Abstract
The scaffold protein p62 (sequestosome 1; SQSTM1) is an emerging key molecular link among the metabolic, immune, and proliferative processes of the cell. Here, we report that adipocyte-specific, but not CNS-, liver-, muscle-, or myeloid-specific p62-deficient mice are obese and exhibit a decreased metabolic rate caused by impaired nonshivering thermogenesis. Our results show that p62 regulates energy metabolism via control of mitochondrial function in brown adipose tissue (BAT). Accordingly, adipocyte-specific p62 deficiency led to impaired mitochondrial function, causing BAT to become unresponsive to β-adrenergic stimuli. Ablation of p62 leads to decreased activation of p38 targets, affecting signaling molecules that control mitochondrial function, such as ATF2, CREB, PGC1α, DIO2, NRF1, CYTC, COX2, ATP5β, and UCP1. p62 ablation in HIB1B and BAT primary cells demonstrated that p62 controls thermogenesis in a cell-autonomous manner, independently of brown adipocyte development or differentiation. Together, our data identify p62 as a novel regulator of mitochondrial function and brown fat thermogenesis.
Introduction
Obesity and its comorbidities, such as cardiovascular disease and type 2 diabetes, are increasingly challenging health threats in modern societies. Obesity, defined as a pathologically exaggerated accumulation of body fat, is the direct consequence of a prolonged positive energy balance that occurs when energy intake chronically exceeds energy expenditure. So far, pharmacological weight loss therapies, frequently aiming to minimize energy intake, have been hampered by limited efficacy or considerable side effects. Therefore, novel strategies to safely and efficiently combat the obesity and diabetes epidemic are urgently required.
In small mammals and infants, brown adipose tissue (BAT) plays an important role in energy metabolism due to its ability to burn energy by dissipating heat in response to sympathetic nerve activity (1, 2). In adult humans, however, BAT was long believed to play, at most, a minor role in energy metabolism. More recently, this historical view has been called into question by the observations that adult humans possess considerable amounts of BAT (3–6) and that the amount of BAT is decreased in obese compared with lean individuals (3, 5). These observations, in addition to recent advances in understanding the complex processes of brown adipocyte differentiation and function (7–12), have resulted in a rekindled scientific interest in BAT as a therapeutic target for the treatment of obesity.
Scaffold proteins are important mediators ensuring efficient and selective cell signal transduction. Enzymatic signal specificity mediated by scaffold proteins is thereby achieved through specific protein-protein interactions between shared motifs located in both the target enzyme and the scaffold protein. p62 (sequestosome 1; SQSTM1) was originally identified as a signal adaptor for isoforms of the atypical PKC subfamily (aPKCs) (13, 14). p62 is a multimodular adaptor protein involved in a number of signaling pathways affecting important biological processes, such as inflammation, cell differentiation, cell growth, and tumorigenesis (15–17). We have previously shown that global ablation of p62 in mice results in obesity and systemic glucose intolerance and insulin resistance (18, 19). The main goal of the present study was to rigorously establish the in vivo target tissue accounting for the role of p62 in the control of metabolic homeostasis and obesity and also to unravel the mechanistic details of p62 in those processes.
Results
Generation of tissue-specific p62–/– mice.
To establish the p62 target tissue responsible for its role in whole-body metabolic control and obesity in vivo, we used the Cre-lox system to target key tissues implicated in systemic energy metabolism control, namely CNS, liver, skeletal muscle, adipose tissue, and cells of the myeloid lineage. To that end, we generated the tissue-specific p62–/– mouse lines shown in Supplemental Figure 1 (supplemental material available online with this article; doi: 10.1172/JCI64209DS1). Briefly, the coding exon 1 of the murine p62 isoform 1 (NM_011018.2) was deleted by mating chimeric p62 floxed mice with mice expressing Cre recombinase (Cre) under the control of the promoter for either nestin (Nes, CNS specific) (20), albumin (Alb, liver specific) (21), myosin light chain 1f (Mlc1f, muscle specific) (22), or adipocyte protein 2 (aP2 also known as Fabp4, adipocyte-specific) (23). As aP2 is also expressed in macrophages (24), we also generated mice that lack p62 in the myeloid lineage using lysozyme M (LysM, also known as Lyz2) cre mice. Validation of tissue-specific p62 deletion was assessed using Western blot analysis (Supplemental Figure 1).
CNS-specific deletion of p62 in mice does not affect systemic metabolism.
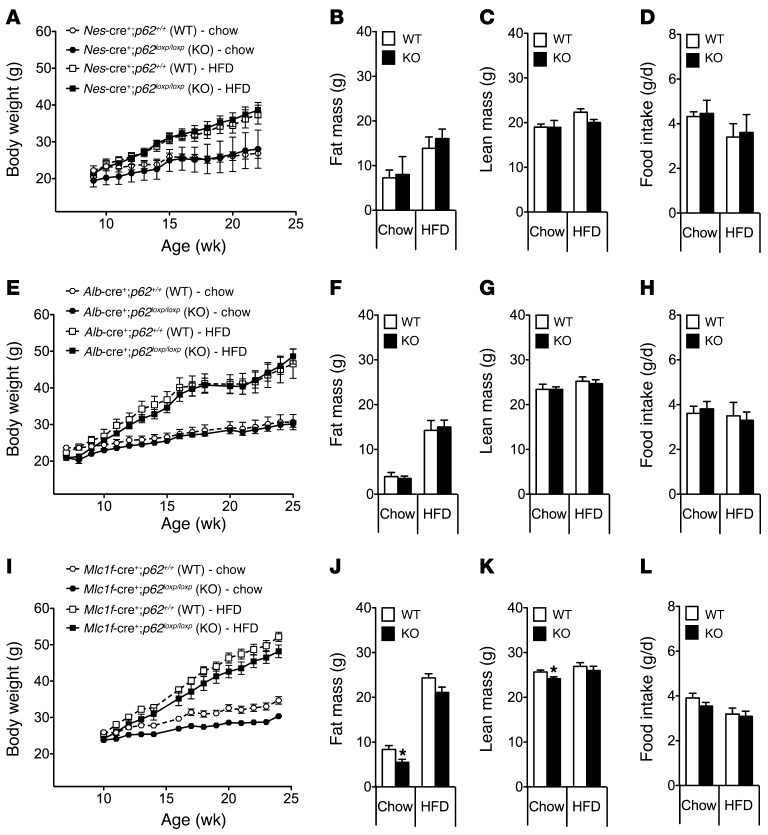
The metabolic phenotype of mice with exclusive deletion of p62 in the CNS was assessed under 2 different feeding regimes. Some mice were fed a regular standard chow diet (5.6% fat), whereas a parallel cohort was fed a high-fat diet (HFD) (58% fat). Surprisingly, we observed no difference in body weight (Figure 1A), body composition (Figure 1, B and C), or food intake (Figure 1D) in mice that lack p62 selectively in the CNS (Nes-cre+;p62loxp/loxp) as compared with WT controls (Nes-cre+;p62+/+), irrespective of the diet. Accordingly, we concluded that obesity in the global p62–/– mice was mediated by peripheral mechanisms and independent of p62 signaling in the brain.
