parenchyma (Figure 3B) [19–22]. These transporters largely belong to the ABCC transporters (e.g., MRP2, MDR1) that are diverse in the substrates they transport out into the blood vessel lumen. For example, paclitaxel is generally retained in low concentration in the brain largely because it is a substrate for the MDR1 gene product (p-glycoprotein) [22]. MDR1 is also regulated at the level of gene transcription and is also under the influence of orphan nuclear receptor systems such as pregnane X receptor (PXR) [23,24]. Therefore, there are potential mechanisms that can alter MDR1 and create drug interactions [25].
The development of agents to inhibit P-glycoprotein at the cellular level and, thus, increase intracellular concentrations of toxic chemotherapy agents is a viable strategy. Inhibitors tested clinically include verapamil, quinidine, cyclosporine, PSC-833 (valspodar), GF120918 (elacridar) and XR9576 (tariquidar). Clinical trials in both solid and hematologic malignancies testing P-glycoprotein inhibitors with cytotoxic P-glycoprotein substrates to overcome cancer cell resistance have been disappointing. Promising Phase II trials were followed by negative Phase III trials, at times with trials being stopped early owing to unacceptable toxicities [26,27]. The molecular mechanisms underlying such interactions are complex and do not follow simple kinetics. For P-glycoprotein, an ATP-binding cassette (ABC) transporter, the inhibition could result from either competition between drug and inhibitor on substrate-binding sites or ATP-binding sites or from the blockage of ATP hydrolysis (e.g., verapamil inhibits P-glycoprotein in a competitive manner but does not affect the ATP hydrolysis). In the substrate-induced fit model, several substrates could bind through specific shifts in the transmembrane segment. These interactions are usually inhibitory but stimulation of P-glycoprotein efflux transport has also been observed (e.g., flavonoids). There are several variations in the allosteric-binding mechanisms for other transporters (e.g., MRP2) that increase the repertoire of transporter drug–drug interactions [28,29].
These negative results have put in doubt the strategy of overcoming cellular drug resistance by the use of P-glycoprotein inhibitors. However, the potential role of P-glycoprotein inhibitors in overcoming the BBB is still an open question. In animal models, the administration of P-glycoprotein inhibitors has been found to increase intracranial concentrations of chemotherapy agents [27,30]. In mice given PSC-833 with paclitaxel, increased concentrations of paclitaxel were found in the brain, and these increased concentrations led to a higher tumor response in these mice [31,32]. Further studies using animal models have found increased concentrations of vinblastine as well as morphine-6-glucuronide when co-administered with PSC-833 and GF 120918 [33,34]. Further evaluation of the use of these inhibitors and their potential role in inhibiting P-glycoprotein at the BBB and, thus, increasing drug levels in the CNS, is warranted. These avenues of research are important given the FDA guidelines on drug interaction studies with p-glycoprotein [35,36].
In humans, these observations of drug interactions leading to poor delivery of drugs (e.g., irinotecan) to the brain have been attributed to systemic metabolism of the drug (e.g., CYP450-inducing drugs and irinotecan in patients with brain cancer). However, it is equally likely that the agents inducing CYP450 peripherally (e.g., valproate, steroids, phenytoin) also act in the brain capillaries through nuclear receptors such as PXR inducing enzymes and transporters. Therefore, strategies to limit PXR activation both systemically and in the brain could assure delivery of drugs to the right compartment at therapeutic concentrations. Of course, this could also be achieved at higher doses of the drug, but due to the duality of metabolism (e.g., in any given case we do not yet understand contributions of systemic versus BBB metabolism that limits drug delivery to the brain) there remains a risk of underestimating delivery to the brain even at toxic systemic doses.
In another example of drug interactions predicted to occur through inhibition of CYP450s, it has become increasingly clear that such interactions are not as ‘pure’ as described in the literature. Ketoconazole is a potent inhibitor of CYP3A4 (and other CYPs) and such has been used as an inhibitor of CYP3A4 in the clinic. Drug–drug interaction (PK) studies have been conducted using ketoconazole to verify and validate the role of CYP3A4 metabolism in vivo. We have been interested in the study of drug metabolism through PXR and have shown that ketoconazole is a moderate inhibitor of activated nuclear receptors (aside from GR and AR, predominantly orphan receptors have been studied in this context). We have shown that whereas ketoconazole weakly activates PXR, it is a moderately strong inhibitor (IC50 ~ 28 μM) of activated PXR. In the context of activating ligands of PXR (e.g., paclitaxel), ketoconazole is able to inhibit the induction of PXR target genes (e.g., CYP450s and ABCC transporters) (Figure 2) [37,38]. Therefore, ketoconazole can affect drug PK through mechanisms beyond CYP450 inhibition. This feature is relevant for newer drugs such as the epothilones that as yet do not have a fully characterized metabolic pathway.
2.2 Enzyme inhibition increasing drug toxicity
In this category of drug–drug interactions, many chemo-therapeutic drugs show altered PK profiles. In some cases, this leads to alterations in PD. For example, inhibition of the CYP450 family of enzymes can significantly alter the metabolism of chemotherapeutic drugs such as the microtubule-binding drugs (e.g., CYP3A4 and vinca alkaloids). To better understand how these interactions may take place, I focus this section on two well described mechanisms based interactions. One involves drugs, sorivudine and 5-FU, and the other ketoconazole and ixabepilone.
2.3 Sorivudine and 5-FU
Sorivudine (1-β-D-arabinofuranosyl-E-5-[2-bromovinyl] uracil; BV-araU; SQ32,756) (Figure 3C) [39–41] is a pyrimidine nucleoside analogue with potent clinical activity against varicella–zoster and other viral infections (e.g., herpes). However, recent studies have shown that there is a potent drug interaction with 5-FU (severe bone marrow suppression). The reason for this is that the sorivudine metabolite, bromovinyluracil, is a potent inhibitor of dihydropyrimidine dehydrogenase (DPD). It has been shown that the metabolite levels after a single dose of sorivudine completely suppress DPD enzyme activity in peripheral blood mononuclear cells. DPD is the first rate-limiting step in the catabolism of 5-FU to dihydrofluorouracil (Figure 3D) and is the only pathway for detoxification of 5-FU. Inhibition of DPD results in loss of 5-FU catabolism and substantially increases 5-FU levels. These levels can be fatal and cause severe toxicity (GI distress, bone marrow toxicity).
2.4 Ketoconazole and Xempra (ixabepilone)
Xempra® (Ixabepilone (BMS-247550)) (Bristol-Myers Squibb, New York, NY, USA) [42] is an analogue of epothilone B that has been approved by the FDA for the treatment of breast cancer. The epothilones (the name being derived from molecular features: epoxide, thiazole, ketone) were originally isolated from fermentation of a common soil bacteria, Sorangium cellulosum, and later synthesized. These natural products were shown to bind to tubulin and exert potent anti-cancer activity, especially in taxane-resistant tumors.
The limited published data on ixabepilone PK in humans are characterized by linear dose kinetics, which are derived from consistent total body clearance and apparent terminal half-life across doses from 10 to 60 mg/m2. The plasma concentration–time profile of ixabepilone is characterized by a steep log decline during the first hour after the completion of the 1-h infusion. The rapid distributive phase is followed by a more prolonged terminal elimination phase with a mean half-life of 16.8 ± 6.0 to 35 ± 14.5 h. This is similar to the profile of other tubulin-binding agents, such as the vinca alkaloids and taxanes. The mean (± s.d.) end of infusion plasma concentration of BMS-247550 for the first dose on the 6 mg/(m2 d) × 5 days dose level is 93.7 ± 40.3 ng/ml. Over the 5-day treatment course, there is minimal accumulation of drug. Total-body clearance of BMS-247550 is rapid (712 ± 247 ml/(min m2)) and does not seem to be dose-dependent. The large volume of distribution (Vdss, volume of distribution at steady-state) (mean ± s.d., 798 ± 375 l) is consistent with extensive tissue binding of the drug. There was also no correlation between Vdss and body weight or surface area. At the clinically relevant schedule (1 – 3 h infusion every 3 weeks), the same PK parameters are observed. Importantly, there is no obvious dependence on body surface area for accurate dosing (flat dosing may perform equally well). Moderate to severely altered liver function may affect ixabepilone PK and toxicity, thereby, necessitating dose modifications for these patients; however, formal recommendations have not been published (pers.commun., Angela Davies, University of California at Davis, CA, USA).
A human mass balance study with ixabepilone has been published. Although, ixabepilone proves to be very unstable when labeled with a radioactive tracer like [(14)C], eight cancer patients received an intravenous dose of 70 mg, 80 nCi of [(14)C]ixabepilone over 3 h. The “mean recovery of ixabepilone-derived radioactivity was 77.3% of dose; fecal excretion was 52.2% and urinary excretion was 25.1%. Only a minor part of the total radioactivity is accounted for by unchanged ixabepilone in both plasma and urine”. This indicates that metabolism is an important elimination mechanism for this drug. Future studies should focus on structural elucidation of ixabepilone metabolites and characterization of their activities.
