Abstract
MicroRNAs (miRNAs) and methionine adenosyltransferase 1A (MAT1A) are dysregulated in hepatocellular carcinoma (HCC), and reduced MAT1A expression correlates with worse HCC prognosis. Expression of miR-664, miR-485-3p, and miR-495, potential regulatory miRNAs of MAT1A, is increased in HCC. Knockdown of these miRNAs individually in Hep3B and HepG2 cells induced MAT1A expression, reduced growth, and increased apoptosis, while combined knockdown exerted additional effects on all parameters. Subcutaneous and intraparenchymal injection of Hep3B cells stably overexpressing each of this trio of miRNAs promoted tumorigenesis and metastasis in mice. Treatment with miRNA-664 (miR-664), miR-485-3p, and miR-495 siRNAs reduced tumor growth, invasion, and metastasis in an orthotopic liver cancer model. Blocking MAT1A induction significantly reduced the antitumorigenic effect of miR-495 siRNA, whereas maintaining MAT1A expression prevented miRNA-mediated enhancement of growth and metastasis. Knockdown of these miRNAs increased total and nuclear level of MAT1A protein, global CpG methylation, lin-28 homolog B (Caenorhabditis elegans) (LIN28B) promoter methylation, and reduced LIN28B expression. The opposite occurred with forced expression of these miRNAs. In conclusion, upregulation of miR-664, miR-485-3p, and miR-495 contributes to lower MAT1A expression in HCC, and enhanced tumorigenesis may provide potential targets for HCC therapy.
Introduction
Methionine adenosyltransferase (MAT) is an essential enzyme that is responsible for the biosynthesis of S-adenosylmethionine (SAMe), the principal biological methyl donor in all mammalian cells (1). Mammals express 2 different genes, MAT1A and MAT2A, that encode for 2 homologous MAT catalytic subunits, α1 and α2, respectively (2). MAT1A is expressed mostly in adult liver and serves as a marker for normal differentiated liver, whereas MAT2A is expressed in all extrahepatic tissues and is induced during rapid liver growth and liver dedifferentiation (1). In liver, the α1 subunit forms dimer (MATIII) and tetramer (MATI) MAT isoenzymes (2). MAT1A expression is reduced in about 60% of patients with cirrhosis due to various etiologies (3) and is often silenced in human hepatocellular carcinoma (HCC) (3, 4). In addition to reduced MAT1A expression, the activity of the MATI/III isoenzymes is often reduced in patients with chronic liver disease, resulting in chronic depletion of hepatic SAMe level (5). The Mat1a-KO mouse model has illustrated the many consequences of chronic hepatic SAMe depletion, the most important of which is spontaneous development of HCC (6, 7).
The importance of hepatic SAMe depletion in HCC development is supported by several rodent models of HCC using hepatocarcinogens in which hepatic SAMe depletion develops and HCC was prevented by SAMe administration (8–10). The dominant mechanism of SAMe chemopreventive effect was thought to be from preventing hypomethylation of the promoter region of several protooncogenes, as SAMe’s chemopreventive effect was blocked by 5-azacytidine (9, 10). However, in an orthotopic liver cancer model, SAMe administration was also able to inhibit HCC establishment but was ineffective in blocking growth of already existing HCC (11). Part of this was because of a compensatory response of the liver to induce methyltransferases that removed excess SAMe to prevent its accumulation (11). A better strategy to maintain increased SAMe level in liver cancer cells is to induce the expression of MAT1A. Consistent with this, liver cancer cells with forced expression of MAT1A doubled-cellular SAMe levels grew slower in vitro and in vivo and exhibited reduced angiogenesis, ERK, and AKT activation and increased apoptosis in vivo (12). This proof of concept prompted the current work to investigate whether MAT1A expression is regulated by microRNAs (miRNAs) that are dysregulated in HCC, as miRNAs are potentially much better therapeutic targets. In the course of our investigation, we uncovered 3 miRNAs whose expression and function in HCC have not been reported to be upregulated in HCC and contribute to the downregulation of MAT1A, tumorigenicity, invasion, and metastasis.
Results
Expression of miRNAs in HCC and the effect of their knockdown on MAT1A expression in HepG2 and Hep3B hepatoma cell lines.
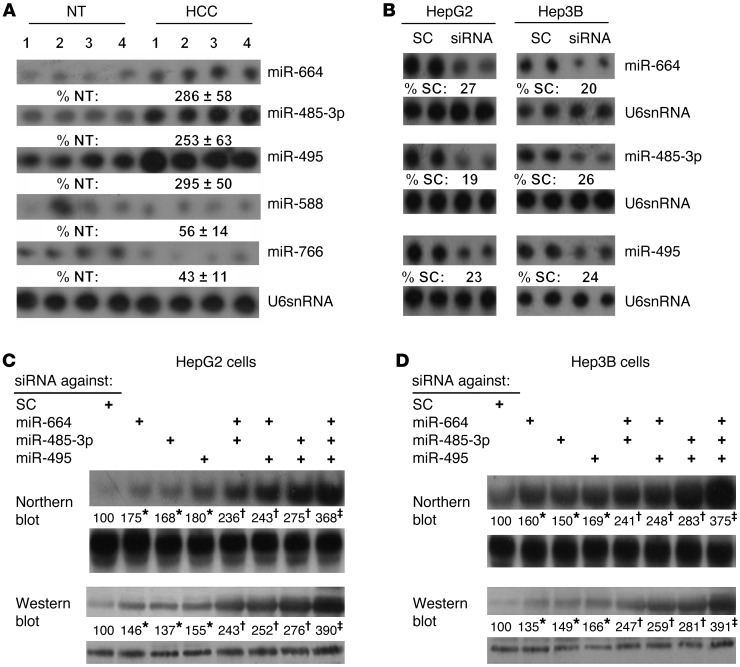
To determine whether miRNAs can potentially regulate MAT1A expression, we used an in silico approach to search for miRNAs that can bind to the 3′ UTR of MAT1A. Using 3 different miRNA prediction target databases (TargetScan, mirDB, miRSVR), many miRNAs were identified (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI63861DS1). We focused on those with high scores (miRNA-664 [miR-664], miR-485-3p, miR-495, miR-588, miR-766) whose expression has not been reported in HCC. Figure 1A shows that miR-664, miR-485-3p, and miR-495 are induced in HCC, while miR-588 and miR-766 are downregulated. Since the dominant mechanism of miRNA regulation is that of downregulation of mRNA/protein levels (13), we focused only on the 3 that are upregulated. To see whether these miRNAs regulate MAT1A expression and to determine the level of regulation, HepG2 and Hep3B cells were treated with siRNA targeting these miRNAs. The efficiency of knockdown is shown in Figure 1B, which is comparable with 75% to 80% reduction after 24 hours. Figure 1, C and D, shows that knockdown of miR-664, miR-485-3p, and miR-495 individually raised MAT1A mRNA and protein levels comparably and combined knockdown exerted nearly an additive effect. This demonstrates that these miRNAs negatively regulate MAT1A expression and the effect is exerted at the mRNA level.
Figure 1. miR-664, miR-485-3p, and miR-495 are induced in HCC and negatively regulate MAT1A expression in liver cancer cell lines.
(A) Northern blot analysis showing expression of select miRNAs in HCC compared with adjacent nontumorous (NT) tissue. (B) Northern blot analysis confirming siRNA knockdown efficiency of miR-664, miR-485-3p, and miR-495 in HepG2 and Hep3B cells as compared with scramble siRNA (SC) control. (C and D) Northern (top) and Western (bottom) blot analyses showing the effect of siRNA knockdown of miR-664, miR-485-3p and miR-495, alone or in combination, on MAT1A expression in HepG2 (C) and Hep3B cells (D). Numbers below the blots represent densitometric values expressed as percentage of respective controls. Representative blots are shown for C and D from 3 experiments, *P < 0.01 vs. SC; †P < 0.05 vs. SC, and triple knockdown; ‡P < 0.001 vs. SC and single knockdown.
