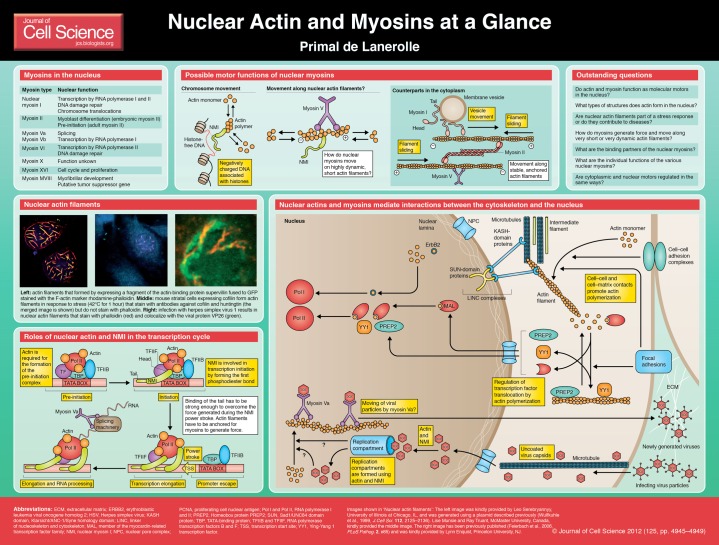
Actin and myosin II form the archetypical molecular motor complex. Myosin II, like all members of the myosin superfamily, is an actin-activated ATPase that uses the energy released when ATP is hydrolyzed to do work. Although we typically associate work with muscle contraction, cell motility and cell division, many nuclear processes require energy. Historically, the notion that the nucleus contains actin or myosin has been highly controversial. Because filaments are central to the biological functions of actin, this controversy was fueled by the absence of actin or myosin filaments in the nucleus. Nevertheless, myosins I, II, V, VI, X, XVI and XVIII have been described in the nucleus, and the presence of actin in the nucleus is now indisputable. However, it has been difficult to recognize structures formed by nuclear myosins and actin that are comparable to actin–myosin structures found in the cytoplasm. Because structure (or ‘form’) is normally a guide to function in biology, the absence of nuclear structures that are obviously similar to those in the cytoplasm has raised the possibility that nuclear motors work in unique ways. This Cell Science at a Glance article will review what is known about actin and myosin in the nucleus and how they differ in form and function from their cytoplasmic counterparts.
Nuclear myosins
One of the best examples of form following function is the sarcomere, the basic contractile unit of striated muscles. The sliding of actin and myosin II organized into filaments in sarcomeres, when ATP is hydrolyzed, is the basis of muscle contraction (Sellers, 2004). However, most members of the myosin superfamily, including nuclear myosin I and most of the other myosins discovered in the nucleus, do not form filaments (Sellers, 2004). Although myosin II has been described in the nucleus (Rodgers, 2005; Li and Sarna, 2009), myosin II filaments are yet to be described in the nucleus. Nuclear myosin Va is found in nuclear speckles, which are sites of RNA processing in transcriptionally active cells (Pranchevicius et al., 2008). By contrast, myosin Vb interacts with actin and RNA polymerase I in nucleoli (Lindsay and McCaffrey, 2009). Myosin VI was implicated in transcription by RNA polymerase II on the basis of chromatin immunoprecipitation (ChIP) assays and the inhibition of transcription by antibodies against myosin VI (Vreugde et al., 2006). Myosin X has also been identified in the nucleus by immunofluorescence, but its role is not known (Woolner et al., 2008). Myosin XVI appears to interact with stress-induced nuclear actin rods and might function in the progression of S-phase (Cameron et al., 2007). Myosin XVIIIb has been found in a subset of nuclei of primary cardiomyocytes and skeletal muscle (Salamon et al., 2003). It is a candidate tumor suppressor gene (Ajima et al., 2007), and mutations in this protein have been observed in a variety of cancers (Bleeker et al., 2009). The above studies implicate myosins in a number of nuclear processes. However, none of these myosins is known to form filaments in the nucleus, and it is unclear how they generate force in the absence of filaments.
