Abstract
Aberrant expression of the homeodomain transcription factor CDX2 occurs in most cases of acute myeloid leukemia (AML) and promotes leukemogenesis, making CDX2, in principle, an attractive therapeutic target. Conversely, CDX2 acts as a tumor suppressor in colonic epithelium. The effectors mediating the leukemogenic activity of CDX2 and the mechanism underlying its context-dependent properties are poorly characterized, and strategies for interfering with CDX2 function in AML remain elusive. We report data implicating repression of the transcription factor KLF4 as important for the oncogenic activity of CDX2, and demonstrate that CDX2 differentially regulates KLF4 in AML versus colon cancer cells through a mechanism that involves tissue-specific patterns of promoter binding and epigenetic modifications. Furthermore, we identified deregulation of the PPARγ signaling pathway as a feature of CDX2-associated AML and observed that PPARγ agonists derepressed KLF4 and were preferentially toxic to CDX2+ leukemic cells. These data delineate transcriptional programs associated with CDX2 expression in hematopoietic cells, provide insight into the antagonistic duality of CDX2 function in AML versus colon cancer, and suggest reactivation of KLF4 expression, through modulation of PPARγ signaling, as a therapeutic modality in a large proportion of AML patients.
Introduction
The homeodomain transcription factor Cdx2 is involved in the formation and maintenance of the trophectoderm lineage (1, 2) and directs anteroposterior axis definition during embryogenesis through modulation of Hox gene expression (3–5). In addition, several studies have indicated a role for Cdx family members in early hematopoietic development (6–8). In adult mammals, Cdx2 expression is confined to the intestinal epithelium, where it controls intestine-specific gene transcription, cell adhesion, columnar morphogenesis, apoptosis, and proliferation (9, 10).
Increased CDX2 activity is important to the pathogenesis of human hematopoietic malignancies (11, 12). We and others have found that CDX2 is aberrantly expressed in the leukemic cells of most patients with acute myeloid leukemia (AML), but not in hematopoietic stem and progenitor cells (HSPCs) derived from normal individuals (13–15). Suppression of CDX2 inhibits the proliferation of AML cell lines and reduces their clonogenic potential (15). Primary murine HSPCs can be transformed in vitro by Cdx2 and generate fully penetrant and transplantable AML in BM transplant (BMT) recipients, consistent with a causal role for increased CDX2 function in myeloid leukemogenesis (15, 16).
The transcriptional effects that mediate the leukemogenic activity of CDX2 remain incompletely understood. Due to the importance of Cdx proteins in the control of Hox gene expression and hematopoiesis during embryonic development, it has been suggested that CDX2 may contribute to AML pathogenesis through dysregulation of HOX genes, a common feature of AML and one implicated in leukemic transformation (17). In support of this hypothesis, aberrant HOX gene programs have been observed in CDX2+ murine and human leukemias as well as in directly transduced HSPCs (14, 15); however, more direct mechanistic evidence linking CDX2 to perturbed HOX gene expression in AML is currently lacking. Furthermore, Cdx2 is known to regulate a wide range of non-Hox genes during embryonic development and in the gastrointestinal tract (1, 2, 18–21). Therefore, it is likely that additional target genes of CDX2 contribute to its leukemogenic potential.
Consistent with its function in maintaining intestinal cell homeostasis, deregulation of CDX2 has also been implicated in colorectal cancer (CRC) development. There is evidence that CDX2 acts as a tumor suppressor due to its ability to reduce the proliferation of CRC cells and its loss in human colon polyps and cancers (22–24), a view that is also supported by gene targeting studies in mice (25, 26). Interestingly, CDX2 may also have oncogenic activity in the colon and the upper gastrointestinal tract (27–29), which suggests that CDX2 possesses both tumor-promoting and tumor-inhibitory properties. The mechanism underlying these context-dependent effects, and the apparently contradictory roles of CDX2 in AML versus colonic epithelial cells, are not well understood.
The objectives of the present study were to examine the consequences of aberrant CDX2 expression in hematopoietic cells using an unbiased approach, requiring no prior knowledge of the normal functions of CDX2, and to gain insight into the opposing effects of CDX2 in AML and CRC. To delineate essential transcriptional features of CDX2-mediated leukemogenesis, we characterized the gene signatures that are activated in mouse HSPCs in response to Cdx2 expression, in a murine model of Cdx2-induced leukemia, and in human AML associated with aberrant CDX2 expression. We identified and validated silencing of the KLF4 transcription factor as a critical oncogenic function of CDX2 and showed that CDX2 differentially regulated KLF4 expression in AML versus CRC cells through a mechanism involving tissue-specific patterns of promoter binding and epigenetic modifications. Furthermore, we used an in silico chemical genomic approach and a murine model to uncover reactivation of KLF4 expression, through modulation of PPARγ signaling, as a therapeutic modality in CDX2-driven AML.
Results
Derivation of a gene signature associated with Cdx2 expression in murine HSPCs.
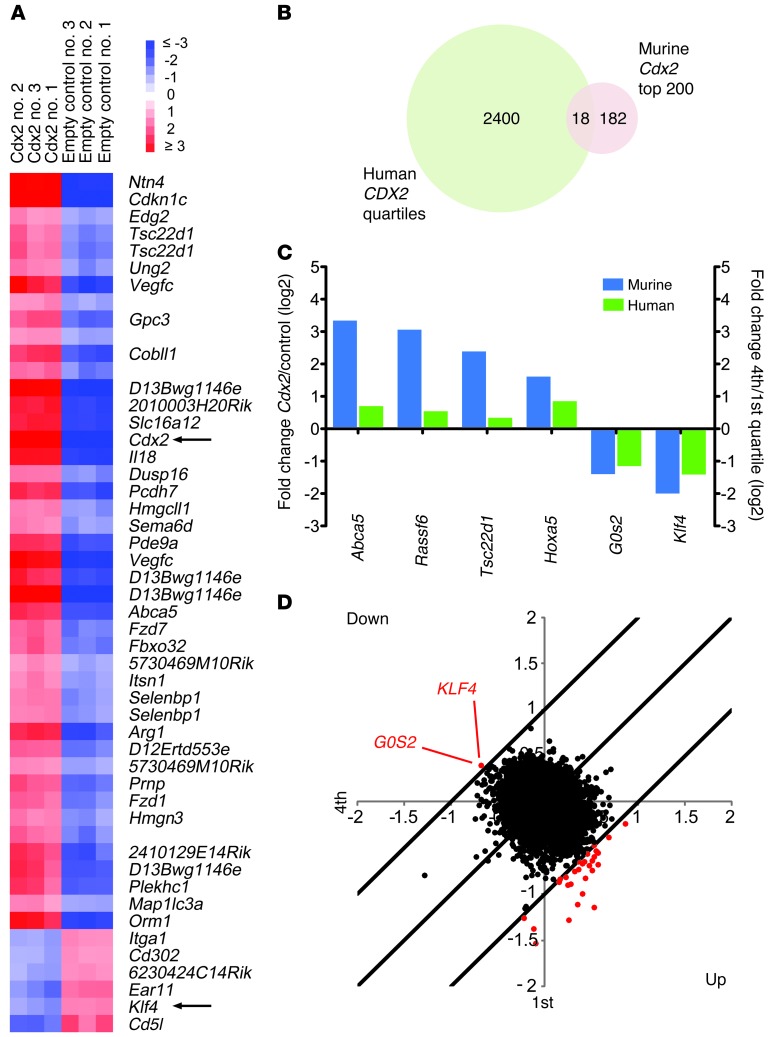
Based on the transforming activity of Cdx2 in mice, we hypothesized that aberrant expression of Cdx2 in murine HSPCs would lead to deregulation of a specific set of transcripts that might be relevant to leukemia development. We transduced c-Kit+Lin– cells from 5-fluorouracil–treated C57BL/6 mice with either a pMSCV-IRES-GFP retroviral construct encoding Cdx2 or an empty control vector and examined gene expression in sorted GFP+ cells from each population after 48 hours using DNA microarrays. Class comparison analysis demonstrated that ectopic Cdx2 expression caused widespread changes in the transcriptome of murine HSPCs. Using a 3-fold cutoff and a P value of 0.001, we found 1,321 genes to be significantly upregulated and 1,385 to be significantly downregulated. The top 50 and 200 deregulated genes are shown in Figure 1A and Supplemental Table 1 (supplemental material available online with this article; doi: 10.1172/JCI64745DS1), respectively. These results showed that Cdx2 induces a distinct pattern of transcriptional changes in immature hematopoietic cells.
Figure 1. Identification of candidate downstream effectors of CDX2 in myeloid leukemogenesis.
(A) Top 50 differentially expressed genes between murine c-Kit+Lin– cells transduced with Cdx2 or an empty control vector. Samples 1–3 indicate biological replicates. (B) Comparison of genes differentially expressed between human AML samples with high (fourth quartile) versus low (first quartile) CDX2 mRNA levels and murine c-Kit+Lin– cells transduced with Cdx2 or an empty control vector. (C) Genes regulated in the same direction between the human and mouse gene expression signatures. (D) Identification of KLF4 and G0S2 outlier expression in human AML samples with high CDX2 mRNA levels using COPA. Outliers at the 95th percentile are indicated in red.
Categorization of human AML based on Cdx2-induced transcriptional changes.
To explore whether the effects induced by Cdx2 in murine HSPCs might be relevant to human leukemogenesis, we evaluated the 200 most highly differentially expressed genes from the murine-derived HSPC Cdx2 signature across the gene expression profiles from 436 unselected AML patients with a wide variety of cytogenetic and molecular abnormalities (available at GEO, accession no. GSE16432). Taking into account probe differences between the human cDNA and mouse oligo-microarray platforms, we found 122 of these 200 genes (represented by 221 clones) to be differentially expressed in the patient cohort. Hierarchical clustering based on this human counterpart of the murine HSPC Cdx2 signature revealed 6 patient groups that differed in their association with karyotypic abnormalities and mutations in the FLT3, NPM1, and CEBPA genes (Supplemental Figure 1), powerful determinants of outcome in patients with AML (30). These data demonstrated that the transcriptional profile driven by Cdx2 in murine HSPCs can be detected in human AML and correlates with clinically relevant markers of disease biology.
