SUMMARY
The consistent observation across all kingdoms of life that highly abundant proteins evolve slowly demonstrates that cellular abundance is a key determinant of protein evolutionary rate. However, other empirical findings, such as the broad distribution of evolutionary rates, suggest that additional variables determine the rate of protein evolution. Here, we report that under the global selection against the cytotoxic effects of misfolded proteins, folding stability (ΔG), simultaneous with abundance, is a causal variable of evolutionary rate. Using both theoretical analysis and multiscale simulations, we demonstrate that the anticorrelation between the pre-mutation ΔG and the arising mutational effect (ΔΔG), purely biophysical in origin, is a necessary requirement for abundance–evolutionary rate covariation. Additionally, we predict and demonstrate in bacteria that the strength of abundance–evolutionary rate correlation depends on the divergence time separating reference genomes. Altogether, these results highlight the intrinsic role of protein biophysics in the emerging universal patterns of molecular evolution.
INTRODUCTION
Understanding the variation of protein structures and sequences in nature is a fundamental problem in biology. Crucial to addressing this question is knowing what determines the rate of protein evolution. A major advance came from the observation that highly expressed proteins consistently evolve slowly in bacteria, yeast, worm, fly, mouse, and humans (Drummond et al., 2005; Drummond and Wilke, 2008; Pál et al., 2001). This anticorrelation between a protein’s cellular abundance and its evolutionary rate (denoted herein as ER) found a unified possible explanation in the global selection against the cytotoxic effect due to misfolded proteins. The primary hypothesis is that misfolded proteins are detrimental to cellular fitness (Drummond et al., 2005). Consequently, more abundant proteins, being prone to produce more toxic molecules (Bucciantini et al., 2002), experience stronger selection pressure, hence will evolve slowly (Drummond et al., 2005). Abundance then is a primary causal variable in determining the ER. However, even for those proteins of comparable expression levels, their ERs still span several orders of magnitude (Drummond and Wilke, 2008). Abundance likewise cannot account for the quasi log-normal distribution of the ER among genes in a genome, a fact observed from bacteria, yeast, worm, fly, mouse, and humans (Lobkovsky et al., 2010). These observations suggest that abundance, although a major determinant of ER, is not its only causal variable.
A candidate for another causal variable is protein folding stability. Most proteins (excluding those that are intrinsically disordered) need to fold to their native 3D structure to function, a property that is thermodynamically determined by its folding free energy (ΔG = Gfolded − Gunfolded). Gross destabilization of proteins by mutations also renders them nonfunctional, and is the etiological basis of several major diseases (Chiti and Dobson, 2006; Serohijos et al., 2008). More importantly, there is intrinsic toxicity associated with protein misfolding because of protein aggregation (Bucciantini et al., 2002).
If indeed stability is a causal variable of protein evolution, what is its systematic effect on ER and on the universally observed abundance-ER anticorrelation? Here, using simulations, theoretical analysis, and bioinformatics, we show that under the global selection against protein misfolding, both abundance and folding stability are causal variables of the ER. We derive an ER surface that defines the coupled role of abundance and folding stability. Further highlighting the role of stability, we demonstrate that the inverse correlation between ΔG and ΔΔG, an intrinsic feature of protein biophysics, is a necessary requirement for highly expressed proteins to evolve slowly. Lastly, we predict from multiscale evolutionary simulations that the strength of the abundance-ER correlation will increase with divergence time. Systematic analysis of species in a class of bacteria confirms this prediction.
RESULTS
Multiscale Evolutionary Model
To systematically determine the role of protein biophysics in the genomic correlation between abundance and ER, we first constructed a monoclonal population of 104 model cells (Figure 1). Each cell consisted of 103 genes that were characterized by their folding stability (ΔG = Gfolded − Gunfolded) and abundance. The abundances of the 103 genes were randomly drawn from the distribution of measured protein abundances in yeast (Ghaemmaghami et al., 2003). Mutations were assumed to occur only in the gene’s coding region, hence only the folding stability of its protein products was evolvable whereas its abundance remained constant throughout the evolution.
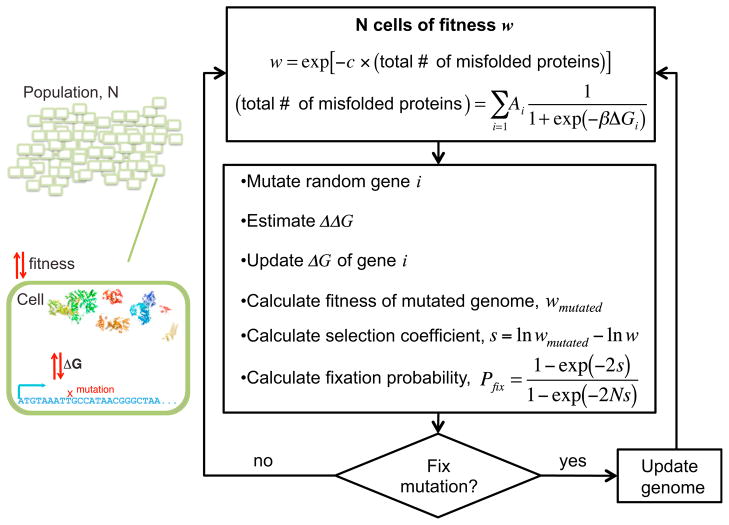
Figure 1. Multiscale Model of Protein Evolution.
