Abstract
An automated phosphopeptide enrichment strategy is described using titanium dioxide (TiO2)-packed, fused silica capillaries for use with liquid chromatography (LC)-mass spectrometry (MS)/MS-based, label-free proteomics workflows. To correlate an optimum peptide:TiO2 loading ratio between different particle types, the ratio of phenyl phosphate-binding capacities was used. The optimum loading for the column was then verified through replicate enrichments of a range of quantities of digested rat brain tissue cell lysate. Fractions were taken during sample loading, multiple wash steps, and the elution steps and analyzed by LC-MS/MS to gauge the efficiency and reproducibility of the enrichment. Greater than 96% of the total phosphopeptides were detected in the elution fractions, indicating efficient trapping of the phosphopeptides on the first pass of enrichment. The quantitative reproducibility of the automated setup was also improved greatly with phosphopeptide intensities from replicate enrichments exhibiting a median coefficient of variation (CV) of 5.8%, and 80% of the identified phosphopeptides had CVs below 11.1%, while maintaining >85% specificity. By providing this high degree of analytical reproducibility, this method allows for label-free phosphoproteomics over large sample sets with complex experimental designs (multiple biological conditions, multiple biological replicates, multiple time-points, etc.), including large-scale clinical cohorts.
Keywords: glycolic acid, liquid chromatography, mass spectrometry, phosphorylation, titanium dioxide
INTRODUCTION
Phosphorylation is a reversible, post-translational modification affecting the folding and function of proteins and is a critical regulator of numerous biological processes, including proliferation, differentiation, and signaling. LC-MS/MS-based phosphoproteomic strategies are widely used for qualitative and quantitative analyses. For relative quantitation, the use of stable isotope labeling by amino acids in cell culture (SILAC) is commonly used.1–3 This approach is largely limited to systems amenable to cell culture and some SILAC-labeled species.4–6 Covalent labeling of peptides with isobaric reporter ion technology (such as isobaric tags for relative and absolute quantitation or tandem mass tags) prior to enrichment is also used but can introduce additional quantitative variation and is limited by cross-talk between coeluting peptides.7 The cost of labeling by reporter ion techniques may be prohibitive, given that the low stoichiometry of phosphorylation requires enrichment of a large amount of proteins, and such labeling costs may be wasteful, as the vast majority of the sample (nonphosphorylated peptides) will not be analyzed. To date, the largest multiplex in a single SILAC experiment has been limited to five; however, recent publications have demonstrated the use of a super-SILAC mix to potentially analyze larger sample sets and overcome the inability to analyze biologically relevant tissues, provided a comparable labeled cell line exists.8–10 Whereas this does serve to broaden the number of SILAC applications, sample dilution is still a challenge, along with the increase in sample complexity (multiple isotope-labeled copies of each peptide in the sample).
Label-free, quantitative proteomics offers the ability to perform relative quantitation, as well as absolute quantitation in some cases, using the area under the curve (AUC) of the accurate mass-selected ion chromatograms without the need to compare this with another signal in the same sample.11–13 Quantitation performed without the need for labeling affords greater flexibility in the experimental design by not limiting the number of samples or treatment groups that can be compared in a single experiment. This is increasingly important for analysis of biological replicates as is necessary to elucidate statistically significant, biologically relevant changes. Label-free experiments provide lower limits of quantitation, as there are no sample losses from cleanups after derivatization/labeling, and the sample dilution that occurs as a result of pooling in multiplexed experiments is avoided. However, in label-free experiments, minimizing the technical variability of the enrichment is critical.
As protein phosphorylation occurs at substoichiometric ratios, an efficient enrichment of phosphorylated peptides is essential. Whereas immobilized metal affinity chromatography (IMAC) was formerly the standard for such enrichments and is still heavily used, there has been an increase in the use of TiO2 materials as a result of its compatibility with reagents frequently used in biological experiments, including alkaline metal salts and EDTA.14–17 TiO2 has also been shown to improve the enrichment of singly phosphorylated peptides over IMAC.18,19 The format of the enrichment also affects performance, and various methods have been used, including spin column and syringe or centrifuge,14,17,18,20 batch format (resin directly added to samples for incubation),21 and packed capillary on-line enrichments.22–24 For the purposes of maintaining quantitative reproducibility and maximizing throughput of the mass spectrometers, our method uses a capillary-based, off-line, automated enrichment strategy. With this automated format, all samples are enriched using the same batch and amount of enrichment material, and the potential for manual operation error is minimized.
