Abstract
Homozygous or compound heterozygous mutations in the GJC2 gene, encoding the gap junction protein connexin47 (Cx47), cause the autosomal recessive hypomyelinating Pelizaeus–Merzbacher-like disease (PMLD1, MIM# 608804). Although clinical and neuroradiological findings resemble those of the classic Pelizaeus–Merzbacher disease, PMLD patients usually show a greater level of cognitive and motor functions. Unpredictably a homozygous missense GJC2 mutation (p.Glu260Lys) was found in a patient presenting with a very severe clinical picture characterised by congenital nystagmus and severe neurological impairment. Also magnetic resonance imaging was unusually severe, showing an abnormal supra- and infratentorial white matter involvement extending to the spinal cord. The novel p.Glu260Lys (c.778G>A) mutation, occurring in a highly conserved motif (SRPTEK) of the Cx47 extracellular loop-2 domain, was predicted, by modelling analysis, to break a ‘salt bridge network', crucial for a proper connexin–connexin interaction to form a connexon, thus hampering the correct formation of the connexon pore. The same structural analysis, extended to the previously reported missense mutations, predicted that most changes were expected to have less severe impact on protein functions, correlating with the mild PMLD1 form of the patients. Our study expands the spectrum of PMLD1 and provides evidence that the extremely severe clinical and neuroradiological PMLD1 form of our patient likely correlates with the predicted impairment of gap junction channel assembly resulting from the detrimental effect of the new p.Glu260Lys mutant allele on Cx47 protein.
Keywords: Pelizaeus–Merzbacher-like disease, GJC2 mutation, magnetic resonance imaging, molecular modelling
Introduction
Pelizaeus–Merzbacher-like disease (PMLD) indicates a disorder virtually identical to Pelizaeus–Merzbacher disease (PMD) (MIM# 312080) but without a mutation in the PLP1 gene.1 Both disorders are characterised by nystagmus, developmental delay, progressive spasticity, ataxia and hypomyelination on brain magnetic resonance imaging (MRI).
Pelizaeus–Merzbacher-like disease-1, PMLD1 (MIM# 608804), also known as leukodystrophy hypomyelinating type 2 (HLD2), is an autosomal recessive disorder caused by mutations in the GJC2 gene, previously named as GJA12, (MIM# 608803), mapping on chromosome 1 (1q42.13).2 The GJC2 sequence encodes the 436-amino-acid gap junction connexin protein, Cx47 (GenBank NP_065168.2), which is highly expressed in oligodendrocytes.3 To date, 25 different GJC2 mutant alleles, harbouring 9 missense, 10 frameshift, 3 nonsense, 1 microinsertion and 1 regulatory mutations have been reported in 54 PMLD1 patients belonging to 32 families.4, 5, 6, 7, 8, 9, 10, 11, 12, 13 In addition, another GJC2 missense mutation causing a milder phenotype, the spastic paraplegia autosomal recessive type 44 (SPG44) (MIM# 613206), has been reported in three members of a single family.14 These findings suggest that this gene may give rise to a spectrum of disorders with different severity, in analogy with the PLP1 gene. Nevertheless, no patients with a very severe clinical picture have been reported so far.
We now describe a young patient with an unusually severe clinical and neuroradiological picture owing to a novel homozygous GJC2 mutation (p.Glu260Lys), predicted to be involved in the gap junction channel assembly by modelling analysis; in addition, we report results of the same structural analysis on other nine previously reported missense mutations. The study, based on a comprehensive comparison of the present clinical, neuroradiological, molecular and computational findings with those of the literature, expands the severity spectrum of PMLD1.
Materials and methods
Clinical features
The proband is the second child of consanguineous healthy parents from Sri Lanka. She underwent clinical examination, laboratory, neurophysiological and neuroradiological investigations (1.5T MRI scanner, Philips Achieva, Best, The Netherlands), as needed in patients with neurological disorders.