Figure 1. Mice with tissue-specific deletion of p62 in the CNS, liver, or skeletal muscles do not show a metabolically relevant phenotype.
Body weight, body composition (fat and lean tissue mass), and food intake of male mice that lack p62 selectively in the CNS (A–D), the liver (E–H), or the skeletal muscles (I–L). Fat and lean tissue mass was measured at an age of 23 weeks (A), 22 weeks (B), or 24 weeks (C). n = 8–10 mice each group. Data represent mean ± SEM. *P < 0.05.
Hepatic deletion of p62 in mice does not affect systemic metabolism.
The phenotypical analysis of mice that lack p62 exclusively in the liver (Alb-cre+;p62loxp/loxp) also failed to reveal any alterations in body weight (Figure 1E), body composition (Figure 1, F and G), or food intake (Figure 1H) when compared with WT controls (Alb-cre+;p62+/+) independently of the diet. In addition, no signs of liver steatosis were observed and no differences in systemic glucose tolerance were observed when comparing liver-specific p62–/– mice and their WT littermate controls (Supplemental Figure 2A). Together, these data do not support a role for hepatic p62 signaling in the regulation of body weight or systemic metabolism.
Deletion of p62 in mouse skeletal muscle does not affect systemic metabolism.
We next analyzed the metabolic phenotype of mice lacking p62 exclusively in skeletal muscle (Mlc1f-cre+;p62loxp/loxp). When compared with WT controls (Mlc1f-cre+;p62+/+), these mutant mice did not show any signs of obesity on either regular chow or HFD (Figure 1I). In fact, the body weight of the muscle-specific p62–/– mice was moderately lower when compared with WT controls fed standard chow diet but not HFD (Figure 1I). The decreased body weight of the chow-fed muscle-specific p62–/– mice was accompanied by a decrease in body fat and lean tissue mass (Figure 1, J and K) without notable changes in food intake (Figure 1L) or glucose tolerance (Supplemental Figure 2B). Indirect calorimetry performed at the age of 20 weeks revealed no detectable differences in energy expenditure, locomotor activity, or substrate utilization between the muscle-specific p62–/– mice and their WT controls (Supplemental Figure 3, A–D). Also, the energy expenditure response to a norepinephrine challenge (1 mg/kg, single s.c. injection) did not differ between both genotypes (Supplemental Figure 3E). Together, these data indicate that lack of p62 signaling in the skeletal muscles does not explain the role of p62 in the systemic control of body weight and adiposity observed in global p62–/– mice.
Adipocyte-specific, but not macrophage-specific, deletion of p62–/– results in obesity and glucose intolerance.
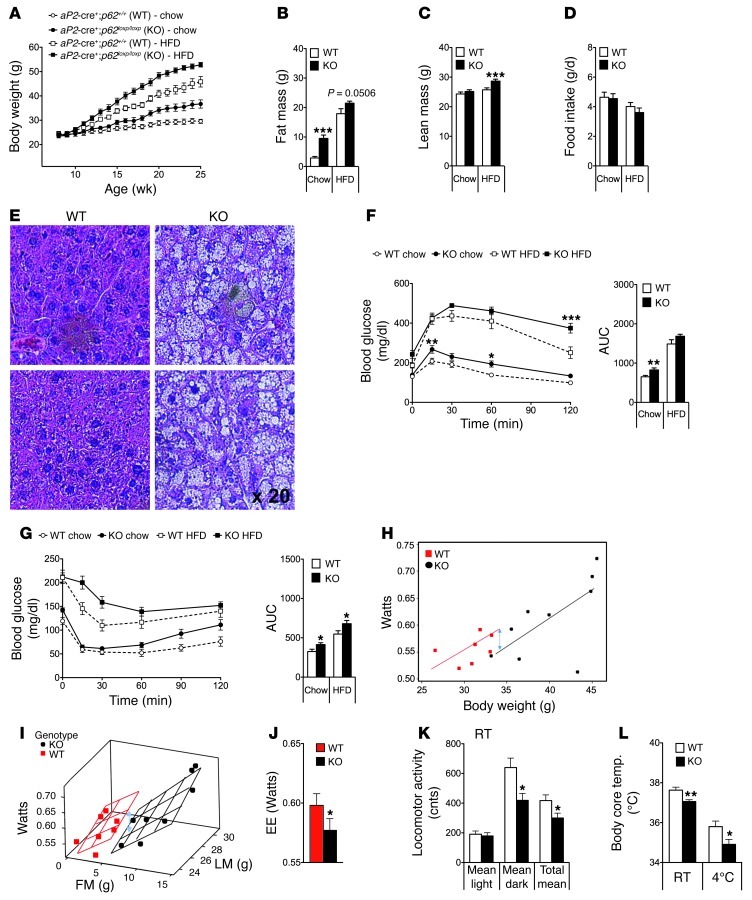
When mice that lack p62 selectively in the adipose tissue (aP2-cre+;p62loxp/loxp) were analyzed, we observed a dramatically enhanced body weight gain as compared with WT controls (aP2-cre+;p62+/+) on both standard chow and HFD (Figure 2A). The obese phenotype of the adipocyte-specific p62–/– mice was comparable to that observed in the global p62–/– mice and was a consequence of increased body fat and lean tissue mass (Figure 2, B and C). Interestingly, as in the case of global p62–/– mice, obesity was not paralleled by changes in food intake (Figure 2D). On either diet, adipocyte-specific p62–/– mice had liver steatosis (Figure 2E), impaired glucose tolerance, and decreased insulin sensitivity (Figure 2, F and G). aP2cre mice have been reported to potentially cause target gene deletion in cells beyond adipocytes, such as CNS neurons or macrophages. However, we had already shown that deletion of p62 in the CNS did not cause a body weight phenotype and consequently could not explain the phenotype of aP2-cre+;p62loxp/loxp mice. Therefore, we next used lysozyme M (LysM) cre mice to selectively knock out p62 in the myeloid cell lineage (LysM-cre+;p62loxp/loxp). Careful phenotype analysis of these mutant mice did not reveal any differences in body weight, body composition, or glucose and insulin sensitivity when compared with the corresponding WT control mice (Supplemental Figure 4, A–E). Collectively, these findings indicate that obesity in aP2-cre+;p62loxp/loxp mice is a consequence of p62 deficiency in adipocytes and does not result from the lack of p62 in myeloid cells.