Nuclear myosin I (NMI) was the first myosin to be discovered in the nucleus (Pestic-Dragovich et al., 2000). It has been studied most extensively and a complicated picture is emerging with regards to its function in the nucleus. On the basis of what we know about actin and myosin in the cytoplasm, it is reasonable to predict that they interact with each other to function as a motor in the nucleus. However, β-actin, the product of one of the six different actin genes found in mammals (Perrin and Ervasti, 2010) and the form that is predominantly found in the nucleus (Hofmann et al., 2004), is necessary for the formation of RNA polymerase II pre-initiation complexes (Hofmann et al., 2004), but NMI is not. By contrast, NMI is required for the formation of the first phosphodiester bond during transcription initiation by RNA polymerase II (Hofmann et al., 2006). These results suggest that actin and NMI can work independently (i.e. without working together or hydrolyzing ATP), perhaps to stabilize protein complexes during the early stages of transcription by RNA polymerase II.
With respect to ribosomal DNA (rDNA) transcription, actin binds to RNA polymerase I, whereas NMI binds to the transcription factor TIF1A (Philimonenko et al., 2004). A mutational approach has revealed that NMI motor activity and polymeric actin are necessary for the efficient transcription of rDNA (Ye et al., 2008). Chuang and colleagues also used a mutational approach to establish that the movement of a chromosome locus is an active process that is powered by actin and NMI (Chuang et al., 2006). This result has been supported by the demonstration that the activation of genes located on different chromosomes by estradiol requires actin and NMI (Hu et al., 2008), as does the repositioning of a gene locus (Dundr et al., 2007) and chromosome repositioning following serum stimulation (Mehta et al., 2010). Thus, actin and NMI appear to have a motor function in the transcription of rDNA and the functional compartmentalization of the nucleus, although the molecular details of their motor activity remain unclear (see Conclusions and future perspectives).
Nuclear actin and transcription
Actin filaments, which are central to biological functions, such as muscle contraction, cell motility, cell division and vesicle transport, are rarely seen in the normal interphase nucleus, and phalloidin, which is commonly used to detect cytoplasmic actin filaments (Cooper, 1987), rarely stains the nucleus. However, this does not exclude the presence of nuclear actin filaments. Sites that are recognized by phalloidin could be buried and inaccessible, and/or nuclear actin filaments could be very short and extremely dynamic, making them difficult to detect using currently available fixation and visualization methods. Another possibility is that the concentration of actin in the nucleus is below its critical concentration for polymerization (0.1–0.6 µM) (Pollard et al., 2000). However, this does not appear to be the case because nuclear actin forms filaments in interphase nuclei under various pathological conditions (see below). Thus, cells either actively limit the formation of actin filaments in the normal interphase nucleus or they form short filaments that turn over very rapidly. Furthermore, the technical difficulties in detecting nuclear actin filaments suggest that any type of filamentous nuclear structure based on actin will be very different from thin filaments found in muscle cells and stress fibers found in the cytoplasm of non-muscle cells. Because form follows function, for example in sarcomeres, not knowing the nature of the nuclear structures formed by actin is a major conceptual barrier to understanding of how actin might work in the nucleus.
Two mechanisms that could regulate the dynamics and filament structure of nuclear actin are post-translational modifications and the import-export of actin (reviewed by de Lanerolle and Serebryannyy, 2011). Although cytoplasmic actin is subject to many post-translational modifications (de Lanerolle and Serebryannyy, 2011), SUMOylation, which increases the nuclear retention of actin, is the only post-translational modification that has been described for nuclear actin (Hofmann et al., 2009). β-actin shuttles between the cytoplasm and the nucleus, with the import and export of actin being mediated by importin 9 (Dopie et al., 2012) and exportin 6 (Stüven et al., 2003), respectively. Dopie and colleagues also reported that nuclear actin levels must be maintained to ensure maximal transcriptional activity, and they suggested that there is a close relationship between nuclear and cytoplasmic actin pools (Dopie et al., 2012). In support of this idea, treating cells with latrunculin B, a drug that disassembles filamentous F-actin in the cytoplasm, results in the formation of nuclear actin filaments that are stained with phalloidin (Pendleton et al., 2003). It has been suggested that the increase in globular G-actin that follows the depolymerization of filamentous F-actin leads to the translocation of actin from the cytoplasm to the nucleus and its organization into nuclear structures that resemble cytoplasmic actin filaments (Pendleton et al., 2003).
