Abstract
Metastasis involves critical interactions between cancer and stromal cells. Intratumoral hypoxia promotes metastasis through activation of hypoxia-inducible factors (HIFs). We demonstrate that HIFs mediate paracrine signaling between breast cancer cells (BCCs) and mesenchymal stem cells (MSCs) to promote metastasis. In a mouse orthotopic implantation model, MSCs were recruited to primary breast tumors and promoted BCC metastasis to LNs and lungs in a HIF-dependent manner. Coculture of MSCs with BCCs augmented HIF activity in BCCs. Additionally, coculture induced expression of the chemokine CXCL10 in MSCs and the cognate receptor CXCR3 in BCCs, which was augmented by hypoxia. CXCR3 expression was blocked in cocultures treated with neutralizing antibody against CXCL10. Conversely, CXCL10 expression was blocked in MSCs cocultured with BCCs that did not express CXCR3 or HIFs. MSC coculture did not enhance the metastasis of HIF-deficient BCCs. BCCs and MSCs expressed placental growth factor (PGF) and its cognate receptor VEGFR1, respectively, in a HIF-dependent manner, and CXCL10 expression by MSCs was dependent on PGF expression by BCCs. PGF promoted metastasis of BCCs and also facilitated homing of MSCs to tumors. Thus, HIFs mediate complex and bidirectional paracrine signaling between BCCs and MSCs that stimulates breast cancer metastasis.
Introduction
An important advance in cancer biology has been the appreciation that, in addition to somatic mutations in oncogenes and tumor suppressor genes within cancer cells, a major mechanism driving disease progression is the interaction of cancer cells with the tumor microenvironment. The tumor stroma consists of extracellular matrix and various mesenchymal cell types, including vascular ECs and pericytes, fibroblasts, myofibroblasts, and various cells of bone marrow origin, including tumor-associated macrophages, bone marrow–derived angiogenic cells, neutrophils, mast cells, myeloid-derived suppressor cells, and mesenchymal stem cells (MSCs), which are recruited to the tumor and enhance primary tumor growth and/or promote metastasis (1). The molecular mechanisms by which stromal cells are attracted to, and communicate with, cancer cells are only understood in a limited number of contexts. For example, breast cancer cell (BCC) production of colony-stimulating factor 1 (CSF1) induces homing of CSF1 receptor–expressing tumor-associated macrophages that secrete epidermal growth factor, which binds to its receptor on cancer cells and stimulates their invasive properties (2, 3).
The combination of cancer cell proliferation and stromal cell recruitment results in an imbalance between O2 consumption and delivery. Tumor vasculature is structurally and functionally abnormal, leading to spatial and temporal heterogeneity in perfusion, even in tumors with active angiogenesis (4, 5). As a result, tissue oxygenation is markedly decreased in the tumor microenvironment. The mean partial O2 pressure in breast cancers is 28 mmHg, compared with 65 mmHg in normal breast tissue (6). As in the case of stromal cells, hypoxia is a critical feature of the tumor microenvironment that promotes invasion and metastasis (7–9). O2 deprivation increases the activity of hypoxia-inducible factors (HIFs) in both BCCs and stromal cells (10–12). HIFs are heterodimeric transcription factors composed of an O2-regulated HIF-1α or HIF-2α subunit and a constitutively expressed HIF-1β subunit (13). Increased levels of HIF-1α or HIF-2α in breast cancer biopsies are associated with metastasis to regional LNs and distant organs and with patient mortality (14–19). HIFs mediate tumor vascularization through the production by cancer cells of angiogenic factors that stimulate ECs and mobilize bone marrow–derived angiogenic cells (20–22). Using both genetic and pharmacologic loss-of-function approaches, HIFs have been shown to play critical roles in breast cancer metastasis to the lungs by activating in BCCs the transcription of genes encoding proteins that play critical roles in establishment of the metastatic niche and in extravasation of BCCs from pulmonary blood vessels (23–28). Treatment of tumor-bearing mice with digoxin or acriflavine, drugs that inhibit HIF activity, resulted in a dramatic reduction in lung metastasis (22, 23). These studies were performed with human cell lines derived from triple-negative breast cancers, which lack expression of estrogen, progesterone, and HER2 receptors and do not respond well to currently available therapies (29).
MSCs are recruited to breast cancers by mechanisms that are not well understood (30, 31). When MDA-MB-231 or MDA-MB-435 human BCCs (referred to herein as MDA-231 and MDA-435, respectively) were mixed with human MSCs and injected subcutaneously into immunodeficient mice, the rate of lung metastasis was increased compared with injection of BCCs alone; this was attributed to MSC production of the chemokine CCL5, which bound to its cognate receptor, CCR5, on BCCs (31). Although these were intriguing observations, the study had several limitations, including the involvement of a heterologous microenvironment (subcutaneous rather than mammary), the large number of MSCs injected (3-fold greater than the number of cancer cells), the long lag time to metastasis (up to 150 days), and the use of CCL5 overexpression in BCCs to implicate this signaling pathway in metastasis. Among the questions raised by the study were the following: What is the mechanism by which MSCs are recruited to primary breast tumors? Do recruited MSCs produce other chemokines that are important for stimulating BCC metastasis? Do BCCs express secreted factors that stimulate MSCs to produce chemokines? What are the molecular mechanisms by which MSC-BCC interaction leads to increased metastasis? Does intratumoral hypoxia modulate this process? To address these issues, we used an orthotopic transplantation model involving injection of cocultured human BCCs and MSCs into the mammary fat pad (MFP) of SCID mice, which results in spontaneous LN and lung metastases within 1 month. Our findings revealed complex bidirectional signaling between MSCs and BCCs that was stimulated by hypoxia in a HIF-dependent manner and promoted metastasis to the LNs as well as the lungs.
Results
Recruitment of MSCs to the primary tumor is HIF dependent, and MSCs enhance metastasis of BCCs to lungs and LNs.
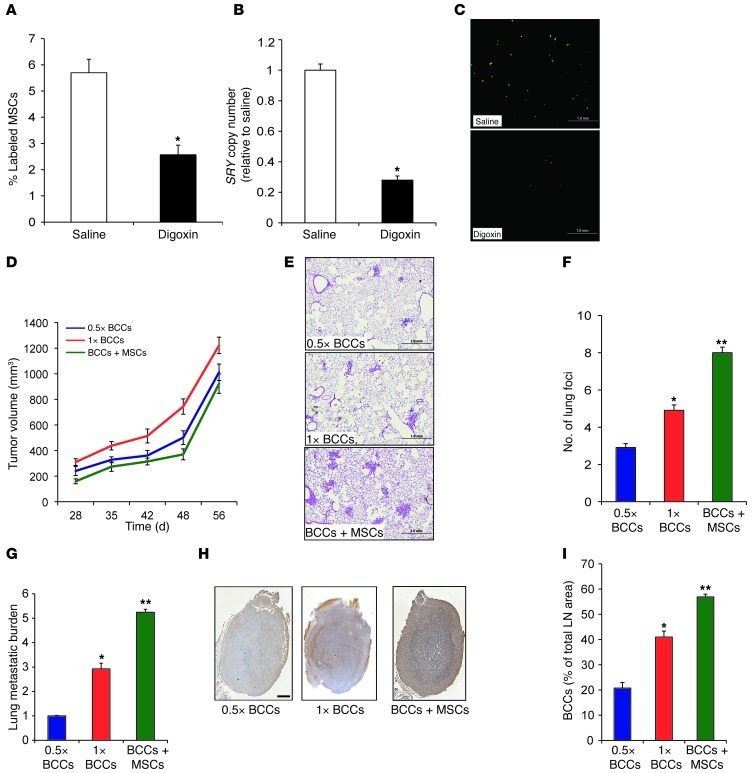
MSCs exhibit homing to sites of tissue injury and tumor growth (31–33). To determine whether HIF activity in the hypoxic tumor microenvironment facilitates recruitment of MSCs to the primary tumor site, MDA-231 human triple-negative BCCs (34) were implanted in the MFP of SCID mice. When the tumors had grown to 200 mm3, the mice received daily intraperitoneal injections of either saline or digoxin (2 mg/kg) to inhibit HIF activity (27, 28, 35). After 1 week of digoxin treatment, CMFDA-labeled human MSCs were injected via tail vein, and primary tumors were harvested 16 hours later. The recruitment of MSCs (derived from a male donor) to the primary tumor (derived from a female donor) was examined by quantitative real-time PCR (qPCR) analysis of genomic DNA using SRY gene primers (to detect Y chromosome sequences) and by FACS analysis of fluorescently labeled MSCs in cell suspensions prepared from primary tumors. Frozen tumor sections were also analyzed for the presence of fluorescently labeled MSCs. All 3 of these assays revealed significantly decreased recruitment of MSCs in tumors from digoxin- versus saline-treated mice (Figure 1, A–C).
Figure 1. MSCs are recruited to breast tumors and enhance lung and LN metastasis.
(A–C) MDA-231 BCCs were implanted into the MFP of SCID mice, which were treated with saline or digoxin (2 mg/kg/d). After 1 week of treatment, CMFDA-labeled human MSCs were injected via tail vein, and tumors were harvested 16 hours later. (A and B) Percent CMFDA+ MSCs in the primary tumor was analyzed by FACS (A) and by qPCR for SRY copy number (B). Data are mean ± SEM (n = 5). *P < 0.05, Student’s t test. (C) Labeled MSCs in tumor sections were detected by fluorescence microscopy. Scale bar: 1 mm. (D–I) SCID mice received MFP injection of 0.5 × 106 (0.5×) or 1 × 106 (1×) MDA-231 BCCs alone or 1 × 106 cells from 1:1 coculture of BCCs+MSCs. Mice were euthanized 56 days later, and tumors, lungs, and ipsilateral axillary LNs were harvested. (D) Tumor volume plotted against time (mean ± SEM; n = 5). (E) H&E staining of lung sections. Scale bar: 1 mm. (F) Metastatic foci in lung sections (≥3 random fields per section) were counted under ×20 magnification. (G) Metastatic burden was determined by qPCR using human HK2 gene primers. (H) Immunohistochemical analysis of LN sections with human-specific vimentin antibody. Scale bar: 0.5 mm. (I) Percent total LN area occupied by BCCs (mean ± SEM; n = 5). *P < 0.05, **P < 0.01 vs. 0.5 × 106 BCCs.
To investigate the functional consequences of interaction between MSCs and BCCs, we cocultured MSCs with MDA-231 BCCs at a 1:1 ratio for 48 hours, followed by MFP implantation of 1 × 106 of the cocultured BCCs+MSCs. Controls included mice injected with BCCs alone — either the same number of BCCs (0.5 × 106) or the same total number of cells (1 × 106). BCCs+MSCs did not accelerate growth of the primary tumor (Figure 1D). However, the lungs and ipsilateral axillary LNs of mice carrying breast tumors derived from BCCs+MSCs displayed a marked increase in metastases (Figure 1, E–I). The number of lung foci, as determined by H&E staining, and the total lung metastatic burden, as determined by qPCR of human-specific HK2 sequences, were significantly increased in mice injected with BCCs+MSCs (Figure 1, E–G). The area occupied by BCCs in the ipsilateral axillary LN was also increased in mice bearing BCCs+MSCs (Figure 1, H and I). In contrast, intravenous injection of BCCs+MSCs or MFP coinjection of BCCs and MSCs (without prior coculture) did not result in increased lung foci compared with injection of BCCs alone (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI64993DS1). These data suggest that coculture of BCCs+MSCs increases metastasis by promoting steps prior to BCC entry into the circulation, such as tissue invasion or vascular intravasation.
HIFs regulate expression of CXCL10 in MSCs and CXCR3 in BCCs in response to coculture and hypoxia.