Model: a constant population of Ne = 104 cells. Each cell contains 103 genes defined by its cellular abundance and the folding stability of its protein product. Because of intrinsic cytotoxicity (Bucciantini et al., 2002), protein misfolding is assumed to be the major determinant of organismic fitness (Drummond and Wilke, 2008). Assuming a two-state protein folding model (Privalov and Khechinashvili, 1974; Shakhnovich and Finkelstein, 1989), the number of misfolded proteins contributed by gene i (with abundance Ai and folding stability ΔGi) is a product of abundance and the probability of being unfolded, 1/(1 + exp(−βΔGi)), where β = 1/kBT. The total number of misfolded proteins in the cell is the sum of all misfolded proteins contributed by each gene. The factor c is the intrinsic fitness cost for every misfolded protein, measured in yeast to be ~32/(total protein concentration) (Geiler-Samerotte et al., 2011). A random mutation in a gene changes its folding stability by ΔΔG (i.e., ΔGmutant − ΔGwild-type), which consequently increases or decreases the fitness of the cell. From classical population genetics, and assuming monoclonality, the fate of a mutation is determined by the selection coefficient s and the fixation probability Pfix (Crow and Kimura, 1970; Sella and Hirsh, 2005). Dynamics: all genes were assigned an initial stability value of −5 kcal/mol, then subjected to mutation, selection, and drift as indicated by the flowchart. The choice of initial ΔG values was irrelevant because the population would eventually reach the dynamic equilibrium imposed by mutation-selection balance (Movie S1). All analyses and calculations of ERs were performed only during the interval that the population is under mutation-selection balance.
To relate the genomic sequence to organismal fitness, we employed the hypothesis posited by Drummond and Wilke (2008) that a major selection pressure underlying coding sequence evolution is the selection against the generic cytotoxicity induced by protein misfolding (Figure 1). Because purported measures of functional selection (e.g., gene essentiality and protein-protein interactions) are weak correlates of the ER at the genome-scale (Pál et al., 2006), functional selection was not taken into account. Assuming a two-state protein folding model (Privalov and Khechinashvili, 1974; Shakhnovich and Finkelstein, 1989), the number of misfolded species contributed by gene i (with abundance Ai and folding stability ΔGi) may be expressed as the product of abundance and the Boltzmann probability of being unfolded, 1/(1 + exp(−βΔGi)), where β = 1/kBT. In principle, mutations may also perturb the folding kinetics; however, we ignored this for now without compromising generality because changes in thermodynamic and kinetic stabilities are highly correlated (Naganathan and Muñoz, 2010). The fitness of a cell w (i.e., the probability of replicating) was then a function of the total number of misfolded protein in the cytoplasm and the intrinsic fitness cost per misfolded protein (denoted as c), measured in Yeast to be ~32/(total protein concentration) (Geiler-Samerotte et al., 2011) (see Figure 1).
Misfolded proteins arise due to either errors in mistranslation or “intrinsic” factors such as genetic mutations. The original proposition of the misfolding hypothesis focused on mistranslation errors (Drummond et al., 2005; Drummond and Wilke, 2008), however, protein misfolding even in the absence of mistranslation also contributes a non-negligible fraction to the total number of misfolded proteins in the cell (Yang et al., 2010). Using the same simulation framework employed by the original proponents of the misfolding hypothesis (Drummond and Wilke, 2008), it was shown that the recapitulation of the abundance-ER covariation is in fact agnostic to the specific source of mis-folding (Yang et al., 2010). The results we present here are also agnostic to the specific source of misfolding (Extended Results).
The monoclonal population of 104 cells was subjected to mutation, drift, and selection (Figure 1). An organism was hit by a mutation at gene i changing the thermodynamic folding stability of the gene’s protein products by ΔΔG (i.e., ΔGmutant − ΔGwild-type), which were randomly drawn from an empirically derived distribution (Equation 1). Measurements of stability changes due to single point mutations in real proteins (the ProTherm database) (Kumar et al., 2006) and computational studies (Tokuriki et al., 2007) showed that the distribution of ΔΔG appears universal across all fold types and protein lengths. In particular, this distribution can be approximated as a Gaussian
| (Equation 1) |
The SD is ΔΔGsd = 1.7 kcal/mol (Zeldovich et al., 2007) and the mean is ΔΔGmean = 1 kcal/mol, both parameters were derived from the ProTherm database.