Identification of candidate downstream effectors of CDX2 in myeloid leukemogenesis.
To nominate specific genes that possibly mediate the activity of CDX2 in AML, we examined the gene expression profiles of 136 patients in which CDX2 mRNA levels had been measured by quantitative RT-PCR (qRT-PCR) (15). Class comparison analysis of cases in the highest (fourth) versus the lowest (first) quartile of CDX2 expression revealed 2,418 differentially regulated cDNA clones (1,335 upregulated, 1,083 downregulated; Figure 1B and Supplemental Figure 2A).
Given the cytogenetic and molecular heterogeneity of human AML, which was also evident in our patient cohort (Supplemental Figure 2, B and C), it seemed likely that the transcriptional pattern associated with high CDX2 mRNA expression was influenced by factors in addition to CDX2, complicating the identification of critical CDX2 target genes. We therefore performed a cross-species comparison between the patient-derived gene signature associated with high CDX2 mRNA expression and the murine HSPC Cdx2 signature. We observed that 13 genes (represented by 18 probe sets) were shared by the human and mouse signatures (Figure 1B), of which 6 were regulated in the same direction (Figure 1C). In murine HSPCs overexpressing Cdx2 and in AML patients with high CDX2 mRNA expression, 2 genes were repressed more than 2-fold: KLF4, encoding Krüppel-like factor 4 (gut), and G0S2, encoding G0/G1 switch 2.
To corroborate these findings, we used cancer outlier profile analysis (COPA), a bioinformatics approach that was developed to discover oncogenes from transcriptome data based on outlier expression in subsets of cancers (31). Application of COPA to the expression profiles of the 136 human AML cases demonstrated that of the putative CDX2 effectors shown in Figure 1C, only KLF4 and G0S2 were among the top-ranking outliers at the 95th percentile, showing markedly decreased expression in patients with high CDX2 mRNA levels (Figure 1D). These observations, together with previous data showing that KLF4 functions as a tumor suppressor in certain gastrointestinal cancers (32–34) and B cell malignancies (35, 36), suggested that suppression of KLF4 might be one mechanism whereby CDX2 promotes leukemogenesis.
Causal relationship between aberrant CDX2 expression and repression of KLF4 in human myeloid leukemia cells.
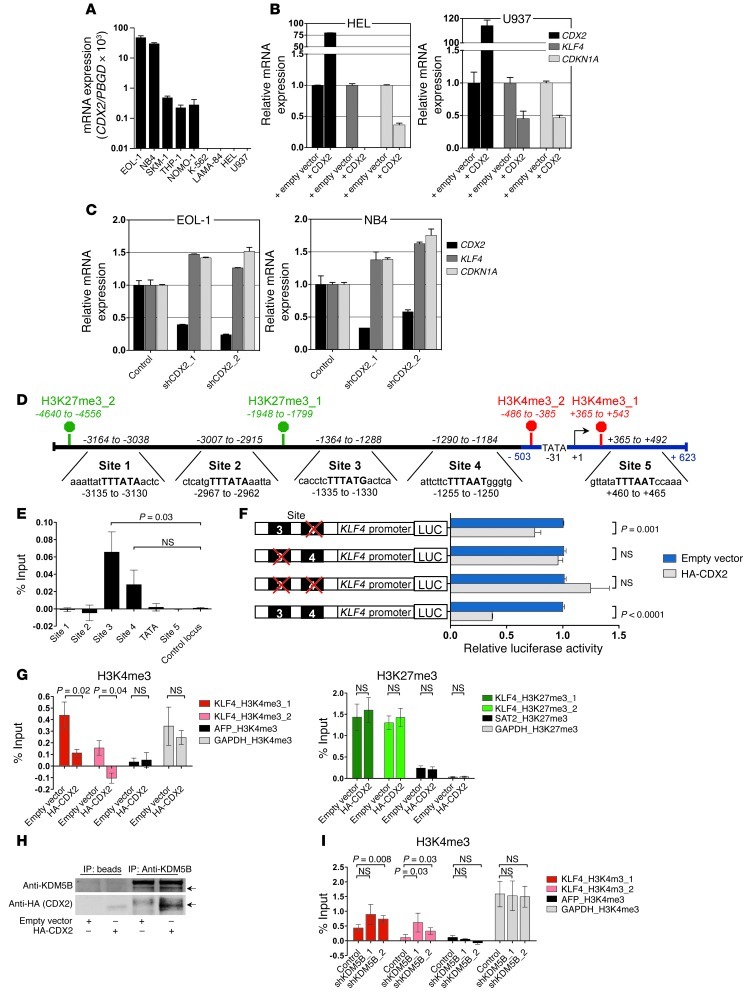
To investigate whether there is a functional link between CDX2 and KLF4 expression, we measured KLF4 mRNA levels by qRT-PCR in response to modulation of CDX2 expression in human myeloid leukemia cell lines (Figure 2A and Supplemental Figure 3A). We first transduced cells that lack CDX2 mRNA with lentiviral vectors encoding full-length CDX2. Confirming the results from the microarray analyses, expression of CDX2 resulted in a decrease in KLF4 transcription that translated into reduced KLF4 protein levels and was accompanied by downregulation of known KLF4 target gene CDKN1A, which encodes cyclin-dependent kinase inhibitor 1A (p21, Cip1) (Figure 2B and Supplemental Figure 3, B and C). Next, we transduced CDX2+ cell lines with lentiviral shRNA constructs targeting CDX2 and observed that stable knockdown of CDX2 led to increased KLF4 and CDKN1A mRNA levels (Figure 2C and Supplemental Figure 3D). These data confirmed our initial observations and supported a causal role for CDX2 in suppression of KLF4 in human myeloid leukemia cells.
Figure 2. Relationship between CDX2 and KLF4 expression and analysis of the KLF4 regulatory region in human myeloid leukemia cell lines.
(A) CDX2 mRNA expression. (B) KLF4 and CDKN1A mRNA levels in response to ectopic CDX2 expression. (C) KLF4 and CDKN1A mRNA levels in response to CDX2 knockdown. (D) Predicted KLF4 promoter (blue) and 4.6 kb upstream of the KLF4 TSS (+1). Possible CDX2 binding sites and the location of the corresponding PCR amplicons (italics) used for ChIP are indicated. The consensus sequence for binding of Cdx proteins is given in bold capital letters. H3K4me3 and H3K27me3 marks, detected in K-562 cells in the ENCODE Histone Modification ChIP-seq project by the Broad Institute, are indicated in red and green, respectively. (E) ChIP showing occupancy of sites 3 and 4 by HA-CDX2 in K-562. GAPDH served as control locus. (F) LUC reporter assays showing that site 3 is required for transcriptional silencing of KLF4 in response to ectopic CDX2 expression in K-562. Mutated sites are indicated by red crosses. (G) ChIP showing reduced H3K4me3 enrichment and unchanged H3K27me3 occupancy in K-562 upon expression of HA-CDX2. AFP and GAPDH served as negative and positive control loci for H3K4me3; GAPDH and SAT2 served as negative and positive control loci for H3K27me3. (H) Co-IP demonstrating interaction of HA-CDX2 with KDM5B in K-562. (I) ChIP showing increased H3K4me3 enrichment in K-562 stably transduced with HA-CDX2 upon KDM5B knockdown. AFP and GAPDH served as negative and positive control loci.
CDX2 binds to the KLF4 regulatory region and causes transcriptional repression of KLF4 in myeloid leukemia cells.
The decrease in KLF4 mRNA associated with aberrant CDX2 expression suggested a direct transcriptional link between CDX2 and KLF4 in myeloid leukemia cells. To investigate this possibility, we examined the KLF4 promoter, as predicted by the MatInspector program ( www.genomatix.de), as well as 3.2 kb of genomic sequence upstream of the KLF4 transcriptional start site (TSS). We identified 5 sites with high homology to the consensus sequence for binding of Cdx proteins, TTTATA/G (Figure 2D and refs. 37, 38). To determine whether CDX2 binds to any of these sites in vivo, we performed ChIP experiments using K-562 cells stably transduced with HA-tagged CDX2 (HA-CDX2). Incubation of whole-cell lysates with an anti-HA Ab followed by PCR-based quantification of IP genomic DNA fragments showed that CDX2 bound to a segment of the KLF4 regulatory region ranging from nt –1,364 to –1,288 (site 3; Figure 2E). A second segment, ranging from nt –1,290 to –1,184 (site 4), was also enriched compared with a control locus (GAPDH), but this difference did not reach statistical significance (Figure 2E). Reporter assays in K-562 cells showed that a segment of the KLF4 regulatory region containing sites 3 and 4 was able to repress luciferase (LUC) activity when cotransfected with CDX2. Mutation analyses demonstrated that disruption of site 3, but not site 4, abolished the ability of CDX2 to repress transcription (Figure 2F).