The balance between G- and F-actin in the cytoplasm can also affect transcription. The nucleus has to respond to changes in the external environment, which are frequently achieved by turning on and off the transcription of specific genes. One mechanism for sending information to the nucleus is through proteins that contain the SUN and KASH domains. Such proteins contribute to the formation of the so-called linker of nucleoskeleton and cytoskeleton (LINC) complexes that physically connect the nuclear lamina to the actin cytoskeleton, microtubules and intermediate filaments (Jaalouk and Lammerding, 2009; Simon and Wilson, 2011). Another mechanism involves regulating the translocation of transcription factors to the nucleus. For instance, the homeobox transcription factor PREP2 (also known as PKNOX2) (Haller et al., 2004) and the transcriptional repressor YY1 (Favot et al., 2005) bind to actin filaments in the cytoplasm and translocate to the nucleus when actin is depolymerized. By contrast, the transcriptional co-activator MAL binds to monomeric actin and translocates to the nucleus upon polymerization of cytoplasmic actin (Vartiainen et al., 2007). Interestingly, the ErbB2 growth factor receptor, which is over-expressed in many human tumors, enhances the transcription of ribosomal genes by binding to β-actin and RNA polymerase I (Li et al., 2011).
Actin related proteins (Arps) also regulate the structure of nuclear actin. For example, in the cytoplasm, the Arp2/3 complex nucleates actin filaments and creates branched actin filaments that are important for extending the leading edge of motile cells (Magdalena et al., 2003). Numerous Arps have been described in the nucleus (reviewed by Oma and Harata, 2011). Actin and Arps recruit chromatin remodeling complexes (Kukalev et al., 2005; Obrdlik et al., 2008; Qi et al., 2011; Blessing et al., 2004), and chromatin remodeling complexes associate with Arp4 and actin (Blessing et al., 2004). The nucleating activity of Arp2/3 is stimulated by Wiskott-Aldrich syndrome family proteins (WASPs), and both Arp2/3 and neural WASP (N-WASP) are found in the nucleus. The polymerization of nuclear actin by Arp2/3 in a N-WASP-dependent manner has been implicated in the regulation of RNA polymerase II (Yoo et al., 2007; Wu et al., 2006). This suggests that the ability of Arp2/3 to regulate actin dynamics is conserved in the nucleus, and that Arp2/3 regulates transcription in manner that is similar to its effects on actin in the cytoplasm, that is, by regulating its polymerization (Yoo et al., 2007).
Nuclear actin filaments in pathology
Although, as stated above, actin filaments are not normally detected in interphase nuclei, they can be detected in certain pathological conditions. Various types of cell stresses, such as DMSO treatment, heat shock, ATP depletion, treatment with latrunculin B (as described above) and viral infections (reviewed by Hofmann, 2009) result in the formation of nuclear actin filaments. Expression of the V163M mutant variant of the α-actin gene, which is only expressed in muscle (Perrin and Ervasti, 2010), also results in the formation of nuclear actin filaments (Domazetovska et al., 2007). V163M is one of a few naturally occurring mutations in the α-muscle actin gene that results in intranuclear rod myopathy, a human disease characterized by the presence of nuclear actin filaments, a decrease in mitotic index and muscle weakness (Domazetovska et al., 2007; Vandebrouck et al., 2010). Mutant huntingtin protein, the causative element in Huntington's disease, also associates with nuclear actin filaments in response to stress (Munsie et al., 2011). However, it remains to be seen whether the nuclear actin filaments have a function in stress responses, or whether they contribute to the pathology associated with any of these or other diseases.
