Abstract
Waxy mutants, in which endosperm starch contains ∼100% amylopectin rather than the wild-type composition of ∼70% amylopectin and ∼30% amylose, occur in many domesticated cereals. The cultivation of waxy varieties is concentrated in east Asia, where there is a culinary preference for glutinous-textured foods that may have developed from ancient food processing traditions. The waxy phenotype results from mutations in the GBSSI gene, which catalyzes amylose synthesis. Broomcorn or proso millet (Panicum miliaceum L.) is one of the world’s oldest cultivated cereals, which spread across Eurasia early in prehistory. Recent phylogeographic analysis has shown strong genetic structuring that likely reflects ancient expansion patterns. Broomcorn millet is highly unusual in being an allotetraploid cereal with fully waxy varieties. Previous work characterized two homeologous GBSSI loci, with multiple alleles at each, but could not determine whether both loci contributed to GBSSI function. We first tested the relative contribution of the two GBSSI loci to amylose synthesis and second tested the association between GBSSI alleles and phylogeographic structure inferred from simple sequence repeats (SSRs). We evaluated the phenotype of all known GBSSI genotypes in broomcorn millet by assaying starch composition and protein function. The results showed that the GBSSI-S locus is the major locus controlling endosperm amylose content, and the GBSSI-L locus has strongly reduced synthesis capacity. We genotyped 178 individuals from landraces from across Eurasia for the 2 GBSSI and 16 SSR loci and analyzed phylogeographic structuring and the geographic and phylogenetic distribution of GBSSI alleles. We found that GBSSI alleles have distinct spatial distributions and strong associations with particular genetic clusters defined by SSRs. The combination of alleles that results in a partially waxy phenotype does not exist in landrace populations. Our data suggest that broomcorn millet is a system in the process of becoming diploidized for the GBSSI locus responsible for grain amylose. Mutant alleles show some exchange between genetic groups, which was favored by selection for the waxy phenotype in particular regions. Partially waxy phenotypes were probably selected against—this unexpected finding shows that better understanding is needed of the human biology of this phenomenon that distinguishes cereal use in eastern and western cultures.
Keywords: diploidization, domestication, granule bound starch synthase, millet, Panicum miliaceum, waxy starch
Introduction
Varieties with a waxy starch phenotype are known in many cereals, including wheat (Triticum spp.), maize (Zea mays), rice (Oryza sativa), barley (Hordeum vulgare), sorghum (Sorghum bicolor), and millets (Panicum miliaceum, Setaria italica, and Coix lacryma-jobi). These varieties have been selected, in societies both ancient and modern, for the altered texture of their endosperm, which results from the absence or near absence of the amylose component of starch. Amylose content in wild-type starch is approximately 20–30%, with amylopectin constituting the other 70–80%. Amylopectin is a branched molecule comprising short (20 to 24-mer) chains of α(1 → 4)-linked α-glucosyl units linked by α(1 → 6) branch linkages. Amylose contains very few branched linkages, and molecules consist of long chains of several thousand α-glucosyl units joined by α(1 → 4)-linkages. As a consequence of these biochemical differences between the starch polymers, waxy and wild-type starches vary in physical properties. Waxy starches lacking amylose gelatinize at lower temperatures and swell more than wild-type starches. On cooking, waxy starches produce a soft paste with a characteristically sticky texture, whereas wild-type starches produce a harder gel that separates easily from the cooking water.
In all species that have been investigated, the waxy phenotype is due to loss of function of the major starch synthase (granule-bound starch synthase [GBSS]) activity in starch granules. GBSS catalyzes the elongation of the amylose chain by transferring adenosine diphosphate (ADP) glucose residues to a glucan substrate and is the sole enzyme responsible for amylose synthesis, in contrast to the complex multienzyme amylopectin biosynthesis pathway (reviewed in Zeeman et al. 2010). The GBSS isoform active in the endosperm is encoded by the gene GBSSI, which also functions in pollen (Yamanaka et al. 2004). In functionally polyploid species, the production of fully waxy types requires the presence of mutant alleles causing loss of function in all homeologs of the GBSSI gene. Broomcorn or proso millet (Panicum miliaceum L.) is an unusual case among cereals with waxy types: it is a polyploid species in which fully waxy types appeared before deliberate recent breeding. In tetraploid and hexaploid wheats, fully waxy lines have only been bred within the last 15 years from partial-waxy types that lacked function in one (or two, in some hexaploid lines) of the GBSSI homeologs. Broomcorn millet is an allotetraploid with 2n = 4x = 36. Its diploid ancestors are unknown but related Panicum species include the wild diploid P. capillare (witchgrass). Graybosch and Baltensperger (2009) demonstrated through crossing experiments the existence of two GBSSI loci in P. miliaceum, consistent with its polyploid constitution. In a previous article (Hunt et al. 2010), we characterized these two loci (GBSSI-L and GBSSI-S) through DNA sequencing of plants from 38 landraces. We found that the GBSSI-S locus has two alleles, a wild-type allele (S0) and a mutant allele (S-15) which contains a 15-bp deletion relative to S0, resulting in the loss of five amino acids from the glucosyl transferase domain GTD1 and the loss of GBSSI-S enzyme activity. We found three GBSSI-L alleles, of which one (LC; GenBank sequence ID ADA61154) was inferred from comparison of the predicted amino acid sequence with those from other GBSSI alleles to be the ancestral allele. Two mutant alleles were discovered. One (LY; GenBank sequence ID ADA61155) differed from the LC allele by a single amino acid substitution from cysteine to tyrosine, at position 153, in exon 7; the other (Lf; GenBank sequence ID ADA61156) differed from the LC allele by a frameshift mutation, specifically the insertion of an additional adenine residue following position 224, in exon 9. Both these mutant alleles result in the loss of functional GBSSI-L protein, as inferred from the loss of endosperm starch synthase activity and amylose in plants that had either of these alleles in combination with the S-15 allele. Among the plants we analyzed, the LC allele occurred in combination with the S0 allele only and therefore we were not able to prove that it encodes a functional version of the GBSSI-L protein. However, the existence of two loci, each with wild-type alleles in P. miliaceum, as inferred by Graybosch and Baltensperger (2009) is consistent with the hypothesis that LC produces a functional protein.
From the data above, the evolution of waxy varieties in broomcorn millet required the coincidence in a single plant of independently arising mutant alleles at the S and L loci. This would necessitate that the mutant alleles were appropriately distributed in populations with respect to both gene pools and geographical location. As in most other cereals, the distribution of waxy types in broomcorn millet is restricted to east Asia, which is thought to reflect their selection by the cultural preference for glutinous-type starchy foods in this region (Sakamoto 1996). Waxy varieties of broomcorn millet have probably existed for at least 2,000 years in China, as indicated by the appearance in classical Chinese texts of the character shu specifying glutinous broomcorn millet (Sakamoto 1996) The cultivation of P. miliaceum in China dates back to at least 8,000 cal BC (Lu et al. 2009), and it is very likely that its domestication occurred in this region, either in the central Yellow River valley or in the upland areas of the Loess Plateau or the Inner Mongolian foothills (Liu et al. 2009). Archaeobotanical records of P. miliaceum are also known from the 6th millennium cal BC in eastern Europe, which has prompted speculation that it may have been domesticated independently in this region (Jones 2004). We recently demonstrated the existence of strong phylogeographic structure among broomcorn millet landraces, based on genotyping data at 16 microsatellite loci. Two major subpopulations exist in Eurasia, one eastern and one western, with the approximate boundary between the two in northwestern China. These data do not resolve the question of whether there were single or multiple centers of domestication: the data could reflect either two independent domestications in the east and west of Eurasia or a single broad domestication in China followed by a founder effect that resulted in the predominance of one gene pool as this crop spread westward (Hunt et al. 2011).
In this study, we investigated the evolution of the waxy phenotype in broomcorn millet in its phylogeographic context. We first sought to determine experimentally whether, as we hypothesized previously, the LC allele produces an active protein. This was to determine whether the waxy endosperm trait in P. miliaceum is controlled by one or two loci. We assessed the functionality of the LC allele in two ways: 1) by studying the GBSSI protein content and activity, and amylose content, in lines with this allele in an S-15 background and 2) by comparing the predicted protein sequence of LC with that of the functional GBSSI in the nonwaxy diploid P. capillare. Second, by comparing the biochemical phenotypes of lines with all combinations of alleles at the GBSSI-L and GBSSI-S loci, we assessed the relative capacity of these two loci for amylose synthesis and their consequent effect on endosperm texture. We also tested whether both alleles of GBSSI were active in pollen grains. This enabled us to clarify which mutations were necessary for the evolution of plants with the waxy phenotype. Third, we analyzed the geographic distribution of alleles at the GBSSI-L and GBSSI-S loci in landrace accessions from across Eurasia and investigated the association of the GBSSI alleles with population structure inferred from microsatellite loci, to determine the likely population history of these mutations. Taking these biochemical and phylogeographic data together, we were able to develop a model for the evolution of the waxy phenotype in broomcorn millet. This model provides some novel findings regarding the evolution of amylose-free starch in polyploid genomes and human selection of waxy endosperm phenotypes.