Based on the coculture data, we hypothesized that MSCs produce paracrine signals that induce BCCs to metastasize. To investigate crosstalk between BCCs and MSCs, an array of antibodies directed against various cytokines and chemokines was incubated with conditioned medium (CM) isolated from BCCs+MSCs that were cocultured at a 1:1 ratio for 48 hours under either nonhypoxic (20% O2) or hypoxic (1% O2) conditions. Levels differed between the 2 conditions for 13 of the 33 cytokines and chemokines analyzed (Supplemental Figure 2A). We focused on the CXCL10 and CCL5 chemokines, which were expressed at higher levels in CM from coculture of BCCs+MSCs under hypoxic conditions.
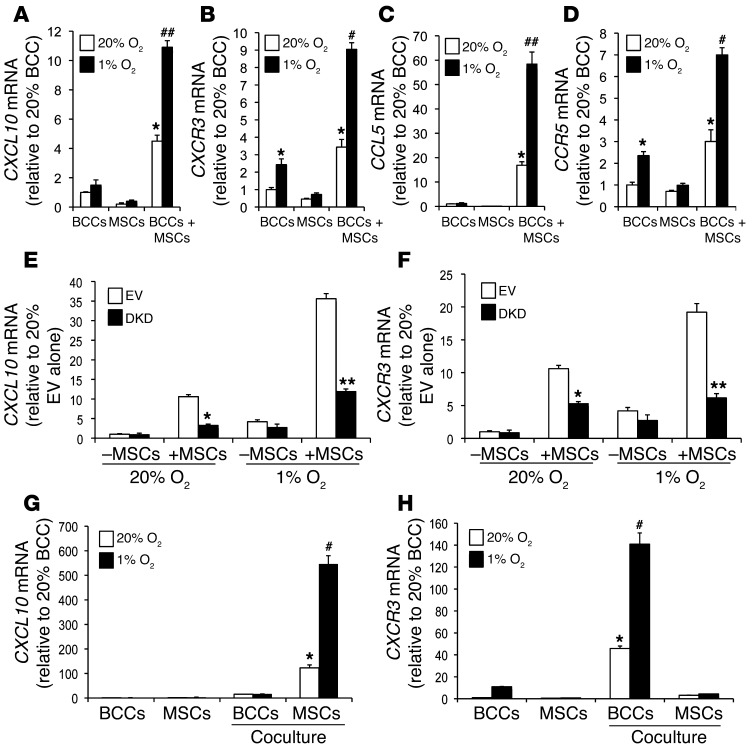
We analyzed the expression of CXCL10 mRNA by RT-qPCR using RNA prepared from coculture of BCCs+MSCs or individually cultured BCCs and MSCs under nonhypoxic or hypoxic conditions. CXCL10 expression was induced in the cocultures, and this effect was further enhanced when the cocultured cells were subjected to hypoxia (Figure 2A). Expression of CXCR3, the cognate receptor for CXCL10 (36), was induced by hypoxia in MDA-231 BCCs alone. BCCs+MSCs increased CXCR3 expression, which was further enhanced when the cocultured cells were subjected to hypoxia (Figure 2B). Similarly, coculture and hypoxia increased the expression of CCL5 and CCR5 (Figure 2, C and D). Our results confirmed a previous report that CCL5 expression was induced in coculture of BCCs+MSCs (31) and extended those findings by demonstrating a synergistic effect of hypoxia.
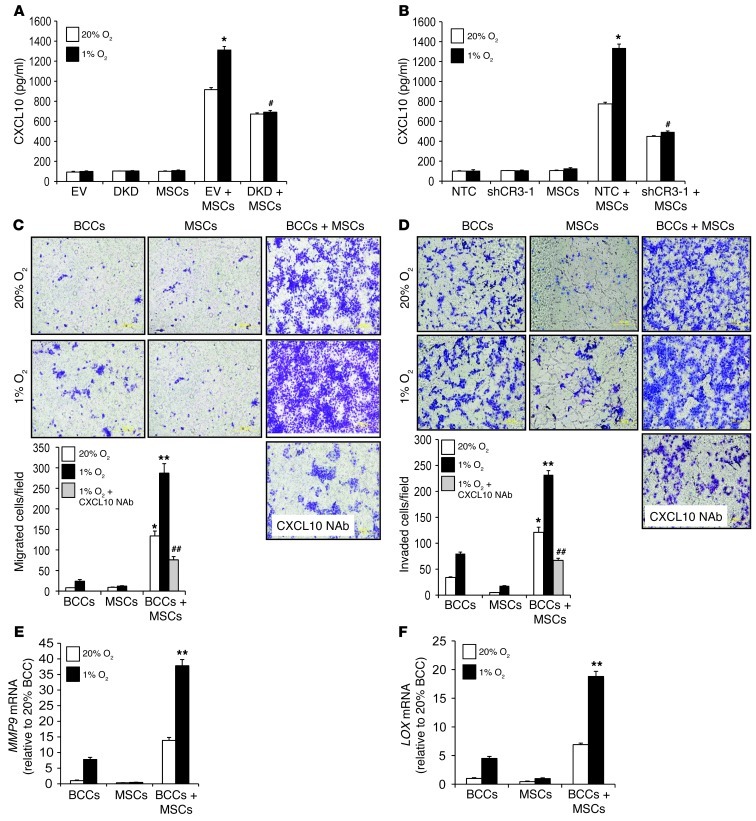
Figure 2. HIFs mediate coculture- and hypoxia-induced CXCL10, CXCR3, CCL5, and CCR5 expression.
(A–D) BCCs, MSCs, or BCCs+MSCs were cultured at 20% or 1% O2 for 48 hours, and CXCL10 (A), CXCR3 (B), CCL5 (C), and CCR5 (D) mRNA levels were determined by RT-qPCR (mean ± SEM; n = 3). Levels were normalized to BCCs at 20% O2. *P < 0.05 vs. 20% BCCs or 20% MSCs; #P < 0.01, ##P < 0.001 vs. 20% BCCs+MSCs, 1-way ANOVA. (E and F) EV cells, DKD cells, EV+MSCs, or DKD+MSCs were cultured at 20% or 1% O2 for 48 hours. CXCL10 and CXCR3 mRNA levels were analyzed by RT-qPCR and normalized to those in EV cells at 20% O2 (mean ± SEM; n = 3). *P < 0.05 vs. 20% EV+MSCs; **P < 0.001 vs. 1% EV+MSCs, 1-way ANOVA. (G and H) GFP-expressing BCCs were cocultured with MSCs at 20% or 1% O2 for 48 hours, then subjected to FACS based on GFP fluorescence for BCCs and CD105 immunofluorescence for MSCs. CXCR3 and CXCL10 mRNA levels were determined in flow-sorted BCCs and MSCs. BCCs and MSCs cultured alone were used as controls. Levels were normalized to BCCs at 20% O2. *P < 0.01 vs. 20% MSCs or BCCs alone; #P < 0.001 vs. 1% MSCs or BCCs alone, 1-way ANOVA.
To investigate the role of HIFs in these phenomena, we used a previously validated double-knockdown subclone of MDA-231 stably transfected with vectors encoding shRNAs directed against HIF1A and HIF2A (MDA-231–DKD; referred to herein as DKD cells) and a control subclone stably transfected with empty vector (MDA-231–EV; referred to herein as EV cells) (26, 27). Expression of CXCR3 and CXCL10 mRNA was significantly decreased in DKD+MSCs compared with EV+MSCs at both 20% and 1% O2 (Figure 2, E and F). CCL5 and CCR5 mRNA levels were also significantly decreased in DKD+MSCs versus EV+MSCs at both 20% and 1% O2 (Supplemental Figure 2, B and C). Expression of RPL13A mRNA, which is not HIF regulated, was not changed in DKD cells and was not induced by coculture or hypoxia (Supplemental Figure 2D). In contrast to the effect of MSCs, coculture of MDA-231 BCCs with human foreskin fibroblasts did not induce increased expression of CXCL10 or CXCR3 (Supplemental Figure 2, H and I), which indicates that the coculture effect is dependent on selective crosstalk between MDA-231 BCCs and MSCs.
To determine the cell types responsible for the production of CXCL10 and CXCR3 under coculture conditions, GFP-expressing MDA-231 BCCs were cocultured with MSCs for 48 hours under nonhypoxic or hypoxic conditions, and FACS was performed using GFP fluorescence and CD105 immunofluorescence to sort for BCCs and MSCs, respectively. RNA was isolated from the sorted cells, and RT-qPCR was performed to analyze CXCR3 and CXCL10 expression. The sorting efficiency was demonstrated using GFP primers to confirm that GFP mRNA was detected only in the BCC population (Supplemental Figure 2E). Analysis of the sorted cells revealed that CXCL10 expression was induced in MSCs, whereas CXCR3 expression was induced in BCCs (Figure 2, G and H). Similarly, induction of CCL5 was observed in MSCs, and CCR5 expression was induced in BCCs (Supplemental Figure 2, F and G). Taken together, the results presented in Figure 2 and Supplemental Figure 2 indicate that HIF-dependent crosstalk between BCCs and MSCs is mediated through the CXCR3-CXCL10 and CCR5-CCL5 signaling pathways in response to coculture and hypoxia.
Acriflavine blocks hypoxia-induced CXCL10-CXCR3 and CCL5-CCR5 expression.
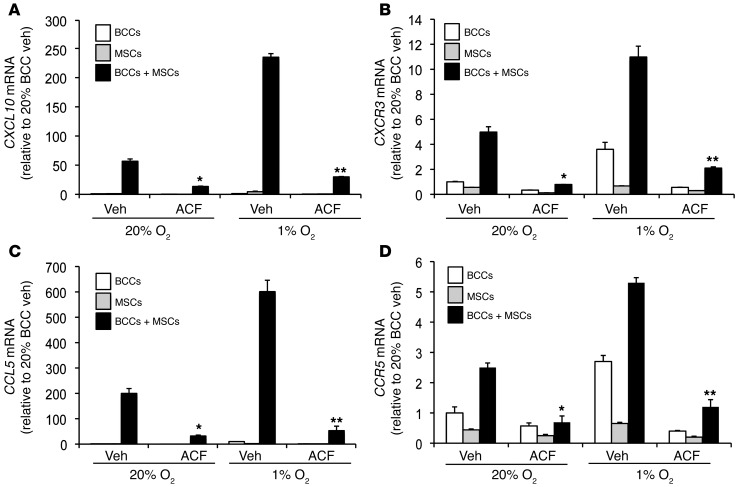
We next explored whether pharmacological targeting of HIF would inhibit coculture-induced gene expression. Acriflavine abrogates HIF activity by inhibiting the dimerization of HIF-1α or HIF-2α with the HIF-1β subunit (22). BCCs+MSCs were treated with vehicle or acriflavine (1 μM) for 48 hours at 20% or 1% O2. The effects of coculture and hypoxia on CXCR3-CXCL10 and CCR5-CCL5 expression were significantly inhibited in the acriflavine-treated cocultures (Figure 3).
Figure 3. Acriflavine blocks coculture-induced expression of CCL5, CCR5, CXCL10, and CXCR3.
BCCs, MSCs, or BCCs+MSCs were treated with 1 μM acriflavine (ACF) or 0.02% DMSO vehicle (Veh) and exposed to 20% or 1% O2 for 48 hours prior to RNA isolation. CXCL10 (A), CXCR3 (B), CCL5 (C), and CCR5 (D) mRNA levels were analyzed by RT-qPCR (mean ± SEM; n = 3). Levels were normalized to vehicle-treated BCCs at 20% O2. *P < 0.01 vs. 20% BCCs+MSCs vehicle; **P < 0.001 vs. 1% BCCs+MSCs vehicle, 1-way ANOVA.
Inhibiting CXCR3 or CXCL10 abrogates CXCR3-CXCL10 crosstalk.