From classical population genetics, the fate of a mutation is determined by its fixation probability (Pfix), itself a function of effective population size (Ne) and the change in fitness due to the mutation (the selection coefficient s) (Figure 1A) (Crow and Kimura, 1970). In general, for mutations that do not affect fitness (i.e., neutral), s = 0 and Pfix = 1/Ne; for beneficial mutations, s > 0 and Pfix > 1/Ne; and for deleterious mutations, s < 0 and Pfix < 1/Ne (Crow and Kimura, 1970). Under the selection against protein misfolding, one could express s as a function of gene properties. Specifically in our model, an organism of fitness w acquired a mutation at gene i that changed its fitness to wmutated (Figure 1A). The change in fitness could be expressed as (Sella and Hirsh, 2005) (see Extended Results for details)
| (Equation 2) |
Because proteins typically exhibit stabilities ΔGi < −3 kcal/mol (Kumar et al., 2006), Equation 2 can be simplified to a more intuitive form
| (Equation 3) |
Equations 2 and 3 are significant because they quantitatively relate the premutation molecular properties of a gene (abundance and stability), the molecular effect of the arising mutation (ΔΔG), and the mutational effect on the organism s. The selection coefficient for protein misfolding due to mistranslation errors is formally equivalent to Equation 3 (see Equation S9 in the Extended Results); hence, without loss of generality, we focused on the effect of intrinsic genetic misfolding. Additionally, it is instructive to note the several special cases of Equation 2. For a neutral mutation (ΔΔG = 0), the fitness effect is trivially zero. However, even for nonzero ΔΔG, near-neutrality (s ≈ 0) is likewise achieved when the premutated gene has low cellular abundance (Ai ≈ 0) and/or is very stable (ΔGi ≪ 0). The latter is due to the flatness of the “Fermi-function” in Equation 2 in very stable ΔG regimes: for very stable proteins, typical ΔΔG values (~1 kcal/mol) (Kumar et al., 2006) exhibit negligible change in the number of unfolded proteins. In general however, for deleterious mutations (ΔΔG > 0), s < 0. Also, in agreement with the standard formulation of the selection against protein misfolding, it is apparent from Equations 2 and 3 that protein abundance A multiplies the detrimental effects of deleterious mutations.
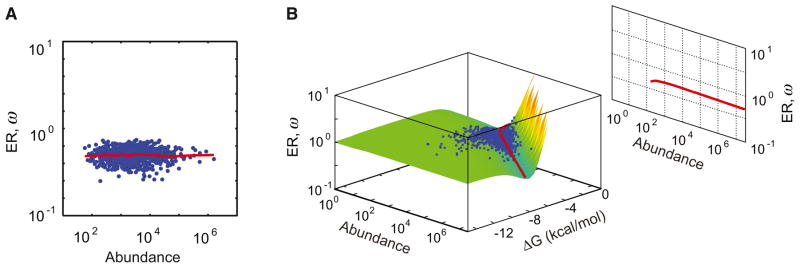
We performed multiscale evolutionary simulations as outlined in Figure 1 then estimated the ER of each gene. Because we knew the full evolutionary history of the population, the ER was simply the total number of accepted substitutions over the divergence time separating two reference genomes (Experimental Procedures). Surprisingly, despite the global selection against the toxic effects of protein misfolding, we did not observe a correlation between abundance and ER (Figure 2A).
Figure 2. Abundant Proteins Do Not Evolve Slowly When ΔΔG Is Independent of ΔG.
(A) Abundance-ER correlation from multiscale evolutionary simulations without ΔΔG versus ΔG correlation (r = 0.01). Red line indicates lowess-smoothed data.
(B) Molecular clock surface without ΔΔG versus ΔG correlation. The molecular clock (Equation 4) can be integrated over possible ΔΔG (Equation 1) to yield a surface that is solely a function of premutation gene properties. The surface heuristically governs the evolution of a gene: a gene in the neutral regime (green) will acquire mutations (on average with ΔΔG > 0) that make it approach the gully (red line). A gene on the verge of misfolding (yellow) will strongly select for beneficial mutations (ΔΔG < 0) that bring it back to the gully or the neutral regime. A gene in the gully will wait the longest time before fixing a mutation. The gully defines the steady-state relationship between abundance, ΔG, and ER. Projection of the gully predicts no abundance-ER correlation. Instantaneous abundance and ΔG values of genes from evolutionary simulations are shown as blue dots (see Experimental Procedures).