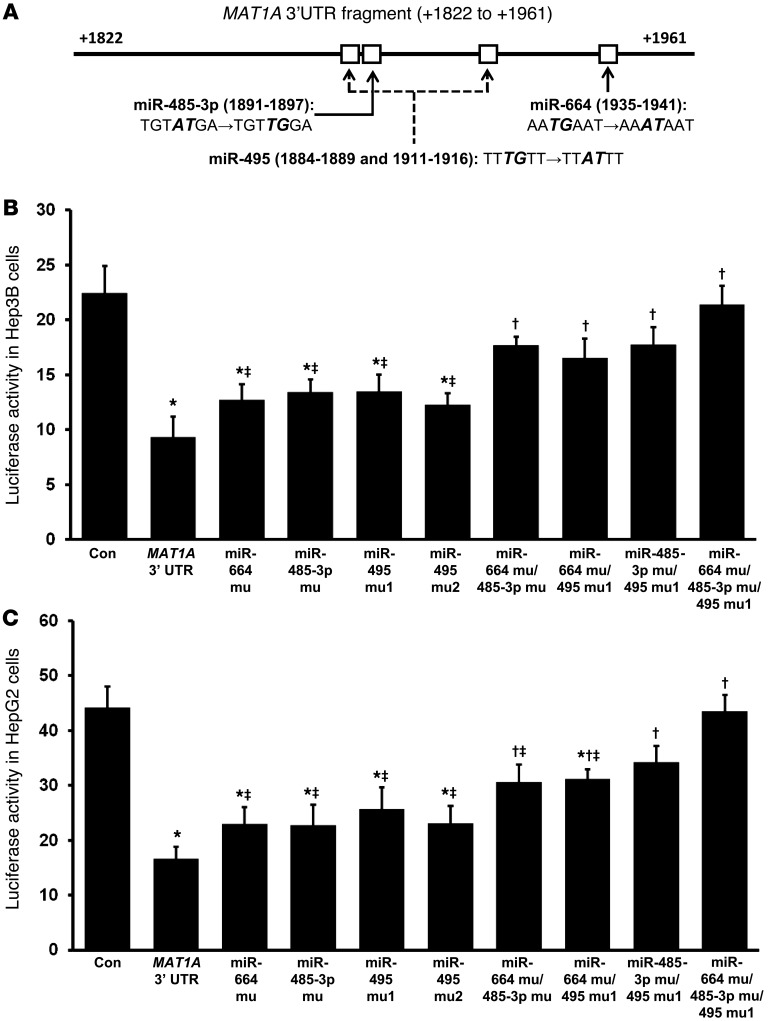
MAT1A 3′ UTR contains functional binding sites for miR-664, miR-485-3p, and miR-495.
Figure 2A shows the MAT1A 3′ UTR sequence containing the putative binding sites for miR-495 (2 sites), miR-664, and miR-485-3p. To determine functionality, each site was mutated alone or in combination and the effect of inserting the WT or mutated MAT1A 3′ UTR on reporter activity was measured in both Hep3B and HepG2 cells after transient transfection. Figure 2, B and C, shows that WT MAT1A 3′ UTR reduced reporter activity by more than 50% and mutation of each miRNA binding site led to slight recovery, with incremental recovery as additional sites were mutated. When miR-664, miR-485-3p, and miR-495 were all mutated, the inhibitory effect of the MAT1A 3′ UTR was completely lost.
Figure 2. MAT1A 3′ UTR-driven reporter activity and the effect of mutating miRNA binding sites.
(A) Diagram of MAT1A 3′ UTR fragment containing the putative binding sites for miR-664, miR-485-3p, and miR-495. Mutations created for each miRNA site are denoted in bold italics. Transient transfection assays were performed using a luciferase reporter system with WT and mutated MAT1A 3′ UTR constructs as described in Methods in (B) HepG2 and (C) Hep3B cells. *P < 0.05 vs. control; †P < 0.05 vs. MAT1A 3′ UTR; ‡P < 0.05 vs. triple miRNA siRNA knockdown. n = 3 experiments, done in triplicate.
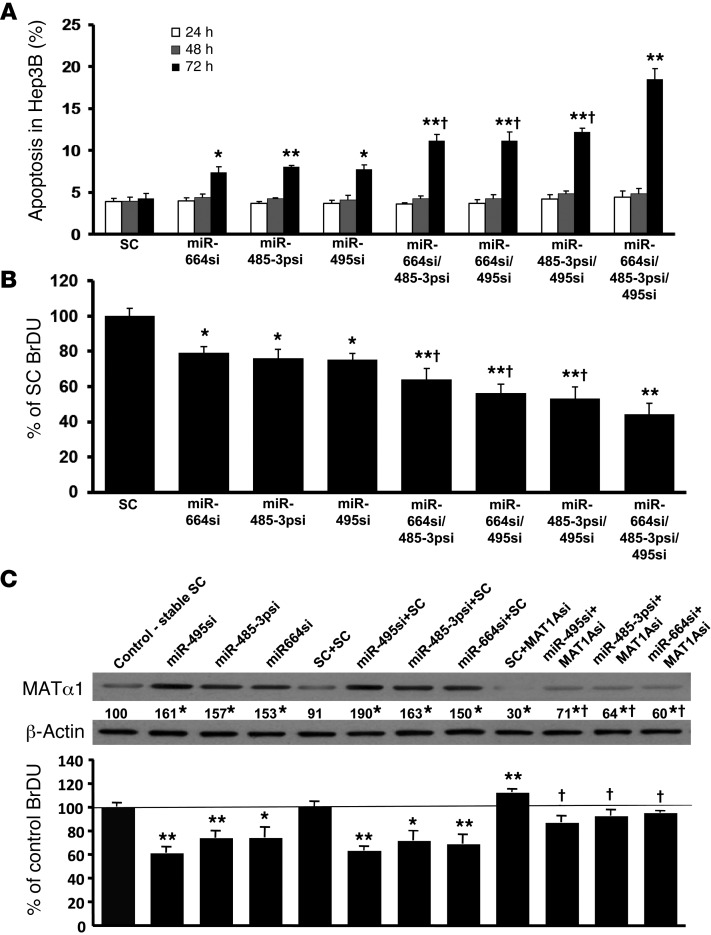
Effect of siRNA against miR-664, miR-485-3p, and miR-495 on apoptosis and cell proliferation.
We previously reported that forced MAT1A expression in liver cancer cells reduced growth while inducing apoptosis (12). To see if inducing MAT1A expression by targeting miRNAs also has the same effect, we measured apoptosis and growth. Figure 3A shows that siRNA treatments (single, double, and triple knockdown) in Hep3B cells induced apoptosis only after 72 hours of transfection and that, similar to the effect on MAT1A expression, the effect on apoptosis was additive with combined knockdown. To avoid toxicity causing nonspecific effects, effect on growth was measured only after 24 hours of siRNA treatment. Figure 3B shows that single knockdown of these miRNAs reduced growth by about 20% and combined knockdown of all 3 reduced growth by about 50%. Similar findings on apoptosis and growth were observed in HepG2 cells (data not shown). To determine the contribution of MAT1A induction on the growth inhibitory effect, Hep3B cells were stably transfected with siRNAs targeting each miRNA. Control Hep3B cells were stably transfected with scramble siRNA. These cells were then transiently transfected with siRNA against MAT1A or scramble siRNA for 24 hours. Figure 3C shows that Hep3B cells with stable knockdown of each miRNA exhibited reduced cell proliferation, but this effect was significantly blunted when MAT1A was also knocked down (Figure 3C).
Figure 3. Increased cellular apoptosis and decreased cell growth in Hep3B cells by miRNA knockdown requires MAT1A induction.
(A) Apoptosis rates were determined by Hoechst staining at 24, 48, and 72 hours after transient miRNA knockdown in Hep3B cells. *P < 0.01, **P < 0.05 vs. SC; †P < 0.05 vs. single or triple siRNA knockdown. n = 3 experiments, each with 12 determinations. (B) BrdU incorporation assay was measured at 24 hours after transient miRNA knockdown in Hep3B cells. *P < 0.05, **P < 0.01 vs. SC; †P < 0.05 vs. single or triple siRNA knockdown. n = 3 experiments, each with 8 determinations. (C) To determine the role of MAT1A induction on growth, Hep3B cells stably transfected with miR-664, miR-485-3p, and miR-495 siRNA or scramble siRNA (stable SC) were transiently transfected with MAT1A siRNA or SC, and BrdU incorporation and MAT1A protein levels were measured 24 hours later. *P < 0.05, **P < 0.01 vs. SC; †P < 0.05 vs. SC+MAT1Asi and each respective miRNA siRNA+SC. n = 3 experiments, each with 8 determinations for BrdU. Numbers below the Western blot represent mean densitometric values expressed as percentage of control.
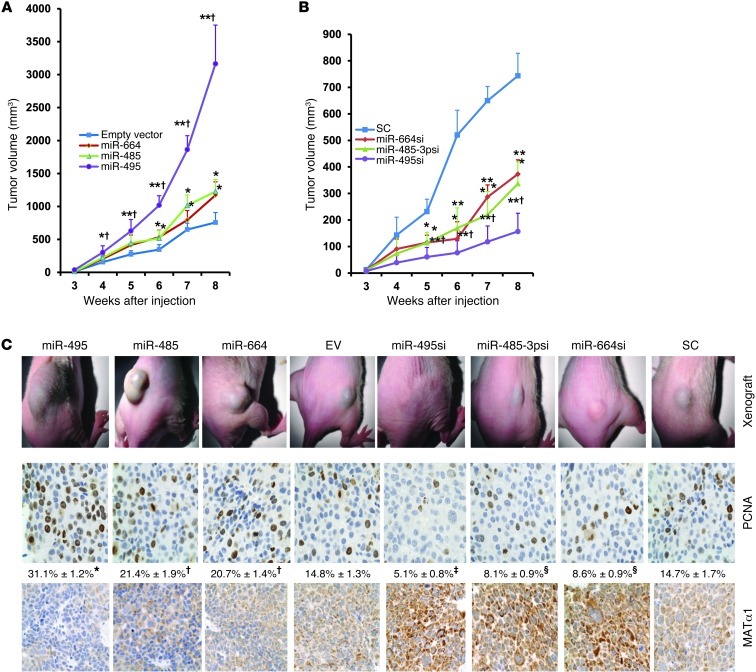
Forced lenti–miR-664, lenti–miR-485, and lenti–miR-495 expression promotes and lenti-siRNAs against these miRNAs inhibit tumorigenesis in xenograft.