Materials and Methods
Identification of LC/S-15 Lines
F4 generation seed was provided for 31 lines derived from the true-breeding wild-type families from the crossing experiments of Graybosch and Baltensperger (2009). We screened the lines to identify those which were homozygous for the LC and S-15 GBSSI alleles as follows. Polymerase chain reactions (PCRs) were carried out for a fragment spanning the 15-bp deletion site in the GBSSI-S locus and labeled with 6-carboxyfluorescein (6-FAM) using the M13 tailing procedure of Boutin-Ganache et al. (2001). Reactions were carried out in 10 μl volumes containing 1 x buffer, 100 nM primer [M13]-int9Sf (5’-[CACGACGTTGTAAAACGAC]-GCCGAATAATCGTCTGATAAATTGAGC-3’), 400 nM primer R11 (5’-CAGGCACACTGCTCCCAATG-3’), and 400 nM primer [FAM]-M13. Cycling conditions were 94°C for 3 min; 30 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 1 min; 10 cycles of 94°C for 30 s, 53°C for 45 s and 72°C for 1 min;, and a final extension step of 72°C for 10 min. Positive controls were included in each set of reactions, using samples that had previously been sequenced across the indel site (Hunt et al. 2010). PCR products were checked on 2% Tris-acetate-EDTA (TAE)-agarose gels and diluted 100-fold in water before analysis by capillary electrophoresis on an ABI3730 instrument (Applied Biosystems). Electropherograms were analyzed in GeneMapper version 4.0 (Applied Biosystems) and scored manually for the S0 or S-15 alleles.
Lines that were monomorphic for S-15 homozygotes were screened at the L locus for the two fragments spanning sites with exon polymorphisms, using a single-base extension method. PCRs for the int5Lf-R3 and M12-R12 fragments, which cover the G/A substitution and frameshift adenine insertion sites, respectively, were carried out essentially as described previously (Hunt et al. 2010). PCR products were checked on TAE-agarose gels and purified using Exonuclease I and Shrimp Alkaline Phosphatase. Cleaned PCR products were then used as the template in SNaPshot™ reactions (Applied Biosystems), which were carried out in 5 μl volumes containing 1 μl cleaned PCR product, 1 μl ABI PRISM® SNaPshot™ Multiplex Ready Reaction Mix, and 500 nM extension primer. The extension primer sequences were 5’GGGAGGATGTCGTGTTCGTCT-3’ for the int5Lf-R3 fragment and 5’-CACGACGTTGTAAAACGACCAGGTACGAGAAGCCTGTGGA-3’ for the M12-R12 fragment. Following this preliminary identification of lines homozygous for the Lc and S-15 alleles, the phenotype of additional grain from these lines was checked by scraping a small amount of endosperm, distal to the embryo, onto a microscope slide and staining with Lugol’s solution (10% (w/v) KI (Sigma-Aldrich Ltd., Gillingham, Dorset, UK), 5% (w/v) I2 (Sigma-Aldrich Ltd., Gillingham, Dorset, UK), diluted 100-fold with water immediately before use). Seed was then sown, and following germination and development of leaf tissue, DNA was extracted, amplified, and sequenced for the exons 2–14 region of the L and S genes, which corresponds to the entire sequence of the mature GBSSI peptide, according to procedures described previously (Hunt et al. 2010).
Other Plant Material
Lines of the five other P. miliaceum genotypes (S0/LC, S0/LY, S0/Lf, S-15/LY, S-15/Lf) were those used previously (Hunt et al. 2010). The six genotypes were compared in experiments to measure GBSSI protein content, GBSSI activity, endosperm amylose concentration, starch swelling power, and visual assessment of the staining with iodine of starch granules from endosperm and pollen grains.
Panicum capillare
Germplasm of P. capillare was provided by the Leibniz Institute of Plant Genetics and Crop Plant Research (Gatersleben, Germany; accession number IPK 781). Grain was phenotyped by iodine staining as described earlier. DNA was extracted from seedlings using a Qiagen Plant DNeasy kit (Qiagen Ltd., Crawley, West Sussex, UK), following the manufacturer’s protocols. The GBSSI locus in this species was amplified using the primers FPSLVVC3 and Rstop3 (supplementary table S1, Supplementary Material online), in 50 µl volumes using 1 x Finnzymes HF buffer (New England Biolabs, Hitchin, Hertfordshire, UK), 200 µM of each deoxynucleotide triphosphate (dNTP), 0.3 µM of each primer, 3% dimethyl sulfoxide (DMSO), and 1 U Finnzymes Phusion™ High-Fidelity DNA Polymerase (New England Biolabs, Hitchin, Hertfordshire, UK). Cycling conditions were 30 s at 98°C; 40 cycles of 10 s at 98°C, and 2 min 30 s at 72°C; final extension step of 10 min at 72°C. PCR products were sequenced for forward and reverse strands using the primers in supplementary table S1, Supplementary Material online. The resulting sequence has been submitted to GenBank (accession number JN587495). This sequence was aligned with those for P. miliaceum GBSSI-S (GU199261) and GBSSI-L (GU199253) in MEGA version 4.0 (Tamura et al. 2007). We updated our previous alignment of GBSS amino acid sequences from a range of monocot and dicot species (Hunt et al. 2010) to include the predicted amino acid sequence for the P. capillare GBSSI and to include all alleles at the P. miliaceum GBSSI-S and GBSSI-L loci. Amino acid alignments were carried out in MEGA 4.0 and formatted in BoxShade 3.31 running on the Mobyle web portal (Néron et al. 2009; http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::boxshade, last accessed 2012 January 25).
Starch Extraction
Starch extraction for SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel) analysis of GBSSI proteins and for enzyme activity assays was performed as described previously (Hunt et al. 2010). Starch extraction for amylose quantification and swelling power tests was performed using a method modified from South and Morrison (1990) and Sulaiman and Morrison (1990). Fifty grains were partially crushed in a pestle and mortar and the husks removed with forceps. Grain was steeped overnight in 5 ml water at 4°C before thorough grinding in a pestle and mortar in a total volume of 10 ml water. The resulting suspension was filtered through Miracloth and the filtrate centrifuged for 20 min at 1,200 × g. The pellet was resuspended in 1 ml water and the suspension layered above 9 ml 80% (w/v) CsCl in a 15 ml centrifuge tube. This was centrifuged for 15 min at 1,200 × g and the supernatant discarded. The pellet was resuspended in 1 ml water and transferred to a 1.5 ml microcentrifuge tube before centrifugation for 5 min at 10,000 × g. The pellet was washed in this way a total of three times with water and then once with ice-cold acetone. Pellets were air dried and stored at −20°C.
GBSSI Protein Content and Activity
Proteins were extracted from purified starch and analyzed by SDS-PAGE. Starch (10 mg) was suspended in 0.5 ml gel sample buffer and heated to 95°C for 3 min. After cooling to room temperature, the samples were centrifuged at 14,000 × g for 10 min and the resulting supernatant recovered. A 10 μl aliquot of supernatant was loaded onto a 7.5% SDS-PAGE (80 × 60 × 0.75 mm), subjected to electrophoresis and then stained with Bio-Safe colloidal Coomassie Blue G-250.
A band of protein at approximately 52 kDa was observed in all samples. The relative amount of protein in each band was estimated from the band density, which was determined by image analysis using ImageJ software (http://rsbweb.nih.gov/ij/, last accessed 2012 September 13).
Starch synthase activity assays were carried out as described previously (Hunt et al. 2010). Three replicates were performed for each sample. Briefly, a suspension of the starch sample was incubated with a reaction mixture including radiolabeled ADP[U14C]glucose. Incorporation of the labeled substrate into the resulting starch was measured by scintillation counting and the rate of uptake calculated by reference to appropriate controls.
Microscopy
Mature seeds of genotype S-15/LC, which had low amylose content, were cut into 1.5 -μm-thick sections, and these were stained with Lugol’s solution to reveal the amylose content of the starch granules. The sections were cut directly from mature endosperm without prior embedding. The advantages of this dry-cut method are that it allows observation of the starch granules in endosperm cells in situ, and because the granules are sectioned, variations in staining intensity and color (i.e., amylose content) within the granule can be discerned. To assess the amylose content of starch in pollen, pollen grains were placed on a microscope slide in a drop of dilute Lugol’s solution, gently squashed under a cover slip, so that some starch was ejected from the ruptured pollen grain, and viewed under a light microscope.
Amylose Quantification
The concentration of amylose in millet starches was estimated using a method modified from Knutson and Grove (1994). Starch (5 mg) was used for each assay (weighed out accurately to 0.01 mg). Three replicate assays were carried out for each sample; 50 µl of 3 M CaCl2 was added, and the samples were vortexed and left to stand for 10 min. Following the addition of 0.5 ml 6 mM I2-DMSO, the samples were stirred and placed in a sonicating bath at 70–75°C for 30 min. A 10 µl aliquot was transferred to a fresh glass tube, and 100 µl 6 mM I2-DMSO and 800 µl water were added. Absorbance at 600 nm was measured using a GENESYSTM 6 spectrophotometer (ThermoSpectronic). Amylose content of samples was determined using a standard curve constructed using millet amylopectin extracted from the waxy line MIL-82 #1 and maize amylose (Sigma-Aldrich, catalog number A7043) in 5 mg total aliquots, over an amylose concentration range of 0–50%.