To further confirm the role of CDX2 in mediating repression of KLF4, we examined the effects of aberrant CDX2 expression on the distribution of histone modifications in the KLF4 regulatory region in myeloid leukemia cells. We performed ChIP-qPCR experiments in K-562 cells transduced with HA-CDX2 or an empty control vector, focusing on domains enriched for trimethylated H3K4 (H3K4me3; associated with active transcription) or trimethylated H3K27 (H3K27me3; associated with repressed transcription) that have been mapped previously in K-562 cells to a genomic region upstream of KLF4 (Figure 2D and Supplemental Figure 3E). A consistent feature associated with exogenous CDX2 expression was the loss of H3K4me3 enrichment around the KLF4 promoter, whereas H3K27me3 occupancy remained unaffected (Figure 2G). CDX2 has been shown to directly bind to the H3K4 demethylase KDM5B (also known as JARID1B; ref. 39). We verified the interaction between CDX2 and KDM5B in K-562 cells (Figure 2H) and observed that depletion of KDM5B using lentiviral shRNA constructs reverted the decrease in H3K4me3 enrichment associated with exogenous CDX2 expression (Figure 2I and Supplemental Figure 3F). Collectively, these data showed that in myeloid leukemia cells, CDX2 occupies a distinct sequence motif in the KLF4 regulatory region to repress KLF4 transcription via epigenetic silencing that involves aberrant H3K4 demethylation by KDM5B.
Growth-inhibitory effect of KLF4 in CDX2+ myeloid leukemia cells.
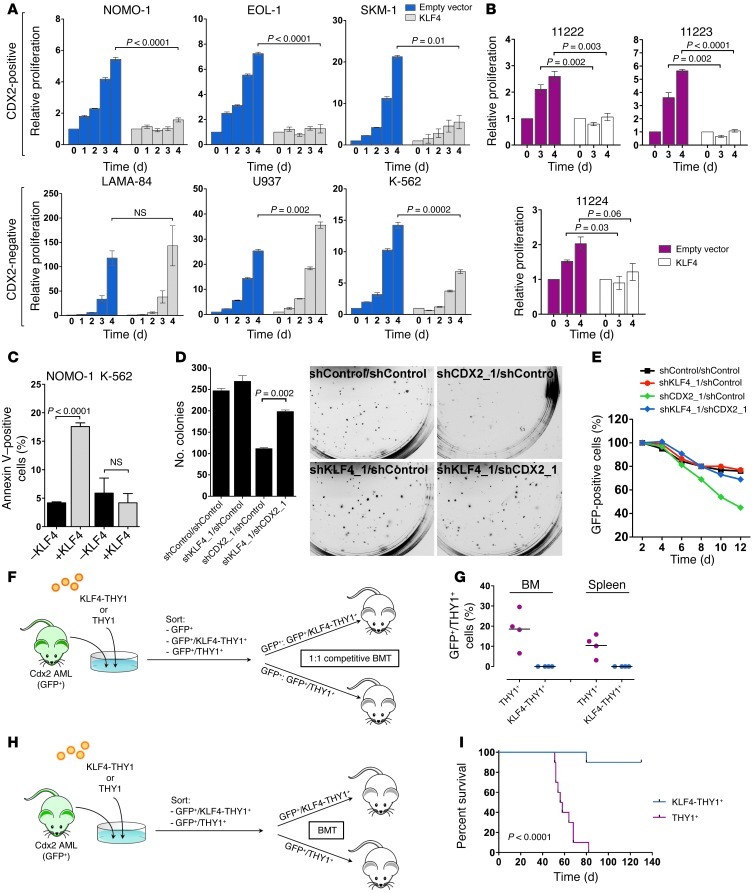
To investigate the functional role of KLF4 in leukemias driven by CDX2, we transduced myeloid leukemia cell lines with a lentiviral vector encoding KLF4 or an empty control vector (Supplemental Figure 4A). Whereas KLF4 inhibited the viability and proliferation of CDX2+ cells, it had no effect or only a marginal one on CDX2– cells (Figure 3A). Likewise, the growth and survival of early-passage cell lines derived from different murine leukemias induced by Cdx2 in a previously described BMT model (15) was also significantly decreased in response to KLF4 expression (Figure 3B).
Figure 3. Growth-inhibitory effect of KLF4 in CDX2+ human myeloid leukemia cell lines and murine AML.
(A) Effects of KLF4 on viability and proliferation of human myeloid leukemia cell lines. (B) Effects of KLF4 on viability and proliferation of early-passage cell lines derived from Cdx2-induced murine leukemias. (C) Apoptosis induction by KLF4 in CDX2+ NOMO-1 cells, but not in CDX2– K-562 cells. (D) Colony formation of AML cells in response to CDX2 and KLF4 knockdown. CDX2+ NOMO-1 cells were stably transduced with combinations of shRNAs and plated in methylcellulose. Knockdown of CDX2 (shCDX2_1/shControl) reduced the number of colonies. Colony formation was partially rescued by concomitant knockdown of KLF4 (shKLF4_1/shCDX2_1). Representative photomicrographs of methylcellulose cultures are shown. Original magnification, ×4.7. (E) CDX2+ NOMO-1 cells stably transduced with an shRNA targeting KLF4 or a nontargeting control shRNA were transduced with a lentiviral vector coexpressing GFP with an shRNA targeting CDX2 or a nontargeting control shRNA. The GFP+ fraction was measured by flow cytometry at the indicated time points. Knockdown of CDX2 alone (shCDX2_1/shControl) depleted GFP+ cells over time. The proportion of GFP+ cells was rescued by concomitant knockdown of KLF4 (shKLF4_1/shCDX2_1). (F) Competitive BMT experiments (n = 4 per group). (G) Proportion of GFP+THY1+ cells in the BM and spleen of mice in F. (H) Noncompetitive BMT experiments (n = 10 per group). (I) Survival of mice in H.
To examine the mechanisms behind this effect, we analyzed apoptosis induction and cell cycle progression in myeloid leukemia cell lines after lentiviral transduction with KLF4. Annexin V staining showed that exogenous KLF4 significantly increased apoptosis in CDX2+ cells, but not CDX2– cells (Figure 3C). Furthermore, flow cytometric analysis of 5-ethynyl-2′-deoxyuridine (EdU) incorporation demonstrated that CDX2+ cells accumulated in the G2/M phase of the cell cycle, whereas CDX2– cells were largely unaffected (Supplemental Figure 4B). These results suggested that KLF4 counteracts the effects of CDX2 in myeloid cells by inducing cell cycle arrest and apoptosis.
To further confirm that the oncogenic effects of CDX2 are mediated through silencing of KLF4, we performed rescue experiments in which we evaluated the effect of suppressing CDX2 in the context of blocked KLF4 expression. We stably transduced CDX2+ NOMO-1 cells with shRNA constructs targeting KLF4 or a nontargeting control shRNA, followed by shRNA knockdown of CDX2. As expected, CDX2 suppression decreased colony formation in methylcellulose of NOMO-1 cells with unimpaired KLF4 expression. In contrast, the effect of CDX2 knockdown was attenuated by prior knockdown of KLF4 (Figure 3D and Supplemental Figure 4, C and E). To extend these results, we introduced a GFP-labeled lentiviral vector that coexpresses an shRNA targeting CDX2 into NOMO-1 cells and monitored viable cells by flow cytometry. Consistent with the colony formation assays, knockdown of CDX2 in the setting of intact KLF4 expression decreased the proportion of GFP+ cells over time, indicative of a toxic effect of CDX2 depletion. In contrast, CDX2 knockdown in the context of blocked KLF4 expression did not result in loss of GFP+ cells (Figure 3E and Supplemental Figure 4D), which indicates that restoration of KLF4 expression is requisite for the antiproliferative effect of CDX2 knockdown.
To corroborate these findings in vivo, we performed competitive BMT with equal numbers of GFP+ leukemic cells derived from Cdx2-induced murine leukemias that were transduced with a retroviral vector expressing KLF4 or an empty control vector (Figure 3F). All mice died of AML within 33–40 days (median, 37.8 d). Flow cytometric quantification of the relative contribution of the different cell populations showed that cells expressing KLF4 were outcompeted by cells transduced with the control vector, as they were undetectable in the BM and spleen of diseased mice (Figure 3G). In a second approach, we transplanted Cdx2+ leukemic cells transduced with KLF4 or empty vector in a noncompetitive fashion (Figure 3H). These experiments showed that only 1 of the mice that received KLF4-expressing cells died, whereas all control mice succumbed to an aggressive leukemia after a median latency of 55 days, resulting in a statistically significant difference in survival (Figure 3I). Together, these observations substantiated the concept that KLF4 inhibits AML development and is transcriptionally silenced in response to aberrant CDX2 expression.
Reversal of CDX2-induced transcriptional changes by PPARγ agonist treatment.
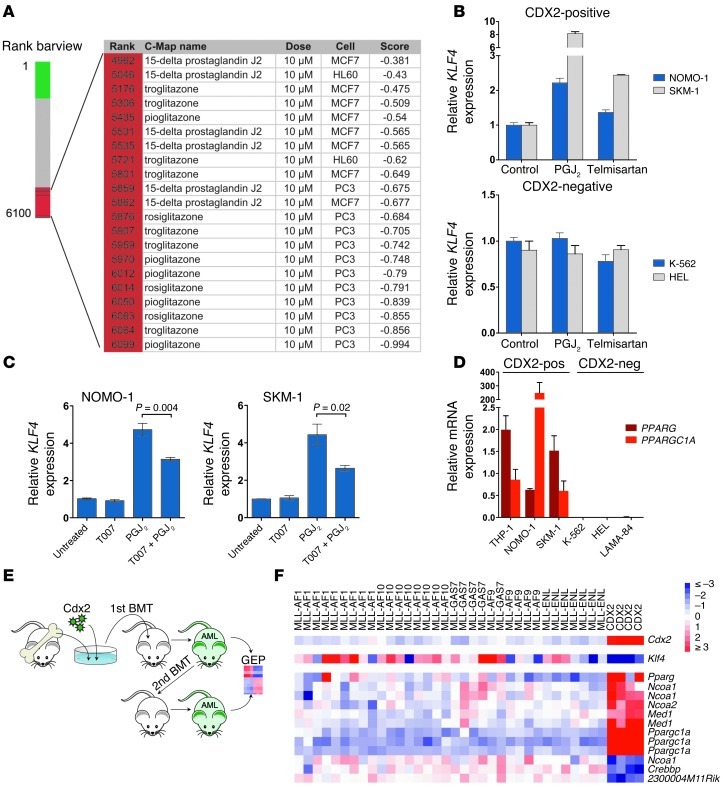
Given that CDX2 is aberrantly expressed in most AML patients and promotes leukemogenesis, interfering with CDX2 or its downstream effectors represents an attractive therapeutic strategy. However, targeting transcription factors is challenging and has not yet been clinically realized. Because of these limitations, we used the Connectivity Map (C-Map), a repository of gene expression profiles from human cell lines treated with a large number of small bioactive molecules (40), to identify modulators of CDX2-induced transcriptional changes. Specifically, we used the human counterpart of the gene signature associated with ectopic expression of Cdx2 in murine HSPCs to query the C-Map for compounds whose activity is linked negatively to this signature. Among the perturbagens with negative enrichment scores (indicative of compounds that may repress the biological state encoded in the Cdx2 signature) were several drugs that stimulate the nuclear receptor PPARγ, a ligand-activated transcription factor involved in the regulation of lipid and glucose homeostasis (41). These drugs included 15-deoxy-Δ12,14-prostaglandin J2 (PGJ2), a natural PPARγ ligand, and 3 thiazolidinediones (rosiglitazone, pioglitazone, and troglitazone), a class of antidiabetic drugs that improve insulin sensitivity (Figure 4A).