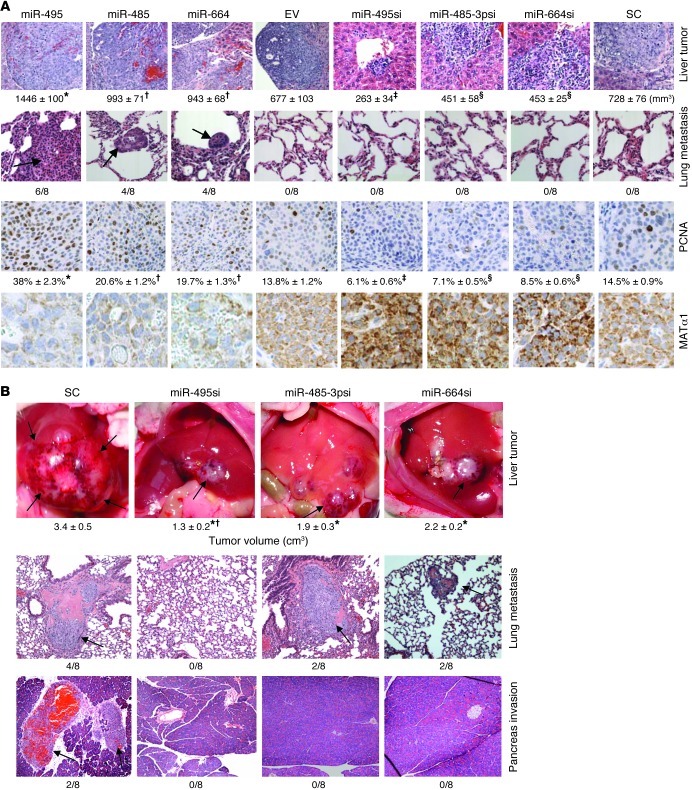
To determine whether miR-664, miR-485-3p, and miR-495 are involved in liver tumorigenesis, stable Hep3B cell lines expressing forced lenti-miRNAs or lenti-siRNAs against these miRNAs were established. Supplemental Figure 1 shows that forced expression of these miRNAs by 3-fold reduced MATα1 levels by 50%, while knockdown of these miRNAs by 80% doubled MATα1 levels. Injection of cells expressing high miRNA levels subcutaneously into the flanks of nude mice resulted in more rapid growth of tumors (Figure 4, A and C), whereas tumor growth was significantly inhibited in cells expressing siRNAs against these miRNAs (Figure 4, B and C). Manipulating the expression of miR-495 resulted in the most dramatic effect. At 8 weeks, tumors from mice injected with forced lenti–miR-664, lenti–miR-485-3p, and lenti–miR-495 showed tumor volumes that were 160%, 170%, and 360% higher as compared with empty vector (EV), respectively (Figure 4A). Tumors from mice injected with cells expressing stable knockdown of miR-664, miR-485-3p, and miR-495 showed tumor volumes that were 50%, 52%, and 79% lower as compared with scramble control, respectively (Figure 4B). Immunohistochemistry staining for proliferating cell nuclear antigen (PCNA) in these tumors showed increased staining in tumors with forced miRNA expression and reduced staining in those with siRNA against these miRNAs (Figure 4C). Stability of the MAT1A expression in tumors is demonstrated using immunohistochemistry, and low MATα1 staining correlated with higher PCNA staining and more rapid growth, whereas high MATα1 staining correlated with lower PCNA staining and slower growth (Figure 4C).
Figure 4. Effect of stably transfected miR-664, miR-485, and miR-495 and their siRNAs on tumor growth in a xenograft mouse model.
Nude mice were injected with Hep3B cells subcutaneously containing either (A) stably transfected miR-664, miR-485, and miR-495 (*P < 0.05, **P < 0.01 vs. EV; †P < 0.05 vs. miR-664 or miR-485; n = 8) or (B) stably transfected siRNAs against miR-664, miR-485-3p, and miR-495 (*P < 0.05, **P < 0.0001 vs. SC; †P < 0.05 vs. miR-664-si or miR-485-3psi; n = 8), and tumor volumes were measured over time. (C) Representative pictures of subcutaneous tumors at 8 weeks following injection of cells containing stably transfected miRNAs (left) and siRNAs (right) (top row), immunohistochemistry stained for PCNA (middle row) and MAT1A protein expression (bottom row) Original magnification, ×200. Numbers below PCNA staining represent percentage of positively stained cells. *P < 0.01 vs. miR-485, miR-664 and EV; †P < 0.05 vs. EV; ‡P < 0.05 vs. miR-485-3psi, miR-664si and SC; §P < 0.05 vs. SC.
Forced lenti–miR-664, lenti–miR-485, and lenti–miR-495 expression promotes and lenti-siRNAs against these miRNAs inhibit tumor invasion and metastasis in an orthotopic liver cancer model.
In order to examine whether these miRNAs play a role in invasion and metastasis, we switched to an orthotopic liver cancer model where cancer cells are directly injected into the left hepatic lobe of nude mice. Figure 5A shows that at the end of day 45, Hep3B cells expressing forced lenti-miRNAs developed larger hepatic tumors as in the xenograft model, higher PCNA staining, lower MATα1 staining, and importantly, exhibited metastasis to the lung in at least 50% of the mice. Hep3B cells stably expressing siRNA against these miRNAs had smaller tumors, lower PCNA staining, and higher MATα1 staining. Since scramble siRNA control cells did not metastasize to the lung at this time point, these results indicate that forced expression of these miRNAs increased invasion and metastatic potential of Hep3B cells.
Figure 5. Effect of varying miR-664, miR-485-3p, and miR-495 expression on tumorigenesis, invasion, and metastasis in an orthotopic liver cancer model.
(A) Hep3B cells stably transfected with lenti–miR-664, miR-485, and miR-495/EV or lenti-siRNA against these miRNAs or SC were injected into the left hepatic lobe, and mice were sacrificed after 45 days. The top row shows H&E staining of liver tumors, and tumor volume at the site of injection are shown below for each condition (*P < 0.05 vs. miR-485, miR-664 and EV; †P < 0.05 vs. EV; ‡P < 0.05 vs. miR-485-3psi, miR-664si, and SC; §P < 0.05 vs. SC). Second row shows H&E staining of lung tissue and incidence of lung metastasis. Arrows point to lung metastasis. Third and fourth rows show immunohistochemistry for PCNA and MAT1A protein. Numbers below PCNA represent percentage of positive cells, *P < 0.01 vs. miR-485, miR-664 and EV; †P < 0.05 vs. EV; ‡P < 0.05 vs. miR-485-3psi, miR-664si and SC; §P < 0.05 vs. SC. Original magnification, ×100 (first row); ×200 (second through fourth rows). (B) HepG2 cells capable of invasion and metastasis were injected into the left hepatic lobe as above, and lentiviral vectors containing miRNA siRNA or SC were injected into the spleen at the time of HepG2 cell injection. Two weeks later, lentiviral siRNA was injected into the tail vein, and this was repeated every 2 weeks until sacrifice at 8 weeks. H&E staining showing the effect of miR-495, miR-485-3p, and miR-664 siRNAs on tumor invasion and metastasis. Arrows point to tumor at the site of injection, and tumor volumes and invasion incidences are shown below each image. *P < 0.01 vs. scrambled siRNA (SC); †P < 0.05 vs. miR-485-3psi and miR-664si. Original magnification, ×100.
Effect of reducing the expression of miR-495, miR-485-3p, and miR-664 in an invasive liver cancer cell line in vivo.
Since Hep3B cells did not metastasize to the lung, we switched to HepG2 cells, which can invade adjacent structures and metastasize to the lung in the same orthotopic liver cancer model (Figure 5B). Mice were treated with siRNA targeting miR-495, miR-485-3p, miR-664, or scramble control for up to 8 weeks. At week 8, there was a significant (35% to 62%) reduction in tumor volume (Figure 5B) and reduced incidence of metastases to the lung, abdominal wall, pancreas, and diaphragm in mice treated with siRNAs (Figure 5B and Supplemental Table 2). Reducing miR-495 expression had the most significant inhibitory impact on tumor growth, invasion, and metastasis.
Role of MAT1A in HCC invasion and antitumorigenic effect of miR-495 siRNA.