Starch Swelling Power
The swelling power of gelatinized starch was measured using a method modified from Konik-Rose et al. (2001). Three replicate assays were carried out for each genotype. Starch (10 mg; accurate to 0.01 mg) was weighed out into preweighed round-bottomed 2 ml Eppendorf tubes; 1 ml H2O was added, and the samples were mixed by thorough shaking. The tubes were placed in a hot block at 90°C in an incubator shaking at 375 revolutions per minute (rpm) for 1 h. Samples were left to cool to room temperature and then centrifuged at 10,000 × g for 10 min. The supernatant was carefully removed with a pipette, and the tubes containing the gelatinized starch pellets were reweighed. Swelling power was calculated as: weight of pellet/dry weight of starch.
Analysis of GBSSI and Microsatellite Genotypes
Landrace accessions of P. miliaceum were provided by the Vavilov Institute, St Petersburg, Russia (VIR), the National Institute of Agrobiological Sciences Genebank, Japan (NIAS), and by the USDA-ARS North Central Regional Plant Introduction Station, Ames, IA, USA. Individual seeds were tested for endosperm starch phenotype as described earlier. A total of 178 individuals from 147 accessions were analyzed (supplementary table S2, Supplementary Material online). Panicum miliaceum is strongly selfing (∼90%; Baltensperger 2002), and for most accessions, only a single individual was analyzed. Up to three individuals were analyzed for some accessions, as waxy phenotype and GBSSI genotype data were already available for these samples from our previous study (Hunt et al. 2010). DNA was extracted from seedlings, and each individual was genotyped for the polymorphic sites in the [M13]int9Sf-R11, int5Lf-R3, and M12-R12 fragments as described earlier. Genotypes at the L and S loci were inferred accordingly from this data. Each sample was also genotyped at 16 of the microsatellite loci characterized by Cho et al. (2010), following the method in Hunt et al. (2011). Microsatellite genotyping data were analyzed in GeneMapper version 4.0 (Applied Biosystems) and scored manually for the diploid genotype at each locus. Multilocus genotypes (MLGs), including both the GBSSI and microsatellite genotype information, were identified by analysis in Microsoft Excel. Where multiple plants from a single accession shared the same MLG, a single representative of each MLG was retained for the subsequent analyses to avoid bias in analysis of associations.
Microsatellite genotype data were used to construct a neighbor-joining tree showing relationships between samples, inferred from a genetic distance matrix using Nei’s distance measure DA (Nei et al. 1983), as calculated by the software PowerMarker version 3.25 (Liu and Muse 2005). Modeling of the number of genetic clusters, based on the microsatellite genotype data, was carried out using a Bayesian clustering algorithm as implemented in the software InStruct (Gao et al. 2007). InStruct uses a Bayesian clustering algorithm similar to the widely used program STRUCTURE (Pritchard et al. 2000) but does not make the assumption of Hardy–Weinberg equilibrium and is therefore more appropriate for analysis of a diploid data set for a strongly inbreeding species, where this assumption is likely to be violated. Ten replicate runs were carried out for each number of clusters (K) from K = 1 to K = 10, with 200,000 burn-in and 1,000,000 Markov chain Monte Carlo reps. The method in Evanno et al. (2005) , as implemented in CorrSieve ver. 1.6-5 (Campana et al. 2011), was used to evaluate the optimal value of K. Correlations between Q-matrices for replicate runs were checked in CorrSieve.
Association of waxy genotypes with genetic clusters inferred from microsatellite loci was evaluated by plotting alleles at the GBSSI-S and GBSSI-L loci on the neighbor-joining DA tree and by performing analyses of variance (ANOVAs) in which waxy alleles were treated as the dependent variable and the proportions of each Q in the model with K = 7 (selected as justified in the Results later as the most informative model) as the independent variable for each sample. ANOVAs were performed separately for the S and L loci and separately for polychotomous and binary codings of the L allele states (the latter equivalent to post hoc t-tests). The two samples that were heterozygous at the GBSSI-L locus were excluded from the analysis. ANOVAs were performed in the R package (R Core Development Team 2005). This method is not perfect, because the values of Q for each sample necessarily sum to 1, so there is redundancy of information in running ANOVAs for all seven clusters. Nonetheless, it provides a clear and quantitative measure of the extent to which the proportional allocation of a sample to each genetic cluster can be explained by its GBSSI genotype.
Maps showing the geographical distribution of samples, genetic allocation to clusters in the K = 7 model in InStruct, and alleles at the GBSSI-L and GBSSI-S loci were plotted in ArcMap 10.1. Precise locations of origin were unknown for many samples. For the purposes of plotting data, these were roughly estimated from the geographic information available using GoogleEarth.
Results
Identification of a GBSSI-L Ortholog in P. capillare
Endosperm starch granules from P. capillare stained dark blue–black with iodine. This suggested that the GBSSI gene in this species confers a wild-type (nonwaxy) phenotype with normal endosperm amylose content, as is the case with all other wild species studied to date (Sakamoto 1996; Shapter et al. 2009).
We characterized the GBSSI sequence in this diploid species. The primers FPSLVVC3 and Rstop3 amplified a single product that yielded unambiguous direct sequence. The presence of a single GBSSI sequence type in P. capillare is consistent with its diploid genome. This sequence was 3,475 bp in length, and alignment with the GBSSI-S and GBSSI-L sequences from P. miliaceum showed that it had very high sequence similarity with GBSSI-L (94.5% including intron sequences or 99.3% considering coding sequence only). Given that alignment of the intron sequences of the GBSSI-S and GBSSI-L homeologs from P. miliaceum is not possible due to their dissimilarity (Hunt et al. 2010), we inferred that the GBSSI sequence in P. capillare is orthologous to GBSSI-L in P. miliaceum. Among the GBSSI-L alleles in P. miliaceum, the predicted amino acid sequence of the P. capillare GBSSI protein is closest to the product of the LC allele. It differs from the latter by three residues: LC has serine (substitution for alanine) at position 298 (supplementary fig. S1, Supplementary Material online), threonine (for methonine) at position 441, and methionine (for valine) at position 499. At all these three sites, the amino acid residue in the P. capillare protein sequence is the same as that in the P. miliaceum GBSSI S0 allele, which is also catalytically active.
Identification of LC/S-15 Lines in P. miliaceum
To evaluate the functionality of the LC allele in P. miliaceum, we needed to identify lines in which this allele was present in an S-15 background. No such lines were present among the total of 147 landrace accessions analyzed either previously (Hunt et al. 2010) or in this study (see later). From the segregation ratio data given by Graybosch and Baltensperger (2009), we inferred that it was highly likely that lines with the genotype S-15/LC would constitute a proportion of the F2-derived families which were true breeding for the nonwaxy phenotype. Thirty-one of these lines, representing the F4 generation, were available for testing. We found that two of these 31 lines—P017-10-2 and P017-10-4—were monomorphic for the genotype S-15/ LC. Both of these lines were derived from the cross “Earlybird” x PI436626. Full sequencing of the L and S loci for these lines confirmed that the predicted protein sequences were identical to those encoded by the LC and S-15 alleles described previously (Hunt et al. 2010).
Amylose Content
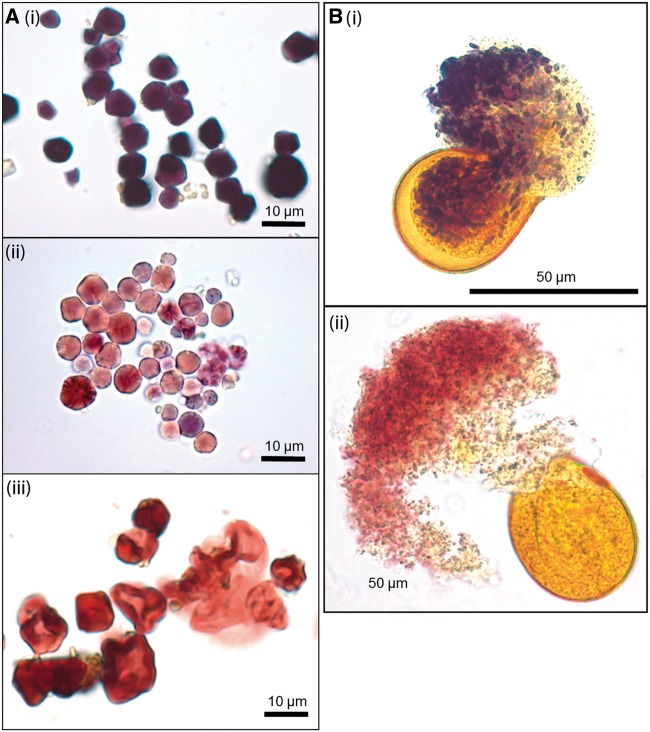
Examination of starch granules stained with Lugol’s solution from the two S-15/LC lines indicated that their phenotype was somewhat different from the previously characterized wild-type lines (genotypes S0/L, where L is any of the genotypes LC, LY, or Lf). A purplish-blue coloration demonstrated the presence of some amylose, but this coloration was less intense than for wild-type granules, and some granules appeared to stain only red rather than blue with iodine (fig. 1A).
Fig. 1.