We and others have also demonstrated that ixabepilone has variable inter-patient PK (CV > 50%). To determine the effect of cytochrome P450s on ixabepilone biotransformation in vitro, a series of studies with liver microsomes were performed together with computational docking studies for ixabepilone using molecular dynamic simulations of CYP3A4 as well as data derived from published crystal structures. First, in vitro data clearly show that ixabepilone is a CYP3A4 substrate (Figure 3E). Modeling ixabepilone and epothilone B to the CYP3A4 catalytic site shows that the interaction energies for the former are far better than for the latter compound, suggesting extensive metabolism by CYP3A4 (Figure 3F). The modeling studies predicted a fairly tight docking into the CYP3A4 catalytic domain and based on docking alone it could be predicted that ixabepilone would probably be metabolized extensively by CYP3A4 (as compared to epothilone B). Because CYP3A4 is known to have homo- and hetero-tropic substrate interactions, the docking with ixabepilone suggested that with this compound such interactions are not likely to occur as the substrates are large enough to occupy the major part of the active site (volume of epothilone B has been estimated to be 709 Å3). However, a cooperative effect involving an external allosteric site cannot be excluded. Furthermore, pure competition effects are expected with ketoconazole as validated for other compounds.
In the clinic, ixabepilone PK and PD are significantly affected by CYP3A4 inhibitors (e.g., ketoconazole) and inducers (e.g., rifampicin). First, ketoconazole (a potent CYP3A4 inhibitor) co-administration reduced the maximum ixabepilone dose administration of 25 mg/m2 compared to single agent therapy of 40 mg/m2. Co-administration of ketoconazole with ixabepilone resulted in a 79% increase in AUC0 –∞ and a relationship of microtubule bundle formation in peripheral blood mononuclear cells to plasma ixabepilone concentration that was well described by the Hill equation (Figure 3G). Microtubule bundle formation in peripheral blood mononuclear cells correlated with neutropenia (Figure 3H). These data directly implicate CYP3A directed metabolism of ixabepilone and its effects on humans. Therefore, any drug with potential effects as a CYP3A4 inhibitor must be used with extreme caution in patients receiving ixabepilone. These dose modifications may be more important in the setting of moderate liver dysfunction, in which case use of CYP3A4 inhibitors may cause fatal drug interactions with ixabepilone. In the reverse example, rifampicin, increases CYP3A4 directed metabolism of ixabepilone (pers.commun., Bristol-Myers Squibb, CT). The plasma exposure of ixabepilone falls dramatically, necessitating an increase in dose to maintain plasma levels. Although the mechanisms of such action seem to be linked to CYP3A4 induction, both rifampicin and ketoconazole have other more general effects on drug metabolism and elimination through ligand action on adopted orphan nuclear receptors controlling xenobiotic metabolism, namely, the PXR and the constitutive androstane receptor. These mechanisms are discussed later.
2.5 Enzyme inhibitors decreasing drug effect
As opposed to enzyme inhibitors increasing drug effect, there are clear examples in oncology in which enzyme inhibitors can decrease drug effect. For example, paroxetine, a commonly used serotonin reuptake inhibitor (SSRI), decreases the conversion of tamoxifen to endoxifen (an active metabolite of tamoxifen) through CYP3A4 directed N-methylation and CYP2D6 directed hydroxylation [43,44]. Other drugs affected by similar mechanisms include irinotecan (CPT-11), iphosphamide, docetaxel and paclitaxel. Again, tamoxifen itself has effects on metabolism through activation of PXR and so in conjunction with paroxetine, the effect on enzyme inhibition becomes complex [45,46]. Also, it is unknown if paroxetine activates or inhibits PXR and other receptors. However, the net effect is clear and more studies are needed to provide a mechanistic basis that comprehensively explains this process.
2.6 Enzyme inducers decreasing drug effect
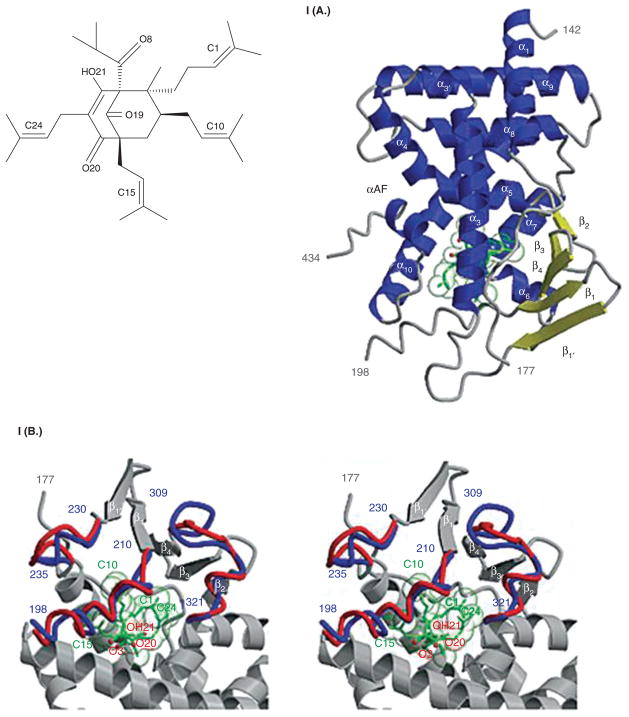
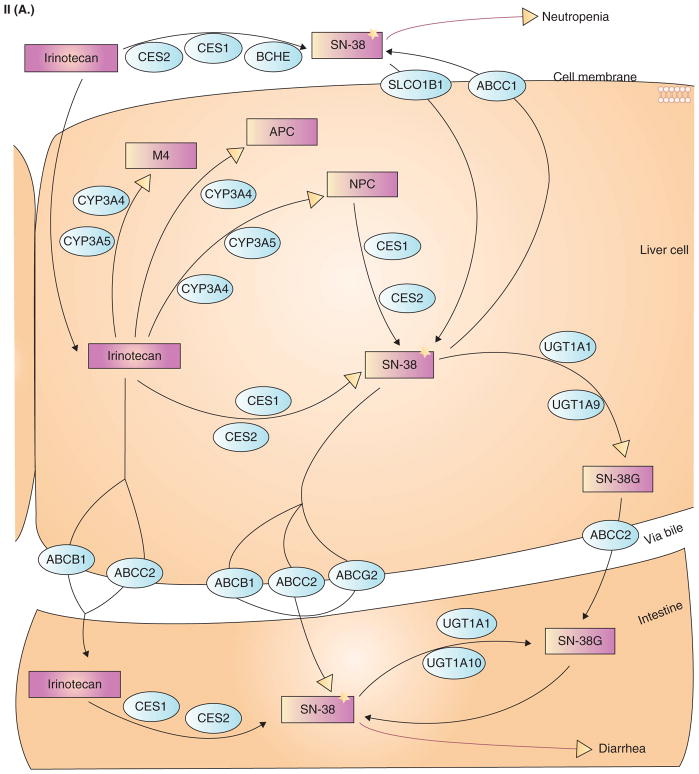
Most drugs in oncology therapeutics are inactivated by enzymes. In this setting, any xenobiotic that induces enzyme expression also reduces drug effect. A well described example is that of hyperforin (an active ingredient in St. John’s wort) and irinotecan (CPT-11) [47–52]. First, hyperforin is a potent PXR ligand (EC50 ~ 23 nM) and binds the PXR ligand-binding pocket (Figure 4.I(A)) [51]. It induces PXR target genes in liver and gut cells. In patients receiving both irinotecan and hyperforin, there is a marked reduction in SN-38 (active metabolite of CPT-11) intracellular accumulation and glucuronidation (inactive metabolite of CPT-11) (Figures 4.I(B), 4.II(A), 4.II(B)). This results in reduced toxicity but also possibly reduced activity of irinotecan [48,53].
Figure 4. Molecular basis for drug interactions.
I. Chemical structure of hyperforin. Crystal structure of human PXR in complex with hyperforin. I(A) 2.1 Å crystal structure of the ligand-binding domain of human PXR bound to hyperforin, an active component of St. John’s wort. PXR residues 142 – 177 and 198 – 434 are shown with α helices in blue and β strands in yellow; hyperforin is rendered in green. The activation function 2 helix, which mediates interactions with transcriptional co-activators and corepressors, is also labeled (αAF). I(B) Stereoview of a superposition between the PXR-hyperforin (red) and PXR-SR12813 (blue) complexes. The α helix 6 novel to the hPXR-LBD/hyperforin structure is labeled in red. Shifts in the loops between residues 198 and 210 and between residues 230 and 235 are also shown (see [52]).
II(A) Metabolism of CPT-11. From http://www.pharmgkb.org/search/pathway/irinotecan/liver.jsp. Copyright 2004 PharmGKB.
II(B) Legend keys to Figure 4II(A).