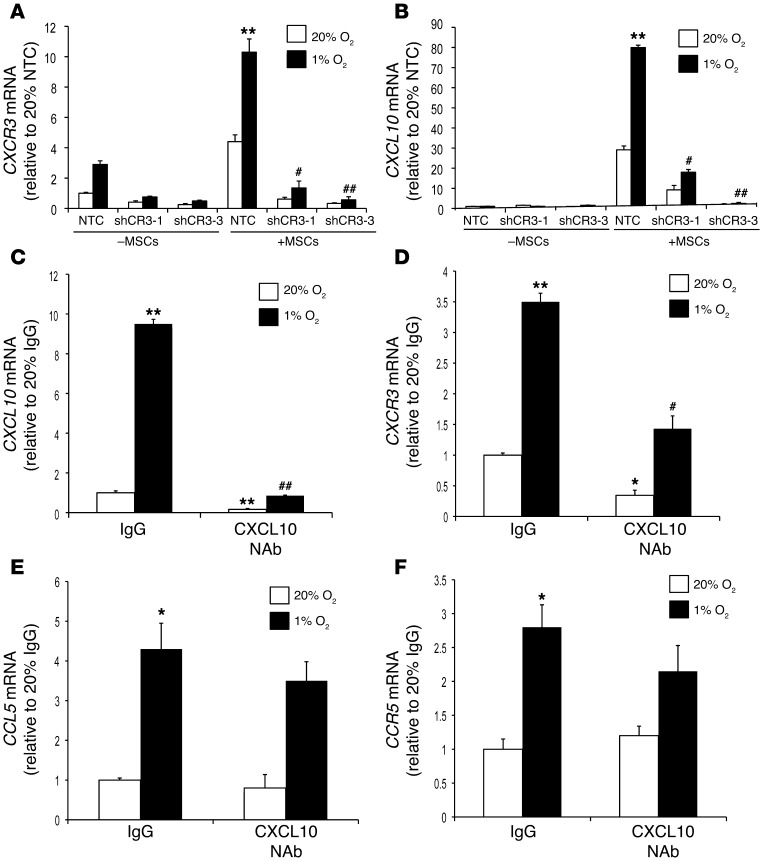
We generated MDA-231 subclones that were stably transfected with vector encoding either of 2 different shRNAs targeting CXCR3 (shCR3-1 and shCR3-3) or a nontargeted control (NTC) shRNA (referred to herein as NTC cells). The knockdown efficiency of different subclones was determined by Western blotting (Supplemental Figure 3A). Expression of CXCR3 and, surprisingly, that of CXCL10 was abrogated when the coculture was performed with the CXCR3-knockdown MDA-231 cells (Figure 4, A and B). MAP kinase signaling correlated with CXCR3 expression and was inhibited when the coculture was performed with CXCR3-knockdown MDA-231 cells (Supplemental Figure 3B). We next added neutralizing antibody (NAb) against CXCL10 or control IgG to cocultures of MSCs with the parental MDA-231 BCCs. Expression of both CXCL10 and CXCR3 mRNA was significantly reduced in the presence of CXCL10 NAb compared with IgG control (Figure 4, C and D). The reduction of CXCL10 and CXCR3 mRNA levels caused by NAb was reversed when an excess of recombinant CXCL10 was added to the culture medium (Supplemental Figure 3, C and D).
Figure 4. Hypoxia augments crosstalk between BCCs and MSCs by promoting CXCL10-CXCR3 signaling.
(A and B) NTC, shCR3-1, and shCR3-3 MDA-231 subclones were cultured alone or cocultured with MSCs and exposed to 20% or 1% O2 for 48 hours. Expression of CXCR3 (A) and CXCL10 (B) mRNA was analyzed by RT-qPCR. Levels were normalized to NTC cells at 20% O2. **P < 0.001 vs. 20% NTC+MSCs; #P < 0.01, ##P < 0.001 vs. 1% NTC+MSCs. (C–F) BCCs+MSCs were treated with CXCL10 Nab or IgG control and exposed to 20% or 1% O2 for 48 hours. Expression of CXCL10 (C), CXCR3 (D), CCL5 (E), and CCR5 (F) mRNA was analyzed by RT-qPCR; levels were normalized to IgG at 20% O2 (mean ± SEM; n = 3). *P < 0.01, **P < 0.001 vs. 20% IgG; #P < 0.05, ##P < 0.001 vs. 1% IgG, 1-way ANOVA.
Similar findings were observed in MDA-435 cells, another triple-negative human BCC line (37), when CXCR3 expression was knocked down (using either of 2 different shRNAs) or when CXCL10 NAb was added to cocultures with MSCs (Supplemental Figure 4, A–D). Taken together, these data indicate that binding of CXCL10 to CXCR3 on BCCs induces BCC→MSC signaling that leads to increased CXCL10 expression in MSCs. When CXCR3 activation is blocked by CXCR3 knockdown or CXCL10 NAb, bidirectional signaling does not occur.
In contrast to its effects on CXCL10 and CXCR3 mRNA expression, CXCL10 NAb did not significantly affect the expression of CCR5 or CCL5 mRNA in cocultures of MSCs with MDA-231 or MDA-435 BCCs (Figure 4, E and F, and Supplemental Figure 4, E and F). These results indicate that loss of CXCL10-CXCR3 signaling did not impede CCL5-CCR5 signaling.
Hypoxia-induced CXCL10 enhances BCC migration and invasion.
We assayed CXCL10 levels in CM from BCCs cultured alone or from BCCs+MSCs. Whereas CXCL10 levels in the CM were very low when BCCs were cultured alone, coculture of EV+MSCs dramatically increased CXCL10 levels, which were further increased under hypoxic conditions (Figure 5A). CM from DKD+MSCs showed decreased CXCL10 levels, which were not induced by hypoxia (Figure 5A). Similarly, CXCL10 levels in CM from cocultures of MSCs with CXCR3-knockdown MDA-231 cells were significantly decreased and not induced by hypoxia (Figure 5B). The increased CXCL10 levels in DKD+MSCs compared with MSCs cultured alone suggests that HIF-independent paracrine signaling also contributes to CXCL10 expression.
Figure 5. CXCL10 enhances BCC migration and invasion.
(A) CM was isolated from cultures of EV cells, DKD cells, MSCs, EV+MSCs, and DKD+MSCs, and ELISA was performed to determine CXCL10 levels (mean ± SEM; n = 3). *P < 0.05 vs. 20% EV+MSCs; #P < 0.01 vs. 1% EV+MSCs, ANOVA. (B) ELISA was performed to determine CXCL10 protein levels (mean ± SEM; n = 3) from NTC cells, shCR3-1 cells, MSCs, NTC+MSCs, and shCR3-1+MSCs. *P < 0.01 vs. 20% NTC+MSCs; #P < 0.01 vs. 1% NTC+MSCs. (C) Naive MDA-231 BCCs were seeded on the top of a Boyden chamber. The number of cells that migrated through the uncoated filter in response to CM from BCCs, MSCs, or BCCs+MSCs (alone or in the presence of CXCL10 NAb) in the lower chamber was counted. The mean number of cells per field was determined from 5 fields per filter (mean ± SEM; n = 3 experiments). *P < 0.05, **P < 0.001 vs. 1% BCCs; ##P < 0.001 vs. 1% BCCs+MSCs. Scale bar: 500 μm. (D) Naive MDA-231 BCCs were seeded on top of Matrigel-coated chamber inserts. The number of cells that invaded through the Matrigel in response to CM from BCCs, MSCs, or BCCs+MSCs (with or without CXCL10 NAb) was counted (mean ± SEM; n = 3). *P < 0.05, **P < 0.001 vs. 1% BCCs; ##P < 0.001 vs. 1% BCCs+MSCs, ANOVA. Scale bar: 500 μm. (E and F) BCCs, MSCs, and BCCs+MSCs were analyzed for MMP9 (E) and LOX (F) mRNA levels, which were normalized to BCCs at 20% O2. **P < 0.001 vs. all other conditions.
CXCL10 is a major chemoattractant for activated T lymphocytes and natural killer cells (38). We therefore evaluated the effect of CXCL10 on the migration and invasion of BCCs. CM from BCCs+MSCs at 20% O2 stimulated increased migration and invasion of naive BCCs, and the effect was augmented when CM from hypoxic cocultures was used (Figure 5, C and D). This increase in migration and invasion was abrogated when CXCL10 NAb was added to the CM (Figure 5, C and D). Treatment of control EV and NTC subclones with recombinant CXCL10 stimulated invasion through Matrigel (tumor-derived extracellular matrix), whereas this effect was significantly decreased in DKD and CXCR3-knockdown (shCR3-1) subclones (Supplemental Figure 5, A and B). These data indicate that CXCL10 produced by MSCs stimulates the HIF-dependent migration and invasion of MDA-231 BCCs bearing the cognate receptor CXCR3.
To identify molecular mechanisms by which coculture of BCCs+MSCs may promote metastasis, we analyzed the expression of prometastatic HIF target genes (21, 22, 39). Expression of matrix metalloproteinase 9 (MMP9) and lysyl oxidase (LOX) mRNAs was induced by coculture and hypoxia (Figure 5, E and F), whereas coculture did not induce expression of LOX-like 4 (LOXL4), angiopoietin-like 4 (ANGPTL4), or L1 cell adhesion molecule (L1CAM) mRNAs (Supplemental Figure 5, C–E).
Coculture of HIF- or CXCR3-knockdown BCCs with MSCs inhibits metastasis.
Inhibition of HIF-1α or HIF-2α expression impairs primary breast tumor growth and lung metastasis after MFP injection (26, 27). To evaluate whether HIF-1α/HIF-2α expression in BCCs is required for cocultured MSCs to stimulate metastasis, EV and DKD cells were cocultured with MSCs at 20% O2 for 48 hours prior to MFP injection. Primary tumor growth was not affected by MSC coculture (Figure 6A). The growth rate of tumors derived from DKD cells was decreased, as previously reported (27). When tumor volume reached 1,300 mm3, mice were euthanized to examine metastasis to lungs and LNs. EV+MSCs, but not DKD+MSCs, significantly increased metastasis to the lungs, as determined by qPCR using human-specific primers or by counting the number of metastatic foci on H&E-stained lung sections (Figure 6, B and C). Human Y chromosome sequences were not detectable by qPCR analysis of genomic DNA isolated from lungs of mice that received MFP injections of BCCs+MSCs, which indicated that MSCs did not colonize the lungs. EV+MSCs, but not DKD+MSCs, also increased metastasis to the ipsilateral axillary LN, as demonstrated by immunohistochemistry using an antibody specific for human vimentin (Figure 6, D and E).
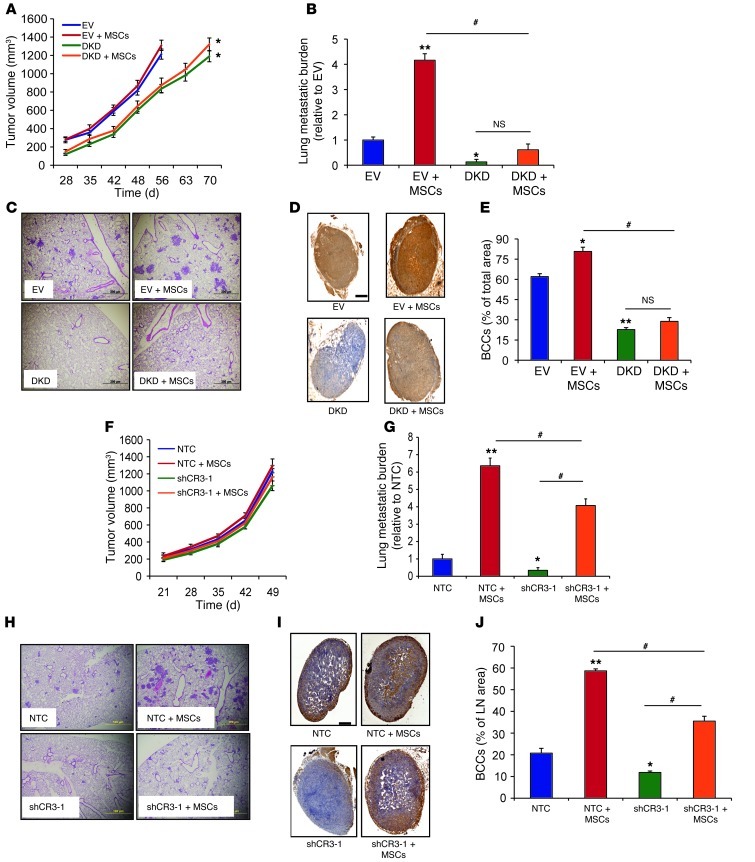
Figure 6. Effect of MSC coculture on metastasis is lost when HIF or CXCR3 expression is inhibited in BCCs.