Revision of literature
Clinical findings of 54 PMLD1 patients belonging to 32 families and 3 SPG44 patients belonging to another family previoulsy reported in the literature were thoroughly reviewed. The clinical severity of PMLD1 patients was scored according to the rating scale for clinical classification ranging from 0 to 4 on the basis of the best motor function, used for PMD patients (Table 1).15
Table 1. Genotype–phenotype correlation: comparison with existing data from the literature.
| Genotype a | Number of patients | Phenotype | Score b | References |
|---|---|---|---|---|
| [p.E260K]+[p.E260K] | 1 | PMLD1 | 0 | Present study |
| [p.Y269D]+[p.R237X] | 1 | PMLD1 | 1 | 2,7 |
| [p.T395I]+[?] | 1 | PMLD1 | 1 | 7 |
| [p.A95G_V96insT]+[?] | 1 | PMLD1 | 2 | 7 |
| [p.P128Rfs]+[p.P128Rfs] | 1 | PMLD1 | 2 | 7 |
| [p.G146S]+[?] | 1 | PMLD1 | 2 | 7 |
| [p.G233R]+[p.G233R] | 1 | PMLD1 | 2 | 7 |
| [p.G233S]+[p.G233S] | 3 (1 fam) | PMLD1 | 2/3 | 4 |
| [p.P87S]+[p.P327Rfs] | 1 | PMLD1 | 3 | 2,7 |
| [p.H129Pfs]+[p.H129Pfs] | 8 (1 fam) | PMLD1 | 3 | 4 |
| [p.Y229X]+[p.Y229X] | 2 | PMLD1 | 3 | 7 |
| [p.A322Pfs]+[p.A322Pfs] | 1 | PMLD1 | 3 | 7 |
| [p.M283T]+[p.M283T] | 3 (1 fam) | PMLD1 | 3 | 2,7 |
| [p.L278Afs]+[ p.L278Afs] | 1 | PMLD1 | 3 | 4 |
| [c.-167A>C]+[c.-167A>C] | 15 (8 fam)c | PMLD1 | 3/4 | 10,11,13 |
| [p.L25Rfs]+[p.L25Rfs] | 2 (1 fam) | PMLD1 | 3/4 | 7 |
| [p.P302Rfs]+[p.P302Rfs] | 3 (2 fam) | PMLD1 | 3/4 | 5,6 |
| [p. R122X]+[p.R122X] | 1 | PMLD1 | 4 | 12 |
| [p.I43M]+[p.I43M] | 1 | PMLD1 | 4 | 9 |
| [p.P70Tfs]+[p.P70Tfs] | 1 | PMLD1 | 4 | 9 |
| [p.P128Rfs]+[?] | 1 | PMLD1 | 4 | 7 |
| [p.E204Gfs]+[p.E204Gfs] | 1 | PMLD1 | 4 | 7 |
| [p.T262A]+[p.T262A] | 1 | PMLD1 | 4 | 7 |
| [p.C315Pfs]+[p.C315Pfs] | 2 (1 fam) | PMLD1 | 4 | 7 |
| [p.T395I]+[?] | 1 | PMLD1 | 4 | 7 |
| [p.I33M]+[p.I33M] | 3 (1 fam) | SPG44 | — | 13 |
Ethical aspects
Following ethical guidelines, the samples were obtained for analysis and storage with the patient's and/or a family member's written informed consent. The consent was sought using a form approved by the local Ethics Committee.
Molecular analysis
Genomic DNA was extracted using standard methods from peripheral blood leukocytes derived from the patient as well as her parents. GJC2 gene exons and exon–intron boundaries were PCR amplified using specific primers designed by reference to the genomic sequence (GenBank accession no. AF014643.1). Sequence analysis was performed by ABI 377 DNA automated sequencer with dye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). The putative mutation was confirmed by sequencing in both directions of the duplicate PCR products. The hypothesis of new possible genetic polymorphisms was also excluded ascertaining that none of the 50 control individuals had this alteration.
Mutation nomenclature
Nucleotide numbers of the mutations are derived from cDNA GJC2 sequence (GenBank-EMBL Accession No. NM_020435.3) considering nucleotide +1 the A of the first ATG translation initiation codon, as recommended by Orthmann-Murphy et al.16
Molecular modelling
For modelling of the GJC2 missense mutations, a homology model of the Cx47 protein was used. This three dimensional model was built by homology using the programme MODELLER17 and, as a template, the crystal structure of the homologous Cx26, a gap junction channel, whose structure has been determined at 3.5A resolution.18 The two proteins share 42% of sequence identity and about 79% of sequence similarity when the cytoplasmatic region is omitted from calculation, thus the model built can be considered as trustable. The obtained structure was then submitted to different energy minimisation cycles using the programme Discover 3 from Insight II suite (Accelrys, San Diego, CA, USA), and the resulting coordinates were then used as a reliable model for visual inspection and graphical representations carried out using the programs Coot19 and Chimera,20 respectively.