The major goals of this work were to improve the reproducibility for relative quantitation and to reduce operator time via automation. For consistency, lysate from a single solubilized rat brain was used throughout these experiments, allowing for direct comparisons between experiments. Multiple enrichment parameters were addressed, including enrichment flow rate and column loading, to ensure a high level of specificity, efficiency, and quantitative reproducibility over previously published, label-free approaches.20,21
MATERIALS AND METHODS
Chemicals
Rat brain tissue was purchased from Bioreclamation (Jericho, NY, USA). MassPREP enhancer (MPE), a 2,5-dihydroxybenzoic acid analog, and RapiGest SF were purchased (Waters, Milford, MA, USA). Phenyl phosphate disodium salt was purchased from VWR (Radnor, PA, USA). Phosphatase inhibitor Cocktails 2 and 3 (Sigma, St. Louis, MO, USA) and Complete EDTA-free protease inhibitor cocktail tablets (Roche, Indianapolis, IN, USA) were used as directed. TiO2 resin was purchased from Protea Biosciences (Morgantown, WV, USA), and bulk 5 μm Titansphere TiO2 particles were purchased from GL Sciences (Tokyo, Japan). The Titansphere particles were dried overnight at 150°C and stored under vacuum until use. Potassium silicate solution, Kasil 2130, was provided by PQ (Valley Forge, PA, USA). All other reagents used were of the highest available purity purchased from commercial sources.
Phenyl Phosphate-Binding Capacity
Previous studies in our lab used TiO2 material purchased from Protea Biosciences in a spin-column enrichment format.20 As the irregular size and shape of this material are not amenable to packing into a capillary, spherical 5 μm Titansphere particles were used, requiring the relative phenyl phosphate-binding capacity of Protea Biosciences TiO2 material to that of GL Sciences TiO2 material to be determined. Two 5-mg aliquots of each material were equilibrated by vortexing with 150 μL Buffer A [80% acetonitrile (ACN), 1% TFA]. After centrifugation, the supernatant was removed, and 150 μL 10 mg/mL phenyl phosphate in Buffer A was added. Slurries were incubated on a rotary shaker for 30 min at room temperature, and the particles were washed twice with 150 μL Buffer A. Elution of bound phenyl phosphate was performed using 100 μL Buffer B (5% aqueous ammonia) and incubation with shaking for 5 min. The supernatant was diluted 1:10 and analyzed by flow injection analysis with a CapLC pump and a 2487 UV detector monitoring 266 nm (Waters). A calibration curve was run before and after sample analysis from 0.0313 mg/mL to 1000 mg/mL phenyl phosphate. The maximum UV absorbance of the injection peak was used for quantitation. The ratio of phenyl phosphate-binding capacities between the two particle types was used in combination with the reported optimal loading of the Protea Biosciences particles of 51.8 μg/mg resin20 as guidelines when packing capillary TiO2 enrichment columns. Validation of the use of this estimate was accomplished by performing duplicate enrichments of 500 μg, 750 μg, 1 mg, and 1.5 mg total digested protein from rat brain lysate on a 250-μm × 10-cm column packed with ∼5 mg Titansphere particles. Optimum protein loading was determined to be where the total phosphopeptide intensity and specificity started to level off at higher loading amounts. The chromatographic peak widths remained constant throughout the loading study (average peak-width at half-height for 50 peaks; 500 μg sample: 26±10 s; 1500 μg sample: 27 s±14 s), indicating that the roll-off was not a result of overloading of the LC column.
Sample Preparation
Rat brain tissue (1.96 g) was solubilized in 20 mL 0.25% RapiGest SF, 50 mM ammonium bicarbonate, in the presence of phosphatase and protease inhibitors using a combination of a tissue tearor (BioSpec Products, Bartlesville, OK, USA) and burst sonication. Insoluble material was pelleted [20,000 relative centrifugal force (rcf)]. The extracted protein yield was 195 mg (mini-Bradford assay calibrated with BSA). This supernatant was aliquoted (5 mg aliquots) and frozen at −80°C prior to digestion. Upon thawing, intact bovine α-casein was used as an internal standard (30 fmol casein/μg lysate). Aliquots were reductively alkylated with 10 mM DTT (80°C for 15 min), followed by 20 mM iodoacetamide (room temperature, dark, 30 min) prior to overnight digestion with trypsin (37°C, 50:1 protein:enzyme ratio). The solution was then acidified (TFA to a final concentration of 1%). After incubation at 60°C for 2 h, samples were centrifuged (20,000 rcf for 5 min) and were dried using a vacuum centrifuge.
TiO2 Column Preparation
TiO2 enrichment columns were prepared using fused-silica capillary (Polymicro Technologies, Phoenix, AZ, USA) with a 360-μm outer diameter and a 250-μm inner diameter (ID). The column-packing procedure was modified slightly from what has been reported previously for self-packed reversed-phase columns.25 Briefly, a 50-cm length of capillary was fritted at the outlet using a glass fiber sol-gel frit, as described previously.26 Columns were slurry packed with 5-μm Titansphere particles, suspended at a concentration of 5 mg/mL in 80:20 ACN:water. Slurries were sonicated for 15 min to reduce aggregates. Packing was initiated at 40 pounds/square inch (psi) and was increased slowly at a rate of ∼500 psi/cm packed bed. The final packing pressure was 6300 psi with a packed bed length of 12 cm. After slowly depressurizing the column, the outlet frit was clipped off, and the column was trimmed to a length of 9.5 cm. The inlet and outlet of the column were secured in fritted unions purchased from Valco Instruments (Houston, TX, USA).
