Summary
Stromal derived growth factor (SDF-1) and gamma-aminobutyric acid (GABA) are two extracellular cues that regulate the rate of neuronal migration during development and may act synergistically. The molecular mechanisms of this interaction are still unclear. Gonadotropin releasing hormone-1 (GnRH) neurons are essential for vertebrate reproduction. During development, these neurons emerge from the nasal placode and migrate through the cribriform plate into the brain. Both SDF-1 and GABA have been shown to regulate the rate of GnRH neuronal migration by accelerating and slowing migration, respectively. As such, this system was used to explore the mechanism by which these molecules act to produce coordinated cell movement during development. In the present study, GABA and SDF-1 are shown to exert opposite effects on the speed of cell movement by activating depolarizing or hyperpolarizing signaling pathways, GABA via changes in chloride and SDF-1 via changes in potassium. GABA and SDF-1 were also found to act synergistically to promote linear rather than random movement. The simultaneous activation of these signaling pathways, therefore, results in tight control of cellular speed and improved directionality along the migratory pathway of GnRH neurons.
Key words: GABA, GIRK, GnRH, Motility, Neuronal migration, SDF
Introduction
Migration of postmitotic neurons to their correct destinations is a cellular event essential for development of appropriate circuitry and function of the nervous system (Rakic, 1990; Cameron et al., 1997; Hatten, 2002). Different modes of neuronal migration have been described, including gliophilic, axophilic and tangential, based on the substrate used for movement and/or orientation to the pial surface. (Marín and Rubenstein, 2003). However, the precise mechanisms by which neurons navigate to their final destination are still unclear. GABA has been implicated both as an acceleratory or a stop signal for gliophilic and homophilic migratory cells (Bolteus and Bordey, 2004; Represa and Ben-Ari, 2005; Cancedda et al., 2007) and appears crucial to positioning of interneurons migrating into the cortex from the ganglionic eminence (Poluch et al., 2008). SDF-1, a chemokine, is involved in primordial germ cell migration (Blaser et al., 2005), migration of neural progenitors (via its receptor CXCR4) and axon growth and branching during development of the nervous system (Tran and Miller, 2003; Pujol et al., 2005). Recent work has shown that GABA and SDF-1 may act synergistically to alter cell movement (Seidel et al., 2007; Bhattacharyya et al., 2008). The molecular mechanisms of this interaction are still unclear, but the possible consequences of this event may have opposite effects on cellular behavior (Seidel et al., 2007; Bhattacharyya et al., 2008).
During axophilic migration, neurons use axons of other cells to reach their final position. Development of neuroendocrine GnRH cells represents the best characterized example of axophilic migration in vertebrates (Wray, 2002; Marín and Rubenstein, 2003). GnRH neurons are essential for reproduction, being components of the hypothalamo-pituitary-gonadal axis (Wray, 2002). To become part of this axis, GnRH cells migrate from the nasal placode into the forebrain in association with olfactory/vomeronasal nerves (Wray, 2002; Schwarting et al., 2004). An important relationship exists between olfactory axons and GnRH neurons. In mice, if olfactory axons do not cross the cribriform plate or do not reach proper brain targets, GnRH neurons remain in the nose or are found in aberrant locations respectively (Murakami and Arai, 1994; Deiner and Sretavan, 1999; Rugarli, 1999; Gao et al., 2000; MacColl et al., 2002). In patients with Kallmann syndrome, malformation of the olfactory system and failure of GnRH neurons to migrate into the brain results in anosmia and delayed or absent pubertal maturation (hypogonadotropic hypogonadism) (Iovane et al., 2004). Among the numerous molecules involved in GnRH neuronal migration, GABA (Fueshko et al., 1998b; Wray et al, 1996; Bless et al., 2000), and SDF-1 (Schwarting et al., 2006; Toba et al., 2008) are potent regulators within the nasal compartment.
The present study characterized the migratory behavior of GnRH neurons and dynamic roles of GABA and SDF-1 in orchestrating their movement, evaluating possible interactions. This study demonstrates a) GnRH neuronal movement is saltatory, b) GABAergic and SDF/CXCR4 systems interact, playing a role in linear movement and c) SDF-1/CXCR4 act via G-protein-activated inward rectifier potassium (GIRK) channels to control GnRH neuronal movement. Further examination of GnRH neuronal migration may provide insight into molecular mechanisms common to all neuronal movement as well as elucidate molecules specific for axophilic processes.
Results
Movement of GnRH neurons
In mouse, GnRH neurons migrate from the nasal region into the forebrain during a 5–6 day time span (Wray et al., 1989; Schwanzel-Fukuda and Pfaff, 1989). In nasal explants generated at E11.5, GnRH neurons also move for 5–6 days (Fueshko and Wray, 1994) and retain other characteristics of GnRH neurons in vivo, including distance and direction of travel and association with olfactory axons (Fueshko and Wray, 1994). By 3 days in vitro (div), GnRH cells move to the explant periphery, allowing visualization of individual neurons with differential interference contrast (DIC) video microscopy (Fig. 1). GnRH cells can be identified in situ by their shape [unipolar body and long leading process (Kusano et al., 1995)] and association with olfactory axons (Kramer and Wray, 2000). In the present study, post hoc verification of the cell phenotype (GnRH, Fig. 1C, brown) and substrate pathway [peripherin, marker of sensory axons, Fig. 1C, blue; (Fueshko and Wray, 1994)] was performed after live imaging.
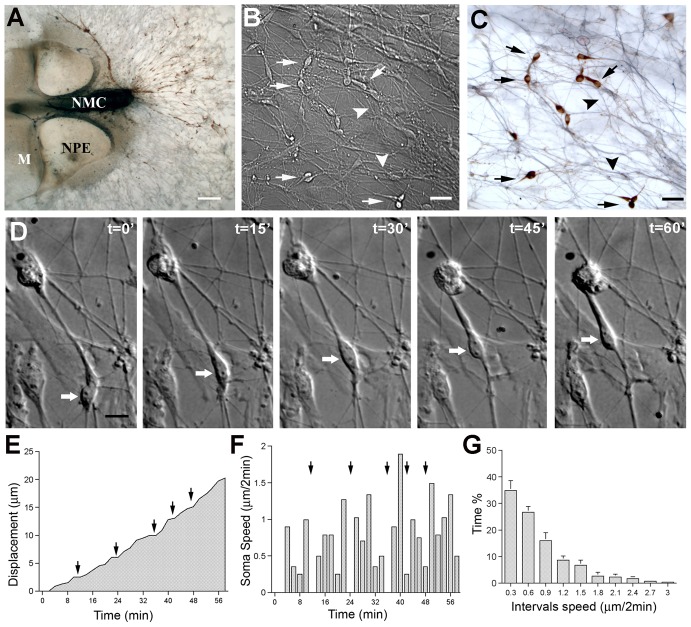
Fig. 1.
In situ GnRH neuron detection. (A) Explant maintained for 4 div, fixed and immunocytochemically stained for GnRH (brown) and peripherin (blue), a marker of olfactory axons. Both GnRH neurons and olfactory axons show directed movement, exiting from the same region of the explant. NMC, nasal midline cartilage; NPE, nasal pit epithelium; M, mesenchyme. (B) DIC image of recording field showing cells exhibiting characteristic uni-bipolar/bipolar shape (arrows) with a long leading process. Cells are located among numerous fiber-like structures, not immediately connected to cell somas (arrowheads). (C) After imaging, explants were stained for GnRH (brown, arrows) and peripherin (blue, arrowheads) to confirm the identity of recorded cells. (D) DIC images 15 min apart showing movement of a GnRH cell in SFM (arrow). (E) GnRH neurons exhibit saltatory movement. Graph showing the speed of cell soma plotted as a function of recording time. The profile of the resulting line shows discontinuous cell movement, with periods of forward movement (upward slope segments) separated by pauses or slower forward movement (horizontal segments, black arrows). (F) Graph showing the speed, every 2 minutes (corresponding to interval pictures), of the same cell shown in E. The height of the bar is proportional to the forward movement, with no bar indicating a pause (black arrows). (G) Frequency distribution showing the percent of time that the cells (n = 12; N = 3) display a particular speed during a 2-hour recording time. GnRH neurons spend ∼35% of their time paused or moving very slowly. Scale bars: 100 µm (A); 20 µm (B,C); 10 µm (D).