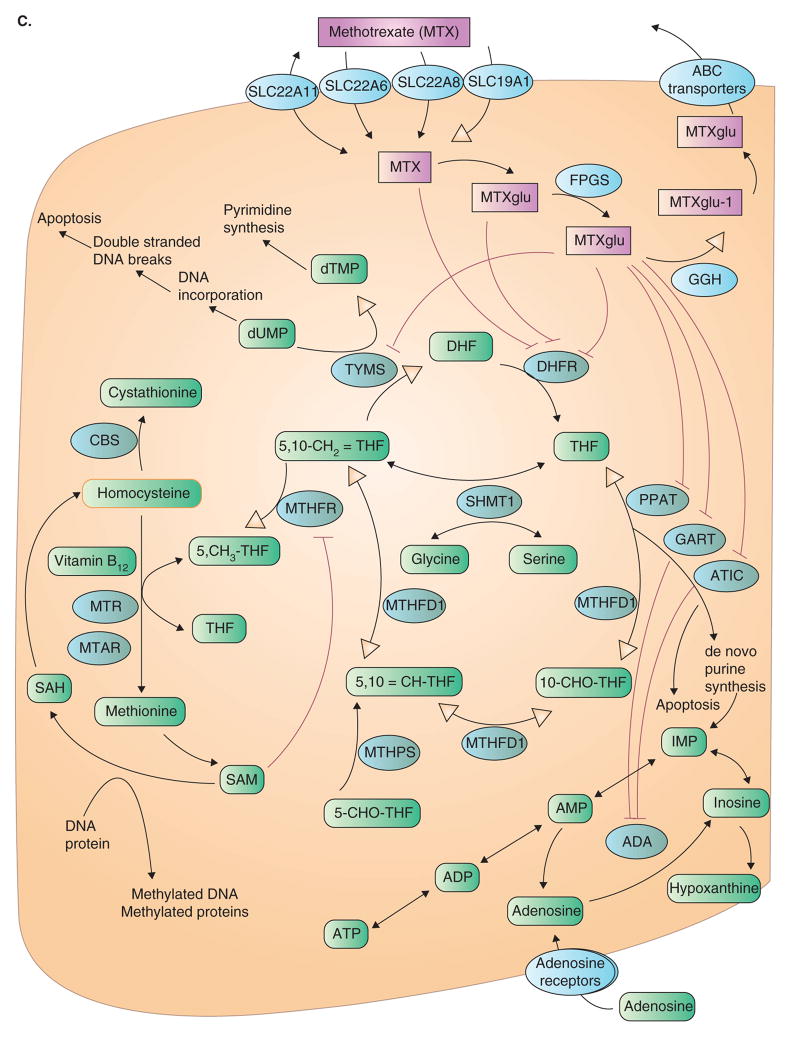
I. Chemical structure of hyperforin. C. This pathway illustrates the genes involved in mediating the effects of the antimetabolite MTX, an analogue of reduced folate, which targets endogenous cellular folate metabolism. At pharmacologic concentrations of 0.1 – 10 mM, cellular entry of MTX is by the reduced folate carrier (SLC19A1). Its main intracellular target is dihydrofolate reductase, inhibition, which results in accumulation of dihydrofolate and a depletion of cellular folates. Cytoplasmic folylpolyglutamyl synthase adds glutamate residues to MTX; these larger and more polar metabolites enhance activity by increasing intracellular retention, as they are not substrates for folate transport systems, and by increasing the affinity for target enzymes (TYMS, PPAT, GART and ATIC). From http://www.pharmgkb.org/search/pathway/mtx/methotrexate.jsp. Copyright 2004 PharmGKB.
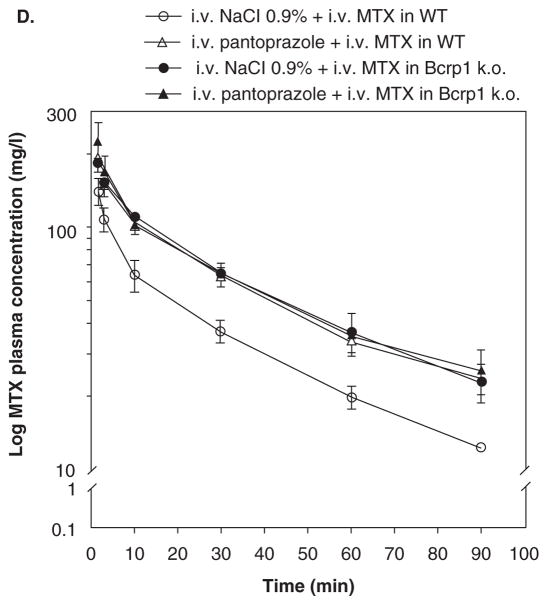
D. Semilogarithmic plot of MTX plasma concentration versus time curves in mice pretreated with i.v. pantoprazole or i.v. NaCl 0.9% (control). Bcrp1 knockout (k.o.) mice (closed symbols) or wild-type (WT) mice (open symbols) were treated with i.v. NaCl 0.9% (circles) or i.v. pantoprazole (40 mg/kg ≈ 120 mg/m2; triangles) 3 min before an i.v. dose of MTX (85 mg/kg ≈ 255 mg/m2). Plasma levels of radiolabeled MTX were determined by liquid scintillation counting at t = 1.5, 3, 10, 30, 60 and 90 min. Results are the means ± s.d. (n = 4). Bars ± s.d. (From Breedveld et al. [60]).
I. Chemical structure of hyperforin. E. (b) From http://en.wikipedia.org/wiki/image:PBB_Protein_TPMT_image.jpg.
MTX: Methotrexate; PXR: Pregnane X receptor.
2.7 Enzyme inducers increasing drug toxicity
Caffeine accelerates the absorption of acetaminophen (APAP) and enhances the analgesic effect of acetaminophen. These effects are seen with higher exposure to acetaminophen after oral loading. Caffeine also induces a member of the CYP450 3A family [54,55]. However, acetaminophen is metabolized to a toxic metabolite by CYP450 (3A4). Oxidation of APAP by P450 3A4, as detected by the formation of its glutathione adduct, has been found to exhibit negative homotropic cooperativity with a Hill coefficient of 0.7. In the presence of caffeine, the observed kinetics are close to classical Michaelis–Menten kinetics with a Hill coefficient approaching 1. Both APAP and caffeine have been found to bind at the active site in proximity to the heme iron using NMR T1. When APAP is incubated with P450 3A4, the acetamido group of APAP was found to be closest to the heme iron consistent with the amide group of APAP weakly associating with the heme iron. The addition of caffeine disrupted the ability of APAP to coordinate with the heme iron of P450 3A4 and enhanced the rate of oxidation to its toxic metabolite. Thus, in this example, enzyme induction is coupled with cooperative enzyme-substrate effects causing facilitated induction of a toxic metabolite of APAP [56]. As much of our caffeine comes from coffee, it would be important to note that another ingredient of coffee, cafestol, is a potent activator of PXR and farnesol X receptor [57,58]. Therefore, this would add to the complexity of interactions when consuming significant amounts of coffee with high doses of acetaminophen daily.
2.8 Altered elimination
Drug interactions leading to progressive accumulation of toxic metabolites and/or parent compound(s) represent another mechanism of toxicity or unanticipated side effects. In this context, one well known interaction is that of methotrexate (MTX) and NSAIDs.
Methrotrexate is an analogue of the natural folate and used widely in the treatment of autoimmune (e.g., rheumatoid arthritis) and cancer (e.g., leukemia) (Figure 4.I(C)). There are numerous reports that indicate that there are significant drug–drug interactions with MTX and NSAIDs (e.g., diclofenac) as well as other drugs such as penicillin and probenecid. These interactions all lead to an increase in the plasma MTX exposure from varying mechanisms. Until now, several potential sites have been proposed for these drug–drug interactions: an increase in the unbound fraction of MTX, a decrease in the urine flow rate resulting from the inhibition of prostaglandin synthesis and inhibition of the renal tubular secretion of MTX [59–62].
Renal elimination (and tubular secretion) of unchanged form of MTX is a major mechanism of regulating its PK. According to inhibition studies using transfectants expressing hOAT1 and hOAT3, NSAIDs have been shown to be potent inhibitors of hOAT3, and inhibition of hOAT3 by certain NSAIDs such as salicylate, indomethacin and phenylbutazone are likely to occur at clinical doses. More recently, it has been shown that renal uptake of MTX through the basolateral membrane is also a potential site of drug–drug interaction, with varying contributions of rOat1, rOat3 and RFC-1 [62–64].
Another prototypical interaction is shown with MTX and benzimidazoles. In cancer patients, co-administration of benzimidazoles and MTX can result in profound MTX-induced toxicity, which correlates with an increase in the serum concentrations of MTX and its main metabolite 7-hydroxymethotrexate [65,66]. In this context, benzimidazoles differentially affect transport of MTX mediated by BCRP and MRP2 [66]. Competition for BCRP may explain the clinical interaction between MTX and benzimidazoles (Figure 4.I(D)) [67].