Results
Clinical features
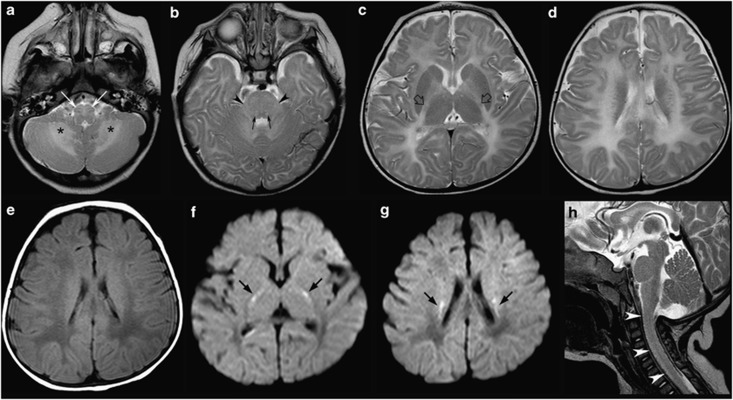
The proband was born at term after uneventful pregnancy and normal delivery. Birth weight and head circumference were normal. Nystagmus was noticed at birth. Developmental delay was observed since the first months of life. At 7 months of life, when she was evaluated for the first time, neurological examination showed nystagmus, normal head circumference, severe axial hypotonia, absent head control, increased muscle tone at lower limbs and brisk tendon reflexes. Blood routine investigations, plasma and urinary amino acids and urinary organic acid were negative or normal. Neurophysiological studies showed: poorly structured background activity without epileptic discharges at EEG, increased I-V truncal conduction time at brainstem auditory evoked potentials, and increased latency at visual evoked potentials, as reported in the literature.12 Brain MRI demonstrated diffuse hyperintensity on T2-weighted images of the cerebral and cerebellar white matter, including the middle cerebellar peduncles and almost the entire brainstem (Figure 1), with partial sparing of the pyramidal tracts at the posterior limb of the internal capsules (PLIC). The white matter of the cervical spinal cord was also abnormal. On T1-weighted images, the cerebral periventricular and lobar white matter was hypointense with a posteroanterior gradient of severity. Cerebral white matter bulk was normal, as well as the basal ganglia and thalami. Interestingly, high signal on diffusion weighted images with correspondingly low apparent diffusion coefficients were detected in the corticospinal tracts through the centra semiovalia and PLIC (Figure 1). Proton MR spectroscopy, performed with a point-resolved proton spectroscopy sequence (PRESS, TE=38 ms) in a region of interest placed in the occipital white matter, showed slightly decreased choline/N-acetyl aspartate (Cho/NAA) ratio compared with that found in age-matched control subjects (data not shown).
Figure 1.
Brain MRI performed at 7 months of age (a–h). (a–d) Axial T2-weighted images show diffuse high signal of almost the entire brainstem, including the hilum of the inferior olivary nuclei (white thin arrows) and the central tegmental tracts (black thin arrows) with only sparing of the most dorsal tegmental portion. The cerebellar white matter is involved (asterisks) as well as the middle cerebellar peduncles (black arrowheads) and supratentorial white matter. Partial myelination of the pyramidal tracts at the PLIC is visible (empty black arrows). (e) Axial T1-weighted image reveals low signal of the lobar white matter with a posteroanterior gradient. (f, g) Axial diffusion weighted images (DWI) images show restricted diffusion within both corticospinal tracts (black thick arrows). (h) Sagittal T2-weighted image reveal diffuse signal changes of the cervical spinal cord (white arrowheads).
Over the years, she had not acquired any motor milestones. At 5 years, neurological examination showed nystagmus, normal head circumference, severe axial hypotonia with still absent head control, increased muscle tone at four limbs, prevailingly at lower limbs, brisk tendon reflexes, bilateral ankle clonus and bilateral Babinski sign. She was able to pronounce only few and isolated words. Despite mild dysphagia for liquids, she was still able to eat semisolid food and drink autonomously. She never suffered from seizures. Nerve conduction velocity studies were normal. Funduscopy showed optic atrophy. A second brain MRI study was not performed because of the relatively poor clinical condition of the girl.