The rate of movement of GnRH neurons was examined from 3 to 7 div (Table 1), with recording periods ranging from 30 min to 2 hours. Most neurons ceased movement by 7div (Fueshko and Wray, 1994). The total distance is the absolute movement a cell exhibits independent of direction. The total distance speed (TDS) was calculated by dividing the value by the recording time. From 3–6 div, the TDS ranged from 12–16 µm/h, with the fastest TDS occurring at 4 div (Table 1). This rate of movement is similar to GnRH neurons in embryonic slices (12.7 µm/h; Bless et al., 2005) and the rate of cortical neuron radial glia-mediated migration during development (Edmondson and Hatten, 1987; Komuro and Rakic, 1995; Nadarajah et al., 2001).
Table 1.
Movement of GnRH neurons is altered by GABAergic signals
| Paradigm | div: | Total distance speed and linear distance speed (µm/h) | ||||
| 3 | 4 | 5 | 6 | 7 | ||
| SFM | TDS | 12.71±0.6 (n = 28, N = 5) | 16.14±0.7a (n = 42, N = 6) | 12.49±0.8 (n = 46, N = 4) | 13.03±0.77 (n = 33, N = 4) | 4.32±0.37 (n = 55, N = 3) |
| SFM | LDS | 8.66±0.62 | 10.93±0.72 | 7.18±0.7 | 7.27±0.69 | |
| Pic | TDS | 16.22±0.6 (n = 26, N = 5) | 22.95±1.49a (n = 19, N = 3) | 16.62±0.95 (n = 36, N = 3) | 15.67±0.55 (n = 66, N = 3) | |
| Pic | LDS | 10.39±0.85 | 10.46±1.58 | 8.98±1.18 | 6.25±0.49 | |
| Mus | TDS | 10.1±0.39 (n = 31, N = 6) | 11.5±0.53 (n = 45, N = 5) | 10.14±0.51 (n = 26, N = 3) | 11.09±0.71 (n = 35, N = 3) | |
| Mus | LDS | 5.66±0.39 | 6.73±0.53 | 5.49±0.62 | 6.15±0.76 | |
| GAD67+/+ | TDS | 14.03±0.88 (n = 52, N = 3) | 11.38±0.9 (n = 22, N = 2) | |||
| GAD67−/− | TDS | 23.72±1.12a (n = 53, N = 5) | 16.84±1.15 (n = 57, N = 3) | |||
| VIAAT−/− | TDS | 22.51±1.24 (n = 53, N = 3) | ||||
Significantly faster compared to speed of cells on other days (one-way ANOVA, P<0.05). n, number of cells recorded; N, number of explants used.
The linear distance traveled by the cells was calculated as the distance between the first picture and the last picture taken during the experiment. Linear distance speed (LDS) was calculated by dividing the value by the recording time. The average LDS from 3 to 6 div was 8.51 µm/h (Table 1). As with TDS, the fastest LDS occurred at 4 div. To quantify the migration efficiency, the ratio between LDS and TDS was calculated. The average LDS/TDS ratio (over all days studied) was 0.63, peaking at 0.68 on both 3 and 4 div. In embryonic slices, with GnRH neuronal migration along stretches of ‘straight’ olfactory axons, a migration efficiency of 0.52 was reported (Bless et al., 2005), decreasing if axonal pathways were severed in slicing. Thus, under basal conditions in nasal explants, robust directional movement of GnRH neurons occurs similar to that found in vivo.
Analysis of cell soma revealed discontinuous movement. At 4 div, the speed of a cell soma plotted as a function of time showed periods of rapid forward movements alternating with periods of slower advancement or complete stop (Fig. 1E). Plotting the speed every 2 minutes showed a similar pattern of cell soma movement (Fig. 1F). Analysis of the frequency distribution of instantaneous speed of soma locomotion (Fig. 1G), indicated that cells spent ∼20 minutes every hour paused or in slow movement (0–0.3 µm/2 min). Together, these data indicate that although GnRH neuronal migration is axophilic, they exhibit saltatory movement similar to that reported for glial-guided cells (Edmondson and Hatten, 1987; O’Rourke et al., 1992; Nadarajah et al., 2001).
GABA increases random movement of neurons
GABAergic neurons inhibit GnRH neuronal movement via GABA receptors (Tobet et al., 1996; Fueshko et al., 1998a). To further understand the mechanisms of this action, 4 div explants were treated during the recording period with GABAA receptor agonist muscimol (Mus, 10−4 M), the GABAA receptor antagonist picrotoxin (Pic, 10−4 M), GABAB receptor agonist baclofen (Bac, 10−4 M), or GABAB receptor antagonist saclofen (Sac, 10−4 M). Application of either ethanol or DMSO vehicle controls did not alter GnRH TDS (data not shown). GABAA receptor stimulation with Mus reduced movement speed of neurons by 22% compared to cells recorded in serum-free medium (SFM) (Table 2). In contrast, blocking GABAA receptors with Pic increased movement speed by 14% (Table 2). There was no significant change in the rate of movement after incubation with Bac or Sac (Table 2). These data indicate that GABAergic signals affecting GnRH neuronal movement are transduced through GABAA receptors.
Table 2.
GABA via GABAA receptors modulate GnRH-1 neuronal movement
| Paradigm | Total distance speed (µm/h) during recording period | n | N | |
| I | II | |||
| SFM, Mus | 18.8±0.7 | 14.6±0.94a | 99 | 5 |
| SFM, Pic | 17.8±1.16 | 20.3±1.23a | 58 | 3 |
| SFM, Bac | 20.37±0.9 | 21.8±0.85 | 87 | 3 |
| SFM, Sac | 16.6±0.9 | 15.24±0.84 | 70 | 3 |
Significantly different from speed during first period (paired t-test, P<0.05). n, number of cells recorded; N, number of explants used.
To determine if GABAergic effects were present throughout the movement period, and what aspect of cell movement was altered, GABAA receptor perturbations were performed as a function of development in vitro. Pic increased the TDS at all ages examined, with an overall increase of 32% (Table 1). In contrast to TDS, Pic did not increase the mean LDS, with the average LDS in the presence of Pic similar to that for cells in SFM. Thus, in the presence of PIC, the LDS/TDS ratio was reduced to 0.5 indicating that neurons exhibited more chemokinesis, increased random and less efficient forward movement when GABAA receptors are blocked. In general, Mus treatment reduced TDS by 21% and also produced a general reduction in the LDS (Table 1). In contrast to the increased chemokinesis seen with Pic, Mus decreased both total and linear movement for neurons with the resulting LDS/TDS similar to controls (0.57 versus 0.63).
Changes in movement speed might be caused by altered speed during movement periods or changes in the amount of pause time. To distinguish between these mechanisms, the speed of the cell soma recorded during Pic treatment was plotted as a function of time (Fig. 2C) and the frequency distribution of the percent of time the cells displayed a particular speed was analyzed (Fig. 2D). The profile of the resulting line for the cell soma showed a decrease in cell pause periods and an increase in movement (Fig. 2C) as compared to cells recorded in SFM (Fig. 1E). Inhibition of endogenous GABAA receptors thus attenuated pause time. Neurons spent ∼35% of their time paused or moving slowly in SFM versus <20% of their time when treated with Pic (Fig. 2D). In contrast, when treated with Mus, there was an increase in cell pause periods (Fig. 2E,F) and a decrease in forward movement as compared to cells recorded in SFM or with Pic. Neurons treated with Mus spent 45% of their time paused or moving slowly. Therefore, stimulating GABAA receptors led to more pauses in GnRH neuron movement, while blocking them led to increased movement by less pause time.