2.9 Genetic polymorphisms in all metabolic processes
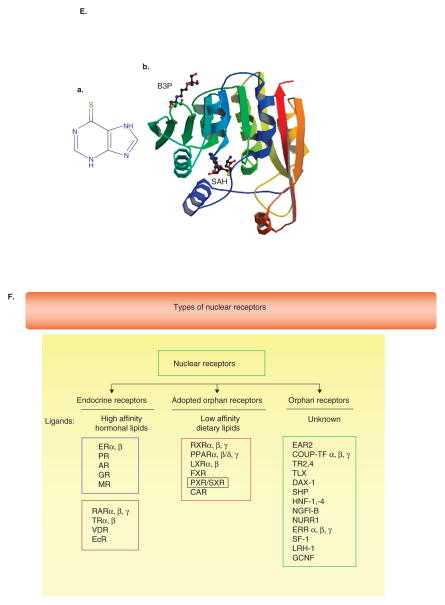
As drugs are metabolized by enzymes and transporters that are largely under the regulation of gene transcription and translation, natural polymorphisms and/or mutations may grossly alter expression and subsequent metabolic pathways. A classical example of this is the evolution and discovery of thiopurine methyltransferase (TPMT) polymorphisms and metabolism of 6-mercaptopurine (Figure 4.I(E), a). The TPMT gene encodes the enzyme that metabolizes thiopurine drugs through S-adenosyl-L-methionine as the S-methyl donor and S-adenosyl-L-homocysteine as a byproduct. Genetic polymorphisms that affect this enzymatic activity are correlated with variations in sensitivity and toxicity to such drugs in individuals. TPMT activity (PDB structure, 2bzg, Figure 4.I(E), b) should be measured before commencing the treatment of patients with thiopurine drugs such 6-mercaptopurine and 6-thioguanine. Patients with low activity (10% prevalence) or especially absent activity (prevalence 0.3%) are at a heightened risk of drug-induced bone marrow toxicity (myelosuppression, anemia, bleeding, infection) due to accumulation of the unmetabolized drug [67–70]. Reuther et al. found that about 5% of all thiopurine therapies will fail due to toxicity [71,72]. There seems to be a great deal of variation in TPMT mutation, with ethnic differences in mutation types accounting for variable responses to 6-mercaptopurine [73]. Although genotyping can identify polymorphisms affecting enzyme function (e.g., polymerase chain reaction assays developed to detect the G238C transversion in TPM*2 and the G460A and A719G transitions in TPM*3 alleles), enzyme function may be better predicted on clinical samples using immunodiagnostic kits[74]. The cost-effectiveness of pretreatment screening has been demonstrated across some but not all conditions [69].
Other classic examples of alteration of drug PK and PD include polymorphisms of UGT1A1 and CPT-11 toxicity, MDR1 polymorphisms in drug PK (e.g., paclitaxel) and disease outcome (e.g., AML) [75–78]. However, these associations have yet to take common place as a pre-condition to therapy. These genetic association studies do not give indication as to dose modifications and as such provide only a propensity towards toxicity. However, it is important to document these polymorphisms in patients because most patients are on several drugs. For example, a brain tumor patient may probably be on an anti-epileptic agent as well as steroid and other cancer drugs. Because transporters are important in brain transport of these drugs (e.g., ABCB1 3435) as well as in regulation of chemotherapy PK (e.g., paclitaxel), it would be important to use a systems approach in evaluating the patient. In this instance, there could be a poor prognosis not only because of alteration in paclitaxel compartment PKs but also owing to anti-epileptic drug resistance.
2.10 Drug interactions through nuclear (adopted or orphan) receptors
A major theme of drug–drug interactions has always focused on enzyme alterations. However, more recently with the cloning of the xenobiotic receptors, several studies have shown that drug interactions may occur through interactions with orphan or adopted orphan receptors.
PXR is a prototypical xenobiotic receptor expressed largely in liver and proximal gut. However, less abundant expression in other tissues such as the BBB (e.g., endothelial cells) and other sites have impact on drug disposition. For example, xenobiotic ligand agonists (comprising ~5 – 10% of human pharmaceutica) of PXR can induce enzyme and transporter expression that alter xenobiotic metabolism and affect its PK. Hyperforin, the main ingredient in St. John’s wort (contrary to clinical data, often used as herbal therapeutics for depression or dysthymia), can significantly alter chemotherapy metabolism, largely through CYP450 metabolism (e.g., irinotecan). Hyperforin is a PXR agonist and induces CYP3A4 expression probably through PXR activation in the liver and gut. In conjunction with irinotecan therapy, hyperforin significantly reduces plasma SN-38 exposure, which reduces both toxicity and efficacy. Another drug, rifampicin, used in the treatment of tuberculosis, significantly induces PXR activation and targets gene expression of metabolizing enzymes [79–106]. In fact, there is a crystal structure defining interactions between rifampicin and PXR as well as hyperforin and PXR [93,107,108]. These structures show that the ligand-binding pocket of PXR is very labile and may be stretched significantly and this may explain why a variety of chemotherapeutics (differing in size and structures) may activate PXR (e.g., paclitaxel). Because PXR is able to accommodate several ligands into its pocket simultaneously, several drug interaction phenotypes may occur during the course of administration of several medications. As an example, herbals such as Wu Wei Zi (Schisandra chinensis Baill) and Gan Cao (Glycyrrhiza uralensis Fisch), which are regularly consumed in the Chinese population for general health, induce PXR activation and rapid clearance of warfarin in rats [109]. Clinical studies proving the effect of PXR activation in vivo have not been developed and such studies are required before claiming drug interactions through PXR. However, there is a preponderance of data suggesting that PXR does indeed have a role in drug interactions that alter PK.
An obvious point to consider is herb–drug interaction. There are several commonly used herbal ‘drugs’ used often for general wellbeing and/or specific ailments that show significant interactions with other allopathic medicine. The exact mechanisms of many of these have not been well described. For example, danshen (Salvia miltiorrhiza) is often used for cardiovascular ailments (e.g., atherosclerosis). However, in conjunction with warfarin (prescribed to most patients with this condition), it causes a decrease in warfarin clearance, although increasing its own bioavailability. The exact mechanisms of drug interaction are unclear [110]. Similarly, Ephedra Sinensis (Cao Mahuang, Ma huang, Yellow astringent) can alter effects of monoamine oxidases, which result in a significant rise in blood pressure. The mechanisms of such interactions are also unclear [111]. Therefore, with herb–drug interactions, although a large number of these could significantly alter drug PK, there might be other mechanisms (e.g., PK independent) that could be involved. As there is a precedent for herb–drug interactions through PXR, it is compelling to invoke its role. Further studies are needed to establish and validate these effects.
Some mechanisms to consider in developing clinical studies include knowledge of the mechanism of transcriptional regulation of orphan nuclear receptor target genes. In this context, PXR mediated transcription depends on the balance of corepressors and co-activators present on ligand activation and subsequent inactivation. We have recently published a review of this subject and call attention to the fact that many of the coregulators of PXR activation have known gene polymorphisms that affect function (e.g., HNF-4α) [112]. Therefore, clinical studies should incorporate the entire range of SNPs to fully understand the individual basis of PXR mediated effects.
As there is significant cross-talk between nuclear receptors, it is worthwhile investigating whether drugs serve as ligands to other X-receptors such as LXR and farnesol X receptor. Constitutive androstane receptor is another receptor that cross-talks with PXR and is also susceptible to xenobiotic mediated activation (e.g., phenobarbital) or inactivation [94]. Therefore, clinical studies must incorporate genetic assessments for the entire range of orphan receptors (Figure 4.I(F)).
2.11 Systems approach to drug–drug interaction problems
The human body is a complex organism comprising physiologic and pathophysiologic states in each organ that affect the PK and PD of xenobiotics.
Environmental effects on drug disposition are widely recognized as salient in determining drug effect. Important factors include food, nutrients, co-mediations and so on, but also include clinical factors such as weight (e.g., cachexia), third-spacing of fluid (increasing volume of distribution) and pathophysiologic conditions such as liver or renal disease [113]. In oncology, cachexia alone causes 22% of cancer deaths. The pathophysiology of cachexia is distinctly different from that of starvation. Resting energy expenditures are elevated, and abnormal intermediary metabolism, proteolysis and lipolysis occur independently of caloric intake. A facilitative interaction among catecholamines, prostaglandins and inflammatory cytokines is responsible for cachexia. Successful treatment requires reduction of energy expenditures, reversal of anorexia, and correction of abnormal intermediary metabolism, lipolysis and proteolysis. Cachexia also decreases the body’s ability to tolerate drugs, at least in part through altered PK and body compartment distribution [113]. Therefore, along with other clinical covariates, this covariate becomes important when building a systems approach towards drug–drug interactions.
One approach to providing a model that would guide our understanding of drug interactions is to develop two nodes: one describing all possible ‘clinically’ feasible interactions between the drug(s) and their ‘on’- and ‘off ’-target(s) and subsequent effects, and the second directly linking drug(s), their concentrations in plasma (or other valid compartment for which measurements exist) and clinical outcome. Finally, one needs to link both nodes. In each of these categories what we want to know is whether the drug(s) interact in an additive, synergistic, antagonistic or ‘no-effect’ manner. Similar analyses have been done for drug–drug surface interactions (Figure 5) and can help classify drug interactions in a manner that now correlates with toxicity (in the second node) [114,115]. The final application of systems biology must also take into account novel bioinformatic approaches such as MetaDrug that: i) predict metabolites for molecules based on their chemical structure, ii) predict the activity of the original compound and its metabolites with various absorption, distribution, metabolism, excretion and toxicity models, iii) incorporate the predictions with human cell signaling and metabolic pathways and networks; and iv) integrate networks and metabolites with relevant toxicogenomic or other high throughput data [116].