The ER Surface as Protein Evolution’s “Free Energy Landscape”
To investigate why highly abundant proteins did not necessarily evolve slowly even when protein misfolding is the only source of evolutionary selection, we performed analytic treatment of the ER. Following earlier works (Nielsen and Yang, 2003), we expressed the normalized ER ω (specifically dN/dS, the ratio between nonsynonymous ER and synonymous ER) as (see Experimental Procedures)
| (Equation 4) |
Equations 2 and 4 provide the crucial link between the premutation gene properties abundance and ΔG, the molecular effect of the arising mutation ΔΔG, and the rate of protein evolution. Equation 4 recapitulates the standard interpretation of ERs—for neutral and near neutral mutations (ΔΔG = 0 and/or ΔG ≪ 0), ω ≈ 1; for deleterious mutations (ΔΔG > 0), ω < 1; and for beneficial mutations (ΔΔG < 0), ω > 1.
More significantly, because the distribution ofmutational effects (ΔΔG) of arising mutations is known (Equation 1), we integrated Equation 4 over all possible ΔΔG to arrive at an ER surface that is solely a function of premutation gene parameters (Figure 2B).
The ER surface is characterized by three regimes (Figure 2B). First, a neutral regime (flat region; colored green), where ω ≈ 1 and the purifying selection due to misfolding is weak because of low abundance and/or high stability. Second, a regime dominated by fixation of beneficial mutations (upward curved leaf; colored red and yellow). In this regime of high abundance and low stability, the protein is closer to the precipice of misfolding, thus there is a strong selection for beneficial mutations, hence the rate is faster than 1. Third, the regime defined by the minimum of the surface (“gully”), where the rate of protein evolution is slower than 1.
Fixation of a nonsynonymous mutation changes ΔG and is a single step on the ER surface; consequently, the full evolution of a gene is essentially a “walk” on this surface. This walk is slowest at the gully (Figure 2B, red line), thus the genes are expected on average to populate this neighborhood. Indeed, when we tracked the location of each gene on the abundance-ΔG plane throughout the simulation, we observed that the genes tend to approach the gully when the population is under the dynamic equilibrium imposed by mutation-selection balance (Figure 2B, blue dots). Thus, the ER surface is conceptually analogous to free energy landscapes in physics, where the minima define the state of the physical system under equilibrium.
This analogy to “energy landscapes” implies that the gully determines the average relationship between the evolutionary variables abundance, folding stability, and ER under mutation-selection balance. When the gully is projected onto the abundance-ΔG plane, we could predict that more abundant proteins will be more stable in agreement with our multiscale evolutionary simulations (Movie S1) and prior computational results (Drummond and Wilke, 2008; Yang et al., 2010). However, when the curvature of the gully is projected on the abundance-ER plane, there is no expected covariation between abundance and ER (Figure 2B, projection).
The explanation to the absence of abundance-ER covariation starts with the realization that the selection coefficient is a function not only of abundance but also of ΔG. Equations 2 and 3 suggest that a low abundance gene can experience the same selection coefficient (hence, the same ER) as a highly abundant gene if the latter is more stable. That is, the role of abundance as a multiplier of the detrimental effects of deleterious mutations can be compensated by greater stability. Indeed, under the selection against protein misfolding, highly abundant proteins evolve to greater stability as shown in this study (Movie S1) and in prior evolutionary simulations by other groups (Drummond and Wilke, 2008; Yang et al., 2010). This specific prediction of the misfolding hypothesis is supported by the observation that highly expressed and slowly evolving proteins share amino acid composition with proteins from thermophilic bacteria (Cherry, 2010). Altogether, these results suggest that the two major predictions of the misfolding avoidance hypothesis—highly abundant proteins evolve slowly and highly abundant proteins are more stable—are apparently mutually inconsistent.
ΔΔG Versus ΔG Anticorrelation Is a Necessary Requirement for Highly Abundant Proteins to Evolve Slowly
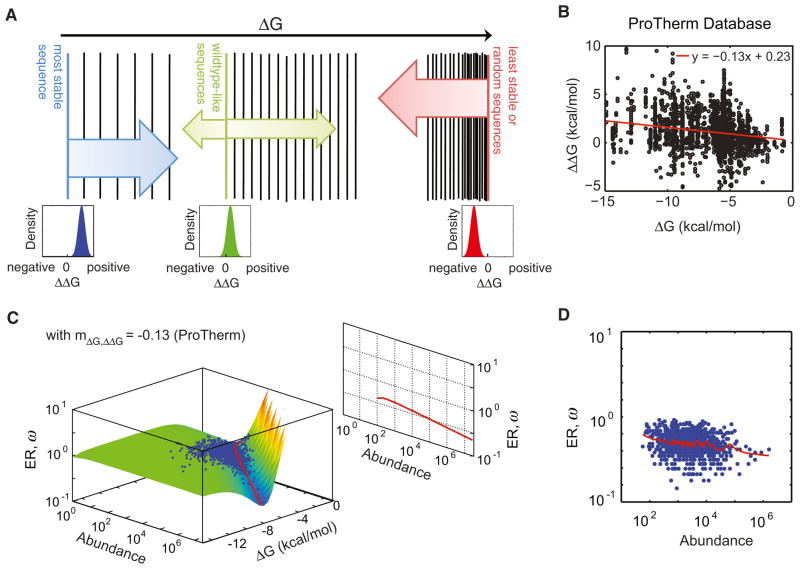
What prevents the exact compensation between abundance and folding stability in nature? We realized that the exact compensation between protein folding stability and abundance could be avoided if the mutational effects (ΔΔG) will couple with premutation gene properties. It is unlikely that ΔΔG couples with its concentration in the cytoplasm. On the other hand, from protein biophysics, there is an expected dependence of ΔΔG on ΔG (Figures 3A and S1). In the limit of the most stable sequence (Figure 3A, blue), all mutations will lead to less stable sequences, hence its ΔΔG distribution will all be destabilizing; for the least stable set of sequences (Figure 3A, red), most mutations lead to sequences that are more stable, hence the distribution of ΔΔG will be biased toward stabilizing mutations. Between these two extremes are wild-type sequences (Figure 3A, green) that are not maximally stable (Kumar et al., 2006; Taverna and Goldstein, 2002; Zeldovich et al., 2007). Indeed, experimental data from ProTherm database confirm this correlation (Figure 3B). We incorporated the empirical statistical correlation between ΔG and ΔΔG into Equation 1 as
Figure 3. ΔΔG versus ΔG Dependence Is Necessary for Abundance-ER Correlation.