Fig. 2.
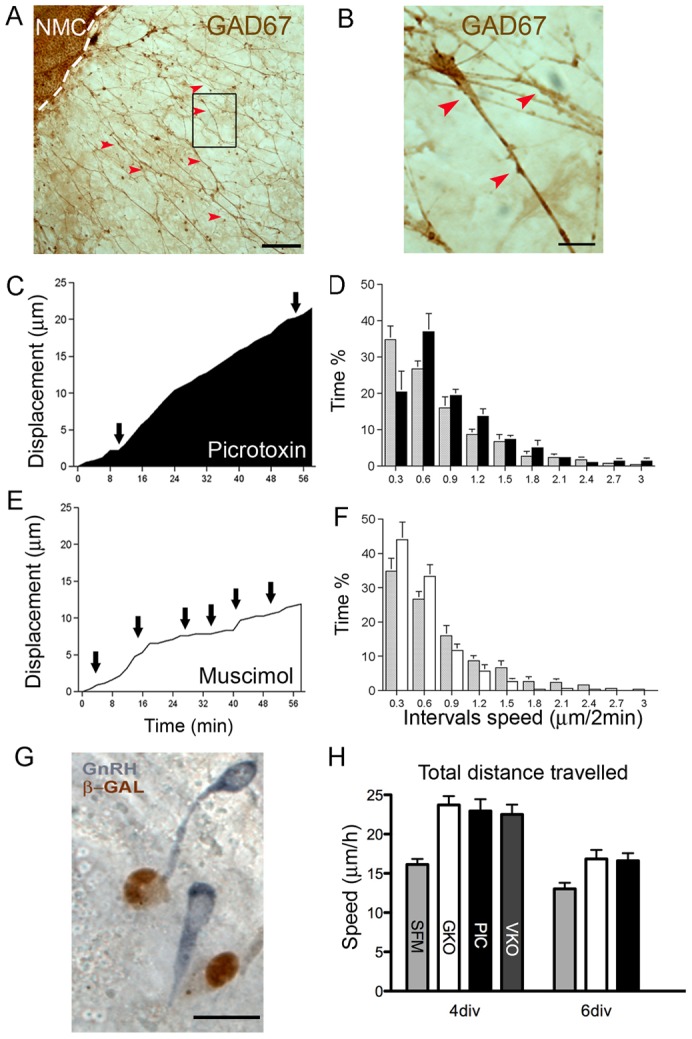
GABA affects neuronal forward movement rate. (A,B) 4 div explant stained with an antibody against GAD67 (brown). Dashed line indicates the border between the nasal midline cartilage (NMC) and periphery of the explant. Red arrows highlight GAD67 positive fibers and cell soma. (B) Magnification of box in A. (C–F) GABAA receptor perturbation changes saltatory movement of neurons. (C,D) Exogenous picrotoxin (Pic, GABAA receptor antagonist) prolongs cell movement. (C) Cell speed plotted as a function of recording time in the presence of Pic. Line shows a decrease in cell pause periods (black arrows, horizontal segments) and an increase in movement (upward slope segments) as compared to cells recorded in serum free medium (SFM, Fig. 1). (D) Frequency distributions showing the percentage of time that the cells (n = 12; N = 3) display a particular speed during a 1 hr recording time in SFM and Pic. Shaded bars, SFM; black bars, Pic. Neurons spent ∼35% of their time paused or moving slowly when recorded in SFM and less than 20% of their time paused or moving slowly when treated with Pic (χ2 test, P<0.05). (E,F) Exogenous muscimol (Mus, GABAA receptor agonist) prolongs pauses in cell movement. (E) Cell speed plotted as a function of time in the presence of Mus shows an increase in cell pause periods (black arrows, horizontal segments) and a decrease in forward movement (upward slope segments) as compared to cells recorded in SFM (Fig. 1E-G). (F) Frequency distribution showing the percentage of time that the cells (n = 12; N = 3) display a particular speed during a 1 hr recording time in SFM vs Mus. Shaded bars, SFM; white bars, Mus. Neurons spent 45% of their time paused or moving slowly when treated with Mus (χ2 test, P<0.05). (G,H). Decrease of GABAergic signaling in GAD67KO mice increases movement rate of neurons. (G) Double immunostaining of a GAD67KO-LacZKIN mouse nasal explant [GnRH (blue) and LacZ (brown)]. (H) The speed of neurons in explants from GAD67−/− (GKO) and VIAAT−/− (VKO) mice is significantly higher (P<0.01, one-way ANOVA with Tukey's post-test) than that of cells in control conditions (WT mouse, SFM), but similar to the speed of neurons in explants (WT) treated with Picrotoxin (Pic). Scale bar: 100 µm (A); 10 µm (B,G).
To directly observe GnRH neuronal movement during a dramatic reduction of endogenous GABA, cells were monitored in explants generated from two different lines of mice with altered GABA synthesis – GAD67 knockout (KO) and VIAAT KO (Oh et al., 2010). GAD67KO animals show a 90% reduction of GABA in the developing brain (Asada et al., 1997) and differences in the distribution of migrating GnRH neurons prenatally (Lee et al., 2008). The TDS of neurons in explants from GAD67+/+ mice was comparable to that of neurons in explants from NIH Swiss mice (Table 1). However, there was a significant increase (∼50%) in TDS of neurons from GAD67−/− mice as compared to GAD67+/+ mice and NIH Swiss mice (Fig. 2H; Table 1). The TDS observed in GAD67−/− mice was similar to measurements in explants from NIH Swiss mice pharmacologically treated with Pic (P>0.05; Fig. 2H). These results were duplicated in VIAAT−/− mice (Table 1; Fig. 2H). GnRH cells in VIAAT−/− explants had a similar TDS to cells in explants from GAD67−/− mice and explants treated with Pic from NIH Swiss mice (Table 1). Importantly, GnRH cells in VIAAT−/− explants slowed in response to muscimol (Table 3). This result indicates that GABAA receptor function is intact in these mutants and that changes in GnRH movement are direct are not secondary to early patterning defects in GnRH neurons. Collectively, these data indicate that 1) endogenous GABAergic signaling slows the speed of GnRH neurons as they move forward, changing the dynamics of saltatory movement to favor pause periods and 2) that another signal(s) in the nasal region (both in vivo and in nasal explants) is present that activates/maintains movement and migrational direction of GnRH cells as they migrate away from the nasal pit.
Table 3.
Midline SDF-1 cues modulate neuronal migration
| Paradigm | Explant | SFM | Drug 1 | Drug 1+2 | n | N | |
| SFM, SDF-1β | TDS | +MNC | 25.79±1 | 25.93±0.91 | 144/146 | 6 | |
| SFM, SDF-1β | TDS | −MNC | 19.87±1.48 | 23.2±0.97d | 49/54 | 3 | |
| SFM, SDF-1β | LDS | −MNC | 16.13±1.9 | 21.27±1.2d | 40 | 2 | |
| SFM, AMD3100 | TDS | +MNC | 16.99±1 | 14.95±1a | 70 | 3 | |
| SFM, AMD3100 | TDS | −MNC | 16±0.65 | 15.8±1.24 | 52 | 2 | |
| SFM, AMD3100 | LDS | +MNC | 14±1.0 | 11±0.92a | 70 | 3 | |
| SFM, Mus, Mus+SDF-1β | TDS | +MNC | 16.2±0.86 | 12.8±0.58a | 17.2±0.69b | 62 | 3 |
| SFM, Mus, Mus+SDF-1β | TDS | −MNC | 19.5±1.06 | 15.6±1.05a | 20.9±1.34b | 63 | 3 |
| SFM, Mus, Mus+SDF-1β | LDS | +MNC | 13.03±0.9 | 10.48±0.7a | 14.3±1.0b | 62 | 3 |
| SFM, Pic, Pic+AMD3100 | TDS | +MNC | 17.8±1.16 | 20.8±1.23a | 15.9±0.98b | 58 | 3 |
| SFM, Pic, Pic+AMD3100 | LDS | +MNC | 15±1.2 | 10.7±0.8b | 58 | 3 | |
| SFM, Mus | TDS | −MNC | 15.5±0.815 | 11.5±0.597a | 67 | 3 | |
| SFM, Pic | TDS | −MNC | 19.9±0.9 | 26.5±1.5a | 47 | 3 | |
| SFM, Mus | TDS | VIAAT−/− | 22.51±1.24 | 13.9±1.09a | 53 | 3 | |
| SFM, Mus | LDS | VIAAT−/− | 15.27±1.3 | 7.2±.86a | 53 | 3 | |
| SFM, Pic, Pic+SDF-1β | TDS | −MNC | 18.2±0.79 | 22.9±0.98a | 21.9±0.8c | 71 | 3 |
Speeds in µm/h. n, number of cells recorded; N, number of explants used. Italics highlight LDS measurements.