Panicum miliaceum starch granules. Starch was stained with Lugol’s solution and observed using a light microscope. The scale is indicated. (A) Material scraped from mature seeds. The genotypes and accessions shown are i: S0/LC, MIL-4 #1 (nonwaxy, dark blue–black staining); ii: S-15/LC, line P017-10-2 (partially waxy, some granules staining red and some granules staining paler blue–purple, indicating the presence of amylose); iii: S-15/Lf, MIL-82 #1 (waxy, granules stain red, some darkly. The characteristic blue of amylose staining is absent). (B) Pollen squashed on a microscope slide to release some of the starch granules within. The genotypes and accessions shown are i: S0/LC, MIL-4 #1 (blue–black staining, amylose present); ii: S-15/Lf, MIL-70 #1 (red staining, amylose free).
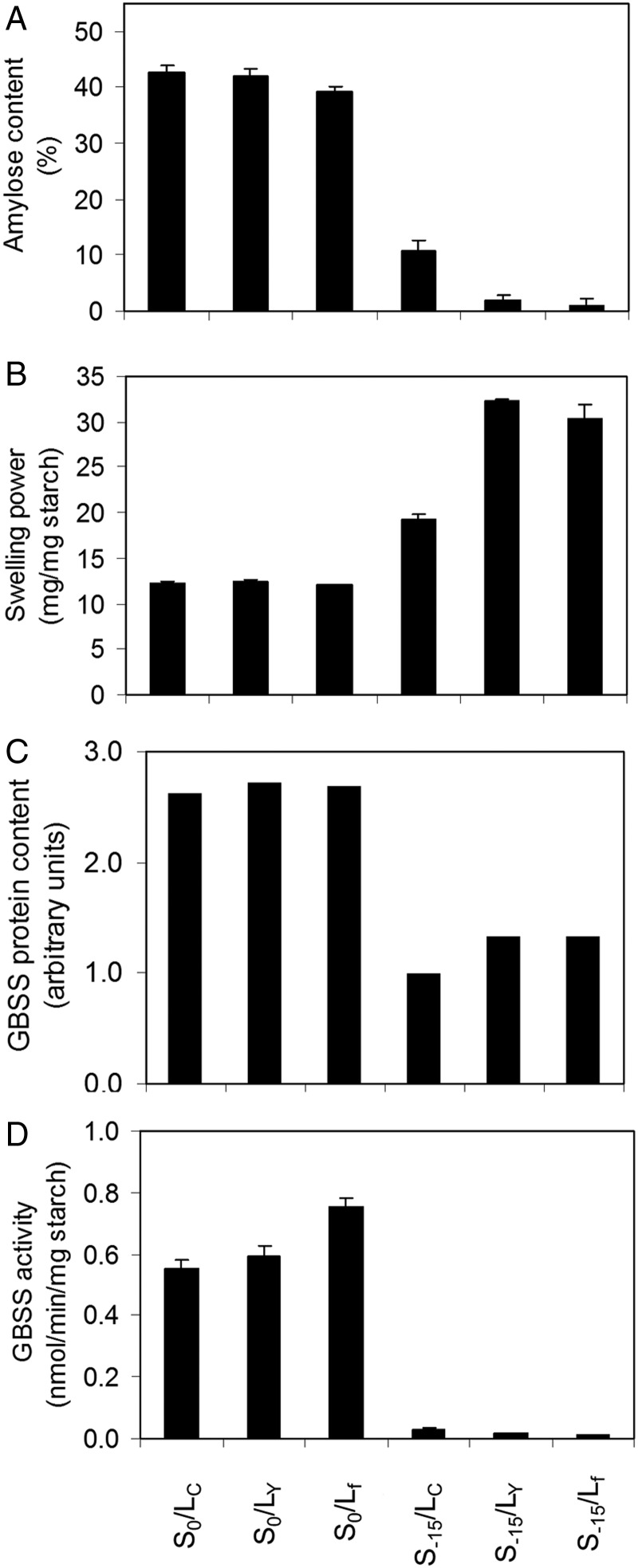
To investigate this result further, we undertook quantitative estimates of the amylose content in all six of the P. miliaceum GBSSI genotypes. The data are shown in figure 2A. The three genotypes previously shown to give wild-type phenotypes (S0/LC, S0/LY, and S0/Lf; Hunt et al. 2010) all contained approximately 35–40% amylose. The two genotypes previously shown to be waxy (S-15/LY and S-15/Lf) both contained approximately 1% amylose. The S-15/LC genotype was confirmed to have an intermediate phenotype with an amylose content of approximately 11%.
Fig. 2.
Biochemical properties of endosperm starch in six GBSSI genotypes of P. miliaceum. (A) Amylose content. (B) Starch swelling power. (C) GBSSI protein content. (D) Starch synthase activity.
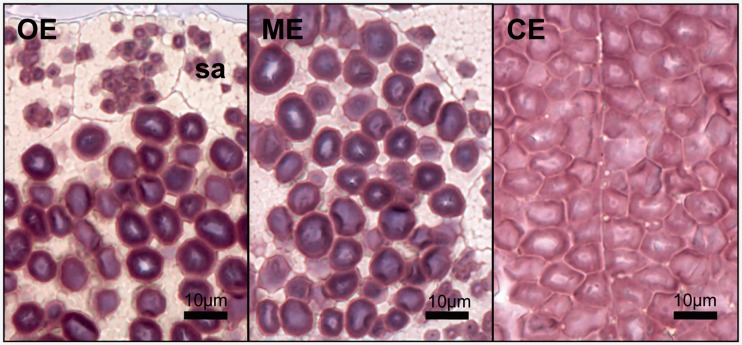
Staining of sections of endosperm from S-15/LC genotype seeds with Lugol’s solution showed that the outer edges of starch granules in the cells in the outer and mid endosperm stained red, indicating the absence of amylose (fig. 3). However, blue staining was visible inside these granules showing that they contained some amylose. In contrast, the starch in the central endosperm cells stained entirely red indicating that these starch granules contained very little, if any, amylose.
Fig. 3.
Sections of grains with low-amylose content. Dry-cut sections of 1.5 μm of mature endosperm of genotype S-15/LC stained with Lugol’s solution. Examples of the outer endosperm (OE; left panel), including the subaleurone cells (sa); mid endosperm further into toward the center of the grain (ME, middle panel); and the central endosperm (CE; right panel) are shown. The scale is indicated.
Starch Swelling Power
To determine the effect of the GBSSI genotype on the functional properties of endosperm starch, we measured the extent to which the starch swelled on gelatinization in the presence of excess water (swelling power). The data are shown in figure 2B. The three wild-type genotypes showed the lowest swelling power, consistently approximately 12%. The two waxy genotypes produced much larger and less dense pellets on gelatinization, with swelling power of approximately 30%. The S-15/LC genotype showed an intermediate swelling power of approximately 19%.
GBSSI Content and Activity
We carried out measurements of protein content and GBSSI activity on mature grains from all six GBSSI genotypes to determine the relative expression of GBSSI alleles and the specific activity of the resulting proteins. This data showed that the S-15/LC genotype had very low GBSSI content and activity, similar to the levels in the waxy genotypes (fig. 2C and D).
Starch Phenotype in Pollen Grains
To test whether both GBSSI alleles were expressed in pollen grains, as in endosperm starch, we used iodine staining to make a qualitative assessment of amylose content in pollen grains of all six GBSSI genotypes. We detected two phenotypes (fig. 1B): a blue-staining starch which we inferred contained amylose, seen in the lines S0/LC, S0/LY, S0/Lf, and S-15/LC, and a red-staining amylose-free starch, seen in the lines S-15/LY and S-15/Lf. Because of the very small amounts of tissue, it was not possible to make quantitative measurements of amylose content or GBSSI content or activity in pollen.
Analysis of GBSSI and Microsatellite Genotypes
Our previous study (Hunt et al. 2010) genotyped 72 plants from 38 accessions for GBSSI-S and GBSSI-L genotypes. In this study, we extended this analysis to a total of 178 plants from 147 accessions, including 69 of the 72 plants analyzed previously. We also genotyped all 178 plants for 16 microsatellite loci with no known connection to the GBSSI loci to analyze the association between GBSSI genotype and phylogeographic clusters. The genotyping of individual plants ensured a rigorous association between waxy genotype and phenotype and microsatellite genotypes. The genotypes at the GBSSI S and L loci for 178 plants, representing landrace accessions from across Eurasia, are shown in supplementary table S2, Supplementary Material online. We found 82 plants with the genotype S0/LC, 29 S0/LY, 17 S0/Lf, 37 S-15/LY, and 11 S-15/Lf. Two plants were heterozygous at the GBSSI-L locus, one with the genotype S0/LC/LY and one with the genotype S0/LC/Lf. No plants were found among the landrace accessions with the genotype S-15/LC.
The full data set of microsatellite genotypes at the 16 simple sequence repeat loci is available in supplementary table S2, Supplementary Material online. There were 151 distinct MLGs among the 178 plants. Excluding multiple plants from the same accession with the same MLG left a total data set of 168 plants, on which subsequent analyses were carried out.