(A–E) EV cells (1 × 106), DKD cells (1 × 106), and EV+MSCs and DKD+MSCs (0.5 × 106 each) were cultured for 48 hours and implanted in the MFP. (A) Primary tumor volumes were measured. Lungs and LNs were harvested when the volume reached 1,300 mm3. (B) Lung DNA was isolated and used to quantify metastatic burden by qPCR with human-specific HK2 primers. (C) H&E-stained lung sections. Scale bar: 200 μm. (D) LN sections were analyzed with human-specific vimentin antibody. Scale bar: 0.5 mm. (E) Percent total LN area occupied by BCCs, determined by image analysis (mean ± SEM; n = 5). *P < 0.05, **P < 0.001 vs. EV; #P < 0.05 as indicated, ANOVA. (F–J) NTC cells (1 × 106), shCR3-1 cells (1 × 106), and NTC+MSCs and shCR3-1+MSCs (0.5 × 106 each) were implanted in MFP. (F) Tumor volume. Lungs and LNs were harvested on day 50. (G) Lung DNA was isolated and used to quantify metastatic burden. (H) H&E-stained lung sections. Scale bar: 100 μm. (I) LN sections were analyzed with human-specific vimentin antibody. Scale bar: 0.5 mm. (J) Percent total LN area occupied by BCCs (mean ± SEM; n = 5). *P < 0.05, **P < 0.001 vs. NTC; #P < 0.05 as indicated, ANOVA.
To further investigate the role of CXCR3 in breast cancer pathogenesis, we orthotopically implanted CXCR3-knockdown BCCs in the MFP of SCID mice. Primary tumor growth and metastasis of 2 independent subclones of CXCR3-knockdown MDA-231 cells (shCR3-1 and shCR3-3) were compared with those of NTC cells in the absence of MSCs. CXCR3 knockdown in MDA-231 cells did not affect growth of the primary tumor (Supplemental Figure 6, A and B). However, mice bearing the CXCR3-knockdown MDA-231 tumors showed a significantly decreased number of metastatic foci and total metastatic burden in the lungs compared with mice bearing NTC tumors (Supplemental Figure 6, C–E). Ipsilateral axillary LNs isolated from mice bearing CXCR3-knockdown breast tumors contained a significantly decreased number of cancer cells, as assessed by immunohistochemical staining for human vimentin (Supplemental Figure 6, F and G).
Next, we cocultured CXCR3-knockdown or NTC control MDA-231 cells with MSCs for 48 hours prior to MFP injection. Inhibition of CXCR3 expression in BCCs decreased the ability of MSCs to enhance BCC metastasis to the lungs and LNs without affecting primary tumor growth (Figure 6, F–J). These results indicated that CXCR3 promotes vascular and lymphatic metastasis of BCCs, which is stimulated by crosstalk between BCCs and MSCs.
Coculture enhances HIF activity.
We next analyzed HIF-1α levels in EV+MSCs and DKD+MSCs. HIF-1α expression was induced in EV+MSCs under nonhypoxic conditions, and was further enhanced with hypoxia (Figure 7A). HIF-1α expression was not induced in DKD+MSCs at 20% or 1% O2 (Figure 7A). Coculture did not induce HIF1A mRNA levels (Supplemental Figure 7). Thus, coculture of BCCs+MSCs specifically increased HIF-1α protein levels under both hypoxic and nonhypoxic conditions.
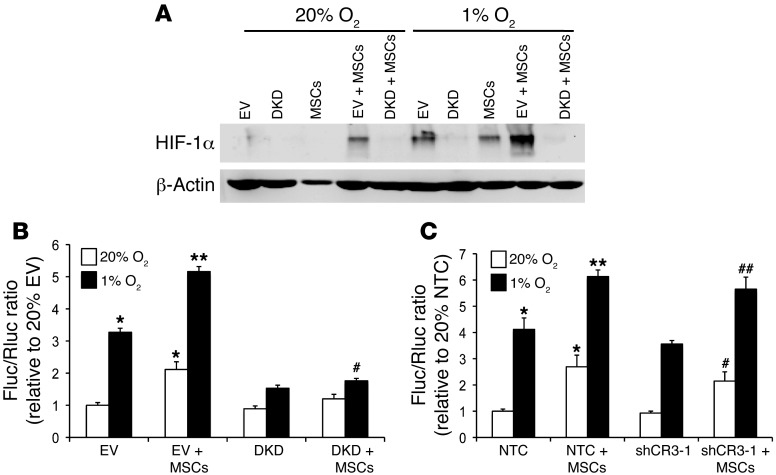
Figure 7. Coculture of BCCs+MSCs enhances HIF-1α expression and HIF transcriptional activity in BCCs.
(A) HIF-1α protein levels in EV cells, DKD cells, MSCs, EV+MSCs, and DKD+MSCs cultured at 20% or 1% O2. β-Actin was used as a loading control. (B) EV and DKD cells were cotransfected with p2.1 and pSV-Renilla and cocultured with MSCs or not for 48 hours at 20% or 1% O2. The Fluc/Rluc ratio was normalized to the value for EV cells at 20% O2. *P < 0.05, **P < 0.001 vs. 20% EV; #P < 0.05 vs. 1% EV, 1-way ANOVA. (C) NTC and shCR3-1 cells were cotransfected with p2.1 and pSV-Renilla and cocultured with MSCs or not for 48 hours at 20% or 1% O2. The Fluc/Rluc ratio was normalized to NTC cells at 20% O2 (mean ± SEM; n = 3). *P < 0.05, **P < 0.001 vs. 20% NTC; #P < 0.05 vs. 20% shCR3-1; ##P < 0.01 vs. 1% shCR3-1, 1-way ANOVA.
To test whether coculture of BCCs+MSCs stimulates HIF transcriptional activity, MDA-231 BCCs were cotransfected with HIF-dependent reporter plasmid p2.1, which contains a hypoxia response element (HRE) from the human ENO1 gene upstream of SV40 promoter and firefly luciferase (Fluc) coding sequences (40), and pSV-Renilla, in which Renilla luciferase (Rluc) expression is driven by the SV40 promoter only. The ratio of Fluc/Rluc activity is a specific measure of HIF transcriptional activity. Transfected cells were cocultured with MSCs and exposed to 20% or 1% O2 for 48 hours. Coculture significantly increased HIF transcriptional activity in EV+MSCs at both 20% and 1% O2 (Figure 7B), consistent with the induction of HIF-1α protein expression (Figure 7A). Coculture of DKD+MSCs significantly reduced HIF transcriptional activity (Figure 7B). In contrast, the effect of coculture on HIF transcriptional activity was not lost when CXCR3 expression was knocked down in MDA-231 BCCs (Figure 7C). Taken together, these results indicate that coculture of BCCs+MSCs enhances HIF transcriptional activity in BCCs by a mechanism independent of CXCR3 signaling that involves increased HIF-1α protein levels.
CXCR3 is a HIF-1 target gene.
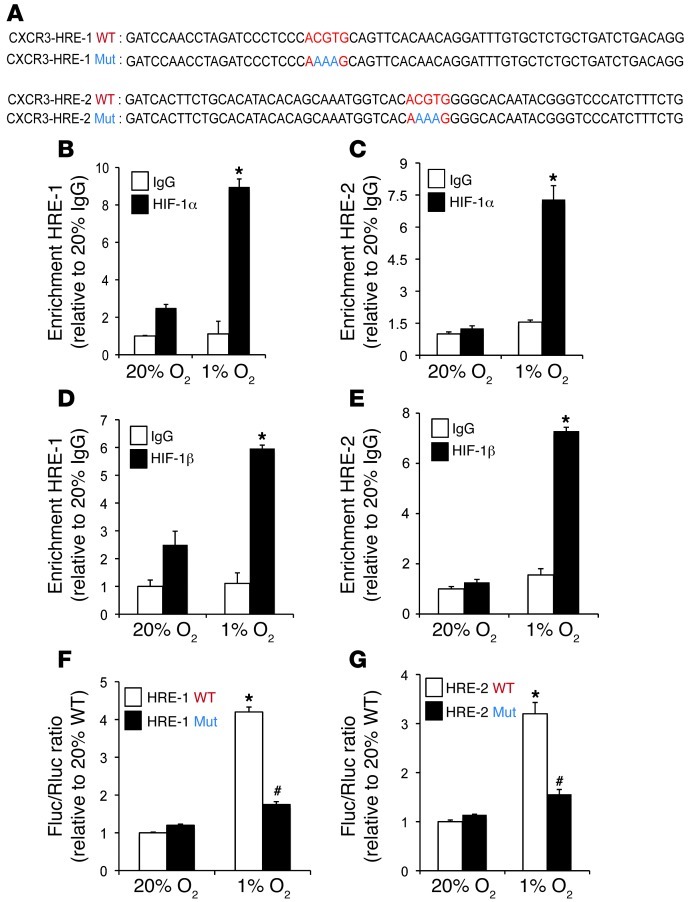
Analysis of the human CXCR3 gene sequence revealed candidate HREs in the 5′-flanking (HRE-1) and 3′-untranslated (HRE-2) regions that contained the HIF binding site sequence 5′-ACGTG-3′ followed by 5′-CACA-3′ (Figure 8A). This bipartite structure was first identified in the human EPO gene (41) and subsequently found in other HREs, including those in the ALDOA (39), COX4I2 (41), PKM2 (42), and ANGPTL4 (27) genes (Supplemental Figure 8). To determine whether HIF-1 binds at these sites, ChIP assays were performed in MDA-231 BCCs, which demonstrated hypoxia-inducible binding of HIF-1α and HIF-1β to both HRE-1 and HRE-2 (Figure 8, B–E). To test whether these putative HREs are functional, a 60-bp fragment encompassing HRE-1 or HRE-2 (Figure 8A) was inserted downstream of SV40 promoter and Fluc coding sequences in the reporter plasmid pGL2 promoter. MDA-231 BCCs were cotransfected with pGL2–CXCR3–HRE-1 or pGL2–CXCR3–HRE-2 and pSV-Renilla and exposed to 20% or 1% O2 for 24 hours. Both HRE-1 and HRE-2 significantly increased Fluc activity in hypoxic MDA-231 BCCs (Figure 8, F and G). Mutation of 5′-CGT-3′ to 5′-AAA-3′ (Figure 8A), which eliminates HIF-1 binding (40, 41), in HRE-1 or HRE-2 significantly decreased hypoxia-induced luciferase activity (Figure 8, F and G). Taken together, the ChIP and reporter data indicated that HIF-1 binds to HREs present in the 5′-flanking and 3′-untranslated regions of the human CXCR3 gene and directly activates its transcription under hypoxic conditions.
Figure 8. CXCR3 is a HIF-1 target gene.