SFM and Drug1 periods significantly different (paired t-test: P<0.05)
Drug1 and Drug1+Drug2 periods significantly different (paired t-test: P<0.05)
Drug1+Drug2 and SFM periods significant different (paired t-test: P<0.05)
SFM and Drug1 periods significantly different (unpaired t-test: P<0.05)
Midline SDF cues
Both in vivo and in nasal explants, SDF-1 producing cells are present, CXCR4 (a SDF-1 receptor) is expressed by GnRH neurons and absence or chronic inhibition of this system decreased GnRH cell movement (Schwarting et al., 2006; Toba et al., 2008). Thus, SDF-1 could be facilitating movement of GnRH neurons in the absence of GABAergic signals. GnRH cells and olfactory axons expressed CXCR4 at 4 div (Fig. 3). SDF-1 producing cells were located bilaterally along the dorsal midline nasal cartilage (MNC) as well as a group of cells clustered at the dorsal MNC tip (Fig. 3). To determine the role of endogenous SDF-1 neuronal movement rate was examined in explants in the absence of MNC (−MNC). Bilateral nasal tissue (+MNC) was cut on either side of the MNC, generating an explant with no or reduced midline cartilage (Fig. 3C). SDF-1 staining was absent (Fig. 3I) but CXCR4 positive fibers and CXCR4 positive GnRH neurons were still present (Fig. 3G,H). In explants with or without MNC, nearly all GnRH cells expressed CXCR4 receptors (Fig. 3F).
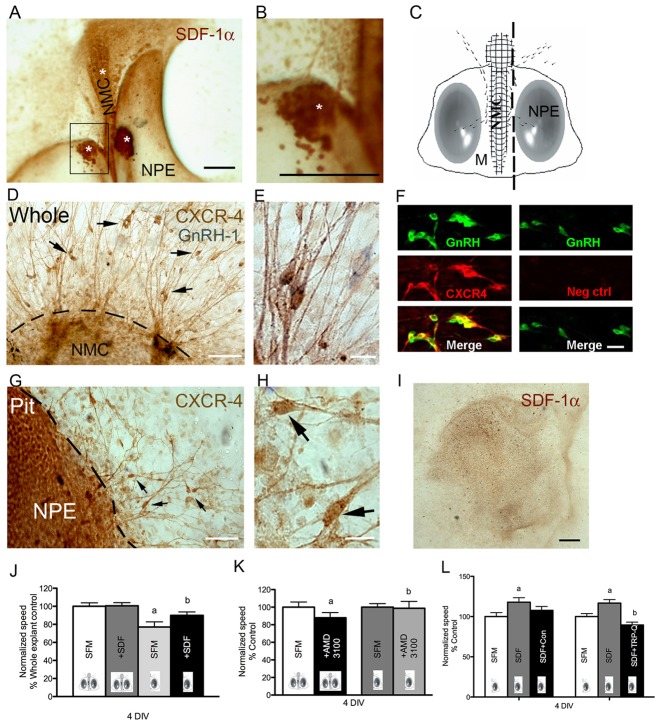
Fig. 3.
The SDF-1/CXCR4 system affects the rate of GnRH neuron movement. (A,B) Explant stained with SDF-1α antibody. SDF-1α is expressed in cells of the nasal midline cartilage (white asterisks). (B) Magnification of box in A. (C) Explant schematic: NMC, nasal midline cartilage; NPE, nasal pit epithelium; M, mesenchyme. Dashed line indicates cutting line to obtain explant without NMC (−NMC). Drawn dots show representative GnRH neurons moving from the main tissue mass into the periphery. (D,E) Explant with NMC (+NMC) stained for CXCR4 (brown) and GnRH (blue) showing large numbers of CXCR4-positive fibers and cells (D, arrows). The majority of cells were labeled for both GnRH and CXCR4 (E). (F) Left: double label fluorescence confirms colocalization of GnRH (green) and CXCR4 (red). Right: negative control omitting CXCR4 primary antibody. (G,H) −MNC explant stained for CXCR4 (brown). (I) Explant −MNC stained with SDF-1α antibody. Staining is markedly reduced compared with whole explants (see A). (J) Exogenous SDF-1β (10 nM) did not change TDS of neurons in +NMC explants (P>0.05). The speed of cells in −NMC explants was significantly lower than that of cells in the +NMC explants (a, t-test, P<0.05). Exogenous SDF-1 rescued the cells' speed to control levels (b, t-test, P>0.05). Results are normalized to whole explant control. (K) AMD3100 significantly decreased TDS of neurons (a, P<0.05) in +NMC explants. The same treatment was ineffective in −NMC explants (b, P>0.05). Speeds are normalized to the control. (L) TRP-Q abolished (b, P<0.05, paired t-test) the SDF-1 induced increase in the speed of neurons in −NMC explants (a, P<0.05, paired t-test). Conotoxin + SDF-1 had no significant effect on TDS when compared to SDF-1 treatment alone (P>0.05, paired t-test). Speeds are normalized to the control period. All images and data from cells in explants maintained for 4 div. Scale bars: 200 µm (A, B, I); 50 µm (D,G); 10 µm (E,H); 20 µm (F).
The movement rate of neurons in −MNC explants in SFM, was variable but averaged 20–35% slower than cells in +MNC explants at 4 div (Fig. 3J, t-test: P<0.005) and 6 div (t-test: P<0.005). Controls, consisting of removal of one NPE and leaving one NPE+MNC, showed cells with movement rates comparable to cells in whole explants [19.17±0.63 µm/h (n = 172, N = 7) vs. 20.45±0.69 µm/h (n = 232, N = 11); unpaired t-test: P>0.05; n, number of cells recorded; N, number of explants used], indicating that the additional dissection did not alter the behavior of the GnRH cells alone. To determine whether removal of SDF-1 was responsible for the decreased TDS of neurons in −MNC explants, the speed of cells with or without MNC was analyzed pre- and post-application of exogenous SDF-1β (10 nM). In explants with MNC, no significant change in TDS was found after SDF-1β administration (Table 3). In contrast, in −MNC explants, TDS during SDF-1β treatment was 17% faster than during control conditions (Table 3; Fig. 3J). The LDS of neurons in −MNC explants was noticeably closer to TDS (LDS/TDS ratio = 0.81). The LDS of neurons in −MNC explants treated with SDF-1β was 30% faster than SFM control condition (Table 3) and the LDS/TDS ratio during SDF-1 treatment increased to 0.90. These data indicate that without MNC, neurons show decreased random movement that is further attenuated by SDF-1.