Molecular and structural analyses
Molecular analysis of GJC2 gene revealed a novel c.778G>A mutation resulting in a Glu260-to-Lys (p.E260K) substitution in homozygosity in the proband and in heterozygosity in her parents. Consistent with its putative pathological authenticity, the underlying nucleotide change was not found in a screen of 100 control alleles, nor listed as polymorphism in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP).
The p.E260K mutation underwent structural analysis by employing the homology model of Cx47.
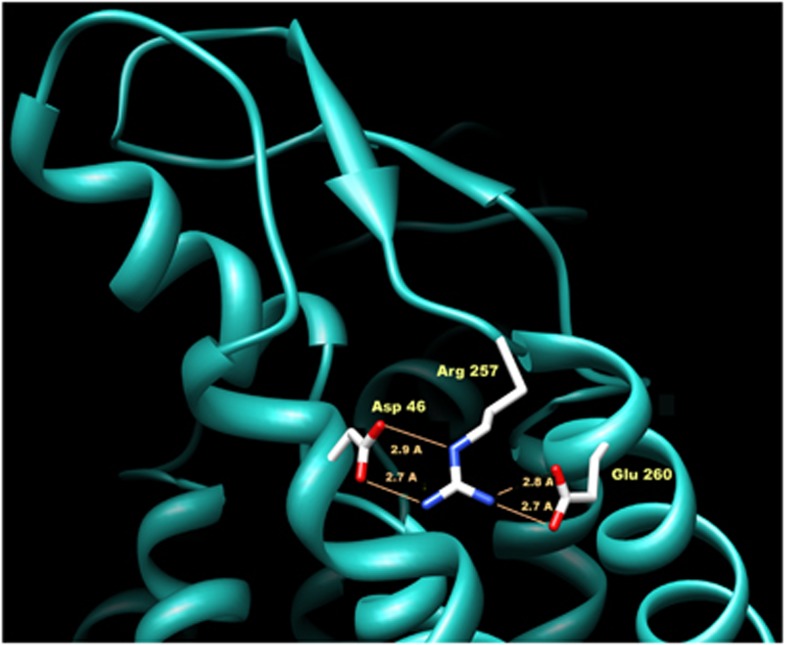
As shown in Figure 2 the replacement of Glu260-to-Lys, introducing a positively charged lysine was predicted to break a salt bridge network, normally formed by residues Asp46, Arg257 and Glu260. The computational analysis, extended to other nine GJC2 missense mutations showing that most of them are conservative or not causative of severe effects on the protein (Table 2).
Figure 2.
Structural modelling of the novel missense mutation p.E260K in the extracellular domain of the Cx47 protein. The newly replaced 260Lys breaks a ‘salt bridge network' normally formed by Asp46, Arg257 and Glu260. This network is important for the correct positioning of the external loop interacting with other connexins to form a connexon. The new 260Lys in this position by interfering with Arg257 is supposed to dislocate the loop, thereby hampering the correct formation of the connexon pore. The distances between adjacent atoms, involved in hydrogen bonds, are indicated in Å (ie, 10−10m).
Table 2. Missense mutation overview for the Connexin47 protein: molecular modelling analysis.
| Mutation a | Position b | Modelling analysis prediction | References c |
|---|---|---|---|
| p.Ile33Met (Ile36Met) | TMD | Possible interference of the long 33Met side chain with the canonical hydrogen bonding between the adjacent helices. 33Met, being in short contact with the Trp3 side chain, might cause the displacement of the N-terminal helix of each Cx47 monomer towards the inner side of the hemichannel thus (partially) blocking the pore. | 14 |
| p.Ile43Met (Ile46Met) | TMD | The replaced 43Met might interact with Arg257, thus perturbing the salt bridge formed between Asp46, Arg257 and Glu260, and possibly misplacing the extracellular loop of the connexon. | 9 |
| p.Pro87Ser (Pro90Ser) | TMD | The new Pro87, inducing a 30° kink in α-helix could alter the transport properties. | 2 |
| p.Gly146Ser (Gly149Ser) | ICD | 146Ser introduces less flexible residues in place of a glycine adding a polar group within the cytoplasmatic region of the protein. | 7 |
| p.Gly233Ser (Gly236Ser) | ECD | 233Ser introduces less flexible residues in place of a glycine adding a polar group between the extracellular and the membrane region of the protein. | 4 |
| p.Gly233Arg (Gly236Arg) | ECD | 233Arg introduces less flexible residues in place of a glycine adding a charged group between the extracellular and the membrane region of the protein. This could mildly interfere with the correct formation of the pore. | 7 |
| p.Glu260Lys | ECD | See legend of Figure 2. | Present study |
| p.Thr262Ala (Thr265Ala) | TMD | The new hydrophobic 262Ala residue, replacing a polar threonine, is supposed to prevent a H-bond formation with Glu235. | 7 |
| p.Tyr269Asp (Tyr272Asp) | TMD | The new 269Asp adds a negative charge in the membrane deleting at the same time, the contribution of the aromatic ring of the tyrosine. | 2 |
| p.Met283Thr (Met286Thr) | TMD | 283Thr changes the local hydrophobic properties of a transmembrane helix by introducing a polar residue. | 2 |
Abbreviations: ECD, Extracellular domain; ICD, Intracellular domain; TMD, Transmembrane domain.