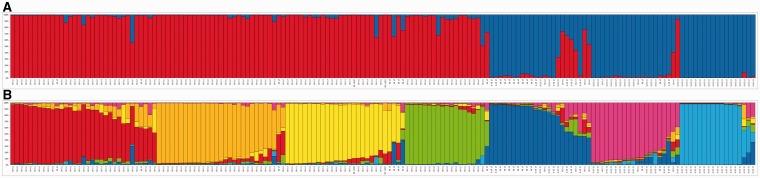
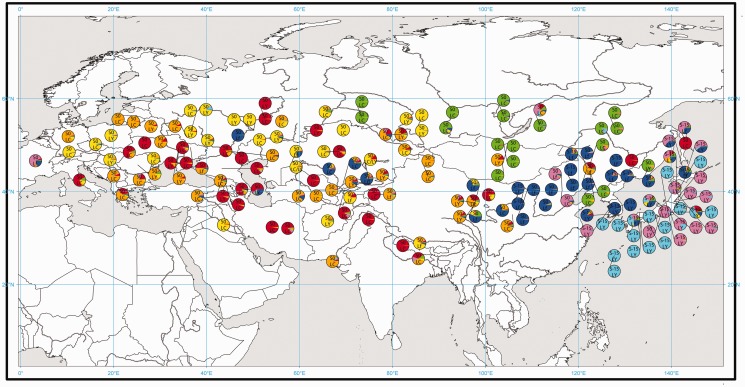
We used both Bayesian clustering analyses, implemented in InStruct, and neighbor-joining phenograms based on Nei's genetic distances (Nei et al. 1983) genetic distances, to evaluate genetic structuring of the microsatellite data set. InStruct output showed no clear value of K where ln P(D) reached a maximum or plateau. The parameter ΔK (Evanno et al. 2005) showed a maximum at K = 2. This split, with two gene pools, divides the samples into eastern and western groups, as found previously (Hunt et al. 2011). In that analysis, a model with six gene pools was also biogeographically meaningful and provided further resolution. In the current analysis, correlations between replicate runs showed that highly stable solutions were obtained up to K = 7 and that the K = 7 model showed a very similar phylogeographic pattern to the K = 6 pattern in Hunt et al. (2011), with an additional subdivision of one of the gene pools. We therefore used the model with seven gene pools as the basis for most of the subsequent analysis. The proportional assignments of each sample to each of the seven gene pools are shown in figures 4 and 5. Under this model, populations 1–4 (shown as red, orange, yellow, and green) fall into the western cluster defined by the K = 2 model, and populations 5–7 (dark blue, pink, and light blue) fall into the eastern cluster of this primary split. The position of the populations 4 (green) under K = 7 within the “western” cluster under K = 2 is in contrast with our previous results (Hunt et al. 2011), in which this group belonged to the “eastern” cluster at the higher level.
Fig. 4.
Microsatellite genotype clusters defined by InStruct. Proportional allocations for each plant sample to each gene pool for the InStruct K = 2 (A) and K = 7 (B) models. Alleles at the GBSSI-S and GBSSI-L loci are shown.
Fig. 5.
Microsatellite genotype clusters and GBSSI allele distribution. For each sample, the pie chart shows the proportional allocation to each gene pool under the K = 7 model. The alleles at the GBSSI-S and GBSSI-L loci are shown superimposed.
In the eastern part of the range, population 5 (shown in dark blue in figures 4, 5, and 6) is largely confined to China and Korea. Population 6 (pink) dominates a small number of samples in northeastern China, and Korea, and approximately half the samples from Japan, predominantly in the northeast. Population 7 (light blue) is confined to Japan, and samples assigned to this population are largely from the southwest of the country. Population 4 (green) has a northerly distribution in Eastern Asia, in North China, Mongolia, Siberia, and the Russian Far East. In the western part of the range, populations 1, 2, and 3 (red, orange, and yellow) all have in a distribution across longitudes ranging from northwestern China to eastern Europe. Of these three populations, number 3 (yellow) appears to have a more northerly center of distribution, at high frequency in northwestern Kazakhstan and the most northwesterly samples from Russia, and in a number of samples in the Novosibirsk region. Populations 1 and 2 (red and orange) show less clear spatial separation in this broad range.
Fig. 6.
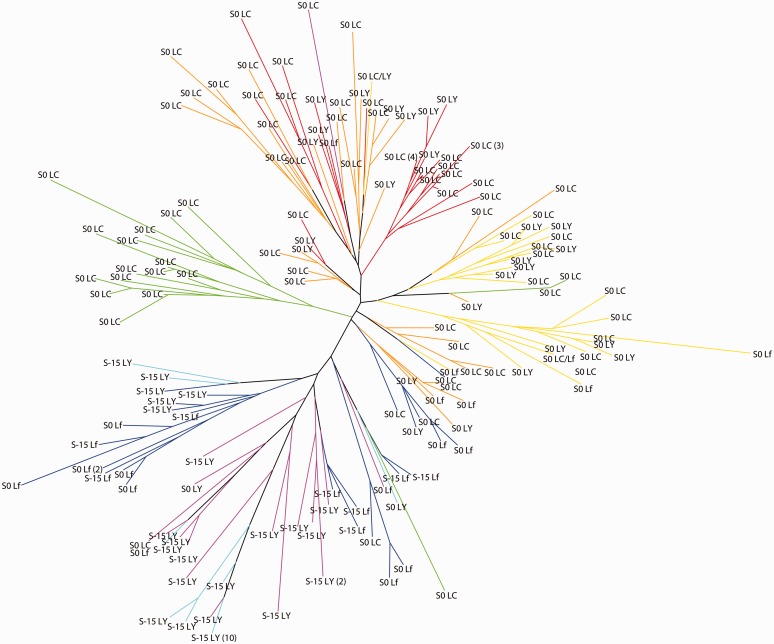
Dendrograms showing microsatellite genotype clusters and GBSSI alleles. Neighbor-joining tree showing relationships among samples based on microsatellite genotypes, using Nei’s genetic distances (Nei et al. 1983). Branches are colored according to the highest proportional allocation to the gene pools identified under the InStruct analysis in the K = 7 model (even where this is <50%). The GBSSI genotype is shown for each sample at the GBSSI-S and GBSSI-L loci. Where multiple individuals share a microsatellite and GBSSI genotype, the number of individuals is indicated in brackets. The genotype of individuals heterozygous for the GBSSI-L locus is shown as both alleles separated by /.
The topology of the neighbor-joining tree (fig. 6) shows broad agreement of the relationships between samples with the InStruct allocations. The branches of the tree are colored consistent with figures 4 and 5, showing the highest InStruct cluster allocation for each sample (even where this is below 50%). Considering the samples with the highest proportional allocation to population 7 (light blue), it appears that these are derived from population 6 (pink). Population 4 (green) forms a clear clade that sits between clades containing populations 1/2/3 (red/orange/yellow) and populations 5/6/7 (dark blue/pink/light blue), respectively. This is consistent with its variable placement in the eastern and western clusters under K = 2 found between the analyses in this article and our previous work (Hunt et al. 2011).
Considering the distribution of the alleles at the GBSSI-S and GBSSI-L loci in relation to the genetic groups identified by the cluster and dendrogram analyses, a number of associations can be seen. Four of the seven InStruct populations—shown in red, orange, yellow, and green—are monomorphic for the wild-type S0 allele. The mutant S-15 allele is at medium–high frequency in populations 5 (dark blue) and 6 (pink) and at 100% frequency in population 7 (light blue). At the GBSSI-L locus, the LC allele occurs at high frequency in populations 1, 2, and 4 (red, orange, and green) and at very low frequency in populations 5 (dark blue) and 6 (pink). It is absent from population 7 (light blue), which is monomorphic for the mutant LY allele. This allele also occurs at moderate to high frequency in populations 6 (pink) and 3 (yellow), at low frequency in populations 5 (dark blue), 1 (red), and 2 (orange), and is absent from population 4 (green). The other mutant L allele, Lf, is at high frequency in population 5 (dark blue), at low frequency in populations 1 (red), 3 (yellow), and 6 (pink), and absent from (orange) and 4 (green). ANOVA tests for association between GBSSI alleles and proportional assignment to each of the populations under the K = 7 model provide statistical support for the observed positive and negative associations between particular GBSSI alleles and genetic clusters inferred from the microsatellite data (table 1).
Table 1.
Results of ANOVA Tests for Association between GBSSI Alleles and Proportional Allocation to each Gene Pool under the K = 7 Model.
| S0 vs. S-15 | LC vs. LY vs. Lf | LC vs. non-LC | LY vs. non-LY | Lf vs. non-Lf | |
|---|---|---|---|---|---|
| 1 Red | *** | ** | ** | ||
| 2 Orange | *** | *** | *** | * | * |
| 3 Yellow | *** | ||||
| 4 Green | ** | *** | *** | *** | |
| 5 Dark blue | * | *** | *** | * | *** |
| 6 Pink | *** | ** | *** | *** | |
| 7 Light blue | *** | *** | *** |
Note.—Statistically significant results can indicate either a positive or negative association.
*P < 0.05.
**P < 0.01.
***P < 0.001.
Discussion
Production of an Active Protein by the GBSSI LC Allele
The identification of lines with the GBSSI LC allele in an S-15 background generated by the crossing program of Graybosch and Baltensperger (2009) enabled us to test the functionality of the LC protein. The data of Graybosch and Baltensperger (2009) implied that the LC allele is sufficient for the production of wild-type endosperm starch. However, these results were based on a large-scale iodine staining screen, and genotype information was not available. The additional investigations we have carried out here have shown that this allele produces a protein which is capable of catalyzing the synthesis of at least some amylose. This corroborates the conclusion of Graybosch and Baltensperger (2009) that endosperm texture in P. miliaceum is under the control of two loci.