Next, the speed of neurons in explants with MNC was recorded pre- and post-application of the CXCR4 receptor antagonist AMD3100 (25 µg/ml) (Toba et al., 2008). AMD3100 significantly decreased (−12%, Table 3; Fig. 3K) the speed of cells in +MNC explants. Similar results were obtained for LDS (−21%; Table 3). The LDS/TDS ratios of SFM and AMD3100 recording periods were respectively 0.82 and 0.74 indicating a decrease in the motility efficiency when CXCR4 receptors are blocked in explants with MNC. Notably, administration of AMD3100 in −MNC explants did not lead to any alteration in neuronal speed (Table 3; Fig. 3K). The average speed of cells from −MNC +SDF-1β explants was then compared to cells in the +MNC ±SDF-1 condition. This analysis revealed that during SDF-1 treatment, cell movement rate in −MNC explants was similar to that of cells maintained in +MNC explants (Table 3; Fig. 3I), suggesting that exogenous SDF-1β could completely rescue cues missing with removal of the MNC. Taken together, these data indicate that endogenous SDF-1β secreted by cells around the MNC has a motogenic effect on GnRH cells through the CXCR4 receptor and suggests that the SDF-1/CXCR-4 system augments the speed of neurons and increases the efficiency of movement by decreasing random chemokinesis.
SDF-1/CXCR4 regulation of GnRH neuronal movement
SDF-1 has been shown to act in the brain as a neurotransmitter (Banisadr et al., 2005) and to be released with GABA from inhibitory neurons (Bhattacharyya et al., 2008). However, work in the hematopoietic system indicates that GABA can block SDF-1α-induced movement of adult mobilized hematopoietic stem and peripheral cells (Seidel et al., 2007). A series of experiments were performed to evaluate if there was synergistic or opposing activity between GABA and SDF-1 to modulate GnRH neuronal movement. The TDS of GnRH cells in ±MNC explants was analyzed pre- and post-application of Mus and then in the presence of both Mus and SDF-1β. Under both explant conditions, there was ∼20% decrease in cell speed recorded during Mus treatment (Table 3), consistent with earlier data (Table 2). When SDF-1β was administered with Mus, there was a significant increase in TDS compared to the Mus period (∼34%; Table 3), but no difference when compared to the SFM period (Table 3). These data reveal that the inhibitory effect of GABA on neuron movement via the GABAA receptor can be reversed by SDF-1. Similar results were found in the LDS measurements. A significant decrease (20%) was detected after Mus application followed by a significant increase (36%) when SDF-1 was added to Mus (Table 3). With Mus+SDF-1, neuron movement reached a LDS also comparable to control (P>0.05). Analysis of the LDS/TDS ratio confirmed previous values and revealed no differences among the recording periods (SFM: 0.80; Mus: 0.81; SDF: 0.83). Together, these data reveal that the inhibitory effect of GABA on neuron movement via the GABAA receptor can be reversed by SDF-1.
To confirm the interaction between GABAergic and SDF-1 signaling, the speed of cells in explants with MNC was analyzed pre- and post-application of Pic and then with both Pic and AMD3100. Consistent with earlier experiments, there was a significant increase in speed after Pic (14%; Table 3). However, addition of AMD3100 in the presence of Pic produced a significant decrease in the speed of cells as compared to Pic only (20%, Table 3). Addition of the CXCR4 antagonist brought the speed of neuronal movement back to a rate similar to that in control conditions (Table 3), suggesting that the main factor increasing cell movement during inhibition of GABAergic signals was SDF-1. Analysis of the LDS of neurons in this group showed that there was a significant decrease (29%) when AMD3100 was added to Pic (Table 3). The LDS/TDS ratio in the presence of PIC and AMD3100 was lower than controls (SFM: 0.79; PIC+AMD: 0.66), indicating that SDF-1/CXCR4 system is needed for the forward movement.
CXCR4 coupling in GnRH neurons
SDF-1 via CXCR4 coupled to Gi/o has been shown to regulate neuronal signaling through inhibition of voltage dependent calcium (Ca2+) currents (Oh et al., 2002), in particular the N-type channel (Simen et al., 2001). Perturbation of the N-type Ca2+ channel has been shown to decrease GnRH neuronal movement rate (Toba et al., 2005). However, SDF-1 also activates GIRK channels (Guyon et al., 2005; Guyon and Nahon, 2007) which GnRH neurons express (Hu et al., 2006; Klenke et al., 2010). To delineate the SDF-1/CXCR4 signaling pathway altering GnRH neuronal movement, experiments were performed using 1) ω-Conotoxin, an inhibitor of N-type Ca2+ channels and 2) tertiapin Q (TPN-Q), an inhibitor of GIRK channels (Klenke et al., 2010).
In +MNC explants, TDS recorded after acute ω-Conotoxin administration (100 nM) was similar to that in the control condition (Table 4). In −MNC explants, cell TDS after ω-Conotoxin was significantly slower (15%) as compared to controls (Table 4). These data indicate that N-Type channels are involved in the movement of GnRH neurons and that the presence of cartilage, possibly SDF-1 producing cells influenced their activity. To assess interactions between N-Type channels and the midline SDF-CXCR4 system, cells in −MNC explants were exposed to SFM, SDF and then SDF+ω-Conotoxin and TDS measured. Consistent with earlier experiments, a significant increase in speed (18%) occurred after SDF application (Table 4). When ω-Conotoxin was added with SDF, no change in the speed of the cells was seen (Table 4; Fig. 3L). Thus SDF-1 blocked the inhibitory activity of ω-Conotoxin alone, but ω-Conotoxin did not block the accelerating action of SDF-1. These data indicate that SDF-1 worked independently of N-type Ca2+ channels to increase cell movement.
Table 4.
SDF-1/CXCR4 signaling via GIRK channels
| Paradigm | Explant | SFM | Drug 1 | Drug 1+2 | n | N | |
| SFM, ω-Conotoxin | TDS | +MNC | 20.4±0.73 | 19.4±0.571 | 105 | 4 | |
| SFM, ω-Conotoxin | TDS | −MNC | 19.1±0.896 | 16.4±0.753a | 74 | 4 | |
| SFM, SDF, SDF+ω-Conotoxin | TDS | −MNC | 16.8±0.824 | 19.8±0.946a | 18.1±0.813 | 82 | 4 |
| SFM, TPN-Q | TDS | +MNC | 23.3±1.43 | 19.8±1.21a | 54 | 3 | |
| SFM, TPN-Q | TDS | −MNC | 20.1±0.93 | 20.7±0.9 | 64 | 3 | |
| SFM, SDF, SDF+TPN-Q | TDS | −MNC | 19.1±0.721 | 22.3±0.835a | 17.1±0.692b | 88 | 4 |
| SFM, Mus, Mus+SDF+TPN-Q | TDS | −MNC | 18.6±0.64 | 14.9±0.64a | 13.9±0.615b | 106 | 4 |
Speeds in µm/h. n, number of cells recorded; N, number of explants used.
SFM and Drug1 periods significantly different (paired t-test: P<0.05)
Drug1 and Drug1+Drug2 periods significantly different (paired t-test: P<0.05)
If SDF-1 acts downstream via GIRK channels, one would predict a change in GnRH neuronal movement in the presence of TPN-Q in +MNC explants (endogenous SDF-1 in midline cells) and no effect in −MNC explants (express little or no endogenous SDF-1). As predicted, a significant decrease in cell speed (15%) was seen when GIRK channels were blocked by TPN-Q (100 nM) in +MNC explants, while no differences in speed were detected after TPN-Q application in NPE-MNC explants (Table 4). These data are consistent with SDF-1 acting via GIRK channels to alter cell movement.
To further test if SDF signaling activates GIRK channels, −MNC explants were analyzed pre- and post-application of SDF-1 or SDF-1+TPN-Q. There was a significant decrease in TDS (23%) when TPN-Q was added with SDF-1, as compared to SDF-1 alone (Table 4; Fig. 3L), but these values were similar to the speed in the control condition (SFM, P>0.05). These data support the hypothesis that SDF-1/CXCR4-mediated movement is signaled via GIRK channels. Finally, to verify independent actions of GABA and SDF-1 signals on cell movement, the effect of Mus, SDF-1 and TPN-Q was investigated in −MNC explants. Earlier results showed that exogenous SDF-1 increased cell movement after application of Mus, suggesting a potential GABA/SDF interaction. To determine the action of GIRK channel inhibition, cells were exposed to SFM, Mus and then Mus+SDF+TPN-Q and TDS measured. No changes in cell TDS were found when SDF-1+TPN-Q were added after Mus (Table 4). Taken together these data indicate that GABA plays a significant but independent role in GnRH cell movement from that of SDF-1, which uses GIRK channels as a downstream signal to alter the membrane potential of moving GnRH cells.