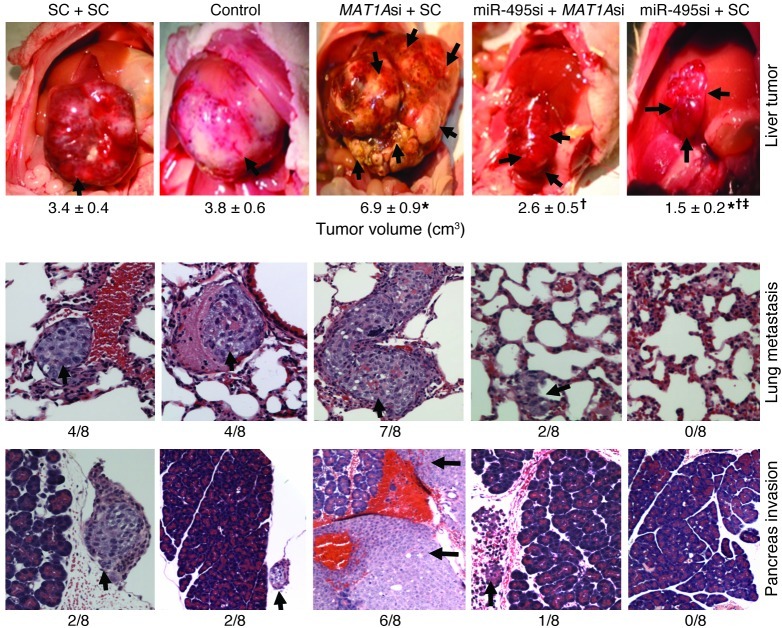
To examine the role of MAT1A directly in tumorigenesis and treatment efficacy of the most potent miRNA siRNA, miR-495 siRNA, mice were treated with siRNA against MAT1A, miR-495, and scramble control alone or together using the same invasive HepG2 cells orthotopic liver cancer model. Figure 6 shows that reducing MAT1A expression with MAT1A siRNA more than doubled the tumor volume after 8 weeks and significantly blunted the treatment efficacy of miR-495 siRNA on tumor growth. MAT1A expression also had a direct effect on invasion and metastasis, as MAT1A siRNA treatment increased invasion to adjacent organs and distant metastasis and blunted the therapeutic efficacy of miR-495 siRNA on the same parameters (Figure 6 and Supplemental Table 3).
Figure 6. Role of MAT1A in tumorigenesis and therapeutic effect of miR-495 siRNA.
HepG2 cells were injected into the left hepatic lobe of male BALB/c nude mice and lentiviral vectors containing MAT1A siRNA (MAT1Asi), miR-495 siRNA (miR-495si), and scramble siRNA (SC), alone or together, were injected into the spleen at the time of HepG2 cell injection (n = 8 per group). Control group received only HepG2 cell injection. Two weeks later, lentiviral siRNAs were injected into the tail vein, and this was repeated every 2 weeks until sacrifice at 8 weeks. First row: arrows point to tumors at the site of injection, and tumor volumes are shown below. *P < 0.005 vs. SC+SC; †P < 0.005 vs. MAT1Asi+SC; ‡P < 0.05 vs. miR-495si+MAT1Asi. Second and third rows show metastasis to lung and pancreas (indicated by arrows) in the various treatment groups, with the incidence shown below. Original magnification, ×200.
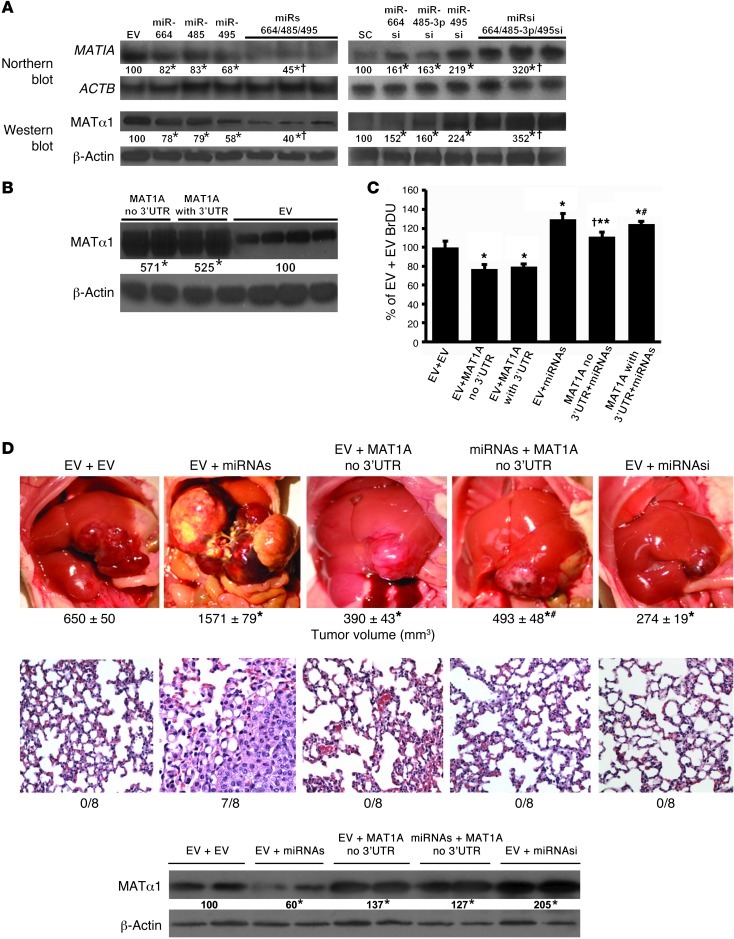
MAT1A is the key mediator of miR-664, miR-485, and miR-495 on tumorigenesis and metastasis.
To further evaluate the role of MAT1A in the tumorigenic effect of miR-664, miR-485, and miR-495, we examined the effect of overexpressing these 3 miRNAs (in 1 vector) in the presence or absence of a MAT1A expression vector that does not have the 3′ UTR so that these miRNAs cannot lower MAT1A expression from this vector. Altering the expression of all 3 miRNAs by combining them or their siRNAs into 1 vector raised or lowered MAT1A expression, respectively, more than altering the expression of single miRNAs (Figure 7A). Absence of the 3′ UTR did not alter MAT1A expression from that of the vector that included it (Figure 7B). In vitro, overexpression of these miRNAs increased, while overexpression of MAT1A (with or without 3′ UTR) decreased cell growth. However, when combined, overexpression of MAT1A without the 3′ UTR was able to reduce the growth-inductive effect of the miRNAs, while overexpression of MAT1A with 3′ UTR was not (Figure 7C). This translated to in vivo tumorigenesis so that overexpression of miRNAs greatly increased tumor size, invasion, and metastasis, while overexpression of either MAT1A or siRNAs targeting all 3 miRNAs inhibited tumor growth by 40% and 58%, respectively. Most importantly, overexpressing the 3 miRNAs failed to increase growth and metastasis in cases in which MAT1A that could not be inhibited by the 3 miRNAs was also overexpressed (Figure 7D). MAT1A protein levels in these tumors confirmed that the miRNAs were not able to inhibit MAT1A expression, as the construct did not contain the MAT1A 3′ UTR (Figure 7D).
Figure 7. MAT1A is the key mediator of miR-664, miR-485, and miR-495 on tumor growth and metastasis.
(A) MAT1A Northern and Western blots of Hep3B cells stably transfected with lenti–miR-664, lenti–miR-485, and lenti–miR-495/EV singly or all 3 miRNAs together (miRs), lenti-siRNA against miRNAs (alone or together, miRsi), or SC. *P < 0.05 vs. respective controls; †P < 0.05 vs. individual miRNA or miRNAsi. (B) MAT1A Western blots of Hep3B cells stably transfected with lenti-MAT1A with or without 3′ UTR. *P < 0.05 vs. EV. (C) BrDU measurement in Hep3B cells stably expressing MAT1A with or without 3′ UTR, then transiently transfected with lentiviral vector containing all 3 miRNAs, or EV for 24 hours and expressed as percentage of control (EV+EV). Results are mean ± SEM from 3 experiments done in triplicate. *P < 0.01 vs. EV+EV; †P < 0.05 vs. EV+MAT1A no 3′ UTR; **P < 0.05 vs. EV+miRNAs and MAT1A with 3′ UTR+miRNAs; #P < 0.05 vs. EV+MAT1A with or without 3′ UTR. (D). Hep3B cells stably transfected with MAT1A without 3′ UTR or EV were injected into the left hepatic lobe and treated with lentiviral vector expressing all miRNAs, siRNA against all 3 miRNAs (miRNAsi), or EV. Tumor volumes at the site of injection 45 days later are shown below for each condition, *P < 0.05 vs. EV+EV; #P < 0.05 vs. EV+miRNAs. MAT1A protein levels and incidence of lung metastasis are shown below. *P < 0.05 vs. EV+EV. Original magnification, ×200. Numbers below all blots refer to densitometric values expressed as percentage of respective controls.