Interestingly, stressed neurons in culture form rod-like actin structures that disappear when the stress is removed (Bernstein et al., 2006). That study also showed the loss of mitrochondrial membrane potential and ATP following stress is lower in cells with actin rods. As the turnover of actin filaments is a major source of energy consumption in neurons (Bernstein and Bamburg, 2003), the authors suggest that the formation of actin rods in the cytoplasm has a protective effect because it increases the amount of ATP that is available for other cellular functions (Bernstein et al., 2006). By analogy, nuclear actin filaments or rods should have a similar protective effect. However, these filaments would have to be readily disassembled when the stress is removed. This does not always appear to be the case. Nuclear actin filaments in Huntington's disease apparently result from cross-links, formed by a transglutaminase, between actin and cofilin (Munsie et al., 2011). How the formation of relatively stable actin filaments, such as those found in Huntington's disease, provides a developmental or survival advantage to a cell is currently unknown.
Actin filaments are also present following viral infections (Taylor et al., 2011; Goley et al., 2006). Viral particles are too large (>125 nm in diameter) to diffuse, and they are usually transported through the cytoplasm on microtubules or actin filaments (Roberts and Baines, 2011). Using cytoplasmic transport as a model, it has been suggested the movement of viral particles in the nucleus is an active process. In support of this hypothesis, viral replication compartments form by moving and merging with each other in a process that requires nuclear actin and NMI (Chang et al., 2011). Baculovirus (Goley et al., 2006), pseudorabies virus and herpes virus infections also result in the formation of actin filaments in the nucleus (Taylor et al., 2011; Roberts and Baines, 2011), and the physical association of nuclear actin filaments with herpes viral capsids appears to be necessary for the correct formation of viral assembly sites and viral transcription (Feierbach et al., 2006).
Conclusions and future perspectives
Although the presence and functional significance of actin and multiple myosins in the nucleus are well established, studies on nuclear actin and myosin paint a complex picture of how they interact, and suggest that they behave in fundamentally different ways in the cytoplasm and nucleus. This body of work also raises many questions with regards to how actin and myosin function in the nucleus at the molecular level.
One of the most enigmatic issues is the absence of filaments in the nucleus. This raises the question of whether actin and any myosin function as a motor complex in the nucleus. Myosins are defined as actin-activated ATPases. Myosin heads have ATP- and actin-binding sites, and ATP is hydrolyzed while myosin heads are detached from actin. When actin and myosin rebind, myosin uses the energy that is released from ATP hydrolysis to perform a power stroke that moves actin relative to myosin. Replacing ADP with ATP detaches the myosin from the actin so that it can repeat the cycle. Myosin II, like myosin I, is a non-processive motor, meaning that one head can detach before the other head binds. Myosin II filaments stay bound to actin filaments because the numerous myosin II heads bind and unbind actin non-synchronously. By contrast, myosin V is a processive motor, in which one head is always bound to actin. Motor proteins also have to be anchored to generate tension or perform work. In muscle contraction, ATP hydrolyzed by myosin II is used to move actin filaments that are anchored to the ends of the sarcomere past the myosin II filaments. The question, then, is whether nuclear actin and myosins are bound by the same kinetic and structural parameters as their cytoplasmic counterparts.
The best evidence for actin and NMI functioning as a motor is in rDNA transcription and chromosome movements (see above). Even though the latter is supported by multiple studies (Chuang et al., 2006; Hu et al., 2008; Dundr et al., 2007; Mehta et al., 2010), it is difficult to visualize how actin and NMI can function as a motor at the molecular level. These studies suggest that chromosomes use NMI to move on a type of actin network in the nucleus that has not been visualized to date. However, NMI, like other class I myosins, is a single headed, non-processive motor (Sellers, 2004). It has to be anchored and a second myosin I molecule has to bind to actin before the first myosin I detaches to function as a motor. Otherwise, it would simply float off the actin filament. One solution to this dilemma is that NMI belongs to the myosin IC sub-family, which has a positively charged region in the tail domain. It has been proposed that myosin IC moves cargoes by simultaneously binding negatively charged lipids with its tail, and actin filaments through the actin-binding site on its head (Mermall et al., 1998). We have previously shown that the NMI tail binds to DNA and proposed a model in which this binding powers the movement of transcriptional complexes by interacting with actin that is bound to the polymerase (de Lanerolle et al., 2005). A crucial element in this model is that the binding of NMI to DNA has to be strong enough so that NMI remains attached to DNA when NMI hydrolyzes ATP and goes through a power stroke. This model also demands a very precise spatial arrangement of actin and NMI relative to each other, the polymerase and the binding of NMI to DNA. A small misalignment in the binding of the NMI tail to DNA could make the NMI head unable to bind to actin.