Although SDF-1 signaling is clearly an important factor for neuronal migration, GnRH cells in −MNC explants still showed movement, though at a reduced rate. To determine whether changes in GABAergic signaling occurred in −MNC explants, the speed of neurons was measured pre- and post-application of Mus or Pic (Table 3). The percent change in −MNC explants treated with Mus (decrease 26%) was similar to the change recorded in explants +MNC (22%), suggesting that GABAA receptors on GnRH cells were intact. No differences were detected in GAD67 staining between ±MNC explants (data not shown). Unexpectedly, application of Pic increased cell movement rate in −MNC (29%, Table 3) even more than that observed in +MNC (17%, Table 2) and produced no change in random movement (LDS/TDS ratio SFM: 0.79, PIC: 0.75). Addition of SDF-1 after a period of PIC alone exposure did not result in any further increase in cell TDS in −MNC explants (Table 3). These results indicate that when GABAA signaling is inhibited, a non-midline factor can stimulate GnRH cell movement in the absence of SDF-1.
Discussion
SDF-1 and GABA are important for appropriate cell migration during central and peripheral nervous system development (Guyon and Nahon, 2007; Tiveron and Cremer, 2008; Represa and Ben-Ari, 2005). The action of GABA is mediated through GABAA receptors while SDF-1 is mediated through receptors CXCR4 and CXCR7 (Tiveron and Cremer, 2008; Sánchez-Alcañiz et al., 2011; Wang et al., 2011; Boldajipour et al., 2008; Odemis et al., 2010; Tiveron et al., 2010). GABAA receptors and CXCR4 receptor are expressed by GnRH neurons (CXCR7 transcript is not found in GnRH neurons, unpublished observations). Both GABA and SDF-1 signals influence migration of GnRH neurons from the nasal pit to the forebrain. In this study, the dynamics of GnRH neurons movement were analyzed and the action of SDF-1 and GABA signaling on neuronal movement determined. In situ analysis revealed that GnRH cells exhibit saltatory movement resembling glial-guided migrating neurons (Nadarajah et al., 2001). In addition, movement of GnRH neurons was accelerated by SDF-1/CXCR-4 signaling, in contrast to the decelerating activity induced by GABA through GABAA receptors. Notably, SDF-1-induced acceleration was distinct from acceleration caused by blocking GABA signaling, as blocking GABA caused increased random movement while SDF-1 facilitated directional movement. The N-type Ca2+ channel, which can be activated by SDF (Simen et al., 2001) and is important in the regulation of neuronal movement (Toba et al., 2005), was not necessary for SDF-1/CXCR4 induced increase in cell movement. Rather, acceleration of GnRH neurons via SDF1/CXCR4 signals was found to operate through activation of GIRK channels.
GABAA signaling slows neuronal movement but improves directional movement
Several studies showed that inhibiting GABAA receptors increased total GnRH neuronal migration while activating GABAA receptors inhibited migration (Fueshko et al., 1998b; Bless et al., 2000; Lee et al., 2008). However, these experiments did not delineate the mechanism whereby activation of GABA resulted in decreased movement. Time-lapse analysis in the present study showed that inhibition of GABA increased total movement but not linear movement of neurons in explants. These data are consistent with inactivation of GABAA receptors resulting in a loss of directional movement and a concomitant increase in random movement. Inhibiting GABA signaling in vivo disrupted association of GnRH neurons with olfactory axon fibers in the brain, causing a disorganized pattern of GnRH neurons (Bless et al., 2000), and altering turning behavior (Bless et al., 2005). Random movements observed in picrotoxin-treated GnRH cells in explants may underlie the disorganization observed when GABAA receptors are blocked during neuronal migration in vivo (Bless et al., 2000). Thus, insufficient GABA signaling seems to accelerate but disorganize neurons; excess GABA signaling maintains neurons on their migratory path but delays their progression.
SDF accelerates neuronal forward movement by activating GIRK channels
In CXCR4 knockout mice, most GnRH neurons fail to migrate away from the vomeronasal organ (Schwarting et al., 2006; Toba et al., 2008). A gradient of SDF-1 mRNA expression along the migratory path of GnRH neurons suggested that SDF-1 might act as a guidance cue (Schwarting et al., 2006). However, SDF-1 was not required for directional movement of GnRH neurons in nasal explants, although blocking SDF-1 signaling reduced movement rates (Toba et al., 2008). These results suggested that the primary function of SDF-1 signaling was linked to neuronal movement per se rather than directional guidance. SDF-1 has been shown to accelerate migration speed of cortical interneurons (López-Bendito et al., 2008). However, acceleration of interneurons by SDF-1 (measured after 36-48 hr) may have been a consequence of reduced cAMP-dependent branching, since branching of the leading process slows cortical interneurons and promotes invasion into the cortical plate (Lysko et al., 2011). Here, by monitoring neurons in situ and capturing images as treatment occurred, SDF-1 was found to accelerate forward movement by promoting saltatory movement of the cell body within minutes. Although effects on saltatory movement and leading process branching are not mutually exclusive, our results suggest SDF-1 accelerates neuronal migration more rapidly than a reduction in leading process branching alone could account for. In addition, the rapid SDF-1 induced changes in saltatory GnRH movement were shown to occur through a GIRK dependent mechanism. GIRK channels open via a signal transduction cascade triggered by the Gβγ dimeric protein released by activated G protein-coupled receptors (Dascal, 1997; Yamada et al., 1998). Opening of the GIRK channel results in permeability to potassium ions and hyperpolarization of the cell. To date, only melanin-concentrating hormone neurons (involved in feeding behavior regulation) have been shown to respond to SDF-1 directly through GIRK current activation and in these cells, SDF-1 influenced neuronal activity (Guyon et al., 2005).
Depolarization has been associated with reduction in GnRH neuronal migration in nasal regions (Kusano et al., 1995; Fueshko et al., 1998b). Administration of TRP-Q, a specific blocker of GIRK channels, likely reduced GnRH neuronal forward movement rates due to depolarization of the cell. The effect of SDF-1 on GnRH forward movement in unilateral explants (lacking endogenous SDF-1) was negated when GIRK channels were blocked with TRP-Q. These data, together with the fact that TRP-Q blocked the ability of SDF-1 to overcome GABA induced reduction in GnRH movement rates, indicates that SDF-1/CXCR4 activate and require potassium extrusion through GIRK channels to regulate neuronal migration. These results provide the first evidence of a molecular link among factors that control neuronal excitability and migration signaling, identifying a new role for GIRK channel activity.
SDF prevents GABA-induced reduction of forward movement rates without increasing random movement
Interacting signaling pathways with opposing effects are frequently utilized by neurons to regulate temporal or spatial development of neural circuits. One well-characterized example is the retinotectal system, in which gradients of branch-inhibiting ephrins and their Eph receptors interact with branch-promoting BDNF signals to confine retinal axon branches to the appropriate tectal locations (McLaughlin and O’Leary, 2005; Rashid et al., 2005; Lim et al., 2008; Marler et al., 2008; Hutchins and Li, 2009). Our results show that SDF-1 and GABA have opposing effects on the forward movement rates of GnRH neurons. GABA and SDF exert inverse effects on membrane potential with Cl− outflow through GABAA receptors (Kusano et al., 1995) versus K+ outflow through GIRK channels (Lüscher and Slesinger, 2010). Previous work showed that inhibiting depolarization-dependent electrical activity in GnRH neurons promoted cell forward movement (Fueshko et al., 1998a), consistent with effects of GABA and SDF-1 being due to direct effects on membrane potential (Fig. 4A). Thus, these two opposing migrational modulators could synergistically confine migration rates, and hence the location, of GnRH neurons along the migratory pathway.