Figure 2. Metabolic phenotype of adipocyte-specific p62–/– mice.
Body weight (A), body composition at an age of 26 weeks (B and C), and food intake (D) of adipocyte-specific p62–/– and control mice fed either with a standard chow diet (5.6% fat) or HFD (58% kcal fat). H&E staining of liver samples from 35-week-old chow-fed adipocyte-specific p62–/– and WT mice (E). Glucose tolerance (F) and insulin sensitivity (G) of 17-week-old chow-fed adipocyte-specific p62–/– and WT mice. Energy expenditure (EE). (H–J) and locomotor activity (K) of 33-week-old chow-fed adipocyte-specific p62–/– and WT mice at room temperature (23 ± 2°C). Body core temperature of 35-week-old chow-fed adipocyte-specific p62–/– and WT mice at room temperature and during acute cold exposure (16 hours at 4°C) (L). n = 7–11 mice each group. Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
White adipocytes of the obese adipocyte-specific p62–/– mice were greatly enlarged (Supplemental Figure 5A) and showed increased basal activity of Mapk1 (also known as ERK) (Supplemental Figure 5B), which is in agreement with previously reported data in total p62-deficient mice (19). Expression of proinflammatory cytokines and chemokines, such as Il6, Tnfa, chemokine cc motif ligand 2 [Ccl2], and integrin α-x [Itgax], as well as markers of macrophage infiltration (Cd68 and Emr1 [also known as F4/80]) were increased as expected, considering the obese phenotype of these mice (Supplemental Figure 5, C and D). Of note, expression of Pparg and Cebpb, well-known master regulators of adipogenesis and fat cell differentiation (2, 25), was not changed in epididymal white adipose tissue (eWAT) of adipocyte-specific p62–/– mice (Supplemental Figure 5E). Gene programs controlling lipoprotein metabolism, lipogenesis, fatty acid transport, or lipolysis were also not affected by adipocyte-specific p62 deficiency (Supplemental Figure 5, F–I). Finally, there was no difference in plasma-free fatty acids and free glycerol between genotypes following a challenge with norepinephrine (1 mg/kg) (Supplemental Figure 5, J and K). We conclude that the obese phenotype of adipocyte-specific p62–/– mice does not result from changes in fatty acid mobilization or lipolysis.
Impaired nonshivering thermogenesis in adipocyte-specific p62–/– mice.
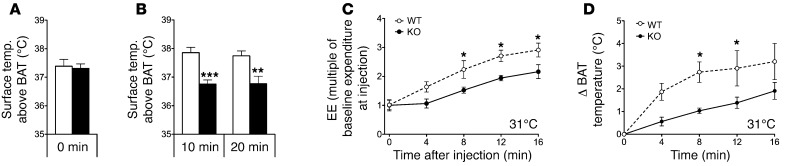
Indirect calorimetry, performed at an age of 33 weeks, revealed that energy expenditure was decreased in adipocyte-specific p62–/– mice as compared with WT controls (Figure 2H). Importantly, this finding was confirmed when energy expenditure raw data were analyzed without normalizing for body size using ANCOVA, with body fat and lean tissue mass as covariants, as proposed recently (ref. 26 and Figure 2, I and J). The decrease in energy expenditure was accompanied by a significant reduction in locomotor activity (Figure 2K) and considerably lower body core temperature (Figure 2L). Based on these observations, we hypothesized that the decreased energy metabolism of the adipocyte-specific p62–/– mice might be a consequence of impaired nonshivering thermogenesis in BAT. Accordingly, to test whether p62 links β-adrenergic stimulation with mitochondrial uncoupling in vivo, we measured the surface temperature above BAT in response to the selective β-3 adrenergic receptor agonist CL-316,243. In line with our hypothesis, single i.p. treatment with CL-316,243 (0.6 mg/kg) significantly increased the surface temperature above BAT in WT mice, but had no measurable effect in adipocyte-specific p62–/– mice (Figure 3, A and B). These findings were corroborated by significantly impaired energy expenditure (Figure 3C) and BAT surface temperature (Figure 3D) responses to systemic norepinephrine (1 mg/kg) challenge in adipocyte-specific p62–/– mice compared with WT controls.
Figure 3. Impaired response to β-adrenergic receptor stimulation in adipocyte-specific p62–/– mice.