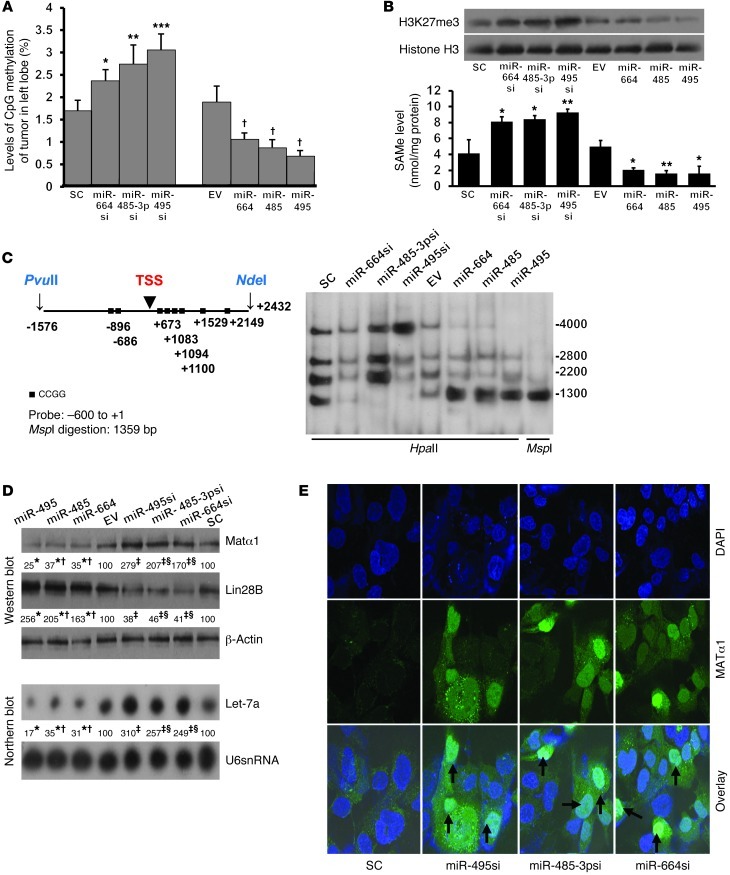
Molecular mechanism of MAT1A-dependent effect on tumorigenesis, invasion, and metastasis.
Increasing MAT1A expression is expected to increase SAMe levels (14), which may alter DNA methylation. Consistent with this, tumors derived from Hep3B cells expressing lower miRNAs (hence higher MAT1A) have higher global DNA methylation and vice versa (Figure 8A). Increased MAT1A expression resulted in higher nuclear levels of H3K27me3 and SAMe (Figure 8B). Although many genes are deregulated in HCC, LIN28B/let-7 (where LIN28B indicates lin-28 homolog B [Caenorhabditis elegans]) axis is of particular interest because LIN28B is overexpressed in HCC and it can repress let-7, a well-studied tumor suppressor often repressed in cancers including HCC (15). There are multiple CCGG sites in the promoter region of LIN28B (Figure 8C). Southern blotting following digestion of DNA with HpaII or MspI shows that tumors expressing high levels of miR-664, miR-485, or miR-495 have a hypomethylated LIN28B promoter region as compared with EV (much stronger band at 1300, weaker or absent band at 4000), while those expressing reduced levels of these miRNAs have a hypermethylated LIN28B promoter region as compared with scramble control (absent band at 1300) (Figure 8C). This correlated with LIN28B expression, so that hypomethylation of LIN28B (with forcing miRNA and lower MAT1A) resulted in higher LIN28B expression, while hypermethylation of LIN28B (with miRNA knockdown and higher MAT1A) resulted in lower LIN28B expression (Figure 8D). LIN28B can repress let-7a (15) and, consistent with this, reducing LIN28B expression resulted in higher let-7a expression and vice versa (Figure 8D). Most interestingly, increasing MAT1A expression by reducing miR-495, miR-485-3p, or miR-664 increased nuclear content of MAT1A protein (MATα1) dramatically (Figure 8E).
Figure 8. Possible mechanism of miR-664, miR-485-3p, miR-495, and MAT1A involvement in tumorigenesis, invasion and metastasis.
(A) 5-mC levels in tumors derived from Hep3B cells stably expressing lower (with siRNA or si) or higher levels of miRNAs. *P < 0.05; **P < 0.01; ***P < 0.0001 vs. SC; †P < 0.05 vs. EV. n = 8 per condition. (B) Effect of varying miRNA expression on nuclear H3K27me3 levels in tumors (Western blots) and SAMe levels. Results are mean ± SEM from 8 per condition. *P < 0.05; **P < 0.01 vs. respective controls. (C) Diagram shows HpaII and MspI sites between PvuII and NdeI in human LIN28B promoter. Black squares, CCGG sites; TSS, transcriptional start site. Numbers are relative to TSS. Southern blot analysis of LIN28B promoter region between –1576 and +2432 (right). DNA samples from liver tumor derived from stably transfected Hep3B containing miR-664, miR-485, and miR-495 and their siRNAs were digested as indicated. MspI digestion results in a band size of 1359 bp as control for HpaII digestion. (D) Effect of overexpressing miR-664, miR-485, and miR-495 and their siRNAs on MAT1A and LIN28B protein expression (top) and let-7a mRNA expression (bottom). Numbers below the blots are densitometric values expressed as percentage of respective controls. *P < 0.01 vs. EV; †P < 0.05 vs. miR-495; ‡P < 0.01 vs. SC; §P < 0.05 vs. miR-495si. (E) Increased nuclear localization of MAT1A protein is seen after knockdown of miR-664, miR-485-3p, and miR-495. Original magnification, ×630 (oil immersion).
Discussion
HCC ranks as the fifth most common cancer and the third most frequent cause of cancer death worldwide (16). Most patients (70% to 85%) with HCC are diagnosed with advanced disease, and the overall 5-year survival rate is less than 12% (17). Even in those HCC patients that undergo surgical resection, the recurrence rate is about 50% at 3 years (18). Sorafenib, a multikinase inhibitor, represents a major breakthrough in treatment of advanced HCC, which was shown to increase the median overall survival from 7.9 to 10.7 months in a randomized, placebo-controlled phase III trial (SHARP) (19). However, sorafenib did not delay time to symptomatic progression, and its cost can be prohibitive. Thus, the search continues for better treatment strategies against HCC.
MAT1A is predominantly expressed in normal liver, and it is often silenced in human HCC (3, 4, 20, 21).
In both animal models and human HCC, decreased MAT1A expression correlates with more aggressive disease and worse prognosis (21). Consistent with an important role of MAT1A in HCC pathogenesis, mice deficient in Mat1a develop spontaneous HCC (7). Liver cancer cells forced to express MAT1A grew slower in vitro and when tested in xenograft model in vivo (3, 12). In animal models of HCC, increasing MAT1A expression achieved stable higher tumor SAMe levels than exogenous SAMe treatment, since chronic administration of SAMe led to an increase in the expression of methyltransferases that prevented SAMe accumulation (11, 12). Thus, developing strategies to increase MAT1A expression in HCC may be effective in slowing HCC growth.
Both transcriptional and posttranscriptional mechanisms have been reported to help explain reduced MAT1A expression in HCC. Promoter hypermethylation correlated with reduced MAT1A expression in cirrhotic patients and HCC (3, 21). In addition, methylation of the coding region near the translational start site can also inhibit MAT1A transcription (22). Recently we showed that the MAT1A 3′ UTR binds to the AU-rich RNA binding factor 1 (AUF1), which is one of the hnRNP proteins known to destabilize target mRNAs (23). HCC specimens express higher AUF1 protein levels, and knockdown of AUF1 increased MAT1A mRNA level (23). However, these mechanisms are difficult to target, since they can affect numerous other genes. This prompted us to consider the possibility that miRNAs might also regulate MAT1A expression.
miRNAs are small noncoding RNAs that regulate gene expression by targeting the 3′ UTR of mRNAs, leading to reduced protein translation and/or increased mRNA degradation in most cases (13). Dysregulation of miRNA expression plays an important role in the pathogenesis of HCC, and miRNA signatures may serve as biomarkers for HCC classification and prognostic risk stratification as well as therapy (24, 25). We used the miRWalk website ( http://www.ma.uni-heidelberg.de/apps/zmf/mirwalk/index.html) to simultaneously search 8 different established miRNA prediction programs to see which miRNAs have the most positive prediction matches. Using the most popular search algorithms — miRSVR (uses miRANDA), TargetScan and mirDB — we generated a list of miRNAs that gave the best scores for MAT1A. Interestingly, while some of these are known to have altered expression in HCC, the expression of most of these miRNAs in HCC is unknown. Of the 5 best-matched miRNAs whose function in HCC is unknown, 3 are induced in 4 sets of HCC samples as compared with adjacent nontumorous liver, and we focused on these for the current work, since the majority of miRNAs downregulate expression of their targets. Indeed, knockdown of miR-664, miR-485-3p, or miR-495 in both HepG2 and Hep3B cells raised MAT1A mRNA and protein levels comparably (Figure 1), which is consistent with the notion that the mechanism of these miRNAs on MAT1A is to increase its mRNA degradation. Reporter assay confirmed the presence of functional miRNA-binding sites in the MAT1A 3′ UTR (Figure 2). Each miRNA exerted a significant influence on growth and apoptosis in vitro and tumor growth in vivo; however, although the miRNAs all have comparable effects on MAT1A expression, miR-495 exerted the most pronounced effect on tumor growth, invasion, and metastasis. This suggests miR-495 has other important targets besides MAT1A. Nevertheless, knocking down miR-664 or miR-485-3p reduced tumor growth by over 50% as well as invasion and metastasis, supporting an important role for MAT1A. Consistent with this, lowering MAT1A expression increased the tumorigenicity, invasion, and metastatic potential of HCC and blunted the therapeutic efficacy of miR-495 siRNA on these parameters. Furthermore, forced expression of MAT1A that cannot be inhibited by these 3 miRNAs completely eliminated the inductive effect of these miRNAs on growth and metastasis. Taken together, these results indicate that MAT1A is the key target for these miRNAs in exerting their influence on HCC tumorigenesis.