Phosphopeptide Enrichment
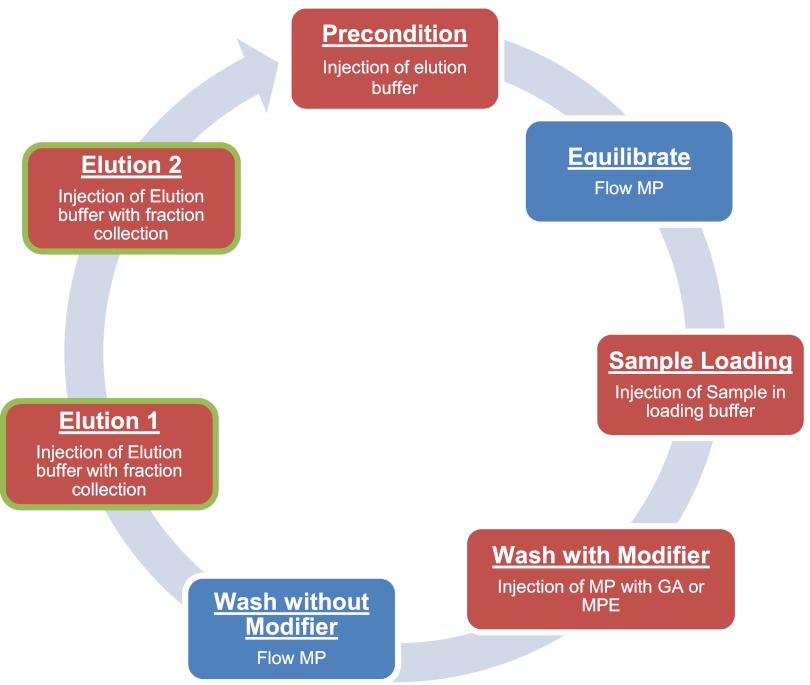
Samples were reconstituted in 40 μL loading buffer (80% ACN, 1% TFA), plus modifier [50 mg/mL MPE in the loading study or 1 M glycolic acid (GA) for all other experiments]. Enrichment of phosphopeptides was performed using a CapLC pump, autosampler, and 2487 UV detector (Waters) and a HoneyComb fraction collector (BASi, West Lafayette, IN, USA). Based on the phenyl phosphate-binding capacity, a 10-cm × 250-μm ID capillary packed with TiO2 particles was used. The mobile phase was kept constant throughout the enrichment at 80% ACN, 1% TFA, and the flow rate was set at 5 μl/min or 10 μL/min. The enrichment process was performed via five injections (Fig. 1) of both sample and various buffers. The first step pre-elutes the column using 100 μL elution buffer (20% ACN, 5% aqueous ammonia). The run time was set to allow for 100 μL (50 column volumes) of mobile phase to flow through the column after the elution buffer to equilibrate the column. The second step injects 40 μL sample in loading buffer. The third step injects 100 μL loading buffer, followed by a delay time to allow for 200 μL mobile phase (100 column volumes) to remove the modifier. The fourth and fifth steps are sequential, 100-μL injections of elution buffer. Elution volumes were chosen to elute and collect 99% of the phosphopeptides based on downstream LC-MS intensity (see Supplemental Table 1). The entire enrichment process was monitored on-line prior to the fraction collector by UV absorption at 215 nm. To re-acidify the elution fraction, each 120-μL fraction was collected into vials containing 30 μL 33% formic acid. An example UV trace of the enrichment process is shown in Supplemental Fig. 1. The total time for the automated enrichment of a single sample performed at a flow rate of 5 or 10 μL/min was 3 h and 37 min, or 2 h, respectively. The two elution fractions were pooled prior to dry-down in a vacuum centrifuge. Samples were reconstituted in 20 μL 2% ACN, 0.1% TFA, with 10 mM citrate for LC-MS/MS analysis.
FIGURE 1.
Experimental workflow for automatic enrichment system. Steps in red involve injections from the autosampler, whereas those in blue correspond to length of time where mobile phase (MP) is allowed to flow through the column.
Phosphopeptide Enrichment Profile
To determine how efficiently phosphopeptides were trapped on the TiO2 column, three fractions were collected, in addition to the pooled elution fractions, assessing break-through/loss: one during the injection of sample as the flow-through fraction, one during the subsequent wash with 1 M GA, and one during the wash without any modifier. All fractions were dried down. The elution fraction and wash fractions were reconstituted with LC loading buffer. The first wash fraction was processed further using a p10 C18 ZipTip (Millipore, Bellerica, MA, USA) to remove excess GA. The flow-through fraction was reconstituted in enrichment buffer with 1 M GA and taken through the automated enrichment process a second time, only collecting the elution fractions. An equivalent percentage (20%) of each fraction was injected on-column for LC-MS/MS analysis.