Body surface temperature above the BAT, measured with an IR camera, of obese adipocyte-specific p62–/– mice before (A) and after (B) i.p. treatment with CL-316,243 (0.6 mg/kg). Measurement of energy expenditure (expressed as multiple of baseline expenditure at injection) (C) and increase of BAT temperature (measured using implanted temperature sensors) at different time points after s.c. treatment with norepinephrine (1 mg/kg) (D). n = 7–11 mice per group (A and B); n = 4 mice each group (C and D). Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Impaired mitochondrial function in BAT of adipocyte-specific p62–/– mice.
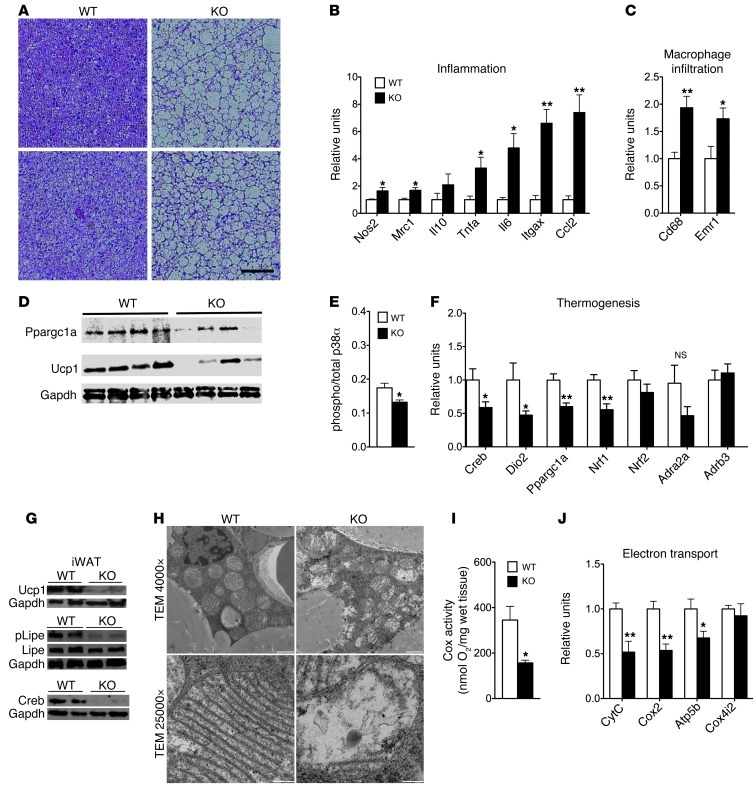
Histological analysis revealed enhanced lipid deposition with increased lipid droplet sizes in BAT of adipocyte-specific p62–/– mice (Figure 4A). Accordingly, expression profiles reflecting inflammation (inducible nitric oxide synthase [Nos2], mannose receptor C-type 1 [Mrc1], Tnfa, Il6, Itgax, Ccl2) and macrophage infiltration (Cd68, Emr1) were increased (Figure 4, B and C). Consistent with our hypothesis that nonshivering thermogenesis is impaired in adipocyte-specific p62–/– mice, protein levels of PPARGC1A (also known as PGC1A) and uncoupling protein 1 (UCP1) were decreased in these mice (Figure 4D). Importantly, we discovered that the loss of p62 in adipocytes decreased activation of p38α in BAT (Figure 4E). Consistent with that observation, we found that mRNA expression of p38 downstream targets and key uncoupling regulators was significantly decreased in adipocyte-specific p62–/– mice compared with WT controls, including cAMP response element binding protein 1 [Creb], diodinase 2 [Dio2], Pgc1a, and nuclear respiratory factor 1 [Nrf1] (Figure 4F). This finding was corroborated by lower protein levels of CREB and UCP1, as well as less phosphorylated LIPE (also known as HSL) in brown adipocyte rich (Brite) inguinal adipose tissue (Figure 4G and ref. 27). Notably, expression of the β-3 adrenergic receptor (Adrb3) and the α2A adrenergic receptor (Adra2a) was not changed in adipocyte-specific p62–/– mice compared with WT controls (Figure 4F), indicating a role for p62 at the molecular interface between β-adrenergic input and intracellular control of uncoupling in BAT.
Figure 4. Impaired mitochondrial function in BAT of chow-fed adipocyte-specific p62–/– mice.
H&E staining of BAT (A). Expression of genes related to inflammation (B) and macrophage infiltration (C) in BAT. Western blot analysis of Ppargc1a and Ucp1 (D) and ratio between levels of total and phosphorylated levels of p38α in BAT (E). Expression of genes related to thermogenesis (F). Western blot analysis of Ucp1, Lipe, and Creb in the inguinal WAT (G). Electron micrograph of brown adipocytes (H), Cox activity (I), and mRNA expression levels of genes related to mitochondrial electron transport (J) in BAT of adipocyte-specific p62–/– and WT mice. Cox4i2, COX subunit 4b. Measurement of gene expression was performed in n = 7–8 mice of each genotype. Cox activity and protein levels of p38α were assessed in n = 4 mice of each genotype. Scale bars: 25 μm (A); 1 μm (H, original magnification ×4,000); 200 nm (H; original magnification, ×25,000. Data represent mean ± SEM. *P < 0.05; **P < 0.01.
Electron microscopy analyses revealed considerable differences in mitochondrial structure as a consequence of p62 deficiency in adipocytes (Figure 4H). Brown adipocytes from adipocyte-specific p62–/– mice contained mitochondria with fewer cristae, which were also shorter and displaced to the periphery with varying degrees of disorientation (Figure 4H). In line with this observation, we found decreased oxidative capacity, as quantified by measuring cytochrome c oxidase (Cox) activity, in BAT of adipocyte-specific p62–/– mice (Figure 4I). Moreover, mRNA expression levels of gene programs relevant to electron transport, such as cytochrome C (CytC), Cox2, and ATP synthase mitochondrial F1 complex subunit β (Atp5b), were likewise decreased (Figure 4J). Together, these data indicate that p62 is essential for the structural integrity and proper function of BAT mitochondria.
Cell autonomous role of p62 in mitochondrial function.