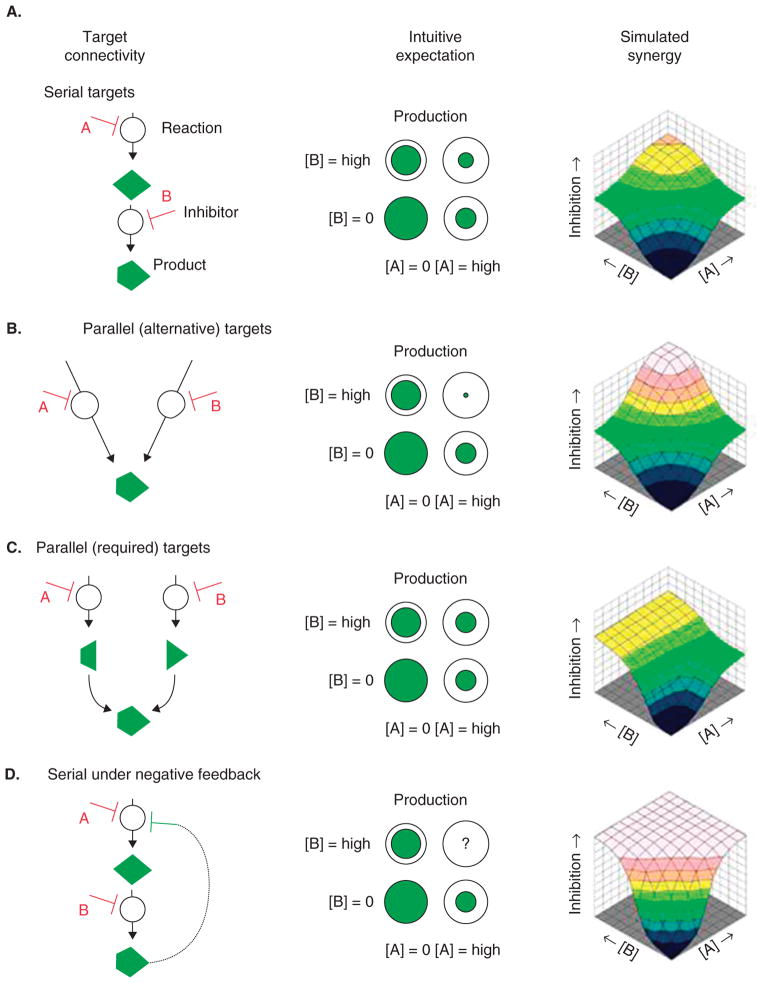
Figure 5. The connection between target relationships and synergy for paired inhibitors.
The underlying association schema for two inhibitors ‘A’ and ‘B’ is shown along with intuitive expectations for their combined effect and the model simulated dose-dependent response surfaces. A. If their targets are serial in a pathway, A and B should help each other reduce production. If they inhibit parallel pathways (B, C), the combination effect ought to reflect the rate-limiting reaction for required (‘AND’) junctions but should be very synergistic for alternative (‘OR’) pathways. D. More complex network topologies without an intuitive expectation can still be simulated. Lehár et al. [114] sought to categorize and link the set of possible response surfaces to underlying network connectivity.
The figure and legend are from [115].
3. Conclusions
Drug interactions that alter drug PK are complex and it is quite clear that mechanistic studies in this area are supplemented with extensive systems biology approaches that take into consideration all targets, physiology, PK and PD. Developing such a model would be an enormous task and as such requires multi-disciplinary committees (computational biologists, pharmacologists, toxicologists, clinical trials, etc.) that tackle each aspect of the problem, and then come together for a final model. The examples of drug interactions provided all have a mechanistic basis and, therefore, are amenable to the systems approach.
4. Expert opinion
Nearly 1 in 25 individuals are at risk for a major drug–drug interaction [117]. Mechanisms describing drug interaction have so far been studied as single gene or target effect(s). The classical examples cited are enzyme inhibition or induction and these account for well described drug interactions in oncology. However, our lack of a systems approach severely reduces the ability with which we can predict drug interactions in oncology (this also extends to other fields). To do this, I propose that individual target based interactions be uncovered for all drugs in the market with the intent of defining new interactions (for example, as I have discussed for the drug ketoconazole and PXR). Second, these data should be captured onto a database from which a systems analysis approach should be performed to unveil important covariates of such interactions. In section 3, I have elaborated my view of systems approach. The ultimate goal is that of reducing unwanted interaction(s) in a patient population in which the cost of such interactions can be enormous to society. This is in addition to the loss of quality of life during a time when every bit of it is needed. I propose that we take cues from the study of the application of systems biology for drug targets and outcomes and apply these to the study of drug targets and ‘toxic’ outcomes (Figure 5).
A systems approach will take into account theoretical model development, which is strengthened by experimental data and clinical outcomes data. A model is developed that predicts interactions when drug A is present and then you sequentially add drugs to perturb the system. In this kind of model, the input functions will constantly change over time to adjust for real-time effects. The net outcome parameters are binomial: is the interaction ‘clinically’ significant to the patient or not? Until such a database is created, we will have to exercise due diligence in recording all clinical adverse events and probing mechanisms of drug–drug interactions, especially as they relate to PK and PD. This means that we will need to continue to report biochemical studies and collaborate with computational biologists early in our efforts to look for models of drug interactions. An example of such an effort is shown with our work on the interaction of ketoconazole and ixabepilone. The greater question is whether clinically used CYP3A4 inhibitors would have the same effect and it would be very helpful to have a model that could predict this. This would prevent efforts of trying to collect such data after adverse events have transpired.
Non-classical data on drug effect must be studied, made publicly available and included in the overall analysis of drug interactions. For example, genotoxic information on compounds, especially those with concentration-dependent effects, are data worthy to include and analyze in such a database. For example, information from bacterial SOS assays can predict genotoxic effects in vivo, especially if several compounds are involved. Transcriptome studies in lower eukaryotes (e.g., yeast) have shown the power of systems biology is deciphering drug targets; yet, this is difficult to perform in higher eukaryotics. An approach that is amenable to complex cell systems is the use of siRNA libraries and concentration-dependent drug screen data would be extremely useful to plug into the systems approach to study drug toxicity (in this case ‘cytotoxicity’).
Footnotes
Declaration of interest
S Mani has received research funding from Bristol-Myers Squibb, Eli Lilly, Onconova, Kosan, Hoffman-La Roche, Amgen, Enzon, Hybridon, AMRI and TLC. The other authors have nothing to declare.
Contributor Information
Sridhar Mani, Email: smani@aecom.yu.edu.
Mohammed Ghalib, Email: mhghalib@montefiore.org.
Imran Chaudhary, Email: ICHAUDHA@montefiore.org.
Sanjay Goel, Email: sgoel@montefiore.org.