LC-MS/MS Analysis
Peptides were separated on a Waters nanoACQUITY ultra performance LC. The trap column was 180 μm × 20 mm (5 μm Symmetry C18), and the analytical column was a 75-μm ID × 250-mm capillary (1.7 μm BEH130 C18). Peptides were trapped and washed for 10 min at 10 μL/min in 99.9% Buffer A. After switching the analytical column in-line, the mobile phase was held at 3% Buffer B for 5 min, followed by a linear ramp to 30% Buffer B over 90 min at 300 nL/min. MS and MS/MS analysis of the eluting peptides was performed on a Waters Synapt high-definition MS G1 mass spectrometer. Data-directed acquisition (DDA) was performed using a single precursor MS scan, followed by three MS/MS product ion scans [0.6-s scans over the range of mass:charge ratio (m/z) 50:1990]. The threshold for MS/MS triggering was set at an individual ion rising above 20 counts/s. Peptide fragmentation was performed using a charge state-dependent, collision-induced dissociation energy versus m/z profile with dynamic exclusion (120 s).
Data Processing and Analysis
Qualitative peptide identifications were made by searching against a forward-reverse National Center for Biotechnology Information rattus protein database, appended with bovine α- and β-casein protein entries (50,558 total entries; downloaded 10/25/10) using Mascot Server 2.2 (Matrix Science, Boston, MA, USA). Precursor and product ion mass tolerances were 20 ppm and 0.04 Da, respectively. Searches were performed with trypsin specificity, allowing up to two missed cleavages; carbamidomethylation was a fixed modification; and oxidation of methionine, deamidation of asparagine and glutamine, and phosphorylation of serine, threonine, and tyrosine were variable modifications. Scaffold result files may be found at Proteome Commons (https://proteomecommons.org) as described in Supplemental Information.
Intensity-based, label-free quantitation of identified phosphopeptides was performed as described previously using Rosetta Elucidator.20 DDA analyses were used for relative quantitation and qualitative purposes. Annotation of aligned features was performed [Mascot ion score thresholding at 1% spectral false discovery rate (FDR)]. Relative quantitation was performed by measuring AUC intensities for all annotated features across all LC-MS/MS runs in a manner analogous to the original accurate mass and retention time approach and comparing between analyses.12 Quantitative precision was expressed as the CV of technical replicates (percent ratio between the sd and the mean). Quantitative results for each peptide in each experiment can be found in Supplemental Tables 1–4.
RESULTS
Binding Capacity and the Effect of Column Loading on Specificity
Previous quantitative phosphoproteomic work conducted in our laboratory used a comprehensive matrix experiment to identify the optimal loading ratio and MPE concentration for the enrichment on Protea Biosciences TiO2 resin.20 In the transition to the use of spherical GL Sciences particles, a standard was needed to compare the two materials directly, as well as to allow for future quality control comparisons of columns to ensure equivalent binding capacities. A phosphate group mimetic, phenylphosphate, was used to determine this difference. The use of phenylphosphate as a surrogate for phosphopeptide-binding capacity has been reported previously.27 Flow injection analysis of the phenyl phosphate eluting from 5 mg of each particle type yielded a binding capacity of 47 ± 2 μmol/g for the Protea Biosciences particles and 188 ± 9 μmol/g for the Titansphere particles.