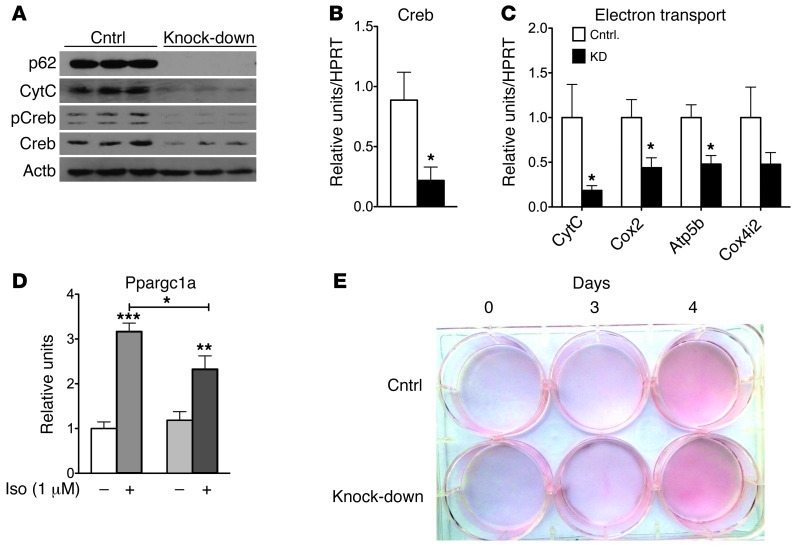
To assess whether the observed effect on mitochondrial function was mediated by p62 in a cell-autonomous manner, we knocked down p62 in the BAT cell line HIB1B using shRNA against p62 (Figure 5). Consistent with our findings from adipocyte-specific p62–/– mice, knockdown of p62 in HIB1B cells led to decreased protein levels of CytC, lower total and phosphorylated levels of Creb, and decreased mRNA levels of the Creb, CytC, Cox2, and Atp5b genes (Figure 5, A–C). In line with this observation, the response of PGC1α to isoproterenol (1 μM) was significantly lower in cells lacking p62 (Figure 5D). On an important note, knockdown of p62 did not affect adipocyte development and differentiation (Figure 5E).
Figure 5. Cell autonomous effect of p62 on mitochondrial function in HIB1B cells.
Western blot analysis of p62, CytC,and total and phosphorylated Creb (A) and mRNA level of Creb (B) and genes related to mitochondrial electron transport (C) in HIB1B cells lacking p62 and controls. mRNA levels of Ppargc1a in p62-deficient HIB1B cells after 6 hours stimulation with isoproterenol (1 μM) (D). Oil red O staining in p62-deficient HIB1B cells (E). *P < 0.05; **P < 0.01; ***P < 0.001.
To validate the results obtained in both the adipocyte-specific p62–/– mice and the HIB1B cells, we assessed the expression profile of the thermogenic genes in BAT primary cells obtained from WT or global p62–/– mice over the time course of adipocyte differentiation (Supplemental Figure 6). In line with our previous findings, expression of Ucp1, Ppargc1a, and Cox2 was decreased in the p62-deficient cells from day 4 with no notable difference compared with WT controls within the first 3 days of differentiation. Cytochrome c expression was decreased from day 3 onwards without any difference between genotypes within the first 2 days of differentiation. No difference at any time point was observed for Atp5b and Cox4 (Supplemental Figure 6, A–F).
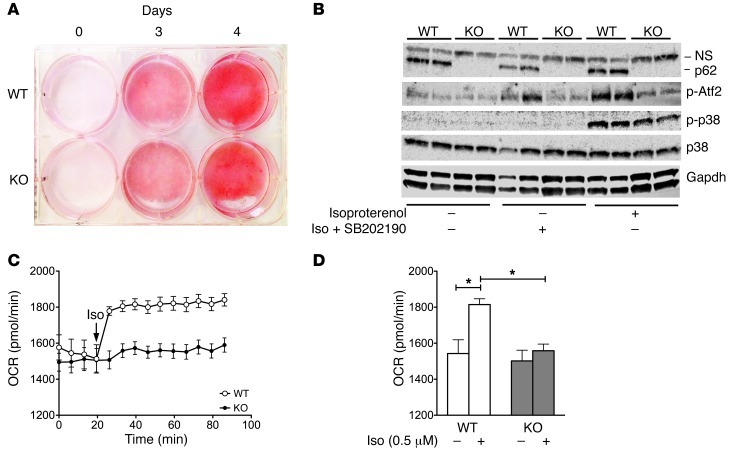
Consistent with our findings in HIB1B cells, we likewise did not observe a difference in adipocyte development and differentiation between genotypes in the BAT primary cells (Figure 6A). Aligned with our in vivo data, phosphorylated levels of both p38 and its direct downstream target Atf2 were decreased in p62-deficient cells upon stimulation with isoproterenol (Figure 6B). These observations support our hypothesis that p62 affects thermogenesis through the p38 MAPK pathway.
Figure 6. Cell autonomous effect of p62 on mitochondrial function in BAT primary cells.
Oil red O staining in p62-deficient BAT primary cells obtained from global p62–/– and WT control mice (A). Western blot analysis of p62, p38, and phosphorylated levels of p38 and Atf2 in 5-day–differentiated BAT primary cells treated for 30 minutes with isoproterenol (0.5 μM) or pretreated for 30 minutes with the p38 inhibitor SB202190 (10 μM) followed by 30 minutes treatment of isoproterenol (0.5 μM) plus SB202190 (10 μM) (B). Measurement of OCR of 2-day–differentiated BAT primary cells obtained from global p62–/– mice and WT controls in response to isoproterenol (0.5 μM) (C and D). *P < 0.05.