Bibliography
- 1.Fety R, Rolland F, Barberi-Heyob M, et al. Clinical impact of pharmacokinetically-guided dose adaptation of 5-fluorouracil: results from a multicentric randomized trial in patients with locally advanced head and neck carcinomas. Clin Cancer Res. 1998;4(9):2039–45. [PubMed] [Google Scholar]
- 2.Fety R, Rolland F, Barberi-Heyob M, et al. Clinical randomized study of 5FU monitoring versus standard dose in patients with head and neck cancer: preliminary results. Anticancer Res. 1994;14(6A):2347–52. [PubMed] [Google Scholar]
- 3.Gupta E, Mick R, Ramirez J, et al. Pharmacokinetic and pharmacodynamic evaluation of the topoisomerase inhibitor irinotecan in cancer patients. J Clin Oncol. 1997;15(4):1502–10. doi: 10.1200/JCO.1997.15.4.1502. [DOI] [PubMed] [Google Scholar]
- 4.Calvert AH, Newell DR, Gumbrell LA, et al. Carboplatin dosage: prospective evaluation of a simple formula based on renal function. J Clin Oncol. 1989;7(11):1748–56. doi: 10.1200/JCO.1989.7.11.1748. [DOI] [PubMed] [Google Scholar]
- 5.Petros WP, Hopkins PJ, Spruill S, et al. Associations between drug metabolism genotype, chemotherapy pharmacokinetics, and overall survival in patients with breast cancer. J Clin Oncol. 2005;23(25):6117–25. doi: 10.1200/JCO.2005.06.075. [DOI] [PubMed] [Google Scholar]
- 6.Riechelmann RP, Krzyzanowska MK. Issues of potential drug interactions and prescription duplication among patients with cancer. Am J Hemtol Oncol. 2007;6(9):529–31. [Google Scholar]
- 7.Riechelmann RP, Tannock IF, Wang L, et al. Potential drug interactions and duplicate prescriptions among cancer patients. J Natl Cancer Inst. 2007;99:592–600. doi: 10.1093/jnci/djk130. [DOI] [PubMed] [Google Scholar]
- 8.Grymonpre RE, Mitenko PA, Sitar DS, et al. Drug-associated hospital admissions in older medical patients. J Am Geriatr Soc. 1988;36:1092–8. doi: 10.1111/j.1532-5415.1988.tb04395.x. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton RA, Briceland LL, Andritz MH. Frequency of hospitalization after exposure to known drug-drug interactions in a Medicaid population. Pharmacotherapy. 1998;18:1112–20. [PubMed] [Google Scholar]
- 10.Mannisto PT, Mantyla R, Nykanen S, et al. Impairing effect of food on ketoconazole absorption. Antimicrob Agents Chemother. 1982;21(5):730–3. doi: 10.1128/aac.21.5.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lelawongs P, Barone JA, Colaizzi JL, et al. Effect of food and gastric acidity on absorption of orally administered ketoconazole. Clin Pharm. 1988;7(3):228–35. [PubMed] [Google Scholar]
- 12.Shepard DR, Mani S, Kastrissios H, et al. Estimation of the effect of food on the disposition of oral 5-fluorouracil in combination with eniluracil. Cancer Chemother Pharmacol. 2002;49(5):398–402. doi: 10.1007/s00280-002-0431-9. [DOI] [PubMed] [Google Scholar]
- 13.Schilsky RL, Bukowski R, Burris H, 3rd, et al. A multicenter phase II study of a five-day regimen of oral 5-fluorouracil plus eniluracil with or without leucovorin in patients with metastatic colorectal cancer. Ann Oncol. 2000;11(4):415–20. doi: 10.1023/a:1008356522080. [DOI] [PubMed] [Google Scholar]
- 14.Mani S, Hochster H, Beck T, et al. Multicenter phase II study to evaluate a 28-day regimen of oral fluorouracil plus eniluracil in the treatment of patients with previously untreated metastatic colorectal cancer. J Clin Oncol. 2000;18(15):2894–901. doi: 10.1200/JCO.2000.18.15.2894. [DOI] [PubMed] [Google Scholar]
- 15.Cao S, Baccanari DP, Joyner SS, et al. 5-Ethynyluracil (776C85): effects on the antitumor activity and pharmacokinetics of tegafur, a prodrug of 5-fluorouracil. Cancer Res. 1995;55(24):6227–30. [PubMed] [Google Scholar]
- 16.Baker SD, Khor SP, Adjei AA, et al. Pharmacokinetic, oral bioavailability, and safety study of fluorouracil in patients treated with 776C85, an inactivator of dihydropyrimidine dehydrogenase. J Clin Oncol. 1996;14(12):3085–96. doi: 10.1200/JCO.1996.14.12.3085. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan RH, Weirnik PH. Neurotoxicity of anti-tumor agents. In: Perry MC, Yarbro JW, editors. Toxicity of Chemotherapy. Orlando, FL: Grune and Stratton; pp. 365–374. [Google Scholar]
- 18.Persidsky Y, Ramirez SH, Haorah J, Kanmogne GD. Blood-brain barrier: structural components and function under physiologic and pathologic conditions. J Neuroimmune Pharmacol. 2006;1(3):223–36. doi: 10.1007/s11481-006-9025-3. [DOI] [PubMed] [Google Scholar]
- 19.Gavins F, Yilmaz G, Granger DN. The evolving paradigm for blood cell-endothelial cell interactions in the cerebral microcirculation. Microcirculation. 2007;14(7):667–81. doi: 10.1080/10739680701404903. [DOI] [PubMed] [Google Scholar]
- 20.Cecchelli R, Berezowski V, Lundquist S, et al. Modelling of the blood-brain barrier in drug discovery and development. Nat Rev Drug Discov. 2007;6(8):650–61. doi: 10.1038/nrd2368. [DOI] [PubMed] [Google Scholar]
- 21.Ohtsuki S, Terasaki T. Contribution of carrier-mediated transport systems to the blood-brain barrier as a supporting and protecting interface for the brain; importance for CNS drug discovery and development. Pharm Res. 2007;24(9):1745–58. doi: 10.1007/s11095-007-9374-5. [DOI] [PubMed] [Google Scholar]
- 22.Gerstner ER, Fine RL. Increased permeability of the blood-brain barrier to chemotherapy in metastatic brain tumors: establishing a treatment paradigm. J Clin Oncol. 2007;25(16):2306–12. doi: 10.1200/JCO.2006.10.0677. [DOI] [PubMed] [Google Scholar]
- 23.Bauer B, Hartz AM, Fricker G, Miller DS. Pregnane X receptor up-regulation of P-glycoprotein expression and transport function at the blood-brain barrier. Mol Pharmacol. 2004;66(3):413–9. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 24.Bauer B, Yang X, Hartz AM, et al. In vivo activation of human pregnane X receptor tightens the blood-brain barrier to methadone through P-glycoprotein up-regulation. Mol Pharmacol. 2006;70(4):1212–9. doi: 10.1124/mol.106.023796. [DOI] [PubMed] [Google Scholar]
- 25.Zhou S, Lim LY, Chowbay B. Herbal modulation of P-glycoprotein. Drug Metab Rev. 2004;36:57–104. doi: 10.1081/dmr-120028427. [DOI] [PubMed] [Google Scholar]
- 26.Ferry DR. Testing the role of P-glycoprotein expression in clinical trials: applying pharmacological principles and best methods for detection together with good clinical trials methodology. Int J Clin Pharmacol Ther. 1998;36(1):29–40. [PubMed] [Google Scholar]
- 27.Oza AM. Clinical development of P glycoprotein modulators in oncology. Novartis Found Symp. 2002;243:103–15. discussion 115–8, 180–5. [PubMed] [Google Scholar]
- 28.Rodrigues AD. Drugs and the Pharmaceutical Sciences. 2. Vol. 179. Informa Healthcare USA, Inc; New York, NY 10017: 2008. Drug-Drug Interactions; p. 579. [Google Scholar]
- 29.Robert J. Multidrug resistance in oncology: diagnostic and therapeutic approaches. Eur J Clin Invest. 1999;29(6):536–45. doi: 10.1046/j.1365-2362.1999.00495.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Balmaceda C, Bruce JN, et al. Tamoxifen paradoxically decreases paclitaxel deposition into cerebrospinal fluid of brain tumor patients. J Neurooncol. 2006;76(1):85–92. doi: 10.1007/s11060-005-4171-7. [DOI] [PubMed] [Google Scholar]
- 31.Kemper EM, van Zandbergen AE, Cleypool C, et al. Increased penetration of paclitaxel into the brain by inhibition of P-glycoprotein. Clin Cancer Res. 2003;9(7):2849–55. [PubMed] [Google Scholar]
- 32.Fellner S, Bauer B, Miller DS, et al. Transport of paclitaxel (Taxol) across the blood-brain barrier in vitro and in vivo. J Clin Invest. 2002;110(9):1309–18. doi: 10.1172/JCI15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lötsch J, Schmidt R, Vetter G, et al. Increased CNS uptake and enhanced antinociception of morphine-6-glucuronide in rats after inhibition of P-glycoprotein. J Neurochem. 2002;83(2):241–8. doi: 10.1046/j.1471-4159.2002.01177.x. [DOI] [PubMed] [Google Scholar]
- 34.Drewe J, Ball HA, Beglinger C, et al. Effect of P-glycoprotein modulation on the clinical pharmacokinetics and adverse effects of morphine. Br J Clin Pharmacol. 2000;50(3):237–46. doi: 10.1046/j.1365-2125.2000.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rautio J, Humphreys JE, Webster LO, et al. In vitro p-glycoprotein inhibition assays for assessment of clinical drug interaction potential of new drug candidates: a recommendation for probe substrates. Drug Metab Dispos. 2006;34(5):786–92. doi: 10.1124/dmd.105.008615. [DOI] [PubMed] [Google Scholar]
- 36.Huang SM, Reynolds KS, Strong JM, et al. A regulatory viewpoint on transporter-based drug interactions. Xenobiotica. 2008;38(7–8):709–24. doi: 10.1080/00498250802017715. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, Wang H, Sinz M, et al. Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene. 2007;26(2):258–68. doi: 10.1038/sj.onc.1209788. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Huang H, Li H, et al. Activated pregnenolone X-receptor is a target for ketoconazole and its analogs. Clin Cancer Res. 2007;13(8):2488–95. doi: 10.1158/1078-0432.CCR-06-1592. [DOI] [PubMed] [Google Scholar]