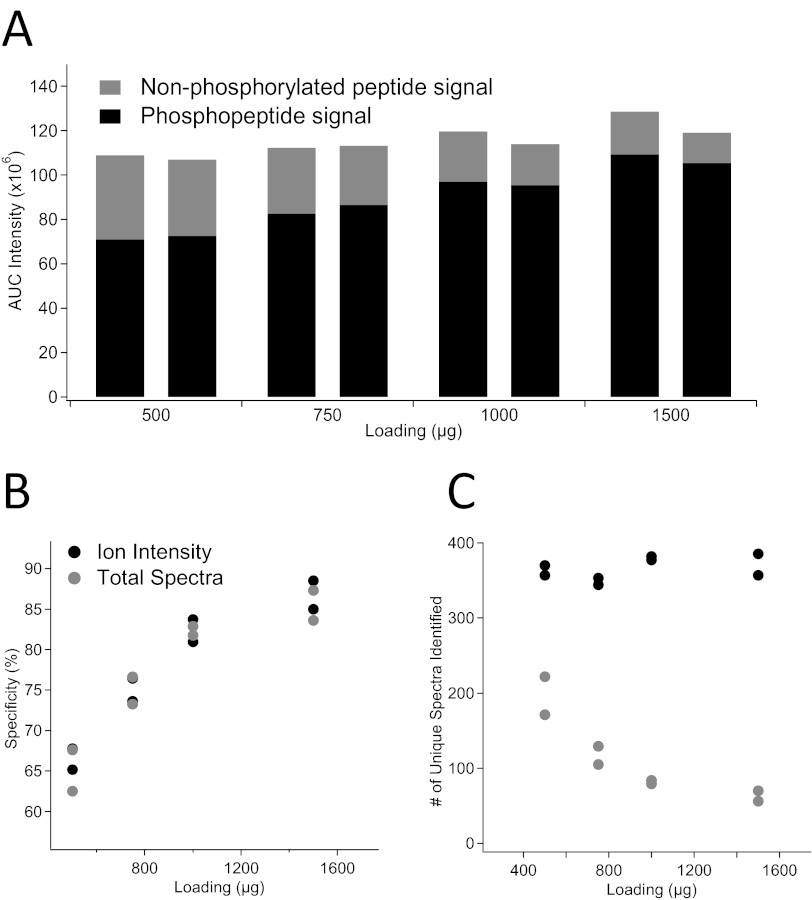
This relative binding was taken into account when determining the ideal column dimensions. Based on optimum loading from previous experiments with Protea Biosciences of 51.8 μg/mg resin, the optimum peptide loading for Titansphere particles was calculated to be 207 μg/mg particles. To allow triplicate LC-MS/MS analysis of the enriched samples, our lab prefers to enrich phosphopeptides from 1-mg quantities of total protein. A 9.5-cm × 250-μm ID capillary column was packed with 5 μm Titansphere material (∼4.89 mg TiO2 material), assuming interparticle porosity of 0.4, giving an optimum load of 1.01 mg digested protein.28 To verify the proper loading, duplicate enrichments of the following amounts of digested rat brain lysate were enriched in the presence of 50 mg/mL MPE: 500 μg, 750 μg, 1.0 mg, and 1.5 mg. To allow for peptide intensity-based comparisons to be made across all sample loading amounts, these data were not intensity scaled. Figure 2A shows the summed ion intensity from phosphopeptides and nonphosphorylated peptides. An increase in phosphopeptide intensity and decrease in nonphosphorylated peptide intensity were seen with increasing loading amount, indicating increased specificity. For the first three loading amounts (Fig. 2B), there is a linear increase in specificity, R2 = 0.94. However, adding in the last loading amount, 1.5 mg, the trend is not maintained (R2=0.83), confirming the optimum loading for this column to be 1.0 mg. The number of unique phosphopeptide spectra is not affected by loading amount (Fig. 2C); therefore, the increase in specificity with higher loading appears to be a result of the decrease in the nonspecific enrichment of nonphosphorylated peptides.
FIGURE 2.
Loading study based on phenyl phosphate-binding capacity. (A) AUC intensity of all signals from phosphopeptides in black and nonphosphorylated peptides in gray. (B) Quantitative specificity of replicated enrichments plotted as a function of loading amount. (C) Number of unique spectra identified to phosphopeptides plotted in black and nonphosphorylated peptides plotted in gray.
Profile of Phosphopeptides throughout the Enrichment Process
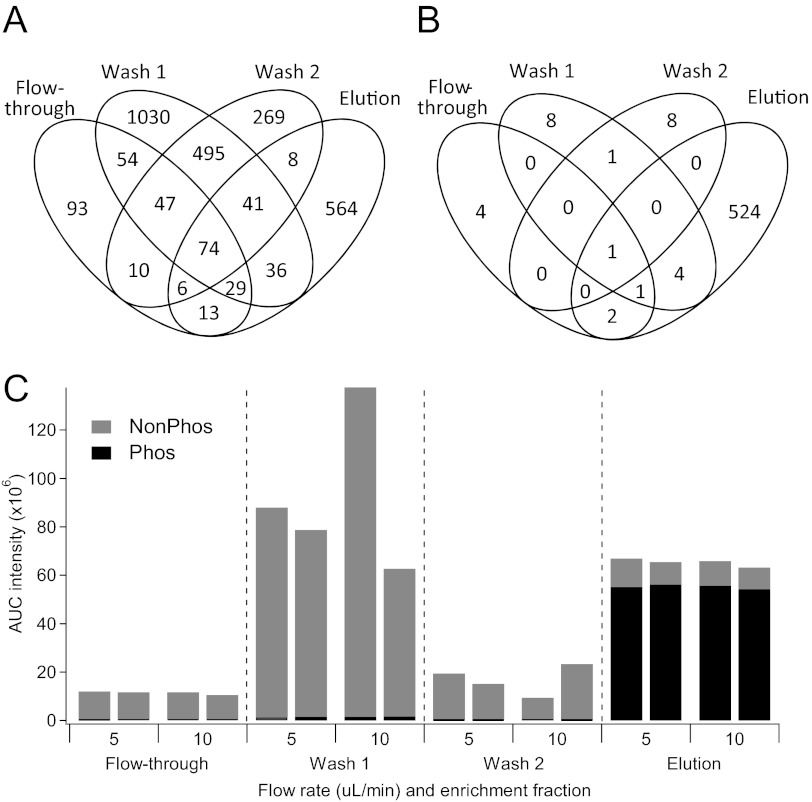
Phosphopeptides were identified and quantified independently for each of the four fractions collected during the enrichment process. The overlap between all peptides identified in each fraction is shown in Fig. 3A, and the overlap of phosphopeptides only is shown in Fig. 3B. The flow-through fraction yielded the fewest number of unique peptides, only four of which were phosphorylated. The wash fractions had a high degree of uniqueness from the elution and re-enriched flow-through fractions in Fig. 3A. However, essentially, all of these were nonphosphorylated peptides (Fig. 3B). Also, important to note is the higher likelihood of false-positives with the addition of variable modifications in database searches. Identifications from individual analyses were made using a forward-reverse database and annotating peptides at a Mascot ion score that yielded a 1% spectral FDR, regardless of the number of modifications on a peptide. The result of this for the elution fractions for each sample was an overall 2.3% peptide-level FDR and a slightly higher phosphopeptide FDR of 2.6%. For Wash Fractions 1 and 2, the overall peptide-level FDRs were 1.5% and 1.2%, respectively. The phosphopeptide FDRs were much higher, at 23.8% and 14.3%, respectively, indicating that presence of phosphopeptides in these wash fractions is likely overestimated.