To functionally validate the results obtained in the HIB1B cells, we measured oxygen consumption in the BAT primary cells, both before and after stimulation with isoproterenol (0.5 μM). Baseline levels of oxygen consumption were comparable between controls and cells lacking p62 (Figure 6, C and D). In primary cells obtained from WT animals, we observed the expected immediate increase in oxygen consumption in response to isoproterenol. However, this effect was completely abrogated in cells lacking p62 (Figure 6, C and D). Based on recent evidence indicating that catecholamines released from macrophages can substantially modulate BAT thermogenesis (28), we next assessed whether the observed changes in mitochondrial function persist after depletion of macrophages from BAT primary cells. We observed a similar lower response to isoproterenol in macrophage-depleted, p62-deficient BAT primary cells (Supplemental Figure 7, B and C). Furthermore, mRNA levels of Ucp1 and Ppargc1a remained decreased in the macrophage-depleted p62-deficient cells both before and after stimulation with isoproterenol (Supplemental Figure 7, D and E). The decreased Ucp1 mRNA expression translated into decreased protein levels of Ucp1 (Supplemental Figure 7F). Notably, no signs of macrophage infiltration were observed after depletion of Itgam-positive cells (Itgam is also known as Cd11b) (Supplemental Figure 7G), which was confirmed by flow cytometry (Supplemental Figure 7A). No differences regarding markers indicative of inflammation, DNA damage, redox balance, or ER stress were observed between genotypes (Supplemental Figure 7, G–K). Together, these data confirm that p62 directly controls basal mitochondrial function of brown adipocytes as well as their responsiveness to β-adrenergic stimulation in a cell autonomous manner, independently of macrophage infiltration and inflammation.
Discussion
Here we show for what we believe is the first time that the lack of p62 exclusively in adipose tissue is sufficient to recapitulate the metabolic syndrome phenotype of total p62-deficient mice, including obesity, glucose intolerance, and insulin resistance. Our results further identify BAT as the main organ responsible for the impaired energy metabolism observed in the adipose-specific p62–/– mice and reveal that p62 controls the mitochondrial function of brown adipocytes in a direct and cell-autonomous manner. The direct effect of p62 deficiency in brown fat thermogenesis includes impaired mitochondrial structure and dysfunction. Interestingly, we observe here that p62 controls activation of p38 in vivo. Several studies have previously shown that p38 MAPK is crucial for sympathetic nervous system–mediated (SNS-mediated) activation of BAT nonshivering thermogenesis and Ucp1 function (7, 29–31). Notably, by screening a HeLa cDNA library, Sudo and colleagues previously identified p62 as a p38-binding protein (32). Following up on that finding, Kawai and colleagues identified 2 p62 binding domains in p38. One domain is located in the N-terminal p38 interaction (NPI) domain, whereas the other is located in the C-terminal p38 interaction (CPI) domain (33). Consistent with our finding that activated levels of p38 are decreased in p62-deficient cells upon stimulation with isoproterenol, Kawai and colleagues found that genetic inactivation of p62 in HeLa cells led to impaired p38 activation upon stimulation with cytokines (33). Using surface plasmon resonance (SPR) imaging experiments, Saito and colleagues showed that the p62 domain comprising amino acids 164–190 can directly bind to p38 (34). However, whereas several lines of evidence support the interaction of p62 and p38 in vitro, using genetically engineered mice, we here show that this interaction appears to be of physiological relevance in vivo. Accordingly, loss of p62 in adipocytes impairs physiological activation of p38 and its downstream pathways, promoting uncoupling in vivo and in vitro. Such programs governing uncoupling have been either directly or indirectly linked to p38 activity and include Atf2, Creb, Ppargc1a, Dio2, Nrf1, Cox2, CytC, Atp5b, and Ucp1. All of these signals were significantly decreased in adipocyte-specific p62–/– mice, suggesting that p62 likely controls mitochondrial function via the p38/Ppargc1a pathway. Notably, this assumption is consistent with reports indicating that p62 binds p38 via its C-terminal domain and that p62 is colocalized with phosphorylated p38 in the nuclei (32–34).
In line with the discovery that p62 controls mitochondrial function of BAT, we found oxidative capacity, measured by Cox activity, to be decreased in BAT of adipocyte-specific p62–/– mice. Of appreciable note, no difference in Cox activity was observed in BAT of mice that specifically lack p62 only in skeletal muscle (Supplemental Figure 3F) or the myeloid lineage (Supplemental Figure 4F).
The observation that lipid metabolism is not changed in the adipocyte-specific p62–/– mice despite the altered morphology of their adipocytes is intriguing and warrants further investigation. As a matter of fact, previously published data showed that global loss of p62 results in increased adipogenesis in vitro and enhanced expression of adipogenic markers such as Pparg in vivo (19). However, using aP2-conditional KO mice, we have been unable to detect these alterations in WAT but rather changes in BAT. A potential explanation for this discrepancy might be that the loss of p62 in a total KO impairs WAT characteristics early during fat development, revealing the role of p62 in WAT. In the conditional model reported here, p62 is depleted at the time point when aP2 is expressed later during the adipocyte development process. WAT differentiation appears to stay intact, but a clear role for p62 governing BAT function is revealed. We conclude that the alterations in BAT may have secondary consequences on WAT in vivo but do not directly affect Pparg-driven differentiation processes.
The maintenance of mitochondrial integrity is central to numerous cellular processes, especially for those regulating the SNS-mediated activation of brown fat thermogenesis. A number of recent studies using cotransfection and shRNA-mediated knockdown of p62 have suggested that it plays an important role in the disposal of dysfunctional mitochondria by autophagy, also known as mitophagy, which is in accordance with p62’s role in the autophagic clearance of polyubiquitinated proteins (35–40). However, the analysis of p62-knockout cells does not support this notion, making this mitophagy function of p62 highly controversial at this moment (40–43). It could be argued that our observation that the mitochondrial structure and function is impaired in the adipocyte-specific p62–/– mice is in line with the reports showing that p62 might be implicated in the regulation of mitophagy. However, our observation that loss of p62 affects the genetic program responsible for mitochondrial biogenesis does not support this model. Together, these data strongly suggest that, although decreased mitophagy cannot be completely ruled out, this would not have a great impact on the p62-deficiency phenotype reported here. We favor the notion that p62 is critical in the regulation of transcriptional programs controlling mitochondrial homeostasis, which underlies the phenotype of adipose-specific p62-KO mice.