(A) Biophysical origin of ΔΔG versus ΔG correlation. In the limit of the most stable sequence (blue), all mutations will lead to less stable sequences, hence its ΔΔG distribution will all be destabilizing; for the set of least stable sequences (i.e., random coils) (red), all mutations lead to sequences that are more stable, hence the distribution of ΔΔG will be biased toward all stabilizing mutations. Between these two extremes are wild-type sequences (green), which are not maximally stable (Kumar et al., 2006; Taverna and Goldstein, 2002; Zeldovich et al., 2007). See also Figure S1.
(B) ΔΔG versus ΔG correlation in real proteins. Compiled empirical measurements of ΔG and ΔΔG values in the Protherm database (Kumar et al., 2006) exhibit a negative correlation (r = −0.2; p value = 10−22). A linear fit (red line) yields a slope of mΔG,ΔΔG = −0.13.
(C) Molecular clock surface with ΔΔG versus ΔG correlation from ProTherm. Abundance and ΔG values of genes from multiscale evolutionary simulation validate the heuristic interpretation (blue dots) (see Movie S1). Two-dimensional projections of the gully predict highly abundant genes will evolve slowly.
(D) Abundance-ER correlation is recapitulated in multiscale evolutionary simulations when ΔΔG versus ΔG correlation is taken into account (Spearman rank correlation r = −0.3; p value = 10−9). Red line indicates lowess-smoothed data.
| (Equation 5) |
In Equation 5 the mean of the Gaussian distribution of ΔΔG becomes more positive as ΔG as proceeds to more stable values (Figure S1), formally reflecting the tendency for stable proteins to exhibit more destabilizing mutations.
When we calculated a new ER surface by integrating Equation 4 over all possible ΔΔG (Equation 5), the gully is deeper at higher abundances where proteins are more stable and mutations are more destabilizing (Figure 3C). Projection of the gully on the abundance-ER plane now predicts that highly expressed proteins will evolve slowly. When we performed evolutionary simulations accounting for the ΔG versus ΔΔG anticorrelation, we recapitulated the abundance-ER anticorrelation (Figure 3D). This new curvature of the gully still predicts that highly abundant proteins are more stable. Thus, in the presence of ΔG versus ΔΔG anticorrelation, the observation of highly abundant proteins being more stable is no longer inconsistent with highly abundant proteins evolving slowly.
Overall, these results suggest that the prevalent interpretation of the selection pressure imposed by protein misfolding—that highly expressed proteins are under stronger selection pressure because they produce greater amount of toxic proteins—is incomplete. A more complete restatement of the hypothesis should be: selection against protein misfolding induces abundant proteins to evolve to greater stability, where the average mutations are more destabilizing; hence, more abundant proteins evolve slowly.
Strength of Abundance-ER Anticorrelation Varies with Divergence Time
The results above demonstrate the causal role of stability on ER. Because ΔG is intrinsically an instantaneous property of a gene (i.e., each sequence has a defined ΔG), the ER ω as defined by Equation 4 is also instantaneous. Nevertheless, estimating ER in practice entails comparing two close, yet sufficiently diverged orthologs. Over the divergence time separating these orthologs, several substitutions would have been fixed, while the gene sampled several ΔG values. We wanted to determine if the Abundance-ER variation likewise exhibits dependence on divergence time.