All mutations are described according to Orthmann-Murphy et al.16 Traditional amino-acid residue numbering has nevertheless also been provided in parentheses. Note that p.Thr395Ile (Thr398Ile), occurring in a region of the model having low homology (see Material and Methods), was excluded from this analysis.
Position is given according to UniProtKB http://www.uniprot.org/.27
Referred to the first report.
Discussion
PMLD1/HLD2 due to GJA12/GJC2 mutations was firstly identified in 2004.2 Since then, molecular analysis of the gene, also extending the screening for copy number variation and for mutations in the non-coding region, has been performed in large PMLD series. The small percentage of molecularly confirmed patients has indicated that GJC2 mutations are a rare cause of PMLD.7, 21 The clinical picture of PMLD1 patients, reported in the literature, is quite homogeneous and is characterised by nystagmus as initial sign, moderate developmental delay, spasticity and ataxia resembling mild forms of PMD. In particular, it is generally accepted that PMLD1 subjects show better cognitive and motor functions when compared with PMD subjects.7
Our patient showed a severe neurological picture from the first months of life. Indeed, she was severely mentally retarded and unable to achieve head control in her childhood (at the age of 5 years), in absence of any clinical deterioration. Thus, the patient may be classified as form 0 (the most severe), according to the previously described rating scale for clinical classification ranging from 0 to 4 on the basis of the best motor function.15 The unexpected severe PMLD1 form led to compare the genotype and the phenotype of our patient with existing data from the literature. As shown in Table 1, the majority of PMLD1 patients (46 out of 54) reported in the literature have a score ranging between 3 and 4, with only six and two patients with score 2 and 1, respectively.
The severity of the clinical features of our patient closely resembles the picture of the most severe form of PMD, that is, the connatal variant, characterised by an early onset of nystagmus, muscle hypotonia, pharyngeal weakness and stridor at birth or within the first few weeks of life, followed by severe mental retardation with absent language and severe spasticity during the follow-up.
On brain MRI, PMLD1 patients usually present homogeneous T2 hyperintensities of the cerebral white matter, often in combination with hypointensity on T1-weighted images. Typical prominent T2 hyperintensity of the pons may be observed with variable myelination of pyramidal tracts. Partial myelination of the PLIC has also been reported,4 as we observed here. The cerebellar white matter may be affected, albeit usually to a lesser extent and with preserved cerebellar volume. The basal ganglia and thalami are normal in most patients. However, Steenweg et al22 recently described a separate cluster of PMLD patients showing T2 hyperintensity of the subcortical white matter contrasting with a lower T2 signal of the remaining cerebral white matter, associated with hypointensity of the anterolateral part of the thalamus.
Our patient demonstrated an unusually severe MRI pattern characterised by extensively abnormal supra- and infratentorial white matter, including the brainstem with additional spinal cord involvement. Such a severe involvement, possibly related to a marked paucity of myelin, likely correlates with the severity of clinical picture. The restricted diffusion within the myelinated pyramidal tracts at the PLIC, previously unreported in any PMLD or PMD patients, may be consistent with intramyelinic oedema.
The novel mutation p.E260K (Glu260Lys) harboured by our patient, occurs in the second extracellular domain and affects a highly conserved residue. Indeed, the analysis of evolutionary conservation by comparing the sequences of orthologous GJC2 proteins from human to zebrafish showed that E260 was invariant at this position as were the flanking residues within the SRPTEK domain (data not shown). It was suggested that this conserved motif (SRPTEK) is important for the mechanism by which two hemichannels dock end-to-end to form junctional channels.23
To shed further light on the deleterious consequences of the novel missense p.E260K mutations at the protein level, we employed the homology model of Cx47 as described in Material and Methods.