miR-485-3p has been shown to mediate topoisomerase IIα downregulation in part via altered regulation of the transcription factor nuclear factor YB and to have a role in drug responsiveness in CEM and CEM/VM-1-5 cells (human leukemic lymphoblastic cells) and in Rh30 and Rh30/v1 cells (human rhabdomyosarcoma cells) (26). miR-495 has been shown to be upregulated in breast cancer stem cells, where its overexpression promoted tumorigenic activity by downregulating E-cadherin and REDD1 (regulated in development and DNA damage responses 1), an inhibitor of mTOR signaling (27). miR-495 is also thought to have a role in liver and pancreas development (28). Increase of miR-664 expression was found in atrial fibrillation patients with rheumatic heart disease (29). However, as of today, the expression and function of these miRNAs in HCC has not been reported. Recently, Koturbash et al. demonstrated that miR-29b might play a role in downregulating Mat1a in the preneoplastic liver tissue of rats treated with the hepatocarcinogen 2-acetylaminofluorene (30). However, miR-29a-c was found to be downregulated in HCC, and it has been shown to promote apoptosis by lowering the expression of Bcl-2 and Mcl-1 (31). Human MAT1A has not been shown to be regulated by miRNA. Our study clearly demonstrates an important role for miR-495, miR-485-3p, and miR-664 in regulating MAT1A expression and that also shows that downregulating their expression inhibited tumor growth, invasion, and metastasis of HCC. The next question is, how does increasing MAT1A expression impact on tumorigenesis, invasion and metastasis?
Since MAT is responsible for SAMe synthesis and MATI/III are the products of the MAT1A gene (1), increasing MAT1A expression would increase steady state SAMe levels (3). Changes in DNA methylation, particularly hypermethylation of tumor suppressors, play a critical role in cancer pathogenesis, including HCC (32). This prompted us to examine whether MAT1A expression in liver cancer cells influences DNA methylation. Indeed, when MAT1A expression was increased by knocking down miR-664, miR-485-3p, or miR-495, nuclear SAMe levels and global DNA methylation increased, and the opposite occurred when MAT1A expression was reduced by forcing the expression of these miRNAs. Of the many genes deregulated in HCC, we focused on LIN28B because of a recent report that demonstrated LIN28B (not LIN28) is overexpressed in HCC and promotes transformation and invasion in HCC in part via repression of let-7 (15). The let-7 family of miRNAs regulates factors that control cell-fate decisions, including oncogenes and cell-cycle factors (33–35). LIN28B (and LIN28) exerts a reciprocal regulation with let-7 (33). let-7 suppresses the expression of LIN28 through let-7–binding sites in the LIN28 3′ UTR, while LIN28/LIN28B suppress the production of mature let-7s at multiple levels as well as enhancing let-7 degradation via 3′ terminal uridylation of let-7 precursors (33). This opposing expression pattern of Lin28 and let-7 can be found throughout development and in oncogenesis and has been compared with a yin-yang balancing act by Ji and Wang (33). This can be illustrated by activation of LIN28 by c-Myc and NF-κB, leading to let-7 repression and cell transformation (33). The LIN28B promoter region has multiple CpG sites, including 3 CpG islands (as determined by CpG island searcher: cpgislands.usc.edu), where Viswanathan et al. correlated the loss of DNA methylation of a downstream CpG island with that of its expression found in HepG2 and K562 erythromyeloblastoid leukemia cells (36). Interestingly, we found that forced miRNAs reduced MAT1A expression and LIN28B promoter methylation and increased LIN28B expression. This correlated with a fall in let-7a expression. Increasing MAT1A expression by knocking down miR-664, miR-485-3p, and miR-495 led to LIN28B promoter hypermethylation, reduced LIN28B expression, and increased let-7a expression (Figure 8D). Thus, enhancing MAT1A expression shifted the balance of LIN28B/let-7 toward let-7 and inhibited tumor growth, invasion, and metastasis.
Recently Reytor et al. reported finding MATI/III in the nuclei (37). The authors speculated that presence of nuclear MAT might provide a continuous source of nuclear SAMe, since SAMe is charged and whether or not it can traverse the nuclear membrane is in debate. In support of this, nuclear accumulation of the active MAT1A protein correlated with higher levels of histone H3K27 trimethylation, an epigenetic modification associated with gene repression and DNA methylation (37). Our findings are consistent with this report. Interestingly, we found that knocking down miR-664, miR-485-3p, and miR-495 increased total cellular MAT1A protein level, particularly the nuclear fraction (Figure 8E). This may explain the dramatic effect that MAT1A expression has on DNA methylation.
In summary, we have identified 3 miRNAs that are increased in HCC that can negatively regulate MAT1A expression at the mRNA level. Reducing the expression of these miRNAs raised MAT1A expression, which we believe is a novel strategy to shift the LIN28B/let-7 balance toward let-7. We suspect this increase in MAT1A expression also has an impact on the expression of many other genes involved in tumorigenesis. This will be the subject of a future investigation. Our results also help to explain why decreased MAT1A expression in HCC is a poor prognostic indicator (21). Few therapeutic options currently exist to treat HCC. These miRNAs are potential therapeutic targets and offer substantial promise in expanding treatment options for patients with HCC.
Methods
Materials and reagents. α-32P-dCTP and γ-32P ATP (3,000 Ci/mmol) were purchased from PerkinElmer.
Antibodies used for either Western blot and/or immunohistochemistry to PCNA, LIN28B, and β-actin were purchased from Cell Signaling Technology. MATα1 antibody was purchased from Novus Biologicals, whereas H3K27me3 and GFP antibody were purchased from Abcam. Lipofectamine 2000 and RNAimax were purchased from Invitrogen, whereas the MethylFlash Methylated DNA quantification kit was purchased from Epigentek. Lenti–miR-664, lenti–miR-485 (lenti–miR-485-3p is not available), lenti–miR-495, lenti–miR-664 siRNA, lenti–miR-485-3p siRNA, lenti–miR-495 siRNA, and a lentiviral purification kit were purchased from SBI System Biosciences. Lenti-MAT1A siRNA was purchased from Applied Biological Material Inc. siRNA to hsa–miR-664 (AGGCTGGGGATAATTGAAT), hsa–miR485-3p (AGAGGAGAGCCGTGTATGAC), and hsa–miR-495 (5′-AGAAGTGCACCATGTTTGTT-3′) were purchased from EXIQON. pMir-Target vector for MAT1A 3′ UTR clone was purchased from OriGene Technologies. GFP expression after injection of lenti-miRNAs or lenti-siRNAs was visualized on paraffin sections by immunohistochemistry with mouse anti-GFP antibody (1:200; Clontech, BD Biosciences) using the ABC method (Vector Laboratories). All other reagents were of analytical grade and obtained from commercial sources.
Source of normal and cancerous liver tissue.
Normal and cancerous liver tissues were obtained as described (38).
Cell lines and stable transfection of lenti-miRNAs and lenti-siRNAs.
293T, Hep3B, and HepG2 cell lines were obtained from Cell Culture Core of the USC Research Center for Liver Diseases and cultured in DMEM supplemented with 10% fetal bovine serum.
To establish stable expression of miRNA or its siRNA, 105 Hep3B cells were seeded in a 24-well plate 1 day prior to infection. To generate cells stably expressing miRNAs or siRNAs, Hep3B cells were transfected with lenti–miR-664, lenti–miR-485, lenti–miR-495, lenti-EV, lenti–miR-664 siRNA, lenti–miR-485-3p siRNA, lenti–miR-495 siRNA, and lenti-scramble siRNA vector for 3 hours by using Lipofectamine 2000 (Invitrogen). Following selection with puromycin (Invitrogen), stable clonal cell lines were established and examined for the expression of miRNA or siRNA and GFP expression by Northern analysis.
In separate experiments, stable cell lines expressing siRNA against miR-495, miR-485-3p, or miR-664 were transiently transfected with scramble siRNA or siRNA against MAT1A (SI00009387; QIAGEN) in Hep3B for 24 hours using RNAimax (Invitrogen). MAT1A expression, apoptosis, and BrdU incorporation were measured as described below.
Construction of vectors and stable cell lines expressing multiple miRNAs or their siRNAs and MAT1A with or without its 3′ UTR.
Single miRNA expression vectors for premiR-485 (PMIRH485PA-1), premiR-495 (PMIRH495PA-1), and preMIR-664 (PMIRH664PA-1) were obtained from SBI System Biosciences. The multiple miRNA expression vector was constructed by sequentially cloning the premiR-485 and premiR-664 insert into the premiR-495 lentiviral expression vector BamHI and EcoRI sites, respectively.