Figure 4. Reversal of CDX2-induced transcriptional changes by PPARγ agonist treatment, and deregulated PPARγ signaling in CDX2-driven AML.
(A) C-Map–based identification of PPARγ agonists among small bioactive molecules negatively linked to a gene signature associated with Cdx2 expression in murine c-Kit+Lin– HSPCs. The red region in the bar plot indicates compounds with a negative enrichment score. Molecules are ordered according to decreasing similarity to Cdx2. (B) Treatment with 5 μM PGJ2 or 50 μM telmisartan for 24 hours increased KLF4 mRNA expression in CDX2+ NOMO-1 and SKM-1 cells, but not in CDX2– K-562 and HEL cells. (C) Pretreatment with 2 μM T0070907 for 2 hours partially reversed the effect of PGJ2 on KLF4 mRNA expression in NOMO-1 and SKM-1 cells. (D) PPARG and PPARGC1A mRNA expression in human myeloid leukemia cell lines. (E) BMT experiments. GEP, gene expression profiling. (F) Relative differential expression of components of BioCarta Pathway “Role of PPAR-gamma Coactivators in Obesity and Thermogenesis” between Cdx2 and MLL murine leukemias. Ppargc1a was the top differentially expressed gene, showing a 30-fold upregulation in Cdx2-induced AML. In addition, overexpression of Cdx2 and repression of Klf4 in Cdx2 leukemias are shown.
In addition to its metabolic actions, PPARγ signaling has also been implicated in the control of cell proliferation, survival, and differentiation. Consistent with these functions, PPARγ ligands inhibit proliferation and induce apoptosis or differentiation in various epithelial and hematologic cancer cell lines and animal models, yet the underlying mechanisms remain incompletely understood (42). In CRC cell lines, PPARγ agonists have been shown to increase KLF4 expression (43–46), which suggests that the tumor-suppressive properties of these drugs may be linked to their ability to upregulate KLF4. Based on our observations that PGJ2 and thiazolidinediones inhibited the gene signature associated with aberrant Cdx2 expression and that suppression of KLF4 was a functionally relevant component of this signature, we reasoned that PPARγ agonists might exert tumor-suppressive effects in AML cells through induction of KLF4.
In support of this hypothesis, treatment of CDX2+ AML cells with 5 μM of the PPARγ agonist PGJ2 for 24 hours led to increased KLF4 mRNA levels (Figure 4B), an effect that was mitigated by a PPARγ antagonist (Figure 4C). To further substantiate that PPARγ agonist treatment leads to induction of KLF4 in CDX2+ AML cells, we also analyzed the effects of telmisartan, an angiotensin II receptor antagonist used for the treatment of hypertension that can function as a partial PPARγ agonist (47). Treatment of CDX2+ cells with 50 μM telmisartan for 24 hours also significantly increased KLF4 transcription, albeit to a lesser extent than did PGJ2. In contrast, PGJ2 or telmisartan treatment were not associated with increased KLF4 mRNA expression in CDX2– cells (Figure 4B).
Upregulation of PPARγ signaling components in CDX2-driven AML.
PPARγ ligands regulate transcription through binding to their cognate receptor, which then undergoes specific conformational changes that allow for coactivator protein recruitment (48). We therefore speculated that the differential effects of PPARγ agonist treatment depending on CDX2 status could be explained by differences in the abundance of PPARγ and its coactivators. Consistent with this hypothesis, qRT-PCR analysis identified PPARG (encoding PPARγ) and PPARGC1A (encoding PPARγ coactivator 1α) transcripts in CDX2+ AML cell lines, whereas CDX2– cells showed no PPARG and PPARGC1A mRNA expression (Figure 4D).
To validate the link among aberrant CDX2 expression, silencing of KLF4, and altered PPARγ signaling in vivo, we generated retrovirally expressed, Cdx2-induced AML in a murine BMT model. We hybridized RNA from spleen cells of diseased primary or secondary transplant recipients to DNA microarrays and compared the resulting gene expression profiles with those of leukemias initiated by 5 different MLL fusion oncogenes that were generated previously using the same experimental strategy (available at GEO, accession no. GSE13690; Figure 4E; and ref. 49). Class comparison analysis identified 9,757 genes that were differentially expressed between leukemias driven by Cdx2 or MLL fusion proteins; the top 200 differentially regulated genes are shown in Supplemental Table 2. We then examined the expression of individual genes in the different datasets and observed that Klf4 was significantly downregulated in Cdx2-induced AML, consistent with our findings in murine c-Kit+Lin– cells and human AML (Figure 4F). These data confirmed that suppression of Klf4 is a characteristic component of the transcriptional program associated with the leukemogenic activity of Cdx2.
Strikingly, gene set enrichment analysis identified the BioCarta Pathway “Role of PPAR-gamma Coactivators in Obesity and Thermogenesis” as the top differentially expressed pathway between Cdx2 and MLL leukemias (Supplemental Table 3). Several components of this pathway were also significantly altered at the single-gene level. For example, Cdx2-driven leukemias were associated with upregulation of Pparg as well as very high expression of Ppargc1a, the top differentially expressed gene between Cdx2 and MLL leukemias (Figure 4F and Supplemental Table 2). These data supported induction of PPARγ signaling components as a mechanism through which CDX2 renders myeloid leukemia cells sensitive to PPARγ agonist treatment.
Functional consequences of PPARγ agonist treatment in CDX2+ myeloid leukemia cell lines.
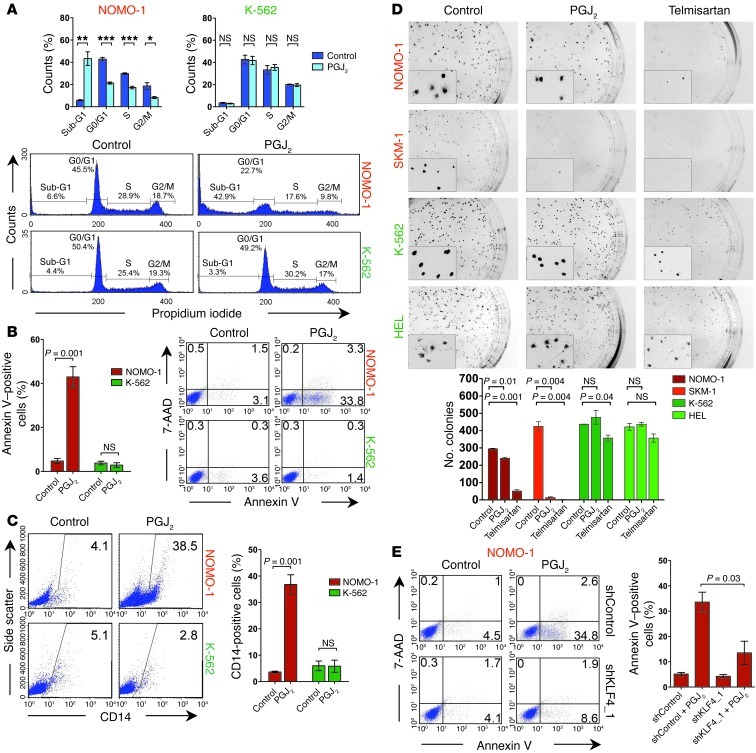
We next investigated whether the transcriptional changes induced by PPARγ agonists translate into a tumor-suppressive effect. In CDX2+ myeloid leukemia cells, PGJ2 treatment induced apoptosis (as evidenced by an increase in annexin V+ cells and a shift to the sub-G1 phase of the cell cycle) as well as myelomonocytic differentiation (as indicated by increased CD14 expression), consistent with previous observations that KLF4 promotes monocyte development (50). In contrast, CDX2– cell lines were largely unaffected by PGJ2 (Figure 5, A–C, and Supplemental Figure 5, A–C). We also tested the effect of PPARγ agonists on the ability of AML cell lines to form colonies in clonogenic methylcellulose assays. For CDX2+ cell lines, we observed a significant reduction in the number of colonies in response to PGJ2 or telmisartan. In contrast, these compounds only marginally decreased colony numbers of CDX2– cells (Figure 5D). Notably, telmisartan reduced colony size in all cell lines (Figure 5D), which indicates that this drug has additional antiproliferative properties that are independent of CDX2 status. To verify that the effects of PPARγ agonist treatment were KLF4 dependent, we incubated CDX2+ NOMO-1 cells with PGJ2 after shRNA knockdown of KLF4 and observed that apoptosis induction by this compound was blunted in the context of impaired KLF4 expression (Figure 5E). Together, these results indicated that PPARγ agonists counteract the effects of aberrant CDX2 expression in HSPCs and suggested upregulation of KLF4 as one mechanism whereby PPARγ agonists inhibit the survival and clonogenic potential of human AML cells.