Another possibility is that myosin Va functions as a motor to transport viral particles in the nucleus. Myosin Va interacts with viral capsid proteins, and a number of viruses promote the formation of nuclear actin filaments (Roberts and Baines, 2011). Nuclear actin filaments and myosin V appear to have important functions in viral replication, and it has been proposed that viruses are moved by myosin V along nuclear actin filaments that are induced by the viruses (Roberts and Baines, 2011). This idea seems feasible because myosin Va is a processive motor so that one head always remains attached to the actin (Mermall et al., 1998), which allows it to walk along actin filaments. Myosin V has a step size of 37 nm (Yildiz et al., 2003), and actin filaments form a double-stranded helix with each node of the helix also repeating every 37 nm (Holmes et al., 1990). Consequently, with each ATP molecule that is hydrolyzed, myosin V can step from one actin to another at the corresponding location in the next turn of the helix (Yildiz et al., 2003). However, actin filaments must have this precise registry, and their myosin-binding sites have to be accessible in order to support the walking of myosin V molecules. It remains to be determined if nuclear actin filaments induced by viruses have this periodicity.
Thus, many questions remain, although we have studied actin and myosin for almost 100 years and know a great deal about these proteins. These studies on cytoplasmic actin and myosins serve as important models for actin and its functions in the nucleus. However, nuclear motors present unusual conceptual and experimental challenges, and they might not behave in ways that we have come to expect on the basis of their cytoplasmic counterparts. Overcoming these challenges will undoubtedly result in exciting new insights into this old family of proteins.
Supplementary Material
Footnotes
Funding
This work was supported by a grant from the NIH [grant number R01GM080587 to P.deL.]. Deposited in PMC for release after 12 months.
A high-resolution version of the poster is available for downloading in the online version of this article at jcs.biologists.org. Individual poster panels are available as JPEG files at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.099754/-/DC1.
References
- Ajima R., Kajiya K., Inoue T., Tani M., Shiraishi–Yamaguchi Y., Maeda M., Segawa T., Furuichi T., Sutoh K., Yokota J. (2007). HOMER2 binds MYO18B and enhances its activity to suppress anchorage independent growth. Biochem. Biophys. Res. Commun. 356, 851–856 10.1016/j.bbrc.2007.03.060 [DOI] [PubMed] [Google Scholar]
- Bernstein B. W., Bamburg J. R. (2003). Actin-ATP hydrolysis is a major energy drain for neurons. J. Neurosci. 23, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. W., Chen H., Boyle J. A., Bamburg J. R. (2006). Formation of actin-ADF/cofilin rods transiently retards decline of mitochondrial potential and ATP in stressed neurons. Am. J. Physiol. Cell Physiol. 291, C828–C839 10.1152/ajpcell.00066.2006 [DOI] [PubMed] [Google Scholar]
- Bleeker F. E., Lamba S., Rodolfo M., Scarpa A., Leenstra S., Vandertop W. P., Bardelli A. (2009). Mutational profiling of cancer candidate genes in glioblastoma, melanoma and pancreatic carcinoma reveals a snapshot of their genomic landscapes. Hum. Mutat. 30, E451–E459 10.1002/humu.20927 [DOI] [PubMed] [Google Scholar]
- Blessing C. A., Ugrinova G. T., Goodson H. V. (2004). Actin and ARPs: action in the nucleus. Trends Cell Biol. 14, 435–442 10.1016/j.tcb.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Cameron R. S., Liu C., Mixon A. S., Pihkala J. P., Rahn R. J., Cameron P. L.2007). Myosin16b: The COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression. Cell Motil. Cytoskeleton 6419–48 10.1002/cm.20162 [DOI] [PubMed] [Google Scholar]