Oligonucleotides used to construct the lenti-vector containing siRNAs targeting miR-664, miR-485-3p, and miR-495 are shown in Supplemental Table 4. miR-664 siRNA oligonucleotide was inserted into the BamHI site of pGreenPuro (SBI System Biosciences). CMV promoter was amplified by PCR using the primers 5′-TAGTTATTAATAGTAATCAATTACGGG-3′ (forward primer) and 5′-GATCTGACGGTTCACTAAACCAG-3′ (reverse primer) from pGreenPuro and cloned into pCR 2.1 vector by TA cloning (Invitrogen). miR-485-3p siRNA oligonucleotide was inserted at the EcoRV, followed by miR-495 inserted at the SacI and SpeI sites. A second CMV promoter was cloned into the HindIII and KpnI sites. The fragment containing the 2 CMV promoters miR-485-3p and miR-495 was subcloned into the miR-664 containing pGreenPuro EcoRI site. Each shRNA and its CMV promoter was confirmed by sequencing.
MAT1A expression vector cloning was described previously (12).
MAT1A 3′ UTR fragment was subcloned into pCDH-CMV-MCS-EF1-copGFP vector (SBI System Biosciences) EcoRI site to create MAT1A without 3′ UTR. Lenti-reporter-luciferase vector containing only the MAT1A 3′ UTR (ABM) was excised and subcloned into the BamHI site of pCDH-CMV-MCS-EF1-copGFP vector to generate MAT1A with 3′ UTR. Stable Hep3B cell lines expressing these vectors were established as described above.
DNA constructs and dual luciferase assay.
A partial forward and reverse MAT1A 3′ UTR fragment from +1822 bp to +1961 bp and mutant fragments containing Sgf1 and MluI linkers were synthesized. Two base pair mutants were performed for miR-664 (AATGAAT→AAATAAT, where italics represent the targeted nucleotides within the miRNA sequence for mutation), miR-485-3p (TGTATGA→TGTTGGA), and miR-495 (TTTGTT→TTATTT) putative target site(s) in MAT1A 3′ UTR (Figure 2A). The annealed fragments and pMirTarget vector were digested with Sgf1 and MluI. WT and mutant MAT1A 3′ UTR were cloned into the pMirTarget vector containing a luciferase reporter (OriGene Technologies). Hep3B cells were placed in 24-well plates the day before transfection. The WT, mutated 3′ UTR of MAT1A pMirTarget vector, or pMirTarget EV (200 ng) and a control Renilla luciferase expression vector (2.5 ng) were cotransfected into Hep3B cells with Superfect (QIAGEN) following the manufacturer’s instructions. Luciferase assays were performed 24 hours later using the Dual Luciferase Reporter Assay System (Promega) as directed by the manufacturer’s suggested protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Xenograft model.
Sixty-four 4-week-old male BALB/c nude mice from Jackson ImmunoResearch Laboratories Inc. were divided equally into 8 groups (n = 8 per group) and given the following Hep3B stable cell line injection: group 1, lenti-EV; group 2, lenti–miR-664; group 3, lenti–miR-485; group 4, lenti–miR-495; group 5, lenti-miRNA scramble siRNA; group 6, lenti–miR-664 siRNA; group 7, lenti–miR-485-3p siRNA; group 8, lenti–miR-495 siRNA. Hep3B cells (1 × 107) in 100 μl PBS were injected subcutaneously into the right flank of each nude mouse. From week 3 on, xenograft tumor size was measured by calipers. The tumor volume was calculated according to the formula: π/6 (length × width2) (39). Animals were sacrificed at week 8. Parts of the tumor tissues were used for RNA and protein analysis; the rest were fixed in 4% formalin for histology and immunohistochemistry.
Orthotopic liver cancer model using Hep3B cells expressing varying levels of miRNAs and MAT1A.
Hep3B cells stably transfected with lentiviral vectors expressing miRNAs or siRNAs against the miRNAs (1.5 × 106 cells/50 μl) were slowly injected into the left hepatic lobe of 4-week-old male BALB/c nude mice (n = 8 per group). Animal groups and number were the same as described for the xenograft model. The tumor size in liver tissues was measured as above at day 45 (pilot experiment showed 50% mice died at day 56 in the miR-495 group, but all mice in different groups survived at day 45) and the tumor volume was calculated. Lung, liver, and adjacent tissues were harvested for DNA, RNA, and protein assays as well as standard pathological studies as described for the xenograft model.
In separate experiments, 4-week-old male BALB/c nude mice (n = 8 per group) were injected in the left hepatic lobe as above with Hep3B cells (1.5 × 106 cells/50 μl) and stably transfected with MAT1A expression vector that does not have the 3′ UTR (MAT1A-no 3′ UTR) or EV. Concurrently, mice were also injected into the spleen with lentiviral vector that expresses either all 3 miRNAs (miRNAs), siRNAs against all 3 miRNAs (miRNAsi), or EV. The packaging was done using the Trans Lentiviral pGIPz Packaging system (TLP4614; Open Biosystems). Viral harvesting was done as described in the Open Biosystems protocol. A total of 1 × 105 Hep3B cells were infected at a multiplicity of 20 PFU/cell for 24 hours. 2 × 109 transducing units (final volume 0.1 ml) were injected into the tail veins of mice. In order to maintain a high level of miRNA knockdown, repeated tail-vein injections were done at week s2 and 4. Tumor volumes in the liver and presence or absence of lung metastasis were documented at day 45 as above.
Orthotopic liver cancer model using HepG2 cells and treatment with siRNAs against miR-495, miR-485-3p, miR-664, or MAT1A.
HepG2 cells have the ability to invade and metastasize (40). To test the effect of knocking down miRNAs, HepG2 cells (1.5 × 106 cells/50 μl) were injected into the left hepatic lobe of 4-week-old male BALB/c nude mice following spleen injection of miRNA siRNA. Then tail-vein injection was done every 2 weeks. Animal groups (n = 8 per group) were as follows: group 1, lenti-scramble siRNA; group 2, lenti–miR-495 siRNA; group 3, lenti-485 siRNA; and group 4, lenti-664 siRNA. Mice were sacrificed at day 56. Lentiviral packaging and harvesting were as described above except HeG2 cells were used and tail vein injection was repeated at week 6. Immunohistochemistry and Western blot were done to assess transduction efficiency using GFP.
To examine the role of MAT1A expression on tumorigenicity and the therapeutic effect of miR-495 siRNA, in separate experiments, mice were injected with HepG2 cells directly into the left lobe as above and treated with lenti-MAT1A siRNA and lenti–miR-495 siRNA alone or in combination as described above. Animal groups (n = 8 per group) were as follows: group 1, lenti-scramble vector and lenti-scramble siRNA; group 2, HepG2 injection; group 3, lenti-MAT1A siRNA+lenti-scramble siRNA; group 4, lenti–miR-495 siRNA+lenti-MAT1A siRNA; and group 5, lenti–miR-495 siRNA+lenti-scramble siRNA. Mice were sacrificed at day 56; liver tumor volume at the site of original injection was measured and tissues (lung, liver, pancreas) were harvested for pathological exam as described above.
Global DNA and LIN28B promoter methylation assay.
Levels of 5-methylcytosine (5-mC) in hepatic tumors derived from Hep3B cells stably expressing miR-664, miR-485, and miR-495 or their siRNAs were measured by the MethylFlash Methylated DNA Quantification Kit.
DNA samples of tumor tissues from mice injected with Hep3B cells expressing forced miR-664, miR-485, and miR-495 and their respective siRNAs were extracted and digested by MspI, HpaII, PvuII, and NdeI. Southern blot was done as described previously (41).
Northern and Western blot analysis.
Northern blotting probes for miR-664, miR485-3p, miR-495, and let-7a were purchased from EXIQON. Total cellular RNA was extracted by using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Equal amounts of total RNA (15 μg) were denatured, fractionated by electrophoresis on a 15% polyacrylamide–8 M urea gel, electroblotted, and cross-linked onto a nylon membrane. Northern blot analysis was performed as described using Ultrahyb-Oligo (Ambion). As a control for normalization of RNA expression levels, blots were hybridized with an oligonucleotide probe complementary to the U6 RNA (5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′).
Xenograft and liver tissues isolated from the different treatment groups were subjected to Western blot analysis. Fifteen micrograms of total protein extract were resolved on 12.5% SDS-polyacrylamide gels. Membranes were probed with antibodies to LIN28B, H3k27me3, and MATα1. To ensure equal loading, membranes were stripped and reprobed with anti–β-actin antibodies. Semi-quantitative analysis was performed for both Northern and Western blots using Quantity One (Bio-Rad).
Histology and immunohistochemistry.
Sections from xenograft, liver, lung, and pancreas were fixed with formalin for 4 hours, embedded in paraffin, sectioned, and stained with H&E, as previously described (12). Staining and counting of PCNA were performed according to the manufacturer’s suggested protocol (Invitrogen), whereas MATα1 antibody was diluted to 1:200. Immunohistochemical staining of MATα1 was performed with the Vector ABC Kit according to the manufacturer’s method. For quantifying immunohistochemical staining, a total of 5 fields at ×100 magnification were randomly selected (minimum of 1000 cells total), and positive nuclei or cells were counted and expressed as a percentage of the total using MetaMorph imaging software. Control with no antibody showed no staining.
Apoptosis and BrdU incorporation.