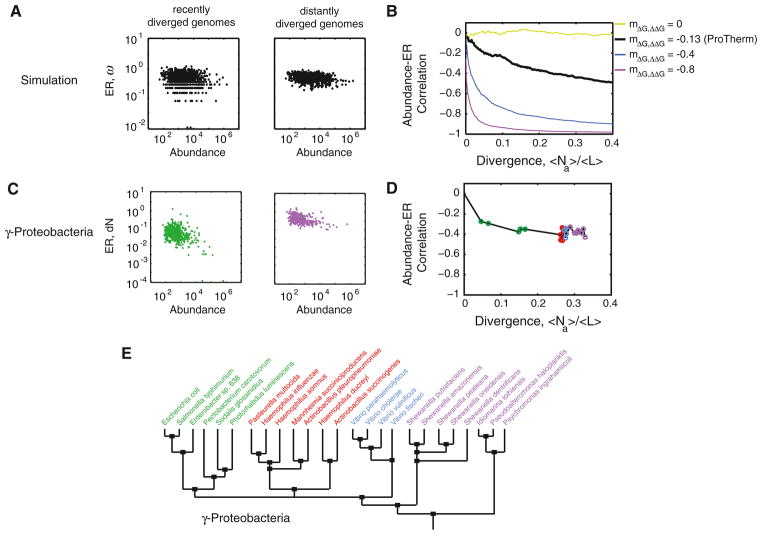
From the multiscale simulations, we estimated the ERs by comparing orthologous pairs of genes between pairs of genomes of various divergences. Simulations predicted that the magnitude of the abundance-ER correlation should be time-dependent (Figures 4A and 4B). Immediately right after divergence, all genes had zero estimated rates by definition, thus the abundance-ER correlation was practically zero. Between very recently diverged species, only the fast evolving genes had fixed mutations, thus the abundance-ER correlation was weak. As divergence time increased, the slow genes started acquiring mutations and the disparity in rate between slow and fast genes intensified, consequently increasing the manifestation of highly abundant genes evolving slowly (Figures 4A and 4B). Strikingly, we found that the nature of the time-dependence of the abundance-ER correlation is strongly influenced by the magnitude of ΔΔG versus ΔG anticorrelation (Figure 4B).
Figure 4. Abundance-ER Anticorrelation Varies with Divergence Time.
ER is measured by counting the number of fixed mutations between two diverged genomes. Divergence time is measured as the number of substitutions per gene (Na) averaged over all genes in the genome (<Na>) normalized by the average length <L> of genes in the genome. See Experimental Procedures.
(A and B) Abundance-ER correlation from multiscale evolutionary simulations. Rates estimated from recently diverged genomes (A, left) exhibit a broader distribution and a weaker correlation with abundance than distantly diverged genome (A, right). (B) The magnitude of abundance-ER correlation varies with divergence time. This variation is influenced by the dependence of ΔG on ΔΔG.
(C and D) Abundance-ER correlation in γ-proteobacteria. Nonsynonymous ER (dN) was calculated between E. coli and orthologous genes in other γ-proteo-bacteria. For example, (C, left) is between E. coli and S. typhimurium and (C, right) is between E. coli and P. ingrahamii. (D) Full time variation of abundance-ER correlation. See Figure S2 for the complete abundance-ER correlations in other γ-proteobacteria.
(E) γ-Proteobacteria phylogeny (adapted from Williams et al., 2010).
We then sought to determine if the predicted time-dependence holds true in real organisms (Figures 4C–4E). We focused on Escherichia coli because its cellular protein abundances have been surveyed (Ishihama et al., 2008) and numerous genomes of its close relatives have been sequenced (Figure 4E), the latter is essential for accurate ER estimation (see Experimental Procedures). We found that the strength of the abundance-rate correlation is weakest when ER is estimated between E. coli and its closest relative, Salmonella typhimurium, than between E. coli and the bacteria Shewanella or Psychromonas ingrahamii. Interestingly, the strength of the correlation asymptotically approaches the value predicted by the ProTherm parameterization of the ΔG versus ΔΔG correlation (Figure 4D). Overall, these results provide yet another proof of the universality of the selection against protein misfolding and of the critical roles protein stability ΔG and protein biophysics play in determining the ER.
DISCUSSION
In agreement with Drummond and Wilke (2008), the protein mis-folding hypothesis is the strongest candidate for a unified explanation to the observed correlation between protein abundance and ER. Moreover, the hypothesis accounts for new genome-wide observations such as indicated in Figure 4 or the finding that proteins in thermophiles share amino acid composition with slowly evolving, highly expressed proteins (Cherry, 2010). These observations suggest that selection against misfolding may have more fundamental implications in shaping the features and architecture of the genome.