(A) Candidate HREs were identified in the 5′-flanking region (HRE-1) and 3′-untranslated region (HRE-2) of the human CXCR3 gene. HREs containing the WT (5′-ACGTG-3′) or mutant (Mut; 5′-AAAAG-3′) HIF-1 binding site sequence were inserted into the Fluc vector pGL2 promoter. (B–E) MDA-231 BCCs were incubated at 20% or 1% O2 for 24 hours, and ChIP assays were performed using IgG, HIF-1α, or HIF-1β antibodies. Specific primers flanking HRE-1 and HRE-2 were used for qPCR, and values were normalized to IgG at 20% O2 (mean ± SEM; n = 3). *P < 0.05 vs. all other conditions, Student’s t test on log-converted values. (F and G) pGL2 promoter containing WT or mutant HRE was cotransfected with pSV-Renilla into MDA-231 BCCs, which were incubated at 20% or 1% O2 for 24 hours. The Fluc/Rluc ratio was normalized to WT at 20% O2 (mean ± SEM; n = 3). *P < 0.05 vs. 20% WT; #P < 0.05 vs. 1% WT, Student’s t test.
HIF regulates PGF expression in BCCs and VEGFR1 expression in MSCs.
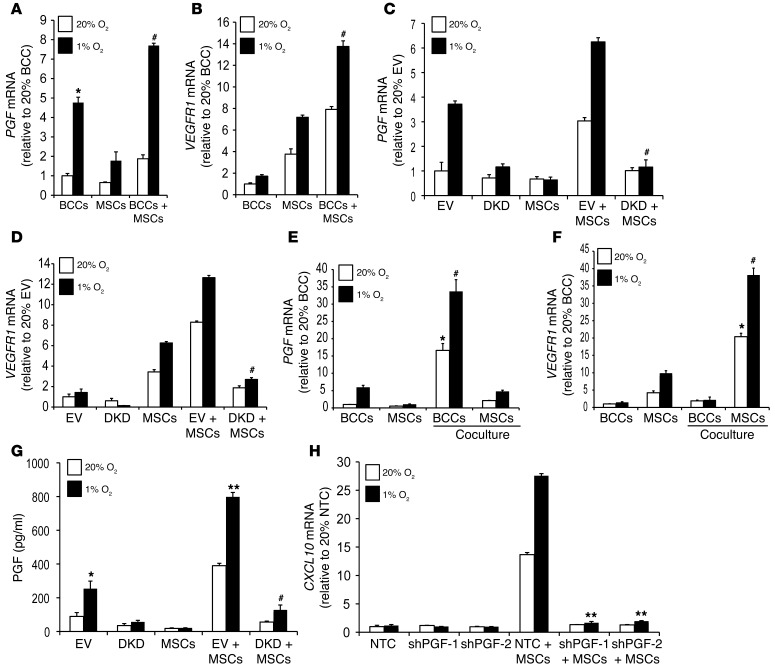
The data presented above demonstrate that production of the chemokines CXCL10 and CCL5 by MSCs enables them to communicate with BCCs, which express the cognate receptors CXCR3 and CCR5, respectively. However, the loss of CXCL10 expression by MSCs when cocultured with BCCs in which CXCR3 was inhibited suggested that BCCs must also communicate with MSCs, i.e., there must be bidirectional signaling. Furthermore, the homing of MSCs to tumors demonstrated in Figure 1 is likely to involve a secreted factor produced by BCCs that binds to a cognate receptor on MSCs. Based on our previous studies of VEGFR1 expression in MSCs (43), we hypothesized that BCCs may communicate with MSCs by producing placental growth factor (PGF), a ligand that binds specifically to VEGFR1. PGF expression in breast cancer biopsies is associated with an increased risk of metastasis and patient mortality (44). PGF mRNA expression was significantly increased in hypoxic MDA-231 BCCs, and VEGFR1 mRNA expression was induced in hypoxic MSCs (Figure 9, A and B). Coculture of BCCs+MSCs induced the expression of PGF and VEGFR1 mRNA. Expression of PGF and VEGFR1 mRNA was markedly decreased in DKD+MSCs compared with EV+MSCs (Figure 9, C and D). Flow sorting of BCCs and MSCs after coculture showed that PGF expression was induced in BCCs, and VEGFR1 expression was induced in MSCs (Figure 9, E and F).
Figure 9. HIF regulates PGF and VEGFR1 expression and facilitates bidirectional signaling.
(A and B) BCCs, MSCs, or BCCs+MSCs were cultured at 20% or 1% O2 for 48 hours. PGF and VEGFR1 mRNA levels, determined by RT-qPCR, were normalized to those in BCCs at 20% O2. *P < 0.05 vs. 20% BCCs; #P < 0.01 vs. 20% BCCs+MSCs, ANOVA. (C and D) EV cells, DKD cells, MSCs, EV+MSCs, and DKD+MSCs were exposed to 20% or 1% O2 for 48 hours. PGF and VEGFR1 mRNA levels were normalized to those in BCCs at 20% O2. #P < 0.01 vs. 1% EV+MSCs. (E and F) GFP+ BCCs were cocultured with MSCs at 20% or 1% O2 for 48 hours, followed by FACS based on GFP fluorescence of BCCs and CD105 immunofluorescence of MSCs. RNA was extracted from flow-sorted cells for analysis of PGF and VEGFR1 expression. *P < 0.05 vs. 20% MSCs or BCCs alone; #P < 0.01 vs. 1% MSCs or BCCs alone. (G) CM was isolated from EV cells, DKD cells, MSCs, EV+MSCs, and DKD+MSCs cultured for 48 hours at 20% or 1% O2. ELISA was performed to determine PGF protein levels in CM (mean ± SEM; n = 3). *P < 0.05, **P < 0.001 vs. 20% EV; #P < 0.01 vs. 1% EV+MSCs, ANOVA. (H) NTC, shPGF-1, and shPGF-2 MDA-231 subclones were cultured alone or with MSCs at 20% or 1% O2 for 48 hours. CXCL10 mRNA expression was analyzed by RT-qPCR (mean ± SEM; n = 3). **P < 0.001 vs. 1% NTC+MSCs, ANOVA.
We assayed secreted PGF levels in CM from BCCs cultured alone or from BCCs+MSCs. PGF levels were induced in the hypoxic CM isolated from MDA-231 or MDA-435 BCCs, which were further increased with coculture and hypoxia (Supplemental Figure 9, A and B). CM from EV+MSCs had dramatically increased PGF levels, which were further increased under hypoxic conditions, whereas PGF levels were markedly decreased in CM from DKD+MSCs (Figure 9G).
To determine whether PGF regulates CXCL10→CXCR3 signaling between MSCs and BCCs, we inhibited PGF expression in MDA-231 BCCs using shRNA. The knockdown efficiency of subclones (shPGF-1 and shPGF-2) was determined by ELISA (Supplemental Figure 9C). Analysis of PGF-knockdown MDA-231 cells cocultured with MSCs revealed a complete loss of CXCL10 mRNA expression and CXCL10 protein secretion (Figure 9H and Supplemental Figure 9D). CXCR3 expression was not changed in PGF-knockdown MDA-231 cells (Supplemental Figure 9E), which ruled out the possibility that autocrine effects of PGF on BCCs regulate CXCR3 expression.
HIF and PGF expressed by BCCs facilitate MSC migration and homing.
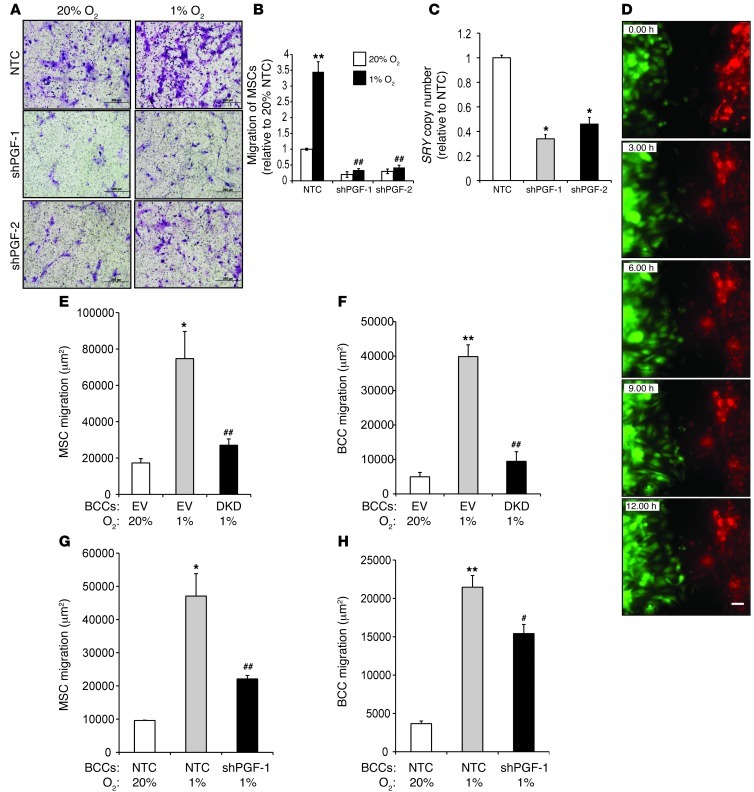
HIF-1 is required for VEGFR1 expression and chemotactic responses to PGF in mouse MSCs (43). To study whether PGF secretion from human BCCs stimulates the motility of human MSCs, we isolated CM from NTC, shPGF-1, and shPGF-2 MDA-231 subclones. Compared with CM from NTC cells, CM from shPGF-1 or shPGF-2 knockdown cells induced significantly less MSC migration (Figure 10, A and B). Furthermore, CM from hypoxic NTC cells markedly increased the chemotactic activity of MSCs, whereas CM from hypoxic shPGF-1 or shPGF-2 cells had no stimulatory effect.
Figure 10. HIF and PGF expressed by BCCs are required for MSC migration and homing.
(A and B) Migration of MSCs in response to CM isolated from NTC, shPGF-1, and shPGF-2 MDA-231 subclones cultured at 20% or 1% O2. MSCs were seeded on the top of the Boyden chamber, and the number of cells that migrated through the filter in response to CM in the lower chamber was counted under light microscopy after staining with crystal violet. Data were normalized to CM isolated from NTC cells at 20% O2. **P < 0.001 vs. 20% NTC; ##P < 0.005 vs. 1% NTC. Scale bar: 200 μm. (C) 1 × 106 NTC, shPGF-1, or shPGF-2 cells were implanted into the MFP of SCID mice. Recruitment of MSCs to the primary tumor was analyzed by qPCR for SRY (mean ± SEM; n = 5). *P < 0.05 vs. NTC, Student’s t test. (D) Representative images acquired by time-lapse photomicroscopy of labeled MDA-231 BCCs (green) and MSCs (red) cocultured in a LiveAssay 2-chamber device coated with fibronectin. Scale bar: 200 μm. (E and F) Migration of MSCs (E) and EV or DKD BCCs (F) after 12 hours of coculture. *P < 0.05, **P < 0.01 vs. 20% EV; ##P < 0.01 vs. 1% EV. (G and H) Migration of MSCs (G) and NTC or shPGF-1 BCCs (H) after 12 hours of coculture. *P < 0.05, **P < 0.01 vs. 20% NTC; #P < 0.05, ##P < 0.01 vs. 1% NTC.
To determine whether PGF secretion from BCCs promotes recruitment of MSCs to the primary tumor site, NTC, shPGF-1 knockdown, and shPGF-2 knockdown MDA-231 cells were implanted in the MFP of SCID mice. When the tumors had grown to 250 mm3, human MSCs were injected via tail vein, and primary tumors were harvested 16 hours later. Recruitment of MSCs to the primary tumor was examined by qPCR analysis of genomic DNA using SRY gene primers. The recruitment of MSCs was significantly decreased in tumors derived from PGF-knockdown MDA-231 cells compared with NTC-derived tumors (Figure 10C).