- Chang L., Godinez W. J., Kim I. H., Tektonidis M., de Lanerolle P., Eils R., Rohr K., Knipe D. M. (2011). Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl. Acad. Sci. USA 108, E136–E144 10.1073/pnas.1103411108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C. H., Carpenter A. E., Fuchsova B., Johnson T., de Lanerolle P., Belmont A. S. (2006). Long-range directional movement of an interphase chromosome site. Curr. Biol. 16, 825–831 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- Cooper J. A. (1987). Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105, 1473–1478 10.1083/jcb.105.4.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lanerolle P., Serebryannyy L. (2011). Nuclear actin and myosins: life without filaments. Nat. Cell Biol. 13, 1282–1288 10.1038/ncb2364 [DOI] [PubMed] [Google Scholar]
- de Lanerolle P., Johnson T., Hofmann W. A. (2005). Actin and myosin I in the nucleus: what next? Nat. Struct. Mol. Biol. 12, 742–746 10.1038/nsmb983 [DOI] [PubMed] [Google Scholar]
- Domazetovska A., Ilkovski B., Cooper S. T., Ghoddusi M., Hardeman E. C., Minamide L. S., Gunning P. W., Bamburg J. R., North K. N. (2007). Mechanisms underlying intranuclear rod formation. Brain 130, 3275–3284 10.1093/brain/awm247 [DOI] [PubMed] [Google Scholar]
- Dopie J., Skarp K. P., Rajakylä E. K., Tanhuanpää K., Vartiainen M. K. (2012). Active maintenance of nuclear actin by importin 9 supports transcription. Proc. Natl. Acad. Sci. USA 109, E544–E552 10.1073/pnas.1118880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Ospina J. K., Sung M. H., John S., Upender M., Ried T., Hager G. L., Matera A. G. (2007). Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 179, 1095–1103 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favot L., Hall S. M., Haworth S. G., Kemp P. R. (2005). Cytoplasmic YY1 is associated with increased smooth muscle-specific gene expression: implications for neonatal pulmonary hypertension. Am. J. Pathol. 167, 1497–1509 10.1016/S0002-9440(10)61236-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierbach B., Piccinotti S., Bisher M., Denk W., Enquist L. W. (2006). Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2, e85 10.1371/journal.ppat.0020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley E. D., Ohkawa T., Mancuso J., Woodruff J. B., D'Alessio J. A., Cande W. Z., Volkman L. E., Welch M. D. (2006). Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314, 464–467 10.1126/science.1133348 [DOI] [PubMed] [Google Scholar]
- Haller K., Rambaldi I., Daniels E., Featherstone M. (2004). Subcellular localization of multiple PREP2 isoforms is regulated by actin, tubulin, and nuclear export. J. Biol. Chem. 279, 49384–49394 10.1074/jbc.M406046200 [DOI] [PubMed] [Google Scholar]
- Hofmann W. A. (2009). Cell and molecular biology of nuclear actin. Int. Rev. Cell Mol. Biol. 273, 219–263 10.1016/S1937-6448(08)01806-6 [DOI] [PubMed] [Google Scholar]
- Hofmann W. A., Stojiljkovic L., Fuchsova B., Vargas G. M., Mavrommatis E., Philimonenko V., Kysela K., Goodrich J. A., Lessard J. L., Hope T. J.et al. (2004). Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 6, 1094–1101 10.1038/ncb1182 [DOI] [PubMed] [Google Scholar]
- Hofmann W. A., Vargas G. M., Ramchandran R., Stojiljkovic L., Goodrich J. A., de Lanerolle P. (2006). Nuclear myosin I is necessary for the formation of the first phosphodiester bond during transcription initiation by RNA polymerase II. J. Cell. Biochem. 99, 1001–1009 10.1002/jcb.21035 [DOI] [PubMed] [Google Scholar]
- Hofmann W. A., Arduini A., Nicol S. M., Camacho C. J., Lessard J. L., Fuller–Pace F. V., de Lanerolle P.2009). SUMOylation of nuclear actin. J. Cell Biol. 186193–200 10.1083/jcb.200905016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K. C., Popp D., Gebhard W., Kabsch W. (1990). Atomic model of the actin filament. Nature 347, 44–49 10.1038/347044a0 [DOI] [PubMed] [Google Scholar]