However, the standard formulation of the hypothesis is incomplete, because the selection coefficient and consequently the ER are determined not only by abundance but also by protein folding stability ΔG, as suggested by previous works (Bershtein et al., 2006; Bloom et al., 2006; Wylie and Shakhnovich, 2011) and systematically proven here. The crux of the misfolding hypothesis is that abundance multiplies the detrimental effect of destabilizing mutations because more abundant proteins will produce more toxic unfolded species. On the other hand, abundant proteins evolve to greater stabilities (Figures 2A and 3C), potentially relieving abundant proteins of the strong selection pressure, thus eliminating the abundance-ER correlation (Figure 2). The exact compensation between abundance and stability is avoided because ΔΔG is correlated with ΔG (i.e., mutations are more destabilizing in more stable proteins) (Figure 3). We advance a more complete statement of the hypothesis: Abundant proteins evolve to greater stability, where the average mutations are more destabilizing; hence, more abundant proteins evolve slowly.
The magnitude of the correlation between ER-abundance is likewise dependent on the strength of the ΔG versus ΔΔG anti-correlation (Figure 3B)—more destabilizing mutations in more stable regime (also high abundance) lead to slower ER. Hence, quantification of this biophysical property is crucial in inferring the strength of the genome-wide abundance-ER correlation. In this work, we parameterized the ΔG versus ΔΔG anticorrelation according to Protherm. Simplified models that fold proteins on a lattice model tend to overestimate the ΔΔG dependence on ΔG because of a small hydrophobic core. Thus prior works (Drummond and Wilke, 2008; Yang et al., 2010) may have overestimated in simulation the strength of abundance-ER correlation.
Previous theoretical analysis of the ER focusing solely on the effect of translational robustness, and excluding the ΔG versus ΔΔG anticorrelation (Wilke and Drummond, 2006), does not recapitulate the shape of the abundance-ER dependence that is observed in simulations using 2D lattice models (Drummond et al., 2005; Drummond and Wilke, 2008; Yang et al., 2010) or from comparative genomics. This finding implies that the ΔG versus ΔΔG dependence is prerequisite to the abundance-ER anticorrelation in both protein misfolding due to errors in mistranslation or misfolding due to genetic mutations.
The distribution of ER from simulations in this model, which is essentially parameter free, spans ~1.5 orders of magnitude, whereas the ER distribution from comparative genomics spans 2–3 orders of magnitude (Lobkovsky et al., 2010). The narrower distribution from simulations is not surprising for several reasons, namely (1) the population is assumed to be monoclonal, (2) all genes have the same length, and (3) other biological factors could impose various selective constraints that will effectively broaden the rate of protein evolution. Real life biology is much more complicated than presented here. Some factors that could influence the rate of evolution of individual genes include protein-protein interactions (e.g., avoidance of nonspecific interactions) (Yang et al., 2012; Zhang et al., 2008), functional selection, and metabolic network topology (Vitkup et al., 2006). Cellular responses (e.g., chaperones and proteases) (Tokuriki and Tawfik, 2009) and functional oligomerization of destabilized proteins (Bershtein et al., 2012) can also attenuate the toxicity of folded proteins. Generalizing, the current framework to include the systematic effect of these constraints is the subject of future work.
EXPERIMENTAL PROCEDURES
Derivation of ω(s) and the Molecular Clock Surface
The rate of the evolutionary clock is a product of the rate at which mutation occurs and the rate at which these mutations will fix (Pfix(s) in Figure 1A) (Nielsen and Yang, 2003). If μ is the mutation rate (number of mutations per base pair per generation) and Ne is the effective population size, the total number of mutations per generation is Neμ. Hence, the nonsynonymous substitution rate dN is
| (Equation 6) |
Assuming that all silent substitutions are neutral, the synonymous rate is dS = Neμ(1/Ne). The clock in Equation 4 is the ratio dN/dS. The molecular clock surface is the integral .
Simulation Protocol
All genes were assigned an initial stability value of −5 kcal/mol, then subjected to mutation, selection, and drift as outlined in Figure 1. The factor kBT = 0.593 kcal/mol. The choice of initial ΔG values was irrelevant because the population would eventually reach the dynamic equilibrium imposed by mutation-selection balance (Movie S1). All analyses and calculations of ERs were performed only during the interval that the population is under mutation-selection balance.
ER Calculation in Simulations
In the multiscale evolutionary simulations, we knew the full history of the evolving population and we recorded all the mutations that was fixed. Thus, the ER was simply the total number of fixed substitutions that occurred over the divergence time separating the two genomes. Because the rates are broadly distributed, they are customarily plotted on a logarithmic scale. We followed (Drummond et al., 2005; Drummond and Wilke, 2008) and used the transformation (ω + 10−4) to include in the analysis the genes with zero estimated rates.
ER Calculation in Bacterial Genomes
To estimate the ER in real proteins, we first determined the ortholog of a gene in the reference genome using the reciprocal smallest distance algorithm implemented in ROUNDUP (Wall et al., 2003). Amino acid alignments of orthologs (generated using CLUSTALW) (Larkin et al., 2007) were used to align their corresponding DNA sequences. Nonsynonymous ERs (dN) were estimated using both Nei and Gojobori (1986) and Maximum Likelihood methods (Yang and Nielsen, 2000) as implemented in CODEML (Yang, 2007). dN values calculated from both methodologies gave nearly identical results because the species are closely related. Thus, the ERs reported in Figures 4 and S2 used only the Nei and Gojobori method (Nei and Gojobori, 1986).