FIGURE 3.
Presence of phosphopeptides throughout each phase of enrichment. Venn overlap of unique spectra for all unique spectra (A) and for unique phosphopeptide spectra (B). (C) Summed AUC intensity for all phosphopeptide species (black) and nonphosphorylated peptides (gray) from fractions taken throughout the enrichment process performed at various enrichment flow rates.
To determine whether different classes of nonphosphorylated peptides were removed at each wash step, the percentage of acidic residues (aspartic acid and glutamic acid) in the nonphosphorylated peptides identified in the first wash or the second wash, but not both, was compared. In the first wash step, the average percentage of acidic amino acids in each peptide was 14.4% and 13.9% in the second wash. Whereas it would be expected that peptides with more acidic residues would need a modifier to competitively inhibit binding to titania, the difference in acidic amino acid content between these two wash steps is not statistically significant [t=0.87; P>0.05; degrees of freedom (d.f.)=757]. LC-MS/MS analysis of an unenriched, digested rat brain sample yields an average percent of acidic residues/peptide of 13.3%, which is statistically different from the Wash 1 peptides (t=2.80; P<0.01; d.f.=1870), indicating that the peptides nonspecifically binding to the column are more likely to contain acidic residues.
The summed intensity for all phosphopeptides and all nonphosphorylated peptides is shown in Fig. 3C. The majority of signals from phosphopeptides (96.1±0.3%) is detected in the elution fractions. The flow-through fraction, which was subjected to a complete re-enrichment, only contained 1% ± 0.1% of the total phosphopeptide signal for a given sample. This indicates that essentially, all phosphopeptides that are going to be enriched are enriched on the first pass through the column. This finding eliminates the need for multiple passes of enrichment.
Effect of Enrichment Flow Rate on Specificity and Reproducibility
Recent publications have focused on the use of batch-mode enrichments, citing difficulties performing reproducible enrichments with spin columns, possibly as a result of difficulty in accurately measuring and controlling flow rate.21,29,30 This type of enrichment format typically involves the incubation of digested lysates in the presence of titania particles (30 min +). One concern when transitioning to an automated protocol with a capillary column was the interaction time that an analyte would have with the titania bed. The spin-column format used in Soderblom et al.20 was operated with a column dead time (the dwell time of an unretained analyte) of 11 s, with a linear velocity of ∼0.4 mm/s. With the use of the 10-cm × 250-μm ID capillary column, the dead time and linear velocity of the enrichment at a flow rate of 5 μL/min are 23.5 s and 4.2 mm/s, respectively. The dead time is twice that of the spin column, and the linear velocity is an order of magnitude larger. To test the effect of changes in flow rate, enrichments were performed at 5 μL/min and 10 μL/min (which would have a comparable dwell time as the spin column).