- 39.Okuda H, Nishiyama T, Ogura K, et al. Lethal drug interactions of sorivudine, a new antiviral drug, with oral 5-fluorouracil prodrugs. Drug Metab Dispos. 1997;25(2):270–3. [PubMed] [Google Scholar]
- 40.Yan J, Tyring SK, McCrary MM, et al. The effect of sorivudine on dihydropyrimidine dehydrogenase activity in patients with acute herpes zoster. Clin Pharmacol Ther. 1997;61(5):563–73. doi: 10.1016/S0009-9236(97)90136-3. [DOI] [PubMed] [Google Scholar]
- 41.Diasio RB. Sorivudine and 5-fluorouracil; a clinically significant drug-drug interaction due to inhibition of dihydropyrimidine dehydrogenase. Br J Clin Pharmacol. 1998;46(1):1–4. doi: 10.1046/j.1365-2125.1998.00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goel S, Cohen M, Cömezoglu SN, et al. The effect of ketoconazole on the pharmacokinetics and pharmacodynamics of ixabepilone: a first in class epothilone B analogue in late-phase clinical development. Clin Cancer Res. 2008;14(9):2701–9. doi: 10.1158/1078-0432.CCR-07-4151. [DOI] [PubMed] [Google Scholar]
- 43.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1734–5. 1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 44.Henry NL, Stearns V, Flockhart DA, et al. Drug interactions and pharmacogenomics in the treatment of breast cancer and depression. Am J Psychiatry. 2008;165(10):1251–5. doi: 10.1176/appi.ajp.2008.08040482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sane RS, Buckley DJ, Buckley AR, et al. Role of human pregnane X receptor in tamoxifen- and 4-hydroxytamoxifen-mediated CYP3A4 induction in primary human hepatocytes and LS174T cells. Drug Metab Dispos. 2008;36(5):946–54. doi: 10.1124/dmd.107.018598. [DOI] [PubMed] [Google Scholar]
- 46.Harmsen S, Meijerman I, Beijnen JH, Schellens JH. Nuclear receptor mediated induction of cytochrome P450 3A4 by anticancer drugs: a key role for the pregnane X receptor. Cancer Chemother Pharmacol. 2008 doi: 10.1007/s00280-008-0842-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 47.Hu Z, Yang X, Ho PC, et al. St John’s wort modulates the toxicities and pharmacokinetics of CPT-11 (irinotecan) in rats. Pharm Res. 2005;22:902–14. doi: 10.1007/s11095-005-4585-0. [DOI] [PubMed] [Google Scholar]
- 48.Mathijssen RH, Verweij J, de Bruijn P, et al. Effects of St John’s wort on irinotecan metabolism. J Natl Cancer Inst. 2002;94:1247–9. doi: 10.1093/jnci/94.16.1247. [DOI] [PubMed] [Google Scholar]
- 49.Frye RF, Fitzgerald SM, Lagattuta TF, et al. Effect of St John’s wort on imatinib mesylate pharmacokinetics. Clin Pharmacol Ther. 2004;76:323–9. doi: 10.1016/j.clpt.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 50.Smith P, Bullock JM, Booker BM, et al. The influence of St John’s wort on the pharmacokinetics and protein binding of imatinib mesylate. Pharmacotherapy. 2004;24:1508–14. doi: 10.1592/phco.24.16.1508.50958. [DOI] [PubMed] [Google Scholar]
- 51.Komoroski BJ, Parise RA, Egorin MJ, et al. Effect of the St John’s wort constituent hyperforin on docetaxel metabolism by human hepatocyte cultures. Clin Cancer Res. 2005;11:6972–9. doi: 10.1158/1078-0432.CCR-04-2488. [DOI] [PubMed] [Google Scholar]
- 52.Watkins RE, Maglich JM, Moore LB, et al. 2.1 A crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry. 2003;42(6):1430–8. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- 53.Hu ZP, Yang XX, Chen X, et al. A mechanistic study on altered pharmacokinetics of irinotecan by St John’s wort. Curr Drug Metab. 2007;8(2):157–71. doi: 10.2174/138920007779815995. [DOI] [PubMed] [Google Scholar]
- 54.DiPetrillo K, Wood S, Kostrubsky V, et al. Effect of caffeine on acetaminophen hepatotoxicity in cultured hepatocytes treated with ethanol and isopentanol. Toxicol Appl Pharmacol. 2002;185(2):91–7. doi: 10.1006/taap.2002.9535. [DOI] [PubMed] [Google Scholar]
- 55.Hazai E, Vereczkey L, Monostory K. Reduction of toxic metabolite formation of acetaminophen. Biochem Biophys Res Commun. 2002;291(4):1089–94. doi: 10.1006/bbrc.2002.6541. [DOI] [PubMed] [Google Scholar]
- 56.Cameron MD, Wen B, Roberts AG, et al. Cooperative binding of acetaminophen and caffeine within the P450 3A4 active site. Chem Res Toxicol. 2007;20(10):1434–41. doi: 10.1021/tx7000702. [DOI] [PubMed] [Google Scholar]
- 57.Wolf KK, Wood SG, Hunt JA, et al. Role of the nuclear receptor pregnane X receptor in acetaminophen hepatotoxicity. Drug Metab Dispos. 2005;33(12):1827–36. doi: 10.1124/dmd.105.005256. [DOI] [PubMed] [Google Scholar]
- 58.Ricketts ML, Boekschoten MV, Kreeft AJ, et al. The cholesterol-raising factor from coffee beans, cafestol, as an agonist ligand for the farnesoid and pregnane X receptors. Mol Endocrinol. 2007;21(7):1603–16. doi: 10.1210/me.2007-0133. [DOI] [PubMed] [Google Scholar]
- 59.Nozaki Y, Kusuhara H, Endou H, Sugiyama Y. Quantitative evaluation of the drug-drug interactions between methotrexate and nonsteroidal anti-inflammatory drugs in the renal uptake process based on the contribution of organic anion transporters and reduced folate carrier. J Pharmacol Exp Ther. 2004;309(1):226–34. doi: 10.1124/jpet.103.061812. [Epub 2004 Jan 13] [DOI] [PubMed] [Google Scholar]
- 60.Breedveld P, Zelcer N, Pluim D, et al. Mechanism of the pharmacokinetic interaction between methotrexate and benzimidazoles: potential role for breast cancer resistance protein in clinical drug-drug interactions. Cancer Res. 2004;64(16):5804–11. doi: 10.1158/0008-5472.CAN-03-4062. [DOI] [PubMed] [Google Scholar]
- 61.van der Heijden JW, Dijkmans ACB, Scheper RJ, Jansen G. Drug Insight: resistance to methotrexate and other disease-modifying antirheumatic drugs—from bench to bedside. Nat Clin Pract Rheumatol. 2007;3:26–34. doi: 10.1038/ncprheum0380. [DOI] [PubMed] [Google Scholar]
- 62.Nozaki Y, Kusuhara H, Endou H, Sugiyama Y. Quantitative evaluation of the drug-drug interactions between methotrexate and nonsteroidal anti-inflammatory drugs in the renal uptake process based on the contribution of organic anion transporters and reduced folate carrier. J Pharmacol Exp Ther. 2004;309(1):226–34. doi: 10.1124/jpet.103.061812. [DOI] [PubMed] [Google Scholar]
- 63.Uwai Y, Taniguchi R, Motohashi H, et al. Methotrexate-loxoprofen interaction: involvement of human organic anion transporters hOAT1 and hOAT3. Drug Metab Pharmacokinet. 2004;19(5):369–74. doi: 10.2133/dmpk.19.369. [DOI] [PubMed] [Google Scholar]
- 64.Takeda M, Khamdang S, Narikawa S, et al. Characterization of methotrexate transport and its drug interactions with human organic anion transporters. J Pharmacol Exp Ther. 2002;303(2):534–9. doi: 10.1124/jpet.102.034330. [DOI] [PubMed] [Google Scholar]
- 65.Joerger M, Huitema AD, van den Bongard HJ, et al. Determinants of the elimination of methotrexate and 7-hydroxy-methotrexate following high-dose infusional therapy to cancer patients. Br J Clin Pharmacol. 2006;62(1):71–80. doi: 10.1111/j.1365-2125.2005.02513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breedveld P, Zelcer N, Pluim D, et al. Mechanism of the pharmacokinetic interaction between methotrexate and benzimidazoles: potential role for breast cancer resistance protein in clinical drug-drug interactions. Cancer Res. 2004;64(16):5804–11. doi: 10.1158/0008-5472.CAN-03-4062. [DOI] [PubMed] [Google Scholar]
- 67.Wu H, Horton JR, Battaile K, et al. Structural basis of allele variation of human thiopurine-S-methyltransferase. Proteins. 2007;67:198–208. doi: 10.1002/prot.21272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yates CR, Krynetski EY, Loennechen T, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997;126(8):608–14. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- 69.Marra C, Esdaile J, Aslam HA. Practical pharmacogenetics: the cost effectiveness of screening for thiopurine s-methyltransferase polymorphisms in patients with rheumatological conditions treated with azathioprine. J Rheumatol. 2002;29:2507–12. [PubMed] [Google Scholar]
- 70.Marra CA, Esdaile JM, Anis AH. Practical pharmacogenetics: the cost effectiveness of screening for thiopurine s-methyltransferase polymorphisms in patients with rheumatological conditions treated with azathioprine. J Rheumatol. 2002;29(12):2507–12. [PubMed] [Google Scholar]
- 71.Reuther LO, Sonne J, Larsen NE, et al. Pharmacological monitoring of azathioprine therapy. Scand J Gastroenterol. 2003;38(9):972–7. doi: 10.1080/00365520310005082. [DOI] [PubMed] [Google Scholar]
- 72.Reuther LO, Vainer B, Sonne J, Larsen NE. Thiopurine methyltransferase (TPMT) genotype distribution in azathioprine-tolerant and -intolerant patients with various disorders. The impact of TPMT genotyping in predicting toxicity. Eur J Clin Pharmacol. 2004;59(11):797–801. doi: 10.1007/s00228-003-0698-8. [DOI] [PubMed] [Google Scholar]
- 73.Wikipedia entry on thiopurine methyltransferase. Available from: http://en.wikipedia.org/wiki/Thiopurine_methyltransferase.