- Hu Q., Kwon Y. S., Nunez E., Cardamone M. D., Hutt K. R., Ohgi K. A., Garcia–Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G.et al. (2008). Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc. Natl. Acad. Sci. USA 105, 19199–19204 10.1073/pnas.0810634105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaalouk D. E., Lammerding J. (2009). Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 10, 63–73 10.1038/nrm2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukalev A., Nord Y., Palmberg C., Bergman T., Percipalle P. (2005). Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 12, 238–244 10.1038/nsmb904 [DOI] [PubMed] [Google Scholar]
- Li Q., Sarna S. K. (2009). Nuclear myosin II regulates the assembly of preinitiation complex for ICAM-1 gene transcription. Gastroenterology 137, 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Y., Chen H., Hsieh Y. H., Wang Y. N., Chu H. J., Chen Y. H., Chen H. Y., Chien P. J., Ma H. T., Tsai H. C.et al. (2011). Nuclear ErbB2 enhances translation and cell growth by activating transcription of ribosomal RNA genes. Cancer Res. 71, 4269–4279 10.1158/0008-5472.CAN-10-3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay A. J., McCaffrey M. W. (2009). Myosin Vb localises to nucleoli and associates with the RNA polymerase I transcription complex. Cell Motil. Cytoskeleton 66, 1057–1072 10.1002/cm.20408 [DOI] [PubMed] [Google Scholar]
- Magdalena J., Millard T. H., Etienne–Manneville S., Launay S., Warwick H. K., Machesky L. M. (2003). Involvement of the Arp2/3 complex and Scar2 in Golgi polarity in scratch wound models. Mol. Biol. Cell 14, 670–684 10.1091/mbc.E02-06-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta I. S., Amira M., Harvey A. J., Bridger J. M. (2010). Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 11, R5 10.1186/gb-2010-11-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermall V., Post P. L., Mooseker M. S. (1998). Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279, 527–533 10.1126/science.279.5350.527 [DOI] [PubMed] [Google Scholar]
- Munsie L., Caron N., Atwal R. S., Marsden I. T., Wild E. J., Bamburg J. R., Tabrizi S. J., Truant R. (2011). Mutant huntingtin causes defective actin remodeling during stress: defining a new role for transglutaminase 2 in neurodegenerative disease. Hum. Mol. Genet. 20, 1937–1951 10.1093/hmg/ddr075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik A., Kukalev A., Louvet E., Farrants A. K., Caputo L., Percipalle P. (2008). The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol. Cell. Biol. 28, 6342–6357 10.1128/MCB.00766-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oma Y., Harata M.2011). Actin-related proteins localized in the nucleus: from discovery to novel roles in nuclear organization. Nucleus 238–46 10.4161/nucl.2.1.14510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton A., Pope B., Weeds A., Koffer A. (2003). Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J. Biol. Chem. 278, 14394–14400 10.1074/jbc.M206393200 [DOI] [PubMed] [Google Scholar]
- Perrin B. J., Ervasti J. M. (2010). The actin gene family: function follows isoform. Cytoskeleton 67, 630–634 10.1002/cm.20475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestic–Dragovich L., Stojiljkovic L., Philimonenko A. A., Nowak G., Ke Y., Settlage R. E., Shabanowitz J., Hunt D. F., Hozak P., de Lanerolle P. (2000). A myosin I isoform in the nucleus. Science 290, 337–341 10.1126/science.290.5490.337 [DOI] [PubMed] [Google Scholar]
- Philimonenko V. V., Zhao J., Iben S., Dingová H., Kyselá K., Kahle M., Zentgraf H., Hofmann W. A., de Lanerolle P., Hozák P.et al. (2004). Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 6, 1165–1172 10.1038/ncb1190 [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Blanchoin L., Mullins R. D. (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 10.1146/annurev.biophys.29.1.545 [DOI] [PubMed] [Google Scholar]