To study migration in a coculture assay, CMFDA-labeled MDA-231 BCCs and CMTPX-labeled MSCs were cocultured in a LiveAssay 2-chamber device coated with fibronectin and allowed to attach overnight. Time-lapse photomicroscopy (Figure 10D), which was performed for 12 hours at 20% or 1% O2, revealed that the migration of MSCs and BCCs toward each other was significantly increased under hypoxic conditions (Figure 10, E–H). Coculture with DKD cells or shPGF-1 cells led to decreased migration of MSCs compared with those cocultured with control BCCs (Figure 10, E and G).
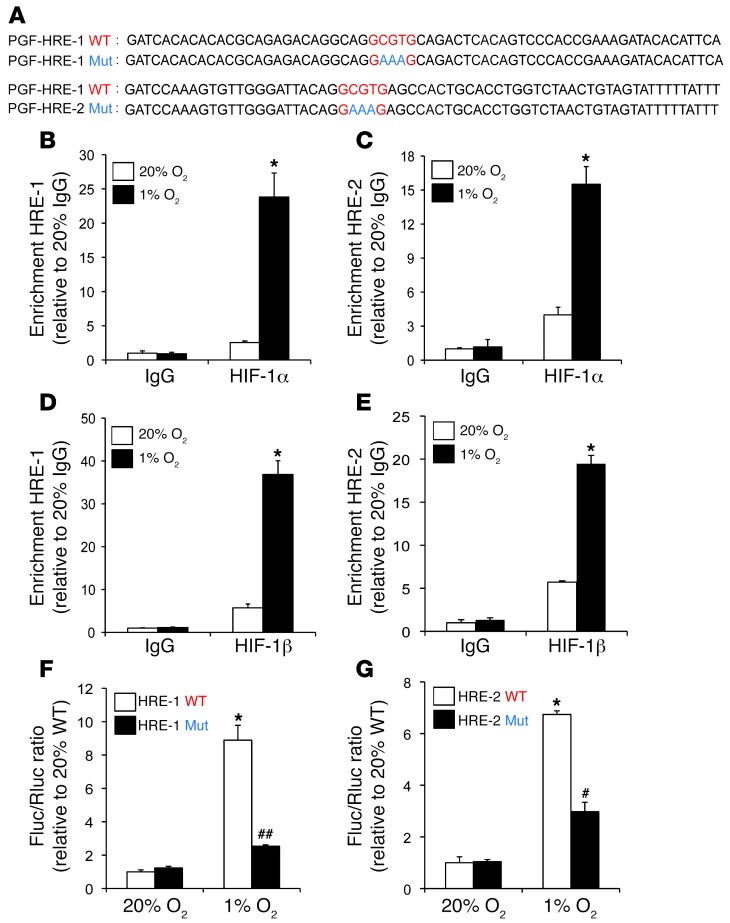
PGF is a HIF-1 target gene.
Analysis of the human PGF gene sequence revealed 2 candidate HIF-1 binding sites in the 5′ flanking region. In PGF, HRE-1 and HRE-2 are located 200 and 2,000 bp upstream of the transcription start site, respectively (Figure 11A). To determine whether HIF-1 binds to these sites, ChIP assays were performed in MDA-231 BCCs, which demonstrated hypoxia-inducible binding of HIF-1α and HIF-1β to both HRE-1 and HRE-2 (Figure 11, B–E), providing evidence for direct regulation of PGF gene transcription by HIF-1. To test whether these putative HREs in the PGF gene are functional, a 60-bp fragment encompassing HRE-1 or HRE-2 (Figure 11A) was inserted downstream of SV40 promoter and Fluc coding sequences in the reporter plasmid pGL2 promoter. MDA-231 BCCs were cotransfected with pGL2–PGF–HRE-1 or pGL2–PGF–HRE-2 and pSV-Renilla and exposed to 20% or 1% O2 for 24 hours. Both HRE-1 and HRE-2 significantly increased luciferase activity in hypoxic MDA-231 BCCs (Figure 11, F and G). Mutation of 5′-CGT-3′ to 5′-AAA-3′ in HRE-1 or HRE-2 significantly decreased hypoxia-induced luciferase activity (Figure 11, F and G). Taken together, the ChIP and reporter data indicated that HIF-1 binds to HREs present in the 5′-flanking region of the human PGF gene and directly activates its transcription under hypoxic conditions.
Figure 11. PGF is a HIF-1 target gene.
(A) Candidate HREs were identified in the 5′-flanking region. HRE-1 and HRE-2 were located 0.2 kb and 2 kb, respectively, from the transcription start site. HREs containing WT (5′-GCGTG-3′) or mutant (5′-GAAAG-3′) HIF-1 binding site sequences were inserted into pGL2 promoter. (B–E) MDA-231 BCCs were incubated at 20% or 1% O2 for 24 hours, and ChIP assays were performed using IgG, HIF-1α, or HIF-1β antibodies. Specific primers flanking HRE-1 and HRE-2 were used for qPCR, and values were normalized to IgG at 20% O2 (mean ± SEM; n = 3). *P < 0.05 vs. all other conditions, Student’s t test on log-converted values. (F and G) WT and mutant HRE sequences were inserted into pGL2 promoter and cotransfected with pSV-Renilla into MDA-231 BCCs, which were incubated at 20% or 1% O2 for 24 hours. The Fluc/Rluc ratio was normalized to WT at 20% O2 (mean ± SEM; n = 3). *P < 0.05 vs. 20% WT; #P < 0.05, ##P < 0.005 vs. 1% WT, Student’s t test.
PGF promotes the metastasis of breast cancer cells.
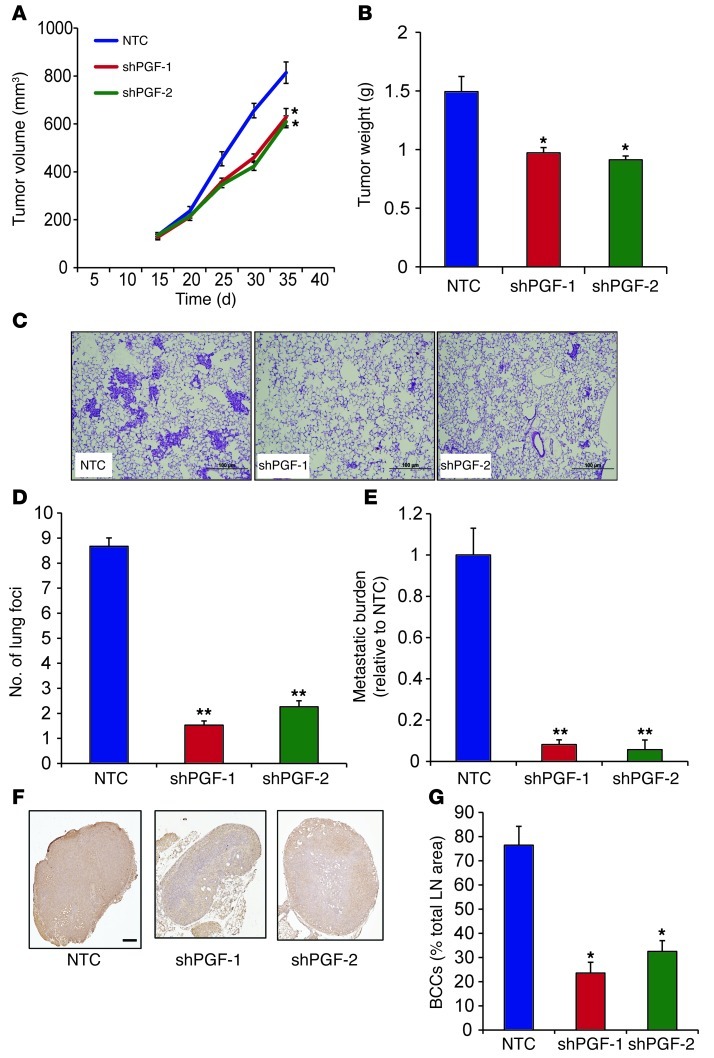
To further investigate the role of PGF in breast cancer pathogenesis, PGF-knockdown BCCs were orthotopically implanted in the MFP of SCID mice. Primary tumor growth and metastasis of the independent shPGF-1 and shPGF-2 MDA-231 subclones were compared with those of NTC control cells. PGF knockdown in MDA-231 BCCs modestly inhibited growth of the primary tumor (Figure 12, A and B). However, mice bearing the PGF-knockdown MDA-231 tumors showed a significantly decreased number of metastatic foci and total metastatic burden in the lungs compared with NTC tumor–bearing mice (Figure 12, C–E). Ipsilateral axillary LNs isolated from mice bearing PGF-knockdown breast tumors contained a significantly decreased number of cancer cells, as assessed by immunohistochemical staining for human vimentin (Figure 12, F and G). Thus, PGF plays a significant role in promoting breast cancer metastasis.
Figure 12. PGF promotes lung and LN metastasis of BCCs.
1 × 106 NTC, shPGF-1, and shPGF-2 MDA-231 subclones were implanted in the MFP of SCID mice. (A) Primary tumor volumes were determined serially. *P < 0.05 vs. NTC, 1-way ANOVA. (B) Primary tumor weights were measured at the end of the experiment. *P < 0.05 vs. NTC, 1-way ANOVA. (C) Photomicrographs of H&E-stained lung sections. Scale bar: 100 μm. (D) Metastatic foci in lung sections. At least 3 random fields were counted per section. **P < 0.005 vs. NTC, 1-way ANOVA. (E) Lung DNA was analyzed by qPCR with human HK2 primers to quantify metastatic burden. **P < 0.005 vs. NTC, 1-way ANOVA. (F) LN sections were stained with human-specific vimentin antibody. Scale bar: 0.5 mm. (G) LN metastasis was quantified by image analysis. *P < 0.05 vs. NTC, 1-way ANOVA.
Discussion
In this study, we demonstrated that HIFs are critical regulators of remarkably complex bidirectional MSC-BCC interactions that promote breast cancer metastasis (Figure 13). Interactions between BCCs and MSCs were mediated by CXCL10→CXCR3, CCL5→CCR5, and PGF→VEGFR1 signaling, which was further augmented by at least 4 feed-forward loops, in which (a) PGF production by BCCs induced expression of CXCL10 by MSCs; (b) CXCL10 secretion by MSCs stimulated CXCR3 expression by BCCs; (c) CXCR3 activity in BCCs stimulated CXCL10 expression in MSCs; and (d) MSCs stimulated HIF-1α expression and HIF transcriptional activity in BCCs through a CXCR3-independent mechanism. Finally, we demonstrated that hypoxia augmented all of these signaling pathways in a HIF-dependent manner. Our data establish CXCR3 and PGF as direct HIF-1 target genes in BCCs, and we have previously demonstrated that VEGFR1 is a direct HIF-1 target gene in MSCs (43). CXCR3 expression has been previously associated with decreased overall survival in breast cancer patients and shown to promote lung metastasis in mice (45).