Figure 5. Effects of PPARγ agonists in CDX2+ human myeloid leukemia cell lines.
(A) Treatment with 5 μM PGJ2 for 48 hours increased the proportion of cells in the sub-G1 phase of the cell cycle, with concomitant loss of cycling cells, in CDX2+ NOMO-1 cells, but not in CDX2– K-562 cells. *P < 0.05; **P < 0.01; ***P < 0.001. (B) Treatment with 5 μM PGJ2 for 48 hours increased the proportion of annexin V+ cells in NOMO-1, but not K-562, cells. Numbers indicate percentages of cells. (C) Treatment with 5 μM PGJ2 for 72 hours induced monocytic differentiation in NOMO-1, but not K-562, cells. Numbers indicate percentages of cells. (D) Treatment with 5 μM PGJ2 or 50 μM telmisartan reduced the number of colonies of CDX2+ NOMO-1 and SKM-1 cells, but not CDX2– K-562 and HEL cells. Representative photomicrographs of methylcellulose cultures are shown. Original magnification, ×4.7. Insets show that telmisartan decreased colony size in all 4 cell lines. Original magnification, ×16. (E) NOMO-1 cells stably transduced with an shRNA targeting KLF4 or a nontargeting control shRNA were treated with 5 μM PGJ2 for 48 hours. PGJ2 increased the proportion of annexin V+ cells. Cell viability was rescued by concomitant knockdown of KLF4 (shKLF4_1/PGJ2). Numbers indicate percentages of cells.
Functional consequences of PPARγ agonist treatment in CDX2+ primary human and murine myeloid leukemias.
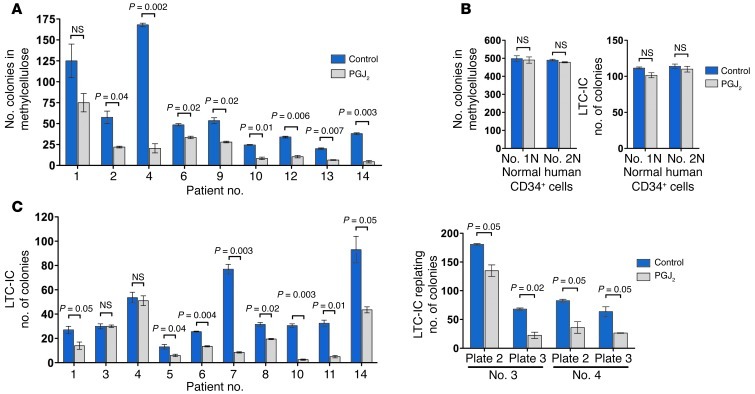
We next sought confirmation in primary human and murine AML. Consistent with the results in myeloid leukemia cell lines, PGJ2 treatment impaired colony formation in methylcellulose of BM mononuclear cells from patients with CDX2+ AML (Figure 6A and Supplemental Table 4). In contrast, PGJ2 had no effect on colony formation of CD34+ cells derived from the BM of healthy individuals (Figure 6B, left). Long-term culture–initiating cell (LTC-IC) assays with stromal cell support for 5 weeks demonstrated that PGJ2 treatment of CDX2+ primary human AML specimens decreased LTC-IC yield in 8 of 10 samples (Figure 6C, left). Subsequent replating of the 2 unaffected LTC-IC cultures in methylcellulose demonstrated reduced colony formation (Figure 6C, right). In contrast, PGJ2 did not reduce LTC-IC frequency in normal CD34+ cells (Figure 6B, right).
Figure 6. Effects of PPARγ agonist treatment in primary human BM samples.
(A) Treatment with 5 μM PGJ2 reduced the number of colonies of CDX2+ AML samples plated in methylcellulose. (B) Treatment with 5 μM PGJ2 had no effect on normal CD34+ cells with respect to colony formation and LTC-IC assays. (C) Treatment with 5 μM PGJ2 reduced LTC-IC frequency in 8 of 10 CDX2+ AML samples (left). PGJ2 decreased the serial replating activity of LTC-IC derived from samples 3 and 4 in secondary and tertiary colony formation assays (right).
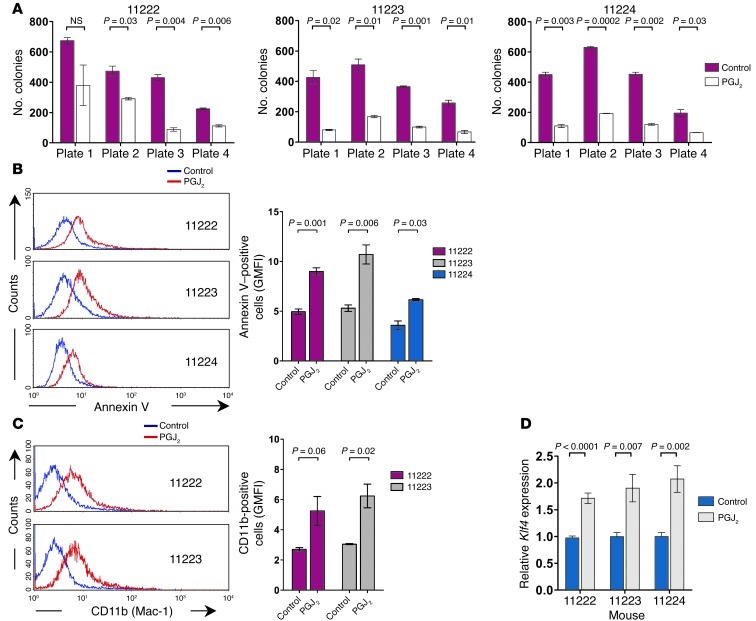
In addition, we investigated the effects of PPARγ agonist treatment in a previously established mouse model of Cdx2-driven AML (15). Incubation of the leukemic cells from different Cdx2-induced murine leukemias with PGJ2 decreased their clonogenic potential in serial replating assays (Figure 7A), promoted apoptosis (Figure 7B), and induced the expression of CD11b, a cell surface marker associated with myelomonocytic differentiation (Figure 7C). As predicted by our findings in human myeloid leukemia cell lines, PPARγ agonist treatment was associated with upregulation of Klf4 mRNA expression (Figure 7D). Together, these data validated our findings in myeloid leukemia cell lines and demonstrated the antileukemic activity of PPARγ agonist treatment in primary CDX2+ AML.
Figure 7. Effects of PPARγ agonist treatment in Cdx2-driven murine leukemias.
(A) Treatment with 5 μM PGJ2 reduced the number of colonies of Cdx2+ murine leukemic cells serially plated in methylcellulose. (B) Treatment with 5 μM PGJ2 for 48 hours induced apoptosis in early-passage cell lines derived from Cdx2-induced murine leukemias. Annexin V staining was analyzed in GFP+ cells. GMFI, geometric mean fluorescence intensity. (C) Treatment with 5 μM PGJ2 for 5 days induced monocytic differentiation of cells from Cdx2-induced murine leukemias. CD11b (Mac-1) surface expression was analyzed in GFP+ cells. (D) Treatment with 5 μM PGJ2 for 24 hours increased Klf4 mRNA expression in early-passage cell lines derived from Cdx2-induced murine leukemias.
Antagonistic regulation of KLF4 expression by CDX2 in CRC versus AML cells.
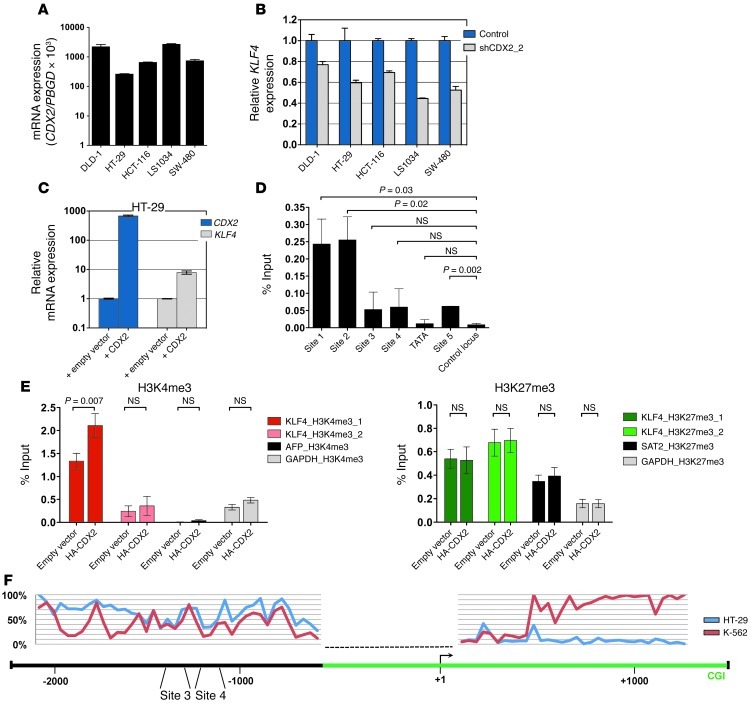
Previous studies have shown that CDX2 acts as a tumor suppressor in the colon and rectum (23–26), a finding diametrically opposed to its function in AML. To investigate whether these context-dependent roles could be explained by opposing effects on KLF4 transcription, we modulated CDX2 expression in human CRC lines and measured KLF4 mRNA levels by qRT-PCR. Depletion of CDX2 by shRNA-mediated knockdown caused a decrease in KLF4 mRNA expression (Figure 8, A and B, and Supplemental Figure 6, A–C). Conversely, overexpression of CDX2 in HT-29, a cell line with low endogenous CDX2 levels, resulted in upregulation of KLF4 mRNA (Figure 8C), consistent with previous data showing that CDX2 activates the KLF4 promoter in CRC cells (51).