As shown in Figure 2 the replacement of Glu260-to-Lys (p.E260K) is predicted to break the salt bridge network, normally formed by residues Asp46, Arg257 and Glu260, which is important to maintain the correct orientation of loop E2 for the connexin–connexin interaction to form a connexon. Indeed, the novel Glu260-to-Lys replacement, introducing a positively charged lysine, would interfere with Arg257 and alter the electrostatic parameters of the stable salt bridge network. This nonconservative change 260Lys most likely induces, the dislocation of the external Cx47 loop, thereby hampering the correct connexon assembly into functional channels.
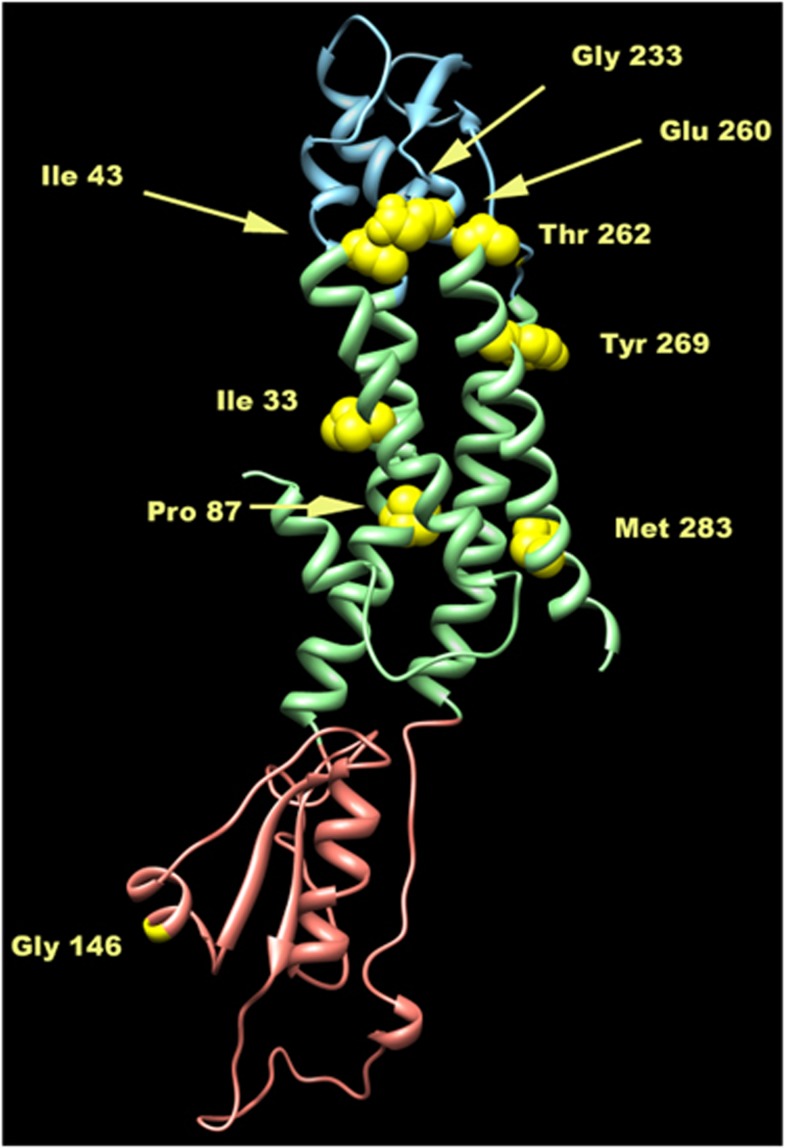
The computational analysis was then extended to nine missense mutations previously reported in the literature in patients with a less severe clinical picture with respect to our patient (Figure 3). The structural analysis of these mutations showed that most of them are conservative or not causative of severe effects on the protein (Table 2). By comparing these findings with the patients' severity score it was observed that the predicted effect of the p.Ile43Met (p.I43M), supposed to potentially perturb a salt bridge, was associated with a mild phenotype, scored as 4. This apparent miscorrelation could be explained by the possibility that the replacement of a branched residue isoleucine-43 with a methionine maintains the hydrophic character of the region. In addition, the flexible methionine side chain, within the protein core, might take a conformation, which does not perturb the correct molecular fold. Another apparent inconsistency might be observed in the case of the ‘in silico' prediction of p.Tyr269Asp (p.Y269D). This mutation, found in combination with a nonsense mutation ([p.Y269D]+[p.R237X]) was expected to have an ameliorative effect on the patient who instead was phenotypically scored as 1. By a targeted examination, the replacement of a polar but uncharged tyrosine with a negatively charged aspartic acid (p.Y269D) occurred in the transmembrane domain. Indeed, the functionally detrimental impact of the interactions between transmembrane aromatic amino acids has already been demonstrated.24 In addition, interactions between the aromatic rings of residues, potentially involving π-stacking, are known to have an important role in the self-assembly, by supplying energy, as well as in the correct orientation, through stacking geometries.25, 26
Figure 3.