- 74.TPMT ImmunoAssay Kit Online Store. Available from: http://www.tpmtassay.com/store/
- 75.Illmer T, Schuler US, Thiede C, et al. MDR1 gene polymorphisms affect therapy outcome in acute myeloid leukemia patients. Cancer Res. 2002;62(17):4955–62. [PubMed] [Google Scholar]
- 76.Siddiqui A, Kerb R, Weale ME, et al. Association of multidrug resistance in epilepsy with a polymorphism in the drug-transporter gene ABCB1. N Engl J Med. 2003;348(15):1442–8. doi: 10.1056/NEJMoa021986. [DOI] [PubMed] [Google Scholar]
- 77.Yamaguchi H, Hishinuma T, Endo N, et al. Genetic variation in ABCB1 influences paclitaxel pharmacokinetics in Japanese patients with ovarian cancer. Int J Gynecol Cancer. 2006;16(3):979–85. doi: 10.1111/j.1525-1438.2006.00593.x. [DOI] [PubMed] [Google Scholar]
- 78.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000;97(7):3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 80.Blumberg B, Sabbagh W, Jr, Juguilon H, et al. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bertilsson G, Heidrich J, Svensson K, et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA. 1998;95:12208–13. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lehmann JM, McKee DD, Watson MA, et al. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7:584–90. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 84.Goodwin B, Moore LB, Stoltz CM, et al. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60:427–31. [PubMed] [Google Scholar]
- 85.Chen Y, Ferguson SS, Negishi M, et al. Induction of human CYP2C9 by rifampicin, hyperforin, and phenobarbital is mediated by the pregnane X receptor. J Pharmacol Exp Ther. 2004;308:495–501. doi: 10.1124/jpet.103.058818. [DOI] [PubMed] [Google Scholar]
- 86.Sonoda J, Xie W, Rosenfeld JM, et al. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR) Proc Natl Acad Sci USA. 2002;99:13801–6. doi: 10.1073/pnas.212494599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie W, Yeuh MF, Radominska-Pandya A, et al. Control of steroid, heme, and carcinogen metabolism by nuclear pregnane X receptor and constitutive androstane receptor. Proc Natl Acad Sci USA. 2003;100:4150–5. doi: 10.1073/pnas.0438010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Falkner KC, Pinaire JA, Xiao GH, et al. Regulation of the rat glutathione S-transferase A2 gene by glucocorticoids: involvement of both the glucocorticoid and pregnane X receptors. Mol Pharmacol. 2001;60:611–9. [PubMed] [Google Scholar]
- 89.Kast HR, Goodwin B, Tarr PT, et al. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem. 2002;277:2908–15. doi: 10.1074/jbc.M109326200. [DOI] [PubMed] [Google Scholar]
- 90.Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- 91.Choudhuri S, Valerio LG. Usefulness of studies on the molecular mechanism of action of herbals/botanicals: the case of St John’s wort. J Biochem Mol Toxicol. 2005;19:1–11. doi: 10.1002/jbt.20057. [DOI] [PubMed] [Google Scholar]
- 92.Moore LB, Goodwin B, Jones SA, et al. St John’s wort induces hepatic drug metabolism through. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watkins RE, Maglich JM, Moore LB, et al. 2.1 A crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry. 2003;42:1430–8. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- 94.Ueda A, Hamadeh HK, Webb HK, et al. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- 95.Wei P, Zhang J, Egan-Hafley M, et al. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–3. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- 96.Kakizaki S, Yamamoto Y, Ueda A, et al. Phenobarbital induction of drug/steroid-metabolizing enzymes and nuclear receptor CAR. Biochim Biophys Acta. 2003;1619:239–42. doi: 10.1016/s0304-4165(02)00482-8. [DOI] [PubMed] [Google Scholar]
- 97.Burk O, Arnold KA, Geick A, et al. A role for constitutive androstane receptor in the regulation of human intestinal MDR1 expression. Biol Chem. 2005;386:503–13. doi: 10.1515/BC.2005.060. [DOI] [PubMed] [Google Scholar]
- 98.Schmiedlin-Ren P, Thummel KE, Fisher JM, et al. Expression of enzymatically active CYP3A4 by Caco-2 cells grown on extracellular matrix-coated permeable supports in the presence of 1alpha, 25-dihydroxyvitamin D3. Mol Pharmacol. 1997;51:741–54. doi: 10.1124/mol.51.5.741. [DOI] [PubMed] [Google Scholar]
- 99.Drocourt L, Ourlin JC, Pascussi JM, et al. Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. J Biol Chem. 2002;277:25125–32. doi: 10.1074/jbc.M201323200. [DOI] [PubMed] [Google Scholar]
- 100.Adachi R, Honma Y, Masuno H, et al. Selective activation of vitamin D receptor by lithocholic acid acetate, a bile acid derivative. J Lipid Res. 2005;46:46–57. doi: 10.1194/jlr.M400294-JLR200. [DOI] [PubMed] [Google Scholar]
- 101.Kostrubsky VE, Lewis LD, Strom SC, et al. Induction of cytochrome P4503A by Taxol in primary cultures of human hepatocytes. Arch Biochem Biophys. 1998;355:131–6. doi: 10.1006/abbi.1998.0730. [DOI] [PubMed] [Google Scholar]
- 102.Luo G, Cunningham M, Kim S, et al. CYP3A4 induction by drugs: correlation between a pregnane X receptor reporter gene assay and CYP3A4 expression in human hepatocytes. Drug Metab Dispos. 2002;30:795–804. doi: 10.1124/dmd.30.7.795. [DOI] [PubMed] [Google Scholar]
- 103.Moore JT, Kliewer SA. Use of the nuclear receptor PXR to predict drug interactions. Toxicology. 2000;153:1–10. doi: 10.1016/s0300-483x(00)00300-0. [DOI] [PubMed] [Google Scholar]
- 104.Xie W, Barwick JL, Downes M, et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–9. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 105.Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259–66. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- 106.Zhou C, Tabb MM, Sadatrafiei A, et al. Tocotrienols activate the steroid and xenobiotic receptor, SXR, and selectively regulate expression of its target genes. Drug Metab Dispos. 2004;32:1075–82. doi: 10.1124/dmd.104.000299. [DOI] [PubMed] [Google Scholar]
- 107.Chrencik JE, Orans J, Moore LB, et al. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol. 2005;19(5):1125–34. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- 108.Watkins RE, Wisely GB, Moore LB, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292(5525):2329–33. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- 109.Mu Y, Zhang J, Zhang S, et al. Traditional Chinese medicines Wu Wei Zi (Schisandra chinensis Baill) and Gan Cao (Glycyrrhiza uralensis Fisch) activate pregnane X receptor and increase warfarin clearance in rats. J Pharmacol Exp Ther. 2006;316(3):1369–77. doi: 10.1124/jpet.105.094342. [DOI] [PubMed] [Google Scholar]
- 110.Chan TY. Interaction between warfarin and danshen (Silvia Miltiorrhiza) Ann Pharmacotherapy. 2001;35(4):501–4. doi: 10.1345/aph.19029. [DOI] [PubMed] [Google Scholar]
- 111.The Herb Research Foundation. Herb Information Greenpaper on the Ephedra site. Available from: http://www.ephedra.nu/medicine_2.html.
- 112.Gollamudi R, Gupta D, Goel S, Mani S. Novel orphan nuclear receptors-coregulator interactions controlling anti-cancer drug metabolism. Curr Drug Metab. 2008;9(7):611–3. doi: 10.2174/138920008785821701. [DOI] [PubMed] [Google Scholar]
- 113.Cova D, Lorusso V, Silvestris N. The pharmacokinetics and pharmacodynamics of drugs in elderly cachectic (cancer) patients. In: Mantovanni G, editor. cachexia and wasting: A modern approach. Springer Milan; 2006. [DOI] [Google Scholar]
- 114.Lehár J, Zimmermann GR, Krueger AS, et al. Chemical combination effects predict connectivity in biological systems. Mol Syst Biol. 2007;3:80. doi: 10.1038/msb4100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yeh P, Kishnoy R. Networks from drug–drug surfaces. Mol Sys Biol. 2007;3:85. doi: 10.1038/msb4100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ekins S, Andreyev S, Ryabov A, et al. A combined approach to drug metabolism and toxicity assessment. Drug Metab Dispos. 2006;34(3):495–503. doi: 10.1124/dmd.105.008458. [Epub 2005 Dec 28] [DOI] [PubMed] [Google Scholar]
- 117.Qato DM, Alexander GC, Conti RM, et al. Use of prescription and over-the-counter medications and dietary supplements among older adults in the United States. JAMA. 2008;300(24):2867–78. doi: 10.1001/jama.2008.892. [DOI] [PMC free article] [PubMed] [Google Scholar]