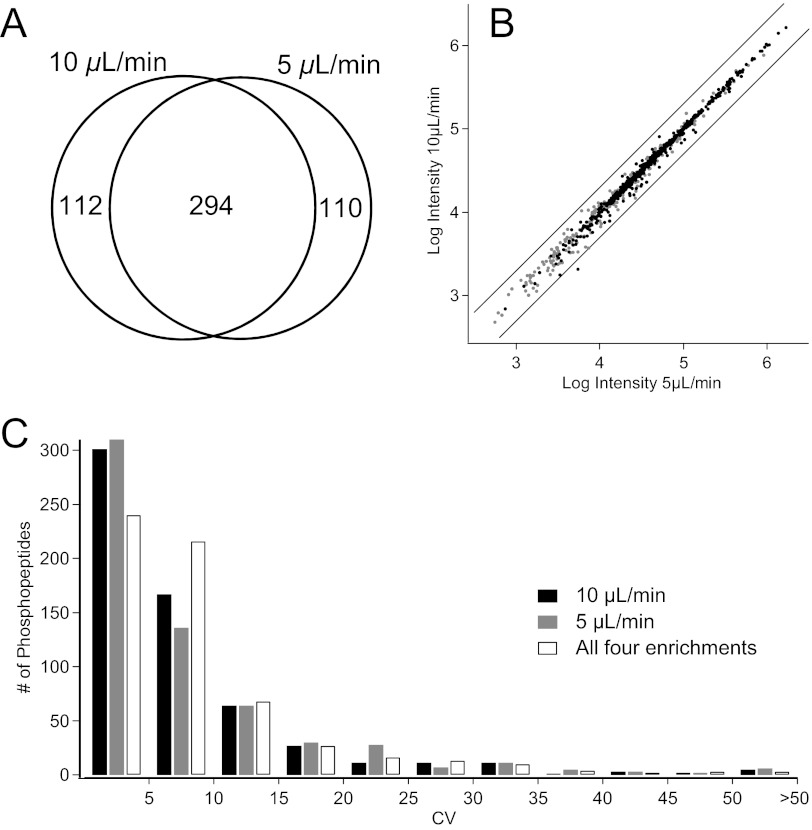
A comparable number of peptides and phosphopeptides were identified in all enrichments. Regardless of enrichment flow rate, the total number of identified spectra was equivalent for all peptides (422±18) and phosphopeptides (340±18). There were 294 phosphopeptide spectra found in both sets of enrichments (Fig. 4A), whereas 112 and 110 phosphorylated spectra were unique to the 10-μL/min enrichment and 5-μL/min enrichment, respectively. With the serendipitous nature of a DDA experiment, this degree of overlap is expected, simply as a result of under-sampling in the LC-MS/MS analysis. Of the peptides that were identified in only one of the conditions, there does not appear to be a bias toward a specific type of phosphopeptide (i.e., multiply phosphorylated).
FIGURE 4.
Reproducibility of phosphopeptide enrichments at various flow rates. (A) Venn overlap of unique phosphopeptide spectra identified from two enrichments performed at 10 μL/min or 5 μL/min. (B) Log/log ratio plot of intensities of phosphopeptides in black and nonphosphorylated peptides in gray after LC-MS/MS analysis of enriched samples. Lines indicate a fold change of two. (C) Distribution of CV as a measure of quantitative reproducibility for each flow rate individually, and all samples combined regardless of enrichment flow rate.
Figure 4B shows the average intensities of peptides enriched at 10 μL/min and at 5 μL/min. Intensity scaling of each run was performed relative to the total phosphopeptide intensity. The phosphopeptides are tightly clustered around y = x (R2=0.997, with a slope of 0.998). To confirm that this correlation is not a result of intensity scaling, unscaled data were plotted, yielding a line with R2 = 0.995 and a slope of 1.010. The variations of intensities were determined within each flow rate separately, as well as across all four enrichments (Fig. 4C). The distributions of CVs for each flow rate follow similar trends, indicating equivalent quantitative reproducibility for both flow rates. Across all four samples, the CV distribution shifts to slightly higher CVs, with a greater percentage of phosphopeptides fitting into the 5–10% CV range. Given that the slope of the line was 0.998 and that there was not a significant increase in CV when combining all four samples, it was determined that there was no difference in performance as a function of flow rate. In addition, the mean and median CVs for the phosphopeptides in this experiment were 8.7% and 5.8%, respectively, with α-casein phosphopeptides having CVs ranging from 1.7% to 6.4%. This is comparable with a recently published, label-free, IMAC-based enrichment31 and is a significant improvement over our previously reported results using spin columns, which obtained a median CV for phosphopeptides of 23.5%.20 A similar batch-format study reported a mean CV of 20.3%.21 For future differential expression experiments, it is recommended that α-casein or another phosphopeptide standard be spiked into each sample to verify enrichment performance.
DISCUSSION
An efficient and selective automated phosphopeptide enrichment strategy has been developed using a TiO2 capillary column-based enrichment system. Initial studies showed that by using a small molecule—phenylphosphate—the relative phosphopeptide-binding capacity of various TiO2 resins could be determined accurately without the need for commercial phosphopeptide standards. This was validated by performing a loading study with a rat brain lysate. Compared with other published protocols, the most significant advantage of the automated enrichment strategy is the improvement in reproducibility. By removing the manual enrichment steps, the average quantitative variability in the enrichment of individual phosphopeptides had a median CV of only 5.8% across all phosphopeptides, which will allow for more confident assignments of differentially expressed phosphopeptides when used in a relative quantitation experiment. For these experiments, increased throughput was not a major objective, and therefore, the number of samples enriched in a 24-h period remains equivalent to that of the spin-column format (12 samples in a 24-h period), albeit in a completely automated platform. However, it is conceivable that with further optimization of the wash conditions and increased flow rates during these steps, the throughput may be improved in the future.
DISCLOSURE
There are no financial conflicts.