Relative Capacities of the GBSSI-S and GBSSI-L Loci for Amylose Synthesis
Our biochemical analyses of the six possible GBSSI genotypes in P. miliaceum allowed us to determine the relative amylose synthesis capacities of the GBSSI-S and GBSSI-L loci. In endosperm, in the absence of the active GBSSI-S allele, S0, the LC allele produces only approximately 25% of the amylose content found in wild-type grain. In contrast, S0 alone (i.e., in combination with a nonfunctional GBSSI-L allele) produces close to 100% of the amylose content of the wild type. The difference between 25% and 100% amylose content relative to wild type is difficult to detect by a simple microscopic examination of iodine-stained crushed grain, accounting for the scoring of S-15/LC genotypes as wild type by Graybosch and Baltensperger (2009). Thus, we infer that the GBSSI-S locus is the major determinant of amylose content in millet endosperm and that the GBSSI-L locus makes only a minor contribution. Our data also show that LC contributes relatively little GBSSI protein compared with the S alleles. It appears that neither GBSSI protein or activity nor amylose content increased in plants with an S0/LC genotype relative to those with S0/LY or S0/Lf genotypes, despite the demonstrated activity of LC. Indeed, GBSSI activity appears to be higher in the S0/Lf than in the S0/LC or S0/LY lines. This could be explained if GBSSI-S has higher specific activity than GBSSI-L: the GBSSI-L protein is absent from the S0/Lf genotype and so all the GBSSI protein in this genotype is the more active S0 form. We conclude that the presence of S0 alone appears to be sufficient for wild-type amylose content.
In pollen, our starch phenotype data showed that GBSSI-S and GBSSI-L both exhibit some activity: pollen grains with either the S0 or LC alleles contain amylose, in the presence of the established mutant alleles LY or Lf, or S-15, respectively. As in the endosperm, pollen grains with mutant alleles at both loci, that is, genotypes S-15/LY and S-15/Lf, are amylose free. Quantitative measurements of amylose contents in pollen grains from different genotypes would be needed to determine whether the relative contributions of GBSSI-S and GBSSI-L in pollen differed from those in endosperm. However, there is no evidence from the present data for substantial differences between the two GBSSI loci in their patterns of expression in these two tissues.
Our finding that the GBSSI-S and GBSSI-L loci contribute unequally to amylose content in endosperm is comparable with data on polyploid wheats. In tetraploid and hexaploid wheats, the different GBSSI homeologs have been shown to make differential contributions to endosperm amylose content, although the extent of the inequality between homeologs in wheat is less than that seen in P. miliaceum. In bread wheat (Triticum aestivum), the Wx-B1 allele contributes most to GBSSI protein levels and amylose content, followed by the Wx-D1 allele, and the Wx-A1 allele contributes least (Yamamori and Quynh 2000). A similar result is found in emmer wheat, with the Wx-A protein making a smaller contribution to total GBSSI than Wx-B (Yamamori et al. 1995).The molecular mechanism responsible for the differential contributions of the Wx homeologs to GBSSI protein and amylose content in wheat is unknown (Yamamori and Quynh 2000).
In millet, we suggest that one or both of two factors are consistent with the reduced contribution of the GBSSI-L locus to GBSSI protein and amylose content in the endosperm. First, sectioning of P. miliaceum grains revealed that the spatial distribution of amylose in the S-15/LC genotypes differed from the wild type both in P. capillare and in S0-bearing genotypes of P. miliaceum. The restriction of amylose to the outer cell layers in S-15/LC lines is a pattern similar to that found in some low-amylose barley lines, in which a mutant wx allele with a 413-bp deletion in the promoter region shows an altered temporal and/or spatial pattern of expression consistent with expression later in endosperm development than normal (Patron et al. 2002; Yanagisawa et al. 2006). The LC allele may show similar alteration in expression patterns relative to its ortholog in P. capillare and to the GBSSI-S protein. Indirect evidence that spatial expression may be altered comes from work on other cereals including barley, maize, and sorghum. These studies indicate that amylose levels in wild-type starch are typically highest in central endosperm and lower in peripheral endosperm (Boyer et al. 1976; Ring et al. 1989; Sullivan et al. 2010), that is, the reverse of the spatial pattern seen in partially waxy barley and broomcorn millet.
Second, as discussed earlier, it is possible that the LC protein may possess lower specific activity relative to its ortholog in P. capillare. We note that the LC allele has three amino acid substitutions relative to both the fully functional P. capillare L-ortholog and the P. miliaceum GBSSI-S0, which might account for reduced specific activity. However, from an alignment (supplementary fig. S1, Supplementary Material online), it could be seen that none of these residues are highly conserved among other functional GBSSIs. Site-directed mutagenesis, which was beyond the scope of this study, would be required to determine whether any or all these three amino acid substitutions significantly affect starch synthase specific activity.
Nonlinearity of the Relationship between Different Measures of “Waxiness”
The biochemical analyses in our article measure several different phenotypic effects of mutations at the GBSSI locus. For some phenotypic measures (swelling power and amylose content), the values for the S-15 /LC lines are intermediate between those of the wild-type and waxy lines, whereas for others (GBSSI activity and GBSSI protein content), the values are very similar to those of the waxy. In respect of iodine staining of seed, the S-15/LC phenotype was very similar to the wild type (Graybosch and Baltensperger 2009). Nonlinear relationships between GBSSI activity/protein content and amylose content are expected for components of multienzyme pathways (Kacser and Burns 1981) and have been described in other species, for example, wheat (Debiton et al. 2010) and potato (Flipse et al. 1996). Low-amylose content is the most frequently employed test for waxiness, because of the ease of screening plant material qualitatively for this trait. However, starch swelling power is likely to be the best proxy measure for the variation in texture of cooked grain, which represents that aspect of the phenotype subject to human selection. The precise relationship between these measures is less important in species, which show only a dimorphism in phenotypes between wild-type and waxy varieties. However, where multiple phenotypic states are known for the waxiness trait, as in broomcorn millet, wheat (Debiton et al. 2010), and rice (Dobo et al. 2010), then the evaluation of selection (in particular, preindustrial selection) on particular genotypes requires consideration of whether the most appropriate measures of "waxiness" have been assessed. In P. miliaceum, we found that for starch swelling power, plants with the S-15/LC genotype showed a clearly intermediate phenotype between waxy and nonwaxy lines.
Partial Diploidization of the GBSSI Locus Responsible for Grain Amylose in Broomcorn Millet and Its Implications for Allele Selection
In summary, our biochemical data suggest that the GBSSI locus responsible for grain amylose is in the process of becoming diploidized in broomcorn millet. Polyploid speciation in plants frequently leads to the loss or silencing of redundant homeologous copies of protein-coding genes or to differences in expression patterns (Chen and Ni 2006). In the endosperm, the GBSSI-L homeolog on its own has a severely reduced capacity for amylose synthesis compared with GBSSI-S, and the presence of S0 appears to be sufficient for wild-type amylose content regardless of the GBSSI-L allele present. Multiple mechanisms may account for this partial diploidization, but on the basis of our data, we cannot determine which of these is most important.
Our biochemical analyses show that distinct, loss-of-function mutations in the GBSSI-L and GBSSI-S loci were needed to give plants with the fully waxy phenotype that has been selected for in east Asia. However, it is also apparent that, given the unequal contributions of the GBSSI-S and GBSSI-L loci to amylose synthesis, the LY and Lf mutations would be selectively neutral in an S0 background. As we inferred previously (Hunt et al. 2010), the S-15 mutation was essential for the evolution of lines with an altered endosperm starch texture. These points help in understanding the spatial and temporal sequence of evolution of alleles at the GBSSI-S and GBSSI-L loci that gave rise to waxy phenotypes.
Inferring the Complex History of Selection and Spread of GBSSI Alleles from Phylogeographic Analyses
The analysis of microsatellite markers gives a phylogenetic context to the distribution of GBSSI alleles and phenotypes that contributes to understanding evolution at this locus. Microsatellite analysis shows that the broomcorn millet gene pool across Eurasia shows strong phylogeographic structure. By screening individual plants from landraces across a wide geographical range for both microsatellite and GBSSI genotypes, we were able to detect associations between phylogeographic clusters and alleles at the two GBSSI loci. Some of these associations were very strong; however, there was also clear evidence for the transfer of mutant alleles among genetic populations, indicating a complex history of spread and selection of GBSSI alleles. The analysis that follows explains these points in detail and allows us to suggest a model for the evolution of waxy phenotypes.
We can assume that the S0 and LC alleles are ancestral to the loss-of-function mutant alleles S-15 and LY/Lf, respectively. The distribution of the LY allele is widespread both geographically and phylogenetically, from which we infer that this mutation probably occurred early in the history of broomcorn millet cultivation. Archaeological and genetic evidence strongly supports northern China as the major center of broomcorn millet domestication and early cultivation as a staple cereal (Hunt et al. 2011). These considerations would suggest that it is likely that the LY mutation arose in this region, perhaps before the divergence of the genetic clusters identified by our microsatellite analysis and then spread both westward to western Russia/eastern Europe and eastward to Japan and Korea. One apparent problem with this model is that the LY allele is very rare among the Chinese samples we analyzed and found only in a single accession (MIL-72) from northwest China. However, it is known that mutations that arise in expanding populations can reach high frequencies in the zone of expansion and remain uncommon in the region of origin (Edmonds et al. 2004; Klopfstein et al. 2006).