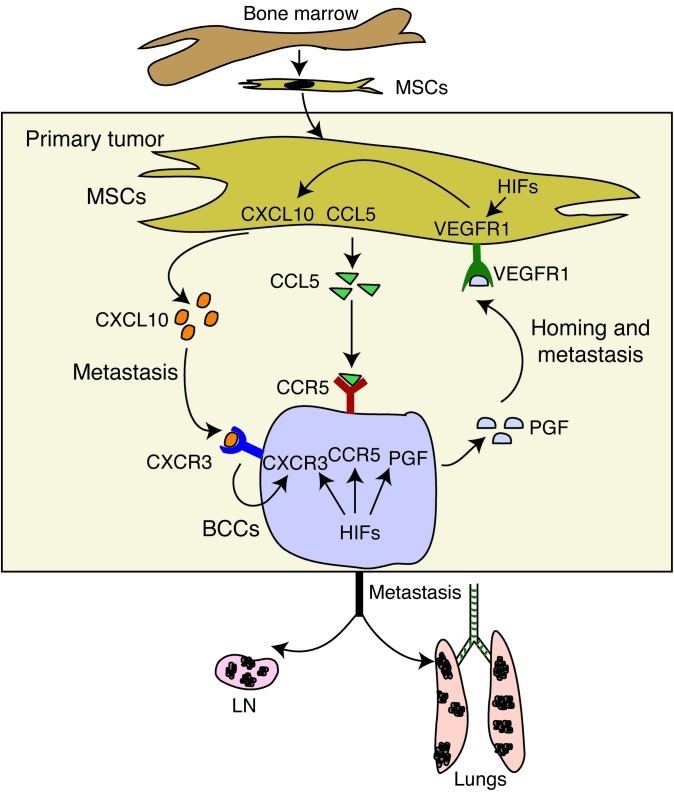
Figure 13. Bidirectional signaling between BCCs and MSCs.
Hypoxia induces recruitment of bone marrow–derived MSCs to the primary tumor site. MSC-BCC interaction induces CXCL10, CCL5, and VEGFR1 expression in MSCs and CXCR3, CCR5, and PGF expression in BCCs. MSC→BCC interaction is mediated by CCL5→CCR5 and CXCL10→CXCR3 signaling. BCC→MSC interaction is mediated by PGF→VEGFR1 signaling, which induces CXCL10 expression in MSCs and thereby establishes a positive feedback loop between the 2 cell types. The PGF→VEGFR1 interaction is important for MSC homing, and CXCL10→CXCR3 and CCL5→CCR5 signaling promote BCC metastasis. The consequence of these interactions is the expression of genes that enhance invasion and the metastasis of BCCs to the lungs and LNs. The expression of CXCR3 and PGF (and probably CCR5) in BCCs as well as VEGFR1 (and perhaps CXCL10 and CCL5) in MSCs are regulated by HIFs.
Although our study was inspired by the work of other investigators who first demonstrated a prometastatic effect of MSCs on BCCs via CCL5→CCR5 signaling (31), we modified the experimental approach and advanced our understanding of the mechanisms and consequences of MSC-BCC interaction in several important aspects. First, MFP injection was performed so that BCCs and MSCs were placed in the breast rather than in a heterologous (subcutaneous) microenvironment. Second, the MSC/BCC ratio was decreased from 3:1 to 1:1, which may better model the cellular composition of human breast cancers. Third, coculture of BCCs+MSCs was performed for 48 hours prior to MFP injection in order to activate bidirectional signaling; moreover, in our model, there was no prometastatic effect of coinjected MSCs in the absence of prior coculture. Fourth, we found that MSC-BCC interaction promoted metastasis via lymphatic and vascular routes to the regional LNs and lungs, respectively. Fifth, we observed that HIF regulated CXCL10→CXCR3 and PGF→VEGFR1 signaling, as well as the previously reported CCL5→CCR5 signaling (31), between MSCs and BCCs. Finally, we established the existence of bidirectional signaling as well as a potential mechanism for the homing of MSCs to the primary tumor (i.e., HIF-dependent PGF expression by hypoxic BCCs).
We have recently demonstrated that hypoxia promotes the metastasis of BCCs to the lungs through the HIF-dependent expression of (a) L1CAM, a cell adhesion molecule that mediates BCC-EC interactions and thereby induces the margination of circulating BCCs in the pulmonary vasculature (27); (b) ANGPTL4, a secreted protein that inhibits EC-EC interactions and thereby facilitates BCC extravasation through the vascular endothelium into the lung parenchyma (27); and (c) LOX and LOXL4, secreted enzymes that mediate collagen crosslinking in the lungs of tumor-bearing mice and thereby initiate metastatic niche formation (26, 28). Coculture of MSCs with MDA-231 BCCs augmented the hypoxia-induced expression of LOX mRNA, whereas ANGPTL4, L1CAM, and LOXL4 mRNA expression was not affected. HIF-dependent expression of the matrix metalloproteases MMP2, MMP9, and MMP14 has been reported (46–48), and coculture of BCCs+MSCs augmented the hypoxia-induced expression of MMP9, but not MMP2 or MMP14. Induction of LOX and MMP9 expression was consistent with the increased invasive properties of BCCs subjected to coculture and/or hypoxia. These results provide specific molecular mechanisms by which MSC coculture may promote BCC metastasis, which appears to involve activation of only a subset of HIF target genes. Further studies are required to establish the mechanism that accounts for this surprising selectivity.
We demonstrated that intravenous injection of BCCs+MSCs did not result in increased lung colonization, compared with injection of BCCs only. This result was consistent with our finding that coculture of BCCs+MSCs increased the expression of genes encoding proteins (e.g., LOX and MMP9) that promote metastasis by mediating tissue invasion and premetastatic niche formation, which involve processes that occur in the primary tumor and are therefore not assayed by intravenous injection of cells. In contrast, the expression of ANGPTL4 and L1CAM, which are HIF-regulated genes that promote extravasation of circulating BCCs into the lung parenchyma (27), was not induced by coculture with MSCs. Thus, our molecular data are consistent with the finding that coculture selectively induces the expression of gene products that mediate steps in the metastatic process that are not interrogated by analysis of lung colonization after intravenous injection.
We recently reported that treatment of mice with the HIF inhibitors digoxin and acriflavine significantly decreases the metastatic dissemination of MDA-231 and MDA-435 triple-negative BCCs from breast to lung (27, 28). Our findings that digoxin treatment blocked the homing of MSCs to the primary breast tumor and that acriflavine blocked coculture-induced CXCL10-CXCR3 and CCL5-CCR5 expression suggest that inhibition of MSC-BCC interaction represents yet another mechanism by which HIF inhibitors prevent breast cancer metastasis and underscore the need for clinical trials of these drugs in women with breast cancer.
Methods
Vectors and cell culture.
Oligonucleotides encoding shRNA targeting HIF2A mRNA (nucleotides 1,992–2,012; GenBank NM_001430) or HIF1A mRNA (nucleotides 2,123–2,141; GenBank NM_181054) were inserted in place of the DNA sequence encoding shRNA against luciferase in lentiviral vector pRRL-LucshRNA-GFP. pLKO.1-puro lentiviral vectors encoding shRNA targeting CXCR3 mRNA (clone NM_001504) or PGF mRNA (clone NM_002632) were purchased from Sigma-Aldrich. The pLKO.1-puro recombinant vectors were cotransfected with plasmid pCMV-dR8.91 and plasmid encoding vesicular stomatitis virus G protein into 293T cells using FuGENE 6 (Roche Applied Science). Viral supernatant was collected 48 hours after transfection, filtered (0.45-μm pore size), and added to MDA-231 or MDA-435 BCCs in the presence of 8 μg/ml polybrene (Sigma-Aldrich). Puromycin (0.5 μg/ml) was added to the medium of cells transduced with pLKO.1-puro vectors for selection.
MDA-231 and MDA-435 BCCs were maintained in high-glucose (4.5 mg/ml) DMEM with 10% FBS and 1% penicillin/streptomycin. CXCR3 and PGF knockdown subclones were maintained in the presence of 0.5 μg/ml puromycin. Human bone marrow–derived MSCs (49) were obtained from the Tulane Center for Gene Therapy. MSCs were maintained in αMEM (Gibco; Invitrogen) supplemented with 20% FBS (Atlanta Biologicals) and 1% penicillin/streptomycin (Cellgro). Cells were maintained at 37°C in a 5% CO2, 95% air incubator (20% O2). Hypoxic cells (1% O2) were maintained at 37°C in a modular incubator chamber (Billups-Rothenberg) flushed with a gas mixture containing 1% O2, 5% CO2, and 94% N2.
Coculture experiments.
Equal numbers of MSCs and MDA-231 or MDA-435 BCCs were seeded in a 1:1 ratio of DMEM with 10% FBS/αMEM with 20% FBS.
RT-qPCR.
Total RNA was extracted from cells and tissue using TRIzol (Invitrogen) and treated with DNase I (Ambion). 1 μg total RNA was used for first-strand DNA synthesis using iScript cDNA Synthesis system (BioRad), and qPCR was performed using human-specific primers and SYBR Green qPCR Master Mix (Fermentas). For each primer pair, annealing temperature was optimized by gradient PCR. The expression of each target mRNA relative to 18S rRNA was calculated based on Ct as 2–Δ(ΔCt), in which ΔCt = Cttarget – Ct18S and Δ(ΔCt) = ΔCtsample – ΔCtcontrol (22). See Supplemental Table 1 for primer sequences.
Immunoblot assays.
Whole cell lysates were prepared in RIPA lysis buffer. Blots were probed with HIF-1α (Novus Biologicals), CXCR3 (R&D), pERK (Santa Cruz), and ERK (Santa Cruz) antibodies. HRP-conjugated anti-rabbit (Roche) and anti-mouse (Santa Cruz) secondary antibodies were used. Signal was detected using ECL Plus (GE Healthcare). Blots were reprobed with a polyclonal anti-actin antibody (Santa Cruz).
Animal studies.
Female 5- to 7-week-old SCID mice (NCI) were studied. Digoxin and saline for injection were obtained from the research pharmacy of the Johns Hopkins Hospital. Cells were harvested by trypsinization, washed in PBS, resuspended at 107 cells/ml in a 1:1 solution of PBS/Matrigel, and injected directly into the MFP. Primary tumors were measured in 3 dimensions (a, b, c), and volume was calculated as abc × 0.52. Primary tumors were harvested from the mammary gland. Lungs were perfused with PBS. One lung was inflated for formalin fixation and paraffin embedding, and the other was used to isolate genomic DNA (22).
Extraction of genomic Dna.
Lungs were digested with lysis buffer and proteinase K at 55°C, and genomic DNA was extracted with phenol-chloroform, precipitated with isopropanol, and washed with ethanol. 200 ng genomic DNA was used for qPCR to quantify human HK2 and mouse 18S rRNA gene sequences (22).
MSC recruitment assay.
2 × 106 MDA-231 BCCs were injected into the MFP. When the tumor reached 200 mm3, the mouse received daily intraperitoneal injections of saline or digoxin (2 mg/kg) for 1 week. MSCs were labeled by 30-minute exposure to 1 μM CMFDA (Invitrogen), 0.5 × 106 MSCs were injected intravenously, and the primary tumor was harvested after 16 hours. The primary tumor was halved for FACS analysis of labeled MSCs and for qPCR analysis of SRY copy number.
CXCL10 and PGF ELISA.
CXCL10 and PGF protein levels in CM were determined using ELISA kits (R&D Systems) according to the manufacturer’s protocol.
Chemokine array.
The levels of different chemokines in the CM were evaluated using a chemokine array (R&D Systems) according to the manufacturer’s protocol.
Migration assay.
BCCs were seeded onto uncoated filters in a 24-well Transwell chamber (8-mm pore size; Costar) and allowed to migrate for 8 hours in the presence or absence of CXCL10 and CXCL10 NAb. The cells that migrated to the underside of the filter were stained with crystal violet and counted under brightfield microscopy.
Invasion assay.
BCCs were seeded onto filters of a 24-well Transwell chamber that were coated with Matrigel (BD Biosciences). Invasion of the cells through the Matrigel to the underside of the filter was assessed 24 hours later by staining with crystal violet and counting under brightfield microscopy.
Analysis of LN metastasis.