REFERENCES
- 1. Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 1999;17:994–999 [DOI] [PubMed] [Google Scholar]
- 2. Ong S-E, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2001;1:376–386 [DOI] [PubMed] [Google Scholar]
- 3. Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 2004;3:1154–1169 [DOI] [PubMed] [Google Scholar]
- 4. Looso M, Borchardt T, Krueger M, Braun T. Advanced identification of proteins in uncharacterized proteomes by pulsed in vivo stable isotope labeling-based mass spectrometry. Mol Cell Proteomics 2010;9:1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sury MD, Chen J-X, Selbach M. The SILAC fly allows for accurate protein quantification in vivo. Mol Cell Proteomics 2010;9:2173–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kruger M, Moser M, Ussar S, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 2008;134:353–364 [DOI] [PubMed] [Google Scholar]
- 7. Lin W-T, Hung W-N, Yian Y-H, et al. Multi-Q: a fully automated tool for multiplexed protein quantitation. J Proteome Res 2006;5:2328–2338 [DOI] [PubMed] [Google Scholar]
- 8. Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods 2010;7:383–385 [DOI] [PubMed] [Google Scholar]
- 9. Geiger T, Wisniewski JR, Cox J, et al. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat Protoc 2011;6:147–157 [DOI] [PubMed] [Google Scholar]
- 10. Molina H, Yang Y, Ruch T, et al. Temporal profiling of the adipocyte proteome during differentiation using a five-plex SILAC based strategy. J Proteome Res 2009;8:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lipton MS, Pasa-Tolic L, Anderson GA, et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci USA 2002;99:11049–11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith RD, Anderson GA, Lipton MS, et al. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics 2002;2:513–523 [DOI] [PubMed] [Google Scholar]
- 13. Zhu W, Smith JW, Huang C-M. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol 2010;2010:840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJD. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics 2005;4:873–886 [DOI] [PubMed] [Google Scholar]
- 15. Thingholm TE, Larsen MR, Ingrell CR, Kassem M, Jensen ON. TiO2-based phosphoproteomic analysis of the plasma membrane and the effects of phosphatase inhibitor treatment. J Proteome Res 2008;7:3304–3313 [DOI] [PubMed] [Google Scholar]
- 16. Thingholm TE, Larsen MR. The use of titanium dioxide micro-columns to selectively isolate phosphopeptides from proteolytic digests. Methods Mol Biol 2009;527:57–66 [DOI] [PubMed] [Google Scholar]
- 17. Thingholm TE, Jorgensen TJD, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc 2006;1:1929–1935 [DOI] [PubMed] [Google Scholar]
- 18. Jensen SS, Larsen MR. Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun Mass Spectrom 2007;21:3635–3645 [DOI] [PubMed] [Google Scholar]
- 19. Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics 2008;7:661–671 [DOI] [PubMed] [Google Scholar]
- 20. Soderblom EJ, Philipp M, Thompson JW, Caron MG, Moseley MA. Quantitative label-free phosphoproteomics strategy for multifaceted experimental designs. Anal Chem 2011;83:3758–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montoya A, Beltran L, Casado P, Rodriguez-Prados J-C, Cutillas PR. Characterization of a TiO2 enrichment method for label-free quantitative phosphoproteomics. Methods (San Diego, CA) 2011;54:370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cantin GT, Shock TR, Park SK, Madhani HD, Yates JR., III Optimizing TiO2 -based phosphopeptide enrichment for automated multidimensional liquid chromatography coupled to tandem mass spectrometry. Anal Chem 2007;79:4666–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pinkse MWH, Uitto PM, Hilhorst MJ, Ooms B, Heck AJR. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem 2004;76:3935–3943 [DOI] [PubMed] [Google Scholar]
- 24. Schlosser A, Vanselow JT, Kramer A. Mapping of phosphorylation sites by a multi-protease approach with specific phosphopeptide enrichment and NanoLC-MS/MS analysis. Anal Chem 2005;77:5243–5250 [DOI] [PubMed] [Google Scholar]
- 25. MacNair JE, Lewis KC, Jorgenson JW. Ultrahigh-pressure reversed-phase liquid chromatography in packed capillary columns. Anal Chem 1997;69:983–989 [DOI] [PubMed] [Google Scholar]
- 26. Maiolica A, Borsotti D, Rappsilber J. Self-made frits for nanoscale columns in proteomics. Proteomics 2005;5:3847–3850 [DOI] [PubMed] [Google Scholar]
- 27. Liang S-S, Makamba H, Huang S-Y, Chen S-H. Nano-titanium dioxide composites for the enrichment of phosphopeptides. J Chromatogr A 2006;1116:38–45 [DOI] [PubMed] [Google Scholar]
- 28. Giddings JC. Dynamics of Chromatography: Principles and Theory. Boca Raton, FL, USA: CRC, 2002 [Google Scholar]
- 29. Kettenbach AN, Gerber SA. Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal Chem 2011;83:7635–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rigbolt KTG, Prokhorova TA, Akimov V, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signaling 2011;4:rs3/1–rs3/17 [DOI] [PubMed] [Google Scholar]
- 31. Wang Y-T, Tsai C-F, Hong T-C, et al. An informatics-assisted label-free quantitation strategy that depicts phosphoproteomic profiles in lung cancer cell invasion. J Proteome Res 2010;9:5582–5597 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.