Collectively, our findings indicate that the adipose-selective inactivation of p62 unveils a new and unexpected function of this important adapter in the direct control of BAT mitochondrial biogenesis and function.
Methods
Animal studies.
For analysis of body weight, body composition, and food intake, mice were kept at an ambient temperature of 23 ± 2°C with constant humidity and a 12-hour light/12-hour dark cycle. Mice had free access to water and were fed ad libitum with either a regular standard chow diet (5.6% fat; LM-485, Teklad) or a HFD (58% kcal fat; Research Diets Inc.). Measurements of energy expenditure were performed using a customized indirect calorimetric system (TSE Systems Gmbh.). After adaptation for 24 hours, recordings were collected over the following 118 hours.
Gene expression analysis.
Gene expression of tissue samples (n = 7–9 mice per group) was profiled with quantitative PCR–based (qPCR-based) techniques using SYBR green, TaqMan Single Probes, or TaqMan Low Density Arrays (Applied Biosystems). TaqMan Low Density Arrays were performed in n = 4 mice of each genotype. The relative expression of the selected genes was measured using the 7900HT Fast Real-Time PCR System (Applied Biosystems). For low-density arrays, the PCR reactions took place on a 384 well reaction card preloaded with the specific primers and probes by the manufacturer. The sequences of primers and probes were designed and validated by Applied Biosystems and were taken from the Assay-on-Demand mouse library. The relative expression levels of each gene were normalized to the housekeeping gene 18S, HPRT, or RPL32.
Western blotting.
Cell extracts for Western blotting were prepared in radioimmunoprecipitation assay buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors). Lysates were separated by SDS–polyacrylamide gel electrophoresis and transferred to Nitrocellulose-ECL membranes (GE Healthcare), where the immune complex was detected by chemiluminescence (GE Healthcare). Antibodies were purchased from Santa Cruz Biotechnology Inc. (Gapdh, CytC, p38, p-p38, Ppargc1a), Abcam plc (Ucp1), Progen Biotechnik (p62), and Cell Signaling Technology Inc. (Creb, P-Creb, Mapk1, P-Mapk1, Lipe, P-Lipe, P-Atf2).
Cell culturing and analysis of BAT primary cells and HIB1B cells.
Culturing of adipocytes was performed according to a protocol provided by C. Ronald Kahn (Joslin Diabetes Center). For isolation of BAT primary cells, the interscapular BAT of 1-day-old global p62–/– and WT mice was dissected and digested for 20 minutes in isolation buffer containing 61.5 mM NaCl, 2.5 mM KCL, 0.65 mM CaCl2, 2.5 mM glucose, 50 mM HEPES, 2% BSA Fraction V, and 1.5 mg/ml collagenase A (Roche Diagnostics Gmbh). Cells were cultured for 2 days in DMEM High-Glucose (Fisher Thermo Scientific) containing 20% FBS and 1% penicillin/streptomycin. After reaching confluency, differentiation was induced by switching to DMEM High-Glucose containing 10% FBS, 20 nM insulin, 1 nM T3, 0.125 mM indomethacin, 0.5 mM IBMX, and 5.1 μM dexamethasone. Two days after start of differentiation, medium was switched to DMEM High-Glucose containing 10% FBS, 20 nM insulin, and 1 nM T3 until start of the experiments. The HIB1B cells were provided from Bruce Spiegelman (Department of Cancer Biology, Dana-Farber Cancer Institute and Department of Cell Biology, Harvard Medical School).
Depletion of macrophages from BAT primary cells.
Separation of macrophages from BAT primary cells was performed by magnetic immunoaffinity isolation using anti-Cd11b antibodies conjugated to magnetic beads (MACS Cell Separation System; Miltenyi Biotec). Following BAT digestion, Itgam-positive (Cd11b) cells were separated using positive selection columns (LD columns; Miltenyi Biotec) according to the manufacturer’s instructions. For validity of cell separation, cell eluates were taken before and after depletion of Cd11b-positive cells as well as from the retained cell fraction. Successful depletion of macrophages was confirmed by flow cytometry and qPCR analysis.
Flow cytometry to determine macrophage frequencies in BAT primary cells.
Dissociated BAT primary cells were suspended in PBS/10% FCS and preincubated with 5 μg/ml anti-Cd16/32 (FcγRII/III block, clone 2.4G2; BD Biosciences). Cells were then stained with FITC-conjugated anti-CD45 (clone 30-F11; eBioscience) and APC-conjugated anti-F4/80 (clone BM8; eBioscience) on ice for 20 minutes. Propidium iodide (1 μg/ml) was used to exclude dead cells. Flow cytometry was performed on a FACS Aria III (BD Biosciences), and results were analyzed using FACS Diva software (BD Biosciences).
Bioenergetics of BAT primary cells.
Primary cells of litters were pooled and independently differentiated and measured. Measurement of oxygen consumption rate (OCR) was performed using an extracellular flux analyzer (XF24, Seahorse Bioscience; Billerica). After differentiation for 2 days on a XF24 well plate, cells were washed in 1× PBS and incubated in 675 μl of DMEM (no. 5030; Sigma-Aldrich) in a non-CO2 incubator for 1 hour. Untreated oxygen consumption was recorded for 20 minutes followed by isoproterenol injection (0.5 μM) and subsequent OCR measurement. OCR measurement in macrophage-depleted primary cells was performed after differentiating for 7 days. Untreated oxygen consumption was recorded for 20 minutes followed by measurement of OCR after injection of isoproterenol (0.5 μM, 35 minutes), oligomycin (2 μg/ml, 21 minutes), FCCP (1 μM, 21 minutes), and rotenon/antimycin A (2.5 μM each, 14 minutes).
Cox activity.