Homoplasy (convergent evolution) of the LY allele in the western populations (shown in yellow/orange/red) and eastern populations (those shown in pink/light blue) is highly unlikely, because examination of the full-length sequences for accessions representing both these groups (MIL-47: GenBank accession number GU199254 and MIL-3o and 3y: GU199257 and GU199258, respectively) shows that exemplars of the LY allele from both these geographic groups also share an intron substitution (G for C at nucleotide position 1408). The parallel character state of this SNP with the cysteine–tyrosine mutation in exon 7 is strong evidence that the LY alleles across the geographic range are identical by descent. In the light of the above, it is also unlikely that the LY allele arose either in the extreme west (eastern Europe) or east (Japan) of its current range and then spread sufficiently widely to become established in both these regions. This spread, which would have had to cross China, would be difficult to reconcile with both the archaeological evidence clearly placing China as the oldest center of broomcorn millet cultivation by some 3 millennia and our data showing the phylogenetic distinctiveness of the western populations (those shown in red, orange, and yellow) and those in Korea and Japan (shown in pink and light blue).
We therefore argue that an origin for the LY allele in China and its outward spread from this region is the most likely explanation of the data. This adds weight on the side of arguments that the phylogeographic patterns observed in P. miliaceum (Hunt et al. 2011) represent a single center of domestication in northern China and that the western genetic cluster arose from a founder effect in westward spread rather than independent domestication in eastern Europe.
Our biochemical data demonstrate that the LC/LY polymorphism would be selectively neutral in the western part of the range, because these populations are monomorphic for the S0 allele. The observed polymorphism at the GBSSI-L locus among the populations shown in red, orange, and yellow is presumably the result of demographic processes. We note that the Ly allele is at higher frequency in population 3 (yellow) than in the closely related populations 1 (red) and 2 (orange), perhaps reflecting founder effects in the splitting and spread of these populations.
The very high frequency of the LY allele in Japan as also reported by Araki et al. (forthcoming) could be accounted for in two different ways. First, it could indicate a founder effect in the spread of population 6 (pink), in which the LY allele was still selectively neutral but approached fixation in Japan by chance. Alternatively, waxy phenotype plants could have arisen through association of the LY allele with the S-15 mutation before, or early in the history of, the spread of broomcorn millet into Japan, which were then subjected to strong positive selection.
In contrast to the LY allele, the Lf allele has a restricted distribution. It is very strongly associated with population 5 (dark blue), which itself is largely restricted to China. The limited geographic spread, and the observation that this allele has not crossed substantially into other genetic groups, suggest that it arose relatively recently. Among the accessions from northeastern China, the Lf allele is the most common (and found in combination with both the S0 and S-15 alleles to give both nonwaxy and waxy phenotypes), but the LC allele is also present in several accessions.
There appear to be two plausible centers of origin for the mutant S-15 allele, namely in Japan or (northeastern) China. Our biochemical data on phenotypes of plants homozygous for the S-15 allele show that this allele would likely have been subject to strong selection for texture regardless of the GBSSI-L allele background in which it appeared. The strongly inbreeding tendency of P. miliaceum (∼90%; Baltensperger 2002) means that the homozygous genotype and therefore the waxy phenotype would be generated rapidly and would have exposed this allele to selection. The S-15 allele is associated with the LY allele in Korea and Japan to produce fully waxy phenotypes, whereas in China these phenotypes result from the association of S-15 with Lf. The microsatellite analysis shows that these populations are genetically differentiated. This suggests that, in whichever of these two populations the S-15 arose, it has crossed between them, likely facilitated by strong positive selection for the waxy texture by human populations in both Japan and northeastern China.
Accounting for the Absence of Partially Waxy Lines in Broomcorn Millet—The Role of Selection
It is striking that partially waxy lines (with the S-15/LC genotype) are either extremely rare in or absent from the broomcorn millet gene pool. This is in apparent contrast to wheat, in which partially waxy landraces are known. Two possible explanations for the absence of partially waxy millet landraces are that 1) the relevant alleles at the two homeologous loci are restricted to distinct geographic or evolutionary clusters, which has limited opportunities for them to come into combination, or 2) there has been selection against this phenotype. Collectively, the microsatellite and GBSSI data demonstrate that gene flow occurs between the differentiated populations. It is thus unlikely that the absence of landraces with the genotype LC/S-15, giving the partially waxy phenotype, can be fully explained by geographic or genetic isolation of populations. We suggest, therefore, that the absence of partially waxy landraces indicates selection against these intermediate phenotypes.
Toward a More Precise Understanding of Cultural Selection for Waxy Phenotypes
Selection against partially waxy phenotypes of broomcorn millet would contrast with the situation in bread wheat, in which partially waxy phenotypes, with mutations in one or two of the three genomes, have been selected in landraces for upon noodle making (Yamamori and Quynh 2000). This highlights the current lack of detailed understanding of the culinary practices and cultural influences that have driven selection for GBSSI genotypes in P. miliaceum. In this regard, we can make several points.
The distribution of waxy types of broomcorn millet in our data set is restricted to
China, Korea, and Japan and one sample from Sakhalin island. Although the geographic
location is imprecise for many of the Chinese samples, it appears that the waxy types are
restricted to the northeastern provinces, whereas lines from northwestern China are
nonwaxy. This is reflected in Chinese-language terms for millet: local farmers in
northeastern China and central Inner Mongolia distinguish between shuzi
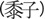 —nonsticky (i.e., nonglutinous or
nonwaxy) broomcorn millet—and mizi
—nonsticky (i.e., nonglutinous or
nonwaxy) broomcorn millet—and mizi
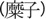 —sticky (i.e., glutinous or waxy)
broomcorn millet, whereas in Gansu province, only shuzi is used, and
mizi is not a recognized term (Liu X, personal communication). This
phenotype geography is similar to that in other cereals, and it is notable that the
western limit of the region in which sticky cereals are found approximately coincides with
both that of the East Asian summer monsoon and the Han Chinese culture (Fuller D, personal
communication). Fuller and Rowlands (2009)
and Yoshida (2002) argue that the
sticky/nonsticky divide, which seems to have developed first in rice, reflects a
fundamental distinction between two different cultures of food processing with different
associated technological artifacts. The first, centered in east Asia, is derived from
Pleistocene exploitation of nuts and tubers (Yoshida 2002) and is based on the boiling and steaming of grain in ceramic
vessels. The second culture, centered on western Eurasia, focuses on the grinding and
baking of grain. The Epipalaeolithic and early Neolithic of this region are characterized
by the presence of grinding stones; pottery postdates the appearance of agriculture by
some 3–4 millennia. However, a number of questions remain. Did the textural
preference for sticky grains relate to the handling properties of the cooked
grain—that is, its cohesiveness in vessels or on eating implements—or its
texture in the mouth? Insufficient attention has also been paid to variation within the
“sticky-grain” zone. This zone is by no means a single cultural unit, either
today or in the past. Variation in usage of sticky grain varieties, and the relative
frequency of cultivation and consumption of sticky and nonsticky types among different
cultural groups, and among different cereals, needs detailed clarification. Discussion of
“preference” for sticky grains seems to imply a psychological choice, but this
may be linked with a physiological component: low-amylose starches are less resistant to
digestion and produce a more pronounced blood sugar spike (Åkerberg et al. 1998; Karlsson et al. 2007). Biochemical and genetic data on a range
of cereal crops demonstrate how selection for the waxy trait has impacted on the plants
themselves. To complete the picture, complementary ethnographic studies are needed that
answer outstanding questions about the human side of this process.
—sticky (i.e., glutinous or waxy)
broomcorn millet, whereas in Gansu province, only shuzi is used, and
mizi is not a recognized term (Liu X, personal communication). This
phenotype geography is similar to that in other cereals, and it is notable that the
western limit of the region in which sticky cereals are found approximately coincides with
both that of the East Asian summer monsoon and the Han Chinese culture (Fuller D, personal
communication). Fuller and Rowlands (2009)
and Yoshida (2002) argue that the
sticky/nonsticky divide, which seems to have developed first in rice, reflects a
fundamental distinction between two different cultures of food processing with different
associated technological artifacts. The first, centered in east Asia, is derived from
Pleistocene exploitation of nuts and tubers (Yoshida 2002) and is based on the boiling and steaming of grain in ceramic
vessels. The second culture, centered on western Eurasia, focuses on the grinding and
baking of grain. The Epipalaeolithic and early Neolithic of this region are characterized
by the presence of grinding stones; pottery postdates the appearance of agriculture by
some 3–4 millennia. However, a number of questions remain. Did the textural
preference for sticky grains relate to the handling properties of the cooked
grain—that is, its cohesiveness in vessels or on eating implements—or its
texture in the mouth? Insufficient attention has also been paid to variation within the
“sticky-grain” zone. This zone is by no means a single cultural unit, either
today or in the past. Variation in usage of sticky grain varieties, and the relative
frequency of cultivation and consumption of sticky and nonsticky types among different
cultural groups, and among different cereals, needs detailed clarification. Discussion of
“preference” for sticky grains seems to imply a psychological choice, but this
may be linked with a physiological component: low-amylose starches are less resistant to
digestion and produce a more pronounced blood sugar spike (Åkerberg et al. 1998; Karlsson et al. 2007). Biochemical and genetic data on a range
of cereal crops demonstrate how selection for the waxy trait has impacted on the plants
themselves. To complete the picture, complementary ethnographic studies are needed that
answer outstanding questions about the human side of this process.