Figure 8. Relationship between CDX2 and KLF4 expression and analysis of the KLF4 regulatory region in human CRC cells.
(A) CDX2 mRNA expression of CRC cell lines. (B) Decreased KLF4 mRNA levels in response to CDX2 knockdown in human CRC cell lines. (C) Increased KLF4 mRNA levels in response to ectopic CDX2 expression in HT-29 cells. (D) ChIP showing occupancy of sites 1, 2 and (to a lesser extent) 5 in the KLF4 regulatory region by HA-CDX2 in HT-29 cells. The location of possible CDX2 binding sites is shown in Figure 2D. GAPDH served as control locus. (E) ChIP showing increased H3K4me3 enrichment and unchanged H3K27me3 occupancy in HT-29 cells upon expression of HA-CDX2. The location of the H3K4me3 and H3K27me3 marks is shown in Figure 2D. AFP and GAPDH served as negative and positive control loci for H3K4me3; GAPDH and SAT2 served as negative and positive control loci for H3K27me3. (F) Quantitative DNA methylation levels at the KLF4 locus in HT-29 cells and K-562 myeloid leukemia cells. The KLF4 TSS (+1) and CDX2 binding sites 3 and 4 are marked, and their relative positions are given. The green bar depicts a CpG island (CGI). The dashed line indicates a region where DNA methylation could not be assessed.
To corroborate these findings, we performed ChIP experiments in HT-29 cells stably expressing HA-CDX2 and observed that exogenous CDX2 associated with 2 genomic regions upstream of KLF4 (sites 1 and 2) that differed from those occupied by CDX2 in myeloid leukemia cells (Figure 2E and Figure 8D). In addition, overexpression of CDX2 in HT-29 cells resulted in an increase in H3K4me3 near the KLF4 TSS, whereas H3K27m3 levels were unchanged (Figure 8E).
To further characterize the mechanism underlying tissue-specific regulation of KLF4 transcription by CDX2, we performed quantitative DNA methylation assessment 2.1 kb upstream and 1.3 kb downstream of the KLF4 TSS using MassARRAY technology (Figure 8F). In total, we interrogated 109 CpG dinucleotides located in 72 addressable CpG units. These experiments revealed prominent differences in DNA methylation levels in an upstream region of intermediate CpG density (nt –1,500 to –2,000), in close vicinity to CDX2 binding sites 3 and 4 (–1,364 to –1,288 and –1,290 to –1,184, respectively) that are occupied by CDX2 in myeloid leukemia cells. While HT-29 CRC cells were almost completely methylated in this region (>75%), K-562 cells exhibited 2 distinct areas of markedly decreased DNA methylation (∼20%). DNA methylation levels at the CpG island near the TSS were low for both cell lines. Further downstream (+0.5 to +1.2 kb, covering the exon 2–intron 2 region), striking DNA methylation differences were again observed: K-562 cells were almost completely methylated, whereas HT-29 cells were virtually unmethylated.
This comparative analysis of the interaction between CDX2 and the KLF4 regulatory region in AML and CRC cells indicated that the divergent properties of CDX2 can be accounted for, at least in part, by differential regulation of KLF4 expression through a mechanism that involves tissue-specific patterns of promoter binding and epigenetic modifications at the KLF4 5′ locus.
Discussion
AML is a genetically and biologically heterogeneous disease. As a consequence, developing broadly applicable targeted AML therapies is challenging. We and others have previously shown that different leukemogenic pathways converge on the homeodomain transcription factor CDX2, which is aberrantly expressed in the leukemic cells of approximately 90% of AML patients (14, 15). Characterization of CDX2 effectors is therefore likely to yield new insights into myeloid leukemogenesis and may uncover common targets for therapy. However, little is known about the consequences of aberrant CDX2 expression in hematopoietic cells (12). Whereas the global gene expression patterns regulated by CDX2 have been determined in embryonic stem cells, intestinal cells, and human CRC cells (9, 19, 21, 52, 53), previous studies of murine and human AML were limited to selected HOX genes that are known transcriptional targets of CDX2 during development (14–16).
We report here the transcriptome changes associated with Cdx2 expression in immature hematopoietic cells, CDX2+ human AML, and Cdx2-driven murine leukemia. To determine the contributions of individual CDX2 target genes to AML development, we focused on KLF4, which was consistently deregulated in the context of aberrant CDX2 expression. There is evidence that KLF4, which encodes a zinc finger transcription factor, can function both as a tumor suppressor and as an oncogene (54). In support of the former, the KLF4 locus is deleted or methylated in human colon and gastric cancers (33, 34), and tissue-specific deletion of Klf4 in mice results in gastric hyperplasia and polyps (32). Furthermore, KLF4 has been identified as a candidate tumor suppressor in classical Hodgkin lymphoma, B cell non-Hodgkin lymphoma, and a mouse model of pre–B cell acute lymphoblastic leukemia (35, 36). On the other hand, KLF4 is often overexpressed in human breast and squamous cell carcinomas and can contribute to oncogenic transformation of cultured cells by mutant RAS (55–57).
Our findings support the view that in the myeloid compartment, KLF4 possesses growth-inhibitory properties: first, KLF4 was repressed in mouse HSPCs following Cdx2 expression ex vivo, in human AML associated with aberrant CDX2 expression, and in Cdx2-driven murine leukemias; second, ectopic expression of KLF4 induced cell cycle arrest and apoptosis in CDX2+ myeloid leukemia cells; third, KLF4 was required for the toxic effect of CDX2 knockdown on AML cells; and fourth, KLF4 blocked the development of AML induced by Cdx2 in a murine model. Thus, given the high prevalence of aberrant CDX2 expression, our data uncover silencing of KLF4 as a common event in AML pathogenesis, although it is likely that CDX2 exerts its leukemogenic effect through additional, as yet unknown mechanisms. Another recent study identified downregulation of KLF4 as a consequence of the JAK2V617F mutation in polycythemia vera (PV) (58). This observation may point to a more general role for reduced KLF4 activity in myeloid leukemogenesis; however, the functional significance of KLF4 repression in PV remains to be determined.
Our findings also provide a mechanistic explanation for the opposing effects of CDX2 in AML and CRC. We observed that KLF4 expression was decreased by CDX2 in myeloid cells, resulting in enhanced survival and proliferation. In contrast, KLF4 was positively regulated by CDX2 in CRC cells, thereby mediating the growth-inhibitory function of APC, a critical inhibitor of Wnt signaling that is commonly inactivated in CRC (22, 51), and possibly other tumor suppressors for colorectal neoplasia. Consistent with these tissue-specific properties, we found that CDX2 bound to distinct sites in the KLF4 regulatory region in AML versus CRC cells, which engendered antagonistic effects on histone modification patterns at the KLF4 promoter that translated into opposing effects on KLF4 expression. DNA methylation analysis indicated that the mechanism underlying context-dependent accessibility of the KLF4 regulatory region likely involves fundamental epigenetic differences between hematopoietic and colonic epithelial cells at the KLF4 5′ locus. Notably, the upstream differentially methylated sequences identified are not located in a CpG island region, but in its less CpG-dense direct neighborhood. Such regions, referred to as “CpG island shores,” have previously been shown to exhibit substantial plasticity of DNA methylation in different tissue and cancer types (59). Therefore, the observed cell type–specific DNA methylation patterns could influence CDX2 occupancy either directly by differential binding affinity or via indirect mechanisms, such as locally altered chromatin conformation, a question that requires further investigation. Another important question to be addressed in the future is how differential binding of CDX2 directs the changes in the chromatin state of KLF4. In any case, our present findings reinforce that CDX2 may be added to the growing list of cancer genes that can affect tumorigenesis both positively and negatively, such as RUNX family members, WT1, TGFB1, and NOTCH1 (60–63).
The leukemogenic activity of CDX2, and its absence in normal hematopoietic progenitors, indicate that blocking the effects of CDX2 may provide a therapeutic window in patients with AML. Direct inhibition of CDX2 is currently not possible; however, our results point to events downstream of CDX2 that could serve as novel targets for treatment. We observed that introduction of KLF4 was selectively toxic to CDX2+ myeloid leukemia cell lines, which indicates that reactivation of KLF4 could be of therapeutic value in AML driven by CDX2. Although reconstituting the function of genes whose expression is lost during carcinogenesis is challenging, there are drugs under development that inhibit tumor growth by inducing KLF4 through a mechanism that involves intracellular zinc depletion (64). While these compounds are primarily being tested in solid tumors, our results suggest that evaluation of their therapeutic utility may also be warranted in myeloid leukemias.
As an alternative approach to devise potential CDX2-directed therapies, we used a chemical genomic strategy based on the C-Map (40), which identified several agonists of the nuclear receptor PPARγ that were negatively correlated with our 200-gene HSPC Cdx2 signature, suggesting that these compounds might antagonize the leukemogenic activity of CDX2. In support of this hypothesis, PPARγ agonist treatment reversed a central feature of the Cdx2 signature, repression of KLF4, and exerted a multifaceted tumor-suppressive effect in CDX2+ myeloid leukemia cell lines and primary human AML samples and in Cdx2-induced murine leukemias, whereas CDX2– cells were unaffected. The unexpected link between CDX2 and altered PPARγ signaling was also demonstrated by the observation that components of the PPARγ pathway, including PPARγ itself, were overexpressed in Cdx2-driven murine leukemias and CDX2+ human AML cells.