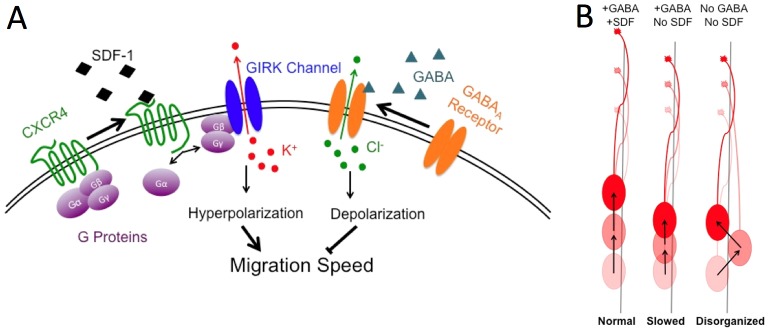
Fig. 4.
Schematic diagram showing the interactions between the SDF-1 and GABA signaling pathways, and subsequent effects on neuronal movement. (A) SDF-1 binding to CXCR4 leads to G-protein mediated activation of GIRK channels and hyperpolarization by potassium efflux. GABA binding to the GABAA receptor leads to opening of the chloride channel and depolarization through chloride efflux. Each pathway antagonizes the other's effect on the depolarization state and thus movement rates. (B) The combined effect of GABA and SDF-1 signaling on the GnRH neuron under normal movement conditions is relatively rapid and organized (‘Normal’, left). Inhibiting SDF-1 while GABA signaling remains intact slows the forward movement rate as neurons become depolarized, but the organizing effect of GABA maintains the forward trajectory of the GnRH neurons (‘Slowed’, middle). Inhibiting both GABA and SDF-1 signaling restores the rate of forward movement to higher levels as the depolarizing effect of GABA is removed, but the organizing effects are lost and movement is more random (‘Disorganized’, right).
Although opposing effects on membrane potential can account for the effects of GABA and SDF-1 on migration speed, it does not explain the fact that SDF-1 had no effect on the directionality imparted by GABA signaling. If SDF-1 completely counteracted the effects of GABA, then augmented random movement should have occurred with SDF-1, similar to the effects of blocking GABA with picrotoxin. The net result of combined SDF and GABA signaling was instead, improved directionality without an apparent reduction of forward movement rates (Fig. 4B). The mechanism by which disruption of GABAergic signaling disorganizes movement is currently unknown. Blocking HGF (Hepatocyte Growth Factor) signaling also led to disorganization of GnRH neuronal migration, with cells appearing to lose their linear orientation, no longer parallel to the olfactory pathway (Giacobini et al., 2007). However, in contrast to GABA, HGF stimulates migratory behavior. Differential effects on total versus linear migration rates have been observed after loss of adhesion (Greciano et al., 2012) or altered Rho GTPase signaling (Wojciak-Stothard and Ridley, 2003; Francis et al., 2006). In addition, manipulation of myosin II, an indirect target of RhoA signaling, can lead to undirected movement of the nucleus orthogonal to the leading process in cortical interneurons (Bellion et al., 2005). Notably, CXCR4 receptors are rapidly internalized after activation (Peled et al., 2012) while GABAA receptors can directly bind to cell adhesion molecules (Zhang et al., 2010) and cytoskeletal adaptor proteins (Wang and Olsen, 2000). If adhesion were strongly enhanced after application of muscimol, a reduction in both total and linear movement might occur. Thus, GABAA receptors could modulate downstream adhesion and cytoskeletal responses that influence directionality. These differences could achieve specificity between the two hyperpolarizing signals, although this hypothesis remains to be tested. However, taken together, the experiments in this study indicate that during neuronal migration, directionality and migration speed can be regulated by independent signaling pathways, each requiring the coordinated action of different sets of extracellular cues. Perturbation of a single signaling pathway would thus have a smaller impact on neuronal migration than if speed and directionality were regulated in tandem.
Materials and Methods
Animals
All mice were killed in accordance with National Institutes of Health-National Institute of Neurological Stroke and Disorders guidelines. Embryos were removed and used for generating nasal explants or in vivo analysis. The Gad1/Gad67 knockout mice and Viaat KO mice have been described previously (Oh et al., 2010).
Nasal explants
Nasal regions were cultured as described previously (Klenke and Taylor-Burds, 2012). Briefly, nasal pits with or without midline nasal cartilage of E11.5 NIH Swiss mice were isolated under aseptic conditions and adhered onto coverslips by a plasma (Cocalico Biologicals, Reamstown, PA)-thrombin (Sigma, St. Louis, MO) clot. Explants were grown in SFM (Fueshko and Wray, 1994) at 37°C with 5% CO2. At 3 div the explants received medium containing fluorodeoxyuridine (8×10−5 M; Sigma, St. Louis, MO). Fresh medium was given again at 6 div.
Embryonic tissue
Embryos were dissected from time-mated females, rinsed in cold PBS, and tissue removed for genotyping (see below) before the head/body was frozen on powdered dry ice and stored at −80°C until sectioning. Serial sections (16 µm, parasagittal plane) were cut (Leica, CM3050 S Cryostat), mounted onto 2X subbed slides (Gold Seal), air-dried (∼min) and stored (−80°C) until use. Embryos were cryosectioned into 4 series.
Immunocytochemistry
Primary antisera: rabbit polyclonal GnRH (1∶3000; SW-1) (Wray et al., 1988) rabbit polyclonal peripherin (1∶2000; Chemicon, Temecula, CA, USA), mouse monoclonal β-galactosidase (1∶1000; Promega, Madison, WI, USA), sheep polyclonal GAD 1440, which recognizes GAD67 with high affinity and to a much lesser extent GAD65 (gift from Dr I.J. Kopin; 1∶14,000), rat monoclonal against CXCR4 (1∶200, BD Biosciences, San Jose, CA, USA), and rabbit monoclonal against SDF-1α (1∶500, Cell Sciences, Canton, MA, USA).
Explants and tissue sections were immunocytochemically stained as described previously (Wray et al., 1994; Toba et al., 2008). Briefly, tissues were fixed (4% formaldehyde, 1 h, rinsed in PBS 3×8 min), blocked (10% normal serum of the species in which the secondary was made 0.3% Triton X-100, 1 hr), and washed in PBS. After washing, tissue was incubated in primary antibody overnight (4°C). The next day tissue was rinsed in PBS, incubated in biotinylated species appropriate secondary antibody (1 hr; 1∶500-1500 in PBS/0.3% Triton X-100; obtained from Vector Laboratories, Burlingame, CA, USA; or 1∶1500 Jackson ImmunoResearch Laboratories, West Grove, PA, USA), and processed using standard procedures for avidin-biotin-conjugated horseradish peroxidase (1∶600, Vectastain Elite; Vector), 3′,3-diaminobenzidine (DAB, Sigma) and glucose oxidase or incubated in a directly conjugated fluorescence secondary (Invitrogen). For double-labeling using chromogens, tissues were rinsed in PBS after DAB reaction (3×8 min), blocked with 10% species appropriate normal serum (30 min), rinsed in PBS (3×8 min), fixed in 4% formaldehyde (1 h), and put in primary antibody for 2 days. After staining with SG (blue, Vector), tissues were counterstained with 0.05% methyl green, washed with distilled water and mounted with crystal mount or Permount (Electron Microscopy Sciences, Hatfield, PA, USA). Controls for staining included omission of the second primary to insure no binding of the second secondary to the first primary/secondary complex occurred.