Immunohistochemical analyses were performed on formalin-fixed, paraffin-embedded LNs. Sections (4 μm thick) were deparaffinized and rehydrated, antigens were retrieved using citrate buffer, and staining was performed using an antibody that specifically recognizes human vimentin (Santa Cruz) and analyzed by Image J software (NIH). The acquired images in RGB color were separated in different color channels by a color deconvolution method (50). The Image J plugin for color deconvolution has a built-in vector for separating hematoxylin and diaminobenzidine (DAB) staining. After color deconvolution, hematoxylin and DAB images were processed separately. By using 5 random test samples stained for vimentin, suitable threshold levels for hematoxylin and DAB were determined. These thresholds were used on both hematoxylin and DAB images and kept constant for analysis of the main image dataset. The extent of staining was calculated as the DAB-positive area divided by the hematoxylin-positive area.
ChIP assays.
MDA-231 BCCs were crosslinked with formaldehyde and lysed with SDS lysis buffer. Chromatin was sheared by sonication, and lysates were precleared with salmon sperm DNA/protein A agarose slurry (Millipore) and incubated with immunoprecipitating antibodies against HIF-1α (Santa Cruz), HIF-1β, and IgG (provided by K. Padgett, Novus Biologicals, Littleton, Colorado, USA). Salmon sperm DNA/protein A agarose slurry was added, and the agarose beads were washed sequentially with low- and high-salt wash buffers (0.1% SDS; 1% Triton X-100; 2 mM EDTA; and 20 mM Tris, pH 8.0; with 0.15 M and 0.5 M NaCl, respectively), LiCl wash buffer (0.25 M LiCl; 1% NP-40; 1% deoxycholate; 10 mM Tris, pH 8.0; and 1 mM EDTA), and TE buffer (10 mM Tris, pH 8.0, and 1 mM EDTA). DNA was eluted in 1% SDS with 0.1 M NaHCO3, and crosslinks were reversed by addition of 0.2 M NaCl. DNA was purified using phenol-chloroform extraction method, suspended in 30 μl TE buffer, and a 2-μl aliquot was used for qPCR.
Luciferase reporter assays.
MDA-231 BCCs were cotransfected with pGL2 promoter reporter containing WT or mutant HRE or pSV-Renilla. Luciferase activities were determined using the Dual-Luciferase Reporter System (Promega).
Time-lapse photomicroscopy.
MDA-231 BCCs were labeled with CellTracker Green CMFDA, and MSCs were labeled with CellTracker Red CMTPX (Invitrogen). The labeled cells were cocultured in a LiveAssay 2-chamber device coated with fibronectin (50 μg/ml) and allowed to attach overnight. After monolayers formed in each chamber, the middle wall was removed, and time-lapse photomicroscopy was performed with a Zeiss Axiovert 2000 phase-contrast microscope using ×20 apochromat objective lens with NA 1.4. Images were acquired using a Cascade 512BII camera (Roper Scientific) and processed using Slidebook 4.2.0 (Intelligent Imaging Innovations). Image analysis was performed by semiautomatic routine, written in MATLAB 2009 (Mathworks). Cell migration over time was calculated for both cell types. For each condition, the longest cell projection for each cell type was calculated manually using Image J (NIH).
Statistics.
Data are expressed as mean ± SEM. Differences between 2 groups and multiple groups were analyzed by 2-tailed Student’s t test and 1-way ANOVA, respectively. A P value less than 0.05 was considered significant.
Study approval.
Animal protocols were in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Johns Hopkins University Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Karen Padgett for providing anti–HIF-1β and IgG antibodies and the Tulane Center for Gene Therapy (supported by PHS grant P40-RR017447 from the NIH) for providing human MSCs. This work was supported by PHS grant U54-CA143868 from the NIH and by funds from the American Cancer Society and Johns Hopkins Institute for Cell Engineering. G.L. Semenza is an American Cancer Society Research Professor and the C. Michael Armstrong Professor at Johns Hopkins University School of Medicine.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(1):189–205. doi:10.1172/JCI64993.
References
- 1.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyckoff J, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64(19):7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 3.Goswami S, et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65(12):5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 5.Cardenas-Navia LI, Mace D, Richardson RA, Wilson DF, Shan S, Dewhirst MW. The pervasive presence of fluctuating oxygenation in tumors. Cancer Res. 2008;68(14):5812–5819. doi: 10.1158/0008-5472.CAN-07-6387. [DOI] [PubMed] [Google Scholar]
- 6.Vaupel P, Hockel M, Mayer A. Detection and characterization of tumor hypoxia using pO2 histography. . Antioxid Redox Signal. 2007;9(8):1221–1235. doi: 10.1089/ars.2007.1628. [DOI] [PubMed] [Google Scholar]
- 7.Chan DA, Giaccia AJ. Hypoxia, gene expression, and metastasis. Cancer Metastasis Rev. 2007;26(2):333–339. doi: 10.1007/s10555-007-9063-1. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan R, Graham CH. Hypoxia-driven selection of the metastatic phenotype. Cancer Metastasis Rev. 2007;26(2):319–331. doi: 10.1007/s10555-007-9062-2. [DOI] [PubMed] [Google Scholar]
- 9.Dayan F, Mazure NM, Brahimi-Horn MC, Pouyssegur J. A dialogue between the hypoxia-inducible factor and the tumor microenvironment. Cancer Microenviron. 2008;1(1):53–68. doi: 10.1007/s12307-008-0006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong H, et al. Overexpression of hypoxia-inducible factor-1α in common human cancers and their metastases. Cancer Res. 1999;59(22):5830–5835. [PubMed] [Google Scholar]
- 11.Talks KL, et al. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIΦ-2α in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157(2):411–421. doi: 10.1016/S0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bos R, et al. Levels of hypoxia-inducible factor-1α during breast carcinogenesis. J Natl Cancer Inst. 2001;93(4):309–314. doi: 10.1093/jnci/93.4.309. [DOI] [PubMed] [Google Scholar]
- 13.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Generali D, et al. Hypoxia-inducible factor-1α expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006;12(15):4562–4578. doi: 10.1158/1078-0432.CCR-05-2690. [DOI] [PubMed] [Google Scholar]
- 15.Schindl M, et al. Overexpression of hypoxia-inducible factor-1α is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clin Cancer Res. 2002;8(6):1831–1837. [PubMed] [Google Scholar]
- 16.Bos R, et al. Levels of hypoxia-inducible factor-1α independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97(6):1573–1581. doi: 10.1002/cncr.11246. [DOI] [PubMed] [Google Scholar]
- 17.Giatromanolaki A, et al. c-erbB-2 related aggressiveness in breast cancer is hypoxia inducible factor-1α dependent. Clin Cancer Res. 2004;10(23):7972–7977. doi: 10.1158/1078-0432.CCR-04-1068. [DOI] [PubMed] [Google Scholar]
- 18.Dales JP, et al. Overexpression of hypoxia-inducible factor HIF-1α predicts early relapse in breast cancer: retrospective study in a series of 745 patients. Int J Cancer. 2005;116(5):734–739. doi: 10.1002/ijc.20984. [DOI] [PubMed] [Google Scholar]
- 19.Helczynska K, et al. Hypoxia-inducible factor-2α correlates to distant recurrence and poor outcome in invasive breast cancer. Cancer Res. 2008;68(22):9212–9220. doi: 10.1158/0008-5472.CAN-08-1135. [DOI] [PubMed] [Google Scholar]
- 20.Du R, et al. HIF-1α induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13(3):206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K, Qian DZ, Rey S, Wei H, Liu JO, Semenza GL. Anthracycline chemotherapy inhibits HIF-1 transcriptional activity and tumor-induced mobilization of circulating angiogenic cells. Proc Natl Acad Sci U S A. 2009;106(7):2353–2358. doi: 10.1073/pnas.0812801106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci U S A. 2009;106(42):17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Erler JT, et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15(1):35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erler JT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440(7088):1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 25.Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1α is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67(2):563–572. doi: 10.1158/0008-5472.CAN-06-2701. [DOI] [PubMed] [Google Scholar]
- 26.Wong CC, et al. Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc Natl Acad Sci U S A. 2011;108(39):16369–16374. doi: 10.1073/pnas.1113483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2011;31(14):1757–1770. doi: 10.1038/onc.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Wong CC, et al. Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. J Mol Med (Berl). 2012;90(7):803–815. doi: 10.1007/s00109-011-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal SK, Childs BH, Pegram M. Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat. 2011;125(3):627–636. doi: 10.1007/s10549-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Haibi CP, Karnoub AE. Mesenchymal stem cells in the pathogenesis and therapy of breast cancer. J Mammary Gland Biol Neoplasia. 2010;15(4):399–409. doi: 10.1007/s10911-010-9196-7. [DOI] [PubMed] [Google Scholar]
- 31.Karnoub AE, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 32.Coffelt SB, et al. The pro-inflammatory peptide LL-37 promotes ovarian tumor progression through recruitment of multipotent mesenchymal stromal cells. Proc Natl Acad Sci U S A. 2009;106(10):3806–3811. doi: 10.1073/pnas.0900244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studeny M, et al. Mesenchymal stem cells: potential precursors for tumor stroma and targeted-delivery vehicles for anticancer agents. J Natl Cancer Inst. 2004;96(21):1593–1603. doi: 10.1093/jnci/djh299. [DOI] [PubMed] [Google Scholar]
- 34.Cailleau R, Young R, Olive M, Reeves WJ., Jr Breast tumor cell lines from pleural effusions. J Natl Cancer Inst. 1974;53(3):661–674. doi: 10.1093/jnci/53.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, et al. Digoxin and other cardiac glycosides inhibit HIF-1α synthesis and block tumor growth. Proc Natl Acad Sci U S A. 2008;105(50):19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colvin RA, Campanella GS, Sun J, Luster AD. Intracellular domains of CXCR3 that mediate CXCL9, CXCL10, and CXCL11 function. J Biol Chem. 2004;279(29):30219–30227. doi: 10.1074/jbc.M403595200. [DOI] [PubMed] [Google Scholar]
- 37.Chambers AF. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res. 2009;69(13):5292–5293. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Z, Xu Y, Cai S. CXCL10 expression and prognostic significance in stage II and III colorectal cancer. Mol Biol Rep. 2010;37(6):3029–3036. doi: 10.1007/s11033-009-9873-z. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 40.Semenza GL, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 41.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12(12):5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okuyama H, Krishnamachary B, Zhou YF, Nagasawa H, Bosch-Marce M, Semenza GL. Expression of vascular endothelial growth factor receptor 1 in bone marrow-derived mesenchymal cells is dependent on hypoxia-inducible factor 1. J Biol Chem. 2006;281(22):15554–15563. doi: 10.1074/jbc.M602003200. [DOI] [PubMed] [Google Scholar]
- 44.Parr C, Watkins G, Boulton M, Cai J, Jiang WG. Placenta growth factor is over-expressed and has prognostic value in human breast cancer. Eur J Cancer. 2005;41(18):2819–2827. doi: 10.1016/j.ejca.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 45.Ma X, et al. CXCR3 expression is associated with poor survival in breast cancer and promotes metastasis in a murine model. Mol Cancer Ther. 2009;8(3):490–498. doi: 10.1158/1535-7163.MCT-08-0485. [DOI] [PubMed] [Google Scholar]
- 46.Manalo DJ, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105(2):659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 47.Petrella BL, Lohi J, Brinckerhoff CE. Identification of membrane type-1 matrix metalloproteinase as a target of hypoxia-inducible factor-2α in von Hippel-Lindau renal cell carcinoma. Oncogene. 2005;24(6):1043–1052. doi: 10.1038/sj.onc.1208305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koh MY, Lemos R, Jr, Liu X, Powis G. The hypoxia-associated factor switches cells from HIF-1α- to HIF-2α-dependent signaling promoting stem cell characteristics, aggressive tumor growth and invasion. Cancer Res. 2011;71(11):4015–4027. doi: 10.1158/0008-5472.CAN-10-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colter DC, Class R, DiGirolamo CM, Prockop DJ. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci U S A. 2000;97(7):3213–3218. doi: 10.1073/pnas.97.7.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.