Divergence Time Metric
Estimating the divergence time between two genomes is very involved (Arbogast et al., 2002); instead, we used a simple metric that allows direct comparison between simulation and bioinformatics results. The mutation rate in our simulations was constant. We likewise assumed that the per base pair mutation rate in the γ-proteobacteria species are comparable because systematic analysis (Anderson et al., 2004) show consistency of mutation rates among bacteria of 0.003 mutations per genome per replication. Thus, divergence time is proportional to the number of substitutions fixed since divergence. A simple metric then is the number of amino acid substitutions accumulated over the time separating two genes. Specifically, divergence time is measured as the number of nonsynonymous substitutions per gene (Na) averaged over all genes in the genome (<Na>), then normalized by the average gene length <L> of genes in the genome. To guard against the potential bias in the composition of gene abundances in the genome, we show that the distribution of abundances for the set of matched orthologs (Figure S2, histograms) are comparable.
Supplementary Material
Acknowledgments
We thank Scott Wiley and Shimon Bershtein for fruitful discussions and comments on the manuscript. This work is supported by NIH grant GM068670 (to E.I.S.).
Footnotes
Supplemental Information includes Extended Results, two figures, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.06.022.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
References
- Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- Arbogast BS, Edwards SV, Wakeley J, Beerli P, Slowinsksi JB. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu Rev Ecol Syst. 2002;33:707–740. [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- Bershtein S, Mu W, Shakhnovich EI. Soluble oligomerization provides a beneficial fitness effect on destabilizing mutations. Proc Natl Acad Sci USA. 2012;109:4857–4862. doi: 10.1073/pnas.1118157109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- Cherry JL. Highly expressed and slowly evolving proteins share compositional properties with thermophilic proteins. Mol Biol Evol. 2010;27:735–741. doi: 10.1093/molbev/msp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An Introduction to Population Genetics Theory. New York: Harper & Row; 1970. [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein mis-folding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci USA. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MD, Bava KA, Gromiha MM, Prabakaran P, Kitajima K, Uedaira H, Sarai A. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 2006;34(Database issue):D204–D206. doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lobkovsky AE, Wolf YI, Koonin EV. Universal distribution of protein evolution rates as a consequence of protein folding physics. Proc Natl Acad Sci USA. 2010;107:2983–2988. doi: 10.1073/pnas.0910445107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganathan AN, Muñoz V. Insights into protein folding mechanisms from large scale analysis of mutational effects. Proc Natl Acad Sci USA. 2010;107:8611–8616. doi: 10.1073/pnas.1000988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol Biol Evol. 2003;20:1231–1239. doi: 10.1093/molbev/msg147. [DOI] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ. An integrated view of protein evolution. Nat Rev Genet. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- Privalov PL, Khechinashvili NN. A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J Mol Biol. 1974;86:665–684. doi: 10.1016/0022-2836(74)90188-0. [DOI] [PubMed] [Google Scholar]
- Sella G, Hirsh AE. The application of statistical physics to evolutionary biology. Proc Natl Acad Sci USA. 2005;102:9541–9546. doi: 10.1073/pnas.0501865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, Riordan JR. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci USA. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhnovich EI, Finkelstein AV. Theory of cooperative transitions in protein molecules. I Why denaturation of globular protein is a first-order phase transition. Biopolymers. 1989;28:1667–1680. doi: 10.1002/bip.360281003. [DOI] [PubMed] [Google Scholar]
- Taverna DM, Goldstein RA. Why are proteins marginally stable? Proteins. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369:1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Vitkup D, Kharchenko P, Wagner A. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 2006;7:R39. doi: 10.1186/gb-2006-7-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- Wilke CO, Drummond DA. Population genetics of translational robustness. Genetics. 2006;173:473–481. doi: 10.1534/genetics.105.051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW. Phylogeny of gammaproteobacteria. J Bacteriol. 2010;192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CS, Shakhnovich EI. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc Natl Acad Sci USA. 2011;108:9916–9921. doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JR, Zhuang SM, Zhang J. Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Mol Syst Biol. 2010;6:421. doi: 10.1038/msb.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JR, Liao BY, Zhuang SM, Zhang J. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc Natl Acad Sci USA. 2012;109:E831–E840. doi: 10.1073/pnas.1117408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Zeldovich KB, Chen P, Shakhnovich EI. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA. 2007;104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Maslov S, Shakhnovich EI. Constraints imposed by non-functional protein-protein interactions on gene expression and proteome size. Mol Syst Biol. 2008;4:210. doi: 10.1038/msb.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.