Ribbon representation of the three dimensional structure of the homology model of wild-type Cx47. The three main regions of the protein are coloured as follows: extracellular in cyan, transmembrane in light green and cytoplasmatic in pink. The wild-type residues analysed are depicted as yellow balls.
Notably, the molecular modelling of p.Ile33Met predicted that the replaced 33Met, being in short contact with the Trp3 side chain, could cause the displacement of the N-terminal helix of each Cx47 monomer towards the inner side of the hemichannel resulting in a potentially partial block of the pore. This ‘in silico' prediction appears to be in line with the functional findings reported in 3 SPG44 patients by Orthmann-Murphy et al,14 who observed—in a cell model system—that the mutant protein (I33M-Cx47) formed gap junction plaques that were indistinguishable from WT-Cx47, but the resulting I33M-Cx47/Cx43 channels showed severely altered voltage-dependent gating. These data further support the reliability of this molecular modelling.
Our studies provide evidence that the extremely severe clinical and neuroradiological PMLD1 form of our patient likely correlates with the impaired formation of the connexon pore resulting from the detrimental effect of the new p.E260K mutant allele on Cx47 protein and that the spectrum of PMLD1 should be widened to include severe forms.
Acknowledgments
The patient sample was obtained from the ‘Cell Line and DNA Biobank from Patients Affected by Genetic Diseases' (G Gaslini Institute)—Telethon Genetic Biobank Network (Project No. GTB07001). The study was also supported by Italian Health Department ‘Finanziamento Ricerca Corrente (contributo per la ricerca intramurale)' and FP7-HEALTH—LeukoTreat no.241622.
The authors declare no conflict of interest.
References
- Garbern JY, Hobson GH.PLP1-Related Disorders. GeneReviews at GeneTests: Medical Genetics Information Resource (database online). Copyright, University of Washington, Seattle 1993 http://www.genetests.org (Updated 16 March 2010).
- Uhlenberg B, Schuelke M, Ruschendorf F, et al. Mutations in the gene encoding gap junction protein α12 (Connexin 46.6) cause Pelizaeus-Merzbacher-like disease. Am J Hum Genet. 2004;75:251–260. doi: 10.1086/422763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleopa KA, Orthmann JL, Enriquez A, Paul DL, Scherer SS. Unique distributions of the gap junction proteins connexin29, connexin32, and connexin47 in oligodendrocytes. Glia. 2004;47:346–357. doi: 10.1002/glia.20043. [DOI] [PubMed] [Google Scholar]
- Bugiani M, Al Shahwan S, Lamantea E, et al. GJA12 mutations in children with recessive hypomielinating leukoencephalopathy. Neurology. 2006;67:273–279. doi: 10.1212/01.wnl.0000223832.66286.e4. [DOI] [PubMed] [Google Scholar]
- Salviati L, Trevisson E, Baldoin MC, et al. A novel deletion in the GJA12 gene causes Pelizaeus-Merzbacher-like disease. Neurogenetics. 2007;8:57–60. doi: 10.1007/s10048-006-0065-x. [DOI] [PubMed] [Google Scholar]
- Wolf NI, Cundall M, Rutland P, et al. Frameshift mutation in GJA12 leading to nystagmus, spastic ataxia and CNS dys-/demyelination. Neurogenetics. 2007;8:39–44. doi: 10.1007/s10048-006-0062-0. [DOI] [PubMed] [Google Scholar]
- Henneke M, Combes P, Diekmann S, et al. GJA12 mutations are a rare cause of Pelizaeus-Merzbacher-like disease. Neurology. 2008;70:748–754. doi: 10.1212/01.wnl.0000284828.84464.35. [DOI] [PubMed] [Google Scholar]