Apoptosis was measured as described (12). BrdU incorporation was measured with BrdU Detection Kit according to the manufacturer’s protocol (BD Biosciences — Pharmingen).
Nuclear SAMe levels.
SAMe levels were measured as described (11) in purified nuclear fractions from tumors expressing varying levels of miRNAs or their siRNAs.
Statistics.
Data are given as mean ± SEM. Statistical analysis was performed using ANOVA followed by Fisher’s test for multiple comparisons. Significance was defined as P < 0.05.
Study approval.
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by Keck School of Medicine University of University of Southern California Health Science Institutional Review Board (Los Angeles, California, USA). All procedure protocols, use, and the care of the animals were reviewed and approved by the Institutional Animal Care and Use Committee at UCLA.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01DK51719 (to S.C. Lu and J.M. Mato), R01AT01576 (to S.C. Lu and J.M. Mato), and Plan Nacional of I+D SAF 2008-04800, and HEPADIP-EULSHM-CT-205 (to J.M. Mato). 293T, HepG2, and Hep3B cells were provided by the Cell Culture Core of the USC Research Center for Liver Diseases (P30DK48522). Pathological sections and staining were done by the Imaging Core of the USC Research Center for Liver Diseases (P30DK48522).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(1):285–298. doi:10.1172/JCI63861.
References
- 1.Lu SC, Mato JM. S-adenosylmethionine in cell growth, apoptosis and liver cancer. J. Gastroenterol Hepatol. 2008;Suppl:S73–S77. doi: 10.1111/j.1440-1746.2007.05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotb M, et al. Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends Genet. 1997;13(2):51–52. doi: 10.1016/S0168-9525(97)01013-5. [DOI] [PubMed] [Google Scholar]
- 3.Avila MA, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33(6):907–914. doi: 10.1016/S0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 4.Cai J, Sun W, Hwang JJ, Stain S, Lu SC. Changes in S-adenosylmethionine synthetase in human liver cancer: molecular characterization and significance. Hepatology. 1996;24(5):1090–1097. doi: 10.1002/hep.510240519. [DOI] [PubMed] [Google Scholar]
- 5.Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45(5):1306–1312. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 6.Lu SC, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A. 2001;98(10):5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez-Chantar ML, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16(10):1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 8.Pascale RM, et al. Chemoprevention by S-adenosyl-L-methionine of rat liver carcinogenesis initiated by 1,2-dimethylhydrazine and promoted by orotic acid. Carcinogenesis. 1995;16(2):427–430. doi: 10.1093/carcin/16.2.427. [DOI] [PubMed] [Google Scholar]
- 9.Pascale RM, et al. Reversal by 5-azacytidine of the S-adenosyl-L-methionine-induced inhibition of the development of putative preneoplastic foci in rat liver carcinogenesis. Cancer Lett. 1991;56(3):259–265. doi: 10.1016/0304-3835(91)90011-6. [DOI] [PubMed] [Google Scholar]
- 10.Pascale RM, et al. Comparative effects of L-methionine, S-adenosyl-L-methionine and 5′-methylthioadenosine on the growth of preneoplastic lesions and DNA methylation in rat liver during the early stages of hepatocarcinogenesis. Anticancer Res. 1991;11(4):1617–1624. [PubMed] [Google Scholar]
- 11.Lu SC, et al. S-adenosylmethionine in the chemoprevention and treatment of hepatocellular carcinoma in a rat model. Hepatology. 2009;50(2):462–471. doi: 10.1002/hep.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, et al. Forced expression of methionine adenosyltransferase 1A in human hepatoma cells suppresses in vivo tumorigenesis in mice. Am J Pathol. 2010;176(5):2456–2466. doi: 10.2353/ajpath.2010.090810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Law PTY, Wong N. Emerging roles of microRNA in the intracellular signaling networks of hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26(3):437–450. doi: 10.1111/j.1440-1746.2010.06512.x. [DOI] [PubMed] [Google Scholar]
- 14.Cai J, Mao Z, Hwang JJ, Lu SC. Differential expression of methionine adenosyltransferase genes influences the rate of growth of human hepatocellular carcinoma cells. Cancer Res. 1998;58(7):1444–1450. [PubMed] [Google Scholar]
- 15.Wang YC, et al. Lin-28B expression promotes transformation and invasion in human hepatocellular carcinoma. Carcinogenesis. 2010;31(9):1516–1522. doi: 10.1093/carcin/bgq107. [DOI] [PubMed] [Google Scholar]
- 16.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden:Globocan 2000. Int J Cancer. 2001;94(2):153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 17.El-Serag HB. Hepatocellular carcinoma. New Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 18.Shah SA, et al. Recurrence after liver resection for hepatocellular carcinoma: risk factors, treatment, and outcomes. Surgery. 2007;141(3):330–339. doi: 10.1016/j.surg.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Llovet JM, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(23):378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Peng JX, Yang JQ, Yang LY. Identification of gene expression profiling in hepatocellular carcinoma using cDNA microarrays. Dig Dis Sci. 2009;54(12):2729–2735. doi: 10.1007/s10620-008-0667-2. [DOI] [PubMed] [Google Scholar]
- 21.Frau M, et al. Role of transcriptional and post-transcriptional regulation of methionine adenosyltransferases in liver cancer progression. Hepatology. 2012;56(1):165–175. doi: 10.1002/hep.25643. [DOI] [PubMed] [Google Scholar]
- 22.Tomasi ML, Li TWH, Li M, Mato JM, Lu SC. Inhibition of methionine adenosyltransferase 1A transcription by coding region methylation. J Cell Physiol. 2012;227(4):1583–1591. doi: 10.1002/jcp.22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vázquez-Chantada M, et al. HuR/Methylated-HuR and AUF1 regulate the expression of methionine adenosyltransferase during liver proliferation, differentiation and carcinogenesis. Gastroenterology. 2010;138(5):1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gramantieri L, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12(6A):2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mott J. MicroRNAs involved in tumor suppressor and oncogene pathways: Implications for hepatobiliary neoplasia. Hepatology. 2009;50(2):630–637. doi: 10.1002/hep.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CF, et al. Novel regulation of nuclear factor-YB by miR-485-3p affects the expression of DNA topoisomerase IIα and drug responsiveness. Mol Pharmacol. 2011;79(4):735–741. doi: 10.1124/mol.110.069633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang-Verslues WW, et al. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30(21):2463–2474. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 28.Simion A, Laudadio I, Prévot PP, Raynaud P, Lemaigre FP, Jacquemin P. miR-495 and miR-218 regulate the expression of the Onecut transcription factors HNF-6 and OC-2. Biochem Biophys Res Commun. 2010;391(1):293–298. doi: 10.1016/j.bbrc.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 29.Lu Y, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122(23):2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 30.Koturbash I, Melnyk S, James SJ, Beland FA, Pogribny IP. Mol Carcinog. Role of epigenetic and miR-22 and miR-29b alterations in the downregulation of Mat1a and Mthfr genes in early preneoplastic livers in rats induced by 2-acetylaminofluorene [Published online ahead of print December 27, 2011]. doi: 10.1002/mc.21861 . [DOI] [PubMed] [Google Scholar]
- 31.Xiong Y, et al. Effects of microRNA- 29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51(3):836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 32.Calvisi DF, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117(9):2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji J, Wang XW. A yin-yang balancing act of Lin28/Let-7 link in tumorigenesis. J Hepatol. 2010;53(5):974–975. doi: 10.1016/j.jhep.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147(5):1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, et al. A mouse model of cholestasis-associated cholangiocarcinoma and transcription factors involved in progression. Gastroenterology. 2011;141(1):378–388. doi: 10.1053/j.gastro.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viswanathan SR, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41(7):843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reytor Y, Pérez-Miguelsanz J, Alvarez L, Pérez-Sala D, Pajares MA. Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J. 2009;23(10):3347–3360. doi: 10.1096/fj.09-130187. [DOI] [PubMed] [Google Scholar]
- 38.Yang HP, Huang ZZ, Zeng ZH, Chen CJ, Wang JH, Lu SC. The role of c-Myb and Sp1 in the up-regulation of methionine adenosyltransferase 2A gene expression in human hepatocellular carcinoma. FASEB J. 2001;15(9):1507–1516. doi: 10.1096/fj.01-0040com. [DOI] [PubMed] [Google Scholar]
- 39.Nyati MK, et al. The potential of 5-fluorocytosine/cytosine deaminase enzyme prodrug gene therapy in an intrahepatic colon cancer model. Gene Ther. 2002;9(13):844–849. doi: 10.1038/sj.gt.3301706. [DOI] [PubMed] [Google Scholar]
- 40.Zhao R, et al. Hepatoma cell line HepG2.2.15 demonstrates distinct biological features compared with parental HepG2. World J Gastroenterol. 2011;17(9):1152–1159. doi: 10.3748/wjg.v17.i9.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Huang ZZ, Zeng Z, Chen C, Selby RR, Lu SC. Role of promoter methylation in increased methionine adenosyltransferase 2A expression in human liver cancer. Am J Physiol Gastrointest Liver Physiol. 2001;280(2):G184–G190. doi: 10.1152/ajpgi.2001.280.2.G184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.