- Pranchevicius M. C., Baqui M. M., Ishikawa–Ankerhold H. C., Lourenço E. V., Leão R. M., Banzi S. R., dos Santos C. T., Roque–Barreira M. C., Espreafico E. M., Larson R. E. (2008). Myosin Va phosphorylated on Ser1650 is found in nuclear speckles and redistributes to nucleoli upon inhibition of transcription. Cell Motil. Cytoskeleton 65, 441–456 10.1002/cm.20269 [DOI] [PubMed] [Google Scholar]
- Qi T., Tang W., Wang L., Zhai L., Guo L., Zeng X. (2011). G-actin participates in RNA polymerase II-dependent transcription elongation by recruiting positive transcription elongation factor b (P-TEFb). J. Biol. Chem. 286, 15171–15181 10.1074/jbc.M110.184374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K. L., Baines J. D. (2011). Actin in herpesvirus infection. Viruses 3, 336–346 10.3390/v3040336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers B. D. (2005). Insulin-like growth factor-I downregulates embryonic myosin heavy chain (eMyHC) in myoblast nuclei. Growth Horm. IGF Res. 15, 377–383 10.1016/j.ghir.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Salamon M., Millino C., Raffaello A., Mongillo M., Sandri C., Bean C., Negrisolo E., Pallavicini A., Valle G., Zaccolo M.et al. (2003). Human MYO18B, a novel unconventional myosin heavy chain expressed in striated muscles moves into the myonuclei upon differentiation. J. Mol. Biol. 326, 137–149 10.1016/S0022-2836(02)01335-9 [DOI] [PubMed] [Google Scholar]
- Sellers J. R. (2004). Fifty years of contractility research post sliding filament hypothesis. J. Muscle Res. Cell Motil. 25, 475–482 10.1007/s10974-004-4239-6 [DOI] [PubMed] [Google Scholar]
- Simon D. N., Wilson K. L. (2011). The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat. Rev. Mol. Cell Biol. 12, 695–708 10.1038/nrm3207 [DOI] [PubMed] [Google Scholar]
- Stüven T., Hartmann E., Görlich D. (2003). Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 22, 5928–5940 10.1093/emboj/cdg565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. P., Koyuncu O. O., Enquist L. W. (2011). Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 9, 427–439 10.1038/nrmicro2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandebrouck A., Domazetovska A., Mokbel N., Cooper S. T., Ilkovski B., North K. N. (2010). In vitro analysis of rod composition and actin dynamics in inherited myopathies. J. Neuropathol. Exp. Neurol. 69, 429–441 10.1097/NEN.0b013e3181d892c6 [DOI] [PubMed] [Google Scholar]
- Vartiainen M. K., Guettler S., Larijani B., Treisman R. (2007). Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316, 1749–1752 10.1126/science.1141084 [DOI] [PubMed] [Google Scholar]
- Vreugde S., Ferrai C., Miluzio A., Hauben E., Marchisio P. C., Crippa M. P., Bussi M., Biffo S. (2006). Nuclear myosin VI enhances RNA polymerase II-dependent transcription. Mol. Cell 23, 749–755 10.1016/j.molcel.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Woolner S., O'Brien L. L., Wiese C., Bement W. M. (2008). Myosin-10 and actin filaments are essential for mitotic spindle function. J. Cell Biol. 182, 77–88 10.1083/jcb.200804062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yoo Y., Okuhama N. N., Tucker P. W., Liu G., Guan J. L. (2006). Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat. Cell Biol. 8, 756–763 10.1038/ncb1433 [DOI] [PubMed] [Google Scholar]
- Ye J., Zhao J., Hoffmann–Rohrer U., Grummt I. (2008). Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 22, 322–330 10.1101/gad.455908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. (2003). Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 10.1126/science.1084398 [DOI] [PubMed] [Google Scholar]
- Yoo Y., Wu X., Guan J. L. (2007). A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J. Biol. Chem. 282, 7616–7623 10.1074/jbc.M607596200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.