- Sartori S, Burlina AB, Salviati L, et al. Increased level of N-acetylaspartylglutamate (NAAG) in the CSF of a patient with Pelizaeus-Merzbacher-like disease due to mutation in the GJA12 gene. Eur J Paediatr Neurol. 2008;12:348–350. doi: 10.1016/j.ejpn.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang H, Wang Y, Chen T, Wu X, Jiang Y. Two novel gap junction protein alpha 12 gene mutations in two Chinese patients with Pelizaeus-Merzbacher-like disease. Brain Dev. 2010;32:236–243. doi: 10.1016/j.braindev.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Osaka H, Hamanoue H, Yamamoto R, et al. Disrupted SOX10 regulation of GJC2 transcription causes Pelizaeus-Merzbacher-like disease. Ann Neurol. 2010;68:250–254. doi: 10.1002/ana.22022. [DOI] [PubMed] [Google Scholar]
- Combes P, Kammoun N, Monnier A, et al. Relevance of GJC2 promoter mutation in Pelizaeus-Merzbacher-like disease. Ann Neurol. 2012;71:146–148. doi: 10.1002/ana.22295. [DOI] [PubMed] [Google Scholar]
- Henneke M, Gegner S, Hahn A, et al. Clinical neurophysiology in GJA12-related hypomyelination vs Pelizaeus-Merzbacher disease. Neurology. 2010;74:1785–1789. doi: 10.1212/WNL.0b013e3181e0f820. [DOI] [PubMed] [Google Scholar]
- Meyer E, Kurian MA, Morgan NV, et al. Promoter mutation is a common variant in GJC2-associated Pelizaeus-Merzbacher-like disease. Mol Genet Metab. 2011;104:637–643. doi: 10.1016/j.ymgme.2011.08.032. [DOI] [PubMed] [Google Scholar]
- Orthmann-Murphy JL, Salsano E, Abrams CK, et al. Hereditary spastic paraplegia is a novel phenotype for GJA12/GJC2 mutations. Brain. 2009;132:426–438. doi: 10.1093/brain/awn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailloux F, Gauthier-Barichard F, Mimault C, et al. Genotype-phenotype correlation in inherited brain myelination defects due to proteolipid protein gene mutations. Clinical European Network on Brain Dysmyelinating Disease. Eur J Hum Genet. 2000;8:837–845. doi: 10.1038/sj.ejhg.5200537. [DOI] [PubMed] [Google Scholar]
- Orthmann-Murphy JL, Enriquez AD, Abrams CK, Scherer SS. Loss-of –function connexin47 mutations cause Pelizaeus-Merzbacher-like disease. Mol Cell Neurosci. 2007;34:629–641. doi: 10.1016/j.mcn.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Maeda S, Nakagawa S, Suga M, et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60 (Pt 12 Pt 1:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ruf N, Uhlenberg B. Analysis of human alternative first exons and copy number variation of the GJA12 gene in patients with Pelizaeus-Merzbacher-like disease. Am J Med Genet Part B. 2008;150B:226–232. doi: 10.1002/ajmg.b.30792. [DOI] [PubMed] [Google Scholar]
- Steenweg ME, Vanderver A, Blaser S, et al. Magnetic resonance imaging pattern recognition in hypomyelinating disorders. Brain. 2010;133:2971–2982. doi: 10.1093/brain/awq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JA, Baker KA, Altenberg GA, Abagyan R, Yeager M. Molecular modeling and mutagenesis of gap junction channels. Prog Biophys Mol Biol. 2007;94:15–28. doi: 10.1016/j.pbiomolbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebermann H, Schoneberg T, Schulz A, et al. A conserved tyrosine residue (Y601) in transmembrane domain 5 of the human thyrotropin receptor serves as a molecular switch to determine G-protein coupling. FASEB J. 1998;12:1461–1471. doi: 10.1096/fasebj.12.14.1461. [DOI] [PubMed] [Google Scholar]
- Gazit E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- Sal-Man N, Gerber D, Bloch I, Shai Y. Specificity in transmembrane helix-helix interactions mediated by aromatic residues. J Biol Chem. 2007;282:19753–19761. doi: 10.1074/jbc.M610368200. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]