Cox activity of interscapular BAT was assessed polarographically using a Clark type electrode (RANK Brothers) and a Powerlab for data processing (ADInstruments). Tissue was weighted and homogenized mechanically in tissue buffer (100 mM potassium phosphate, 2 mM EDTA, 10 mM glutathione, pH 7.2, at 37°C) using QIAGEN TissueLyser. Homogenates were treated with detergent (0.1% n-dodecyl-β-D-maltoside) and subjected to a temperature-controlled reaction chamber containing 130 μM cytochrome c from horse heart (Sigma-Aldrich) and 18 mM ascorbate in measuring buffer (100 mM potassium phosphate, 5 mM EDTA, pH 7.2, at 37°C ).
Measurement of p38α.
Protein levels of total and phosphorylated p38α were assessed from BAT tissue using an InstantOne ELISA (eBioscience Inc.). BAT samples were disrupted using a TissueLyser (Qiagen) and lysed and analyzed according to the manufacturer’s instructions.
Transmission electron microscopy.
Tissue samples were fixed in 2.5% electron microscopy grade glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4 (Science Services), postfixed in 2% aqueous osmium tetraoxide, dehydrated in gradual ethanol (30%–100%) and propylene oxide, embedded in Epon (Merck), and cured for 24 hours at 60°C. Semithin sections were cut and stained with toluidine blue. Ultrathin sections of 50 nm were collected onto 200 mesh copper grids and stained with uranyl acetate and lead citrate before examination by transmission electron microscopy (Zeiss Libra 120 Plus; Carl Zeiss NTS GmbH). Pictures were acquired using a Slow Scan CCD-camera and iTEM software (Olympus Soft Imaging Solutions).
Statistics.
Statistical analyses were performed using the statistical tools implemented in Graph Pad Prism (GraphPad Software). Differences between treatment groups were assessed by 2-way ANOVA followed by Dunnett’s or Bonferroni’s post hoc test, 1-way ANOVA, or Student’s 2-tailed t test. All results are given as mean ± SEM. P < 0.05 was considered statistically significant.
Study approval.
All procedures were approved by the Institutional Animal Care and Use Committee of the University of Cincinnati and were performed in accordance with the NIH principles of laboratory animal care.
Supplementary Material
Acknowledgments
The authors would like to thank Bruce Spiegelman and C. Ronald Kahn for their kind support and for providing the BAT cell lines.
Footnotes
Conflict of interest: John Speakman holds stock in Swedish Orphan Biovitrum, has received grants from AstraZeneca and Pfizer, and is a founder shareholder in Cambridge Biotechnology. Yu-Hua Tseng receives grant support from Chugai Pharmaceutical Co. D. Perez-Tilve has a collaborative association with F. Hoffmann-La Roche Ltd. pertaining to peptide-based therapeutics in metabolism.
Citation for this article: J Clin Invest. 2013;123(1):469–478. doi:10.1172/JCI64209.
References
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11(4):257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293(2):E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 5.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 6.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 7.Cao W, et al. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol. 2004;24(7):3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kajimura S, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460(7259):1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tseng YH, et al. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat Cell Biol. 2005;7(6):601–611. doi: 10.1038/ncb1259. [DOI] [PubMed] [Google Scholar]
- 10.Tseng YH, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454(7207):1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng YH, Kriauciunas KM, Kokkotou E, Kahn CR. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol. 2004;24(5):1918–1929. doi: 10.1128/MCB.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404(6778):652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 13.Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase C zeta with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci U S A. 1997;94(12):6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18(5):3069–3080. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moscat J, Diaz-Meco MT. To aggregate or not to aggregate? A new role for p62. EMBO Rep. 2009;10(8):804. doi: 10.1038/embor.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137(6):1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moscat J, Diaz-Meco MT. Feedback on fat: p62-mTORC1-autophagy connections. Cell. 2011;147(4):724–727. doi: 10.1016/j.cell.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SJ, et al. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 2010;11(3):226–232. doi: 10.1038/embor.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez A, et al. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3(3):211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 21.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274(1):305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 22.Bothe GW, Haspel JA, Smith CL, Wiener HH, Burden SJ. Selective expression of Cre recombinase in skeletal muscle fibers. Genesis. 2000;26(2):165–166. doi: 10.1002/(SICI)1526-968X(200002)26:2<165::AID-GENE22>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 23.He W, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erbay E, et al. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat Med. 2009;15(12):1383–1391. doi: 10.1038/nm.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4(4):263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tschöp MH, et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2011;9(1):57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121(1):96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen KD, et al. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480(7375):104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bordicchia M, et al. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J Clin Invest. 2012;122(3):1022–1036. doi: 10.1172/JCI59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao W, et al. p38 Mitogen-activated protein kinase plays a stimulatory role in hepatic gluconeogenesis. J Biol Chem. 2005;280(52):42731–42737. doi: 10.1074/jbc.M506223200. [DOI] [PubMed] [Google Scholar]
- 31.Cao W, Medvedev AV, Daniel KW, Collins S. beta-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J Biol Chem. 2001;276(29):27077–27082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- 32.Sudo T, Maruyama M, Osada H. p62 functions as a p38 MAP kinase regulator. Biochem Biophys Res Commun. 2000;269(2):521–525. doi: 10.1006/bbrc.2000.2333. [DOI] [PubMed] [Google Scholar]
- 33.Kawai K, Saito A, Sudo T, Osada H. Specific regulation of cytokine-dependent p38 MAP kinase activation by p62/SQSTM1. J Biochem. 2008;143(6):765–772. doi: 10.1093/jb/mvn027. [DOI] [PubMed] [Google Scholar]
- 34.Saito A, Kawai K, Takayama H, Sudo T, Osada H. Improvement of photoaffinity SPR imaging platform and determination of the binding site of p62/SQSTM1 to p38 MAP kinase. Chem Asian J. 2008;3(8–9):1607–1612. doi: 10.1002/asia.200800099. [DOI] [PubMed] [Google Scholar]
- 35.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27(2):306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bulteau AL, Szweda LI, Friguet B. Mitochondrial protein oxidation and degradation in response to oxidative stress and aging. Exp Gerontol. 2006;41(7):653–657. doi: 10.1016/j.exger.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793(9):1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6(6):e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding WX, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285(36):27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 42.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6(8):1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okatsu K, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15(8):887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.