Collectively, these findings suggest a model in which aberrant expression of CDX2 in HSPCs causes silencing of KLF4, which in turn leads to upregulation of PPARγ signaling intermediates, thereby sensitizing the leukemic cells to PPARγ agonist treatment (Figure 9). Given the known relationship between expression of CDX2 and induction of KLF4 in colonic epithelial cells, a similar mechanism may be operative in CRC cells, which are commonly characterized by a loss of CDX2 function. Our observation that deregulation of PPARγ signaling was present in Cdx2-driven murine AML and in established CDX2+ human cell lines, but not in murine HSPCs acutely transduced with Cdx2, argues against the possibility that the increased transcription of components of the PPARγ pathway in CDX2+ leukemic cells is due to direct induction by CDX2. Instead, we reason that altered PPARγ signaling reflects more complex, multilayered changes in gene expression that occur gradually as HSPCs adapt to the presence of CDX2. However, further studies are required to characterize the entire molecular circuitry involving KLF4 suppression and deregulated PPARγ signaling in different tissues characterized by perturbation of CDX2 function.
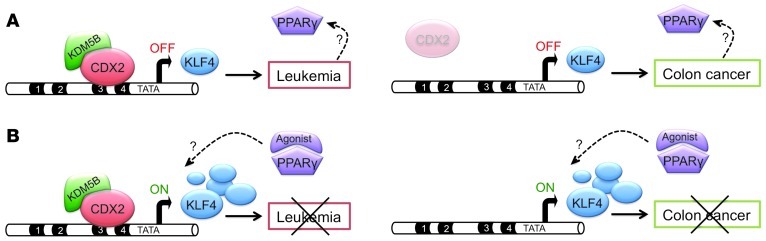
Figure 9. Opposing actions of CDX2 in AML and CRC development.
(A) In AML (left), aberrant CDX2 expression leads to downregulation of KLF4 (OFF) through binding to distinct sites in the KLF4 regulatory region and recruitment of the H3K4 demethylase KDM5B, thereby contributing to leukemogenesis. Aberrant CDX2 expression also causes deregulated PPARγ signaling via an unknown mechanism. In colonic epithelial cells (right), CDX2 induces KLF4 expression through binding to different sites in the KLF4 regulatory region, consistent with previous studies showing that loss of CDX2 and subsequent downregulation of KLF4 promote colorectal carcinogenesis. (B) PPARγ agonist treatment upregulates KLF4 expression (ON) regardless of tissue context, thereby inhibiting the viability and proliferation of AML (left; present study) and CRC cells (right; previous observations). Further studies are required to elucidate the mechanism whereby aberrant CDX2 expression and loss of KLF4 cause deregulated PPARγ signaling (dashed arrows).
In summary, our data demonstrated that the leukemogenic activity of CDX2 was linked to a distinct set of transcriptional changes and that silencing of KLF4 was an essential feature of this aberrant gene expression program. Hence, KLF4 emerges as a previously unrecognized growth-inhibitory gene involved in multiple AML subtypes, an observation that also offers insight into the mechanistic basis for the disparate functions of CDX2 in AML and CRC. Furthermore, gene expression analyses enabled the identification of deregulated PPARγ signaling in myeloid leukemias driven by CDX2. Because PPARγ expression is lower in normal HSPCs compared with AML (65, 66), and PPARγ agonist treatment preferentially inhibits the viability and proliferation of CDX2+ AML cells, a therapeutic window may exist in which PPARγ-directed therapy might attack CDX2+ leukemias while sparing normal cells. These findings provide a potential new lead for the development of targeted therapeutics in CDX2+ AML.
Methods
Isolation and retroviral transduction of mouse HSPCs.
BM cells were harvested from 3x15 C57BL/6 mice, and differentiated cells were removed by incubation with rat Ab against lineage antigens (CD3, CD4, CD8, Gr-1, B220, CD19, IL-7R, and Ter119), followed by depletion with magnetic beads (Dynabeads; Invitrogen). Lineage-depleted cells were stained with APC-conjugated anti–c-Kit and PE-Cy5–conjugated goat anti-rat Ab, and c-Kit+Lin– cells were sorted using a BD FACSAria cell sorter (BD Biosciences). Cells were plated in RetroNectin-coated tissue culture dishes (Takara Bio) and transduced twice with pMSCV-Cdx2-IRES-GFP or pMSCV-IRES-GFP retroviral constructs. After 48 hours, GFP+ cells were sorted using a BD FACSAria cell sorter, and RNA was extracted using the RNeasy Micro Kit (Qiagen).
Gene expression profiling, COPA, and C-Map analysis.
Gene expression profiling of mouse HSPCs and human AML samples, COPA, and C-Map analysis were performed as described previously (see Supplemental Methods). The microarray datasets reported herein were deposited in GEO (accession no. GSE40939).
Cell culture.
Myeloid leukemia cell lines (EOL-1, HEL, HL-60, K-562, LAMA-1, NB4, NOMO-1, SKM-1, and U937), CRC cell lines (DLD-1, HCT-116, HT-29, LS1034, and SW-480), and 293T cells were maintained under standard conditions. PGJ2 and T0070907 were obtained from Cayman Chemical. Telmisartan was obtained from Sigma-Aldrich.
Plasmids and viral transduction.
The mouse Cdx2 cDNA was described previously (15), the human CDX2 cDNA was cloned from FirstChoice Human Colon Tumor Total RNA (Ambion), and the human KLF4 cDNA was obtained from Open Biosystems. For expression in murine and human hematopoietic cells, cDNAs were cloned into the pMSCV-IRES-GFP or pMSCV-IRES-THY1 retroviral vectors or the pLenti6.2/V5-DEST lentiviral vector (Invitrogen). For inducible expression, the CDX2 cDNA was cloned into an IPTG-inducible pLKO.1puro vector (provided by S. Silver, Broad Institute, Cambridge, Massachusetts, USA). For apoptosis and cell cycle analysis in response to KLF4, the pRRL.PPT.SF.hKLF4.Tomato.pre lentiviral vector was used (provided by M. Milsom, Children’s Hospital Boston, Boston, Massachusetts, USA). RNAi experiments were performed using pLKO.1 lentiviral shRNA vectors obtained from the MISSION TRC-Hs 1.0 (Human) shRNA library (shCDX2_1, TRCN0000013685; shCDX2_2, TRCN0000013684; shKLF4_1, TRCN0000005316; shKLF4_2, TRCN0000010934; shKDM5B_1, TRCN0000014761; shKDM5B_2, TRCN0000014762) through Open Biosystems or Sigma-Aldrich. For coexpression of shCDX2_1 and GFP, the puromycin resistance gene of pLKO.1 was replaced with GFP, which was PCR-amplified from pMSCV-IRES-GFP. Generation of viral supernatants and viral transduction were performed as described previously (15, 67).
RNA isolation, cDNA synthesis, and qRT-PCR.
RNA isolation, cDNA synthesis, and qRT-PCR were performed using standard procedures. See Supplemental Methods for details.
ChIP and LUC reporter assays.
ChIP was performed using K-562 and HT-29 cells stably transduced with HA-CDX2, alone or in combination with shRNAs targeting KDM5B, or an empty pLenti6.2/V5-DEST control vector. Expression values were normalized to input chromatin and reported as percent input after background subtraction. For LUC reporter assays, the KLF4 promoter and a fragment of the KLF4 upstream regulatory region containing CDX2 binding sites 3 and 4 were cloned into pGL4.12[luc2CP] (Promega). Mutated binding sites were generated by site-directed mutagenesis. See Supplemental Methods for details.
Quantitative DNA methylation analysis.
DNA methylation was quantitatively assessed at single CpG units using MassARRAY EpiTyper technology (Sequenom) as previously described (68). See Supplemental Methods for details.
Cell viability and proliferation assays.
Viability and proliferation of primary murine leukemic cells and human myeloid leukemia cell lines were determined using the CellTiter96AQueousOne Solution Proliferation Assay (Promega).
Cell cycle, apoptosis, and differentiation analysis.
Cell cycle, apoptosis, and differentiation were analyzed using standard procedures. See Supplemental Methods for details.
Colony formation and LTC-IC assays.
Colony formation and LTC-IC assays were performed using standard procedures. See Supplemental Methods for details.
Murine BMT assays.
Noncompetitive BMT was performed as described previously (15). For competitive BMT, GFP+ cells derived from Cdx2-induced murine leukemias were transduced with a retroviral vector expressing THY1, either alone or together with KLF4, followed by sorting of GFP+, GFP+KLF4-THY1+, and GFP+THY1+ cells using a BD FACSAria cell sorter. Sublethally irradiated mice were injected with equal numbers (2 × 104) of GFP+ cells and either GFP+KLF4-THY1+ or GFP+THY1+ cells, and the proportion of GFP+THY1+ cells in the BM and spleen of diseased mice was quantified by flow cytometry.
Statistics.
Experiments were performed at least 3 times; unless otherwise indicated, 1 representative experiment is shown. Error bars represent mean ± SEM. Statistical analysis was performed using paired or unpaired 2-tailed t test, Kaplan-Meier survival estimates, or log-rank test as appropriate. A P value less than 0.05 was considered significant. Computations were performed using GraphPad Prism (version 5.0a; GraphPad Software).
Study approval.
Human AML samples were obtained under institutional review board–approved protocols following written informed consent. Animal experiments were performed after approval and in accordance with the guidelines of the Subcommittee on Research Animal Care of Children’s Hospital Boston.
Supplementary Material
Acknowledgments
We thank Gary Gilliland for advice and critical review of the manuscript. We thank Ursula Botzenhardt for technical assistance. This work was supported by grant DJCLS R 09/12 from the Deutsche José Carreras Leukämie-Stiftung. L. Bullinger is the recipient of a Heisenberg Fellowship from the German Research Foundation. S.W. Lane is supported by the Leukaemia Foundation of Australia. C. Scholl is the recipient of an Emmy Noether Fellowship from the German Research Foundation. S. Fröhling is the recipient of a European Hematology Association–José Carreras Foundation Young Investigator Fellowship.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(1):299–314. doi:10.1172/JCI64745.