Supplementary Material
Supplementary tables S1 and S2 and figure S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the Vavilov Research Institute (St Petersburg, Russia), the National Museum of Georgia, the USDA-ARS PI station (Ames, Iowa), NIAS Genebank (Japan), IPK Gatersleben (Leibniz, Germany), and Xinyi Liu and Loukas Barton for providing germplasm; the University of Cambridge Botanic Garden for plant growth facilities and support; the John Bingham Laboratory, NIAB (Cambridge, UK), for experimental facilities; the University of Cambridge’s computational grid, CamGRID, on whose resources analyses were performed; and Jenny Barna for computing support. This work was supported by a Wellcome Trust Bioarchaeology Research Training Fellowship (ref. 076815/Z/05/Z) to H.V.H. The research in this article was carried out at the McDonald Institute for Archaeological Research, University of Cambridge, at the John Bingham Laboratory, NIAB, and at the John Innes Centre.
References
- Åkerberg A, Liljeberg H, Björck I. Effects of amylose/amylopectin ratio and baking conditions on resistant starch formation and glycaemic indices. J Cereal Sci. 1998;28:71–80. [Google Scholar]
- Araki M, Numaoka A, Kawase M, Fukunaga K. Origin of waxy common millet, Panicum miliaceum L. in Japan. Genet Resour Crop Evol. Forthcoming doi 10.1007/s10722-011-9755-9. [Google Scholar]
- Baltensperger DD. Progress with proso, pearl and other millets. In: Janick J, Whipkey A, editors. Trends in new crops and new uses. Alexandria (VA): ASHS Press; 2002. pp. 100–103. [Google Scholar]
- Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques. 2001;31:24–28. [PubMed] [Google Scholar]
- Boyer CD, Shannon JC, Garwood DL, Creech RG. Changes in starch granule size and amylose percentage during kernel development in several Zea-Mays-L genotypes. Cereal Chem. 1976;53:327–337. [Google Scholar]
- Campana MG, Hunt HV, Jones H, White J. CorrSieve: software for summarising and evaluating STRUCTURE output. Mol Ecol Resour. 2011;11:349–352. doi: 10.1111/j.1755-0998.2010.02917.x. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Ni Z. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays. 2006;28:240–252. doi: 10.1002/bies.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y-I, Chung J-W, Lee G-A, Ma K-H, Dixit A, Gwag J-G, Park Y-J. Development and characterization of twenty-five new polymorphic microsatellite markers in proso millet (Panicum miliaceum L.) Genes Genom. 2010;32:267–273. [Google Scholar]
- Debiton C, Bancel E, Chambon C, Rhazi L, Branlard G. Effect of the three waxy null alleles on enzymes associated to wheat starch granules using proteomic approach. J Cereal Sci. 2010;52:466–474. [Google Scholar]
- Dobo M, Ayres N, Walker G, Park WD. Polymorphism in the GBSS gene affects amylose content in US and European rice germplasm. J Cereal Sci. 2010;52:450–456. [Google Scholar]
- Edmonds CA, Lillie AS, Cavalli-Sforza LL. Mutations arising in the wave front of an expanding population. Proc Natl Acad Sci U S A. 2004;101:975–979. doi: 10.1073/pnas.0308064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Flipse E, Keetels C, Jacobsen E, Visser RGF. The dosage effect of the wildtype GBSS allele is linear for GBSS activity but not for amylose content: absence of amylose has a distinct influence on the physico-chemical properties of starch. Theor Appl Genet. 1996;92:121–127. doi: 10.1007/BF00222961. [DOI] [PubMed] [Google Scholar]
- Fuller DQ, Rowlands M. Towards a long-term macro-geography of cultural substances: food and sacrifice traditions in East, West, and South Asia. Chinese Rev Anthr. 2009;12:1–37. [Google Scholar]
- Gao H, Williamson S, Bustamante C. An MCMC approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybosch RA, Baltensperger DD. Evaluation of the waxy endosperm trait in proso millet (Panicum miliaceum) Plant Breeding. 2009;128:70–73. [Google Scholar]
- Hunt HV, Campana MG, Lawes MC, Park Y-J, Bower MA, Howe CJ, Jones MK. Genetic diversity and phylogeography of broomcorn millet (Panicum miliaceum L.) across Eurasia. Mol Ecol. 2011;20:4756–4771. doi: 10.1111/j.1365-294X.2011.05318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt HV, Denyer K, Packman LC, Jones MK, Howe CJ. Molecular basis of the waxy phenotype in broomcorn millet (Panicum miliaceum L.) Mol Biol Evol. 2010;27:1478–1494. doi: 10.1093/molbev/msq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK. Between fertile crescents: minor grain crops and agricultural origins. In: Jones MK, editor. Traces of ancestry: studies in honour of Colin Renfrew. Cambridge: Oxbow Books; 2004. pp. 127–135. [Google Scholar]
- Kacser H, Burns JA. The molecular basis of dominance. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson ME, Leeman AM, Björck IME, Eliasson A-C. Some physical and nutritional characteristics of genetically modified potatoes varying in amylose/amylopectin ratios. Food Chem. 2007;100:136–146. [Google Scholar]
- Klopfstein S, Currat M, Excoffier L. The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol. 2006;23:482–490. doi: 10.1093/molbev/msj057. [DOI] [PubMed] [Google Scholar]
- Knutson CA, Grove MJ. Rapid method for estimation of amylose in maize starches. Cereal Chem. 1994;71:469–471. [Google Scholar]
- Konik-Rose CM, Moss R, Rahman S, Appels R, Stoddard F, McMaster G. Evaluation of the 40 mg swelling test for measuring starch functionality. Starch-Stärke. 2001;53:14–20. [Google Scholar]
- Liu K, Muse SV. PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Liu X, Hunt HV, Jones MK. River valleys and foothills: changing archaeological perceptions of North China’s earliest farms. Antiquity. 2009;83:82–95. [Google Scholar]
- Lu H, Zhang J, Liu K, et al. (13 co-authors) Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc Natl Acad Sci U S A. 2009;106:7367–7372. doi: 10.1073/pnas.0900158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Néron B, Ménager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C. Mobyle: a new full web bioinformatics framework. Bioinformatics. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Smith AM, Fahy BF, Hylton CM, Naldrett MJ, Rossnagel BG, Denyer K. The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5' non-coding region. Plant Physiol. 2002;130:190–198. doi: 10.1104/pp.005454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team. 2005. R Version 2.10.1. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ring SH, Akingbala JO, Rooney LW. A study of factors affecting amylose content of sorghum determined by an automated-method. Starch-Stärke. 1989;41:457–461. [Google Scholar]
- Sakamoto S. Glutinous-endosperm starch food culture specific to Eastern and Southeastern Asia. In: Ellen R, Fukui K, editors. Redefining nature: ecology, culture and domestication. Oxford: Berg; 1996. pp. 215–231. [Google Scholar]
- Shapter FM, Eggler P, Lee LS, Henry RJ. Variation in granule-bound starch synthase I (GBSSI) loci amongst Australian wild cereal relatives (Poaceae) J Cereal Sci. 2009;49:4–11. [Google Scholar]
- South JB, Morrison WR. Isolation and analysis of starch from single kernels of wheat and barley. J Cereal Sci. 1990;12:43–51. [Google Scholar]
- Sulaiman BD, Morrison WR. Proteins associated with the surface of wheat starch granules purified by centrifuging through caesium chloride. J Cereal Sci. 1990;12:53–61. [Google Scholar]
- Sullivan P, O'Flaherty J, Brunton N, Gee VL, Arendt E, Gallagher E. Chemical composition and microstructure of milled barley fractions. Eur Food Res Technol. 2010;230:579–595. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Yamamori M, Nakamura T, Nagamine T. Polymorphism of two waxy proteins in the Emmer group of tetraploid wheat, Triticum-Dicoccoides, T-Dicoccum, and T-Durum. Plant Breeding. 1995;114:215–218. [Google Scholar]
- Yamamori M, Quynh NT. Differential effects of Wx-A1, -B1, and -D1 protein deficiencies on apparent amylose content and starch pasting properties in common wheat. Theor Appl Genet. 2000;100:32–38. [Google Scholar]
- Yamanaka S, Nakamura I, Watanabe KN, Sato Y-I. Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor Appl Genet. 2004;108:1200–1204. doi: 10.1007/s00122-003-1564-x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa T, Donion E, Fujita M, Kiribuchi-Otobe C, Takayama T. Starch pasting properties and amylose content from 17 waxy barley lines. Cereal Chem. 2006;83:354–357. [Google Scholar]
- Yoshida S. Wild plant foods and vegeculture. In: Yoshida S, Matthews PJ, editors. Vegeculture in Eastern Asia and Oceania. Osaka: Japan Centre for Area Studies. National Museum of Ethnology; 2002. pp. 31–44. [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.