References
- 1.Niwa H, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123(5):917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 2.Strumpf D, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132(9):2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 3.Charite J, et al. Transducing positional information to the Hox genes: critical interaction of cdx gene products with position-sensitive regulatory elements. Development. 1998;125(22):4349–4358. doi: 10.1242/dev.125.22.4349. [DOI] [PubMed] [Google Scholar]
- 4.Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F. Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci U S A. 2004;101(20):7641–7645. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Akker E, et al. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129(9):2181–2193. doi: 10.1242/dev.129.9.2181. [DOI] [PubMed] [Google Scholar]
- 6.Lengerke C, et al. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2(1):72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Cdx gene deficiency compromises embryonic hematopoiesis in the mouse. Proc Natl Acad Sci U S A. 2008;105(22):7756–7761. doi: 10.1073/pnas.0708951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Yates F, Naveiras O, Ernst P, Daley GQ. Embryonic stem cell-derived hematopoietic stem cells. Proc Natl Acad Sci U S A. 2005;102(52):19081–19086. doi: 10.1073/pnas.0506127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16(4):588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo RJ, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther. 2004;3(7):593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- 11.Fröhling S, Scholl C, Bansal D, Huntly BJP. HOX gene regulation in acute myeloid leukemia: CDX marks the spot? Cell Cycle. 2007;6(18):2241–2245. doi: 10.4161/cc.6.18.4656. [DOI] [PubMed] [Google Scholar]
- 12.Rawat VPS, Humphries RK, Buske C. Beyond HOX: the role of ParaHox genes in normal and malignant hematopoiesis. Blood. 2012;120(3):519–527. doi: 10.1182/blood-2012-02-385898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chase A, et al. Fusion of ETV6 to the caudal-related homeobox gene CDX2 in acute myeloid leukemia with the t(12;13)(p13;q12). Blood. 1999;93(3):1025–1031. [PubMed] [Google Scholar]
- 14.Rawat VP, et al. Overexpression of CDX2 perturbs HOX gene expression in murine progenitors depending on its N-terminal domain and is closely correlated with deregulated HOX gene expression in human acute myeloid leukemia. Blood. 2008;111(1):309–319. doi: 10.1182/blood-2007-04-085407. [DOI] [PubMed] [Google Scholar]
- 15.Scholl C, et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J Clin Invest. 2007;117(4):1037–1048. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rawat VPS, et al. Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc Natl Acad Sci U S A. 2004;101(3):817–822. doi: 10.1073/pnas.0305555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 18.Hinoi T, et al. CDX2 regulates liver intestine-cadherin expression in normal and malignant colon epithelium and intestinal metaplasia. Gastroenterology. 2002;123(5):1565–1577. doi: 10.1053/gast.2002.36598. [DOI] [PubMed] [Google Scholar]
- 19.Nishiyama A, et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell. 2009;5(4):420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suh E, Traber PG. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol Cell Biol. 1996;16(2):619–625. doi: 10.1128/mcb.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verzi MP, et al. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc Natl Acad Sci U S A. 2010;107(34):15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Costa LT, et al. CDX2 is mutated in a colorectal cancer with normal APC/beta-catenin signaling. Oncogene. 1999;18(35):5010–5014. doi: 10.1038/sj.onc.1202872. [DOI] [PubMed] [Google Scholar]
- 23.Hinoi T, Loda M, Fearon ER. Silencing of CDX2 expression in colon cancer via a dominant repression pathway. J Biol Chem. 2003;278(45):44608–44616. doi: 10.1074/jbc.M307435200. [DOI] [PubMed] [Google Scholar]
- 24.Wicking C, et al. CDX2, a human homologue of Drosophila caudal, is mutated in both alleles in a replication error positive colorectal cancer. Oncogene. 1998;17(5):657–659. doi: 10.1038/sj.onc.1201971. [DOI] [PubMed] [Google Scholar]
- 25.Bonhomme C, et al. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut. 2003;52(10):1465–1471. doi: 10.1136/gut.52.10.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386(6620):84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 27.Almeida R, et al. Expression of intestine-specific transcription factors, CDX1 and CDX2, in intestinal metaplasia and gastric carcinomas. J Pathol. 2003;199(1):36–40. doi: 10.1002/path.1246. [DOI] [PubMed] [Google Scholar]
- 28.Dang LH, et al. CDX2 has tumorigenic potential in the human colon cancer cell lines LOVO and SW48. Oncogene. 2006;25(15):2264–2272. doi: 10.1038/sj.onc.1209247. [DOI] [PubMed] [Google Scholar]
- 29.Silberg DG, et al. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 2002;122(3):689–696. doi: 10.1053/gast.2002.31902. [DOI] [PubMed] [Google Scholar]
- 30.Döhner H, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 31.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 32.Katz JP, et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128(4):935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 33.Wei D, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65(7):2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 34.Zhao W, et al. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23(2):395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan H, et al. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classical Hodgkin lymphoma. Blood. 2010;116(9):1469–1478. doi: 10.1182/blood-2009-12-256446. [DOI] [PubMed] [Google Scholar]
- 36.Kharas MG, et al. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood. 2007;109(2):747–755. doi: 10.1182/blood-2006-03-011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivera-Pomar R, Lu X, Perrimon N, Taubert H, Jackle H. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature. 1995;376(6537):253–256. doi: 10.1038/376253a0. [DOI] [PubMed] [Google Scholar]
- 38.Suh E, Chen L, Taylor J, Traber PG. A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol Cell Biol. 1994;14(11):7340–7351. doi: 10.1128/mcb.14.11.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W, Chen H, Zhang L. The PcG protein hPc2 interacts with the N-terminus of histone demethylase JARID1B and acts as a transcriptional co-repressor. BMB Rep. 2009;42(3):154–159. doi: 10.5483/BMBRep.2009.42.3.154. [DOI] [PubMed] [Google Scholar]
- 40.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 41.Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123(6):993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 42.Ondrey F. Peroxisome proliferator-activated receptor gamma pathway targeting in carcinogenesis: implications for chemoprevention. Clin Cancer Res. 2009;15(1):2–8. doi: 10.1158/1078-0432.CCR-08-0326. [DOI] [PubMed] [Google Scholar]
- 43.Cekanova M, Lee SH, McEntee MF, Baek SJ. MCC-555-induced NAG-1 expression is mediated in part by KLF4. Eur J Pharmacol. 2010;637(1–3):30–37. doi: 10.1016/j.ejphar.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drori S, et al. Hic-5 regulates an epithelial program mediated by PPARgamma. Genes Dev. 2005;19(3):362–375. doi: 10.1101/gad.1240705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo J, et al. Peroxisome proliferator-activated receptor gamma-dependent activity of indole ring-substituted 1,1-bis(3’-indolyl)-1-(p-biphenyl)methanes in cancer cells. Cancer Chemother Pharmacol. 2010;66(1):141–150. doi: 10.1007/s00280-009-1144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rageul J, et al. KLF4-dependent, PPARgamma-induced expression of GPA33 in colon cancer cell lines. Int J Cancer. 2009;125(12):2802–2809. doi: 10.1002/ijc.24683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benson SC, et al. Identification of telmisartan as a unique angiotensin II receptor antagonist with selective PPARgamma-modulating activity. Hypertension. 2004;43(5):993–1002. doi: 10.1161/01.HYP.0000123072.34629.57. [DOI] [PubMed] [Google Scholar]
- 48.Yki-Jarvinen H. Thiazolidinediones. N Engl J Med. 2004;351(11):1106–1118. doi: 10.1056/NEJMra041001. [DOI] [PubMed] [Google Scholar]
- 49.Somervaille TC, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feinberg MW, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26(18):4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20(35):4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao N, Kaestner KH. Cdx2 regulates endo-lysosomal function and epithelial cell polarity. Genes Dev. 2010;24(12):1295–1305. doi: 10.1101/gad.1921510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takakura Y, et al. CDX2 regulates multidrug resistance 1 gene expression in malignant intestinal epithelium. Cancer Res. 2010;70(17):6767–6778. doi: 10.1158/0008-5472.CAN-09-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6(1):11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 55.Foster KW, et al. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60(22):6488–6495. [PubMed] [Google Scholar]
- 56.Foster KW, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10(6):423–434. [PubMed] [Google Scholar]
- 57.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7(11):1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 58.Berkofsky-Fessler W, et al. Transcriptional profiling of polycythemia vera identifies gene expression patterns both dependent and independent from the action of JAK2V617F. Clin Cancer Res. 2010;16(17):4339–4352. doi: 10.1158/1078-0432.CCR-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5(5):376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 61.Huff V. Wilms’ tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 2011;11(2):111–121. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Massague J. TGFbeta in cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17(1):52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 64.Huesca M, et al. A novel small molecule with potent anticancer activity inhibits cell growth by modulating intracellular labile zinc homeostasis. Mol Cancer Ther. 2009;8(9):2586–2596. doi: 10.1158/1535-7163.MCT-08-1104. [DOI] [PubMed] [Google Scholar]
- 65.Konopleva M, et al. Peroxisome proliferator-activated receptor gamma and retinoid X receptor ligands are potent inducers of differentiation and apoptosis in leukemias. Mol Cancer Ther. 2004;3(10):1249–1262. [PubMed] [Google Scholar]
- 66.Tsao T, et al. Role of peroxisome proliferator-activated receptor-gamma and its coactivator DRIP205 in cellular responses to CDDO (RTA-401) in acute myelogenous leukemia. Cancer Res. 2010;70(12):4949–4960. doi: 10.1158/0008-5472.CAN-09-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 68.Ehrich M, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.