Video microscopy
Time lapse video microscopy was performed on explants between 3 and 7 div. Nasal explants were placed into a perfusion chamber (Warner Instruments, Hamden, CT) housed in an incubator on top of the microscope stage. Temperature was maintained at 37°C±2 and the CO2 concentration at 5% (Live cell, Pathology Devices, Inc., Westminister, MD, USA). Preheated SFM (37°C, or SFM plus drug) was pumped through the chamber with a peristaltic pump (Spectra Hardware, Inc., Westmoreland City, PA). Experiments were performed on either an inverted Nikon TE2000 or TE200 microscope (Nikon USA, Melville NY), both equipped with Differential Interference Contrast (DIC) optics, a motorized stage (Ludl, Electronic Products Ltd, Hawthorne, NY USA) and an ICCD camera (Retiga, Qimaging, Burnaby, Canada). Both systems were connected to a Macintosh computer and the experiments were piloted by imaging software (IPLab Spectrum, Scanalytics Inc., Rockville, MD). Multiple fields of the same explant were monitored every 2 min for the duration of recording period (30-180 min). Images were taken with an ELWD 20×objective (Nikon).
Speed of movement
Movement of the cells was analyzed a posteriori using ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997-2008). Two measurements were determined; total distance and linear distance. Total distance reflects the entire route that the cell moved while linear distance equals the change between the first position and final position. If total distance and linear distance are equal then the cell moved in a ‘straight’ line. In contrast, if total distance is greater than linear distance, the cell took a circuitous route to its final position. To calculate distance traveled by the cells the following methods were employed. GnRH cell soma exhibits a characteristic uni/bipolar morphology. In ImageJ a circle was drawn around the neuron, with the diameter surrounding the maximum width of the cell and the centroid recorded as the cell's position. The circle was positioned on the cell body over the maximum width at each recorded frame. Distance traveled was calculated from the X and Y coordinates plotted in ImageJ for each frame (total) or first and last frame (linear).
Recording experiments that involved drug treatments were divided into either 2 periods [SFM control period (40-60 min), treatment period (60-120 min)] or 3 periods [SFM control period (40-60 min), first treatment period (60 min), second treatment period (60 min)].
Drugs
Muscimol (Mus, agonist of GABAA receptor); Picrotoxin (Pic, antagonist of GABAA receptor); baclofen (Bac, agonist of GABAB receptor); and saclofen (Sac, antagonist of GABAB receptor) were obtained from Sigma (St. Louis, MO). Murine SDF-1β was obtained from PeproTech Inc. (Princeton Business Park, Rock Hill, NJ); ω-Conotoxin and TPN-Q were obtained from Tocris Bioscience (Ellisville, MO). All stock solutions [made up in ethanol (final concentration ≤0.001% v/v), DMSO (DMSO, final concentration ≤0.001% v/v) or Dulbecco's Modified Eagle Medium (DMEM)] were stored at −20°C, and solutions were prepared before each experiment by diluting stock solutions into SFM used for growing the explants (Fueshko and Wray, 1994).
Statistical analysis
The effects of drug on the forward movement rate of GnRH neurons were calculated by comparing the mean cell speed in control conditions (SFM) with that in the drug-treated explants using custom R scripts (http://www.r-project.org; custom scripts for measuring cell body position by D.D.). Data are given as mean of movement rate ± SEM, and the levels of significance were determined by a two-way ANOVA between multiple groups or paired Student's t-tests (same cells recorded and analyzed pre- and post-application of the drugs) or unpaired Student's t-tests (different cells of the same explant or different explants recorded and analyzed pre- and post-application of the drugs). t-tests were considered significantly different if P<0.05. n = number of cells recorded, and N = number of explants used. Chi-squared tests were used to compare ratios of cells speeding, slowing, or remaining unchanged between different drug applications. For analysis of changes in saltatory movement, frequency distributions of interval speed (µm/2 min) were generated and compared using the chi-squared test. All chi-squared tests were considered significantly different if P<0.05.
Footnotes
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Neurological Disorders and Stroke [grant number ZIA NS002824-21]; and the National Institutes of Health [grant number R01MH064794 to B.G.C.]. B.I.H. was supported by the Pharmacology Research Associate (PRAT) Program of he National Institute of General Medical Sciences, National Institutes of Health. Deposited in PMC for release after 12 months.
References
- Asada H., Kawamura Y., Maruyama K., Kume H., Ding R. G., Kanbara N., Kuzume H., Sanbo M., Yagi T., Obata K. (1997). Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc. Natl. Acad. Sci. USA 94, 6496–6499 10.1073/pnas.94.12.6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banisadr G., Gosselin R. D., Mechighel P., Kitabgi P., Rostène W., Parsadaniantz S. M. (2005). Highly regionalized neuronal expression of monocyte chemoattractant protein-1 (MCP-1/CCL2) in rat brain: evidence for its colocalization with neurotransmitters and neuropeptides. J. Comp. Neurol. 489, 275–292 10.1002/cne.20598 [DOI] [PubMed] [Google Scholar]
- Bellion A., Baudoin J. P., Alvarez C., Bornens M., Métin C. (2005). Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 25, 5691–5699 10.1523/JNEUROSCI.1030-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya B. J., Banisadr G., Jung H., Ren D., Cronshaw D. G., Zou Y., Miller R. J. (2008). The chemokine stromal cell-derived factor-1 regulates GABAergic inputs to neural progenitors in the postnatal dentate gyrus. J. Neurosci. 28, 6720–6730 10.1523/JNEUROSCI.1677-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser H., Eisenbeiss S., Neumann M., Reichman–Fried M., Thisse B., Thisse C., Raz E. (2005). Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J. Cell Sci. 118, 4027–4038 10.1242/jcs.02522 [DOI] [PubMed] [Google Scholar]
- Bless E. P., Westaway W. A., Schwarting G. A., Tobet S. A. (2000). Effects of gamma-aminobutyric acid(A) receptor manipulation on migrating gonadotropin-releasing hormone neurons through the entire migratory route in vivo and in vitro. Endocrinology 141, 1254–1262 10.1210/en.141.3.1254 [DOI] [PubMed] [Google Scholar]
- Bless E. P., Walker H. J., Yu K. W., Knoll J. G., Moenter S. M., Schwarting G. A., Tobet S. A. (2005). Live view of gonadotropin-releasing hormone containing neuron migration. Endocrinology 146, 463–468 10.1210/en.2004-0838 [DOI] [PubMed] [Google Scholar]
- Boldajipour B., Mahabaleshwar H., Kardash E., Reichman–Fried M., Blaser H., Minina S., Wilson D., Xu Q., Raz E. (2008). Control of chemokine-guided cell migration by ligand sequestration. Cell 132, 463–473 10.1016/j.cell.2007.12.034 [DOI] [PubMed] [Google Scholar]
- Bolteus A. J., Bordey A.2004). GABA release and uptake regulate neuronal precursor migration in the postnatal subventricular zone. J. Neurosci. 247623–7631 10.1523/JNEUROSCI.1999-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron R. S., Ruffin J. W., Cho N. K., Cameron P. L., Rakic P. (1997). Developmental expression, pattern of distribution, and effect on cell aggregation implicate a neuron-glial junctional domain protein in neuronal migration. J. Comp. Neurol. 387, 467–488 [DOI] [PubMed] [Google Scholar]
- Cancedda L., Fiumelli H., Chen K., Poo M. M. (2007). Excitatory GABA action is essential for morphological maturation of cortical neurons in vivo. J. Neurosci. 27, 5224–5235 10.1523/JNEUROSCI.5169-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascal N. (1997). Signalling via the G protein-activated K+ channels. Cell. Signal. 9, 551–573 10.1016/S0898-6568(97)00095-8 [DOI] [PubMed] [Google Scholar]
- Deiner M. S., Sretavan D. W. (1999). Altered midline axon pathways and ectopic neurons in the developing hypothalamus of netrin-1- and DCC-deficient mice. J. Neurosci. 19, 9900–9912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson J. C., Hatten M. E. (1987). Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J. Neurosci. 7, 1928–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis S. A., Shen X., Young J. B., Kaul P., Lerner D. J. (2006). Rho GEF Lsc is required for normal polarization, migration, and adhesion of formyl-peptide-stimulated neutrophils. Blood 107, 1627–1635 10.1182/blood-2005-03-1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fueshko S., Wray S. (1994). LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev. Biol. 166, 331–348 10.1006/dbio.1994.1319 [DOI] [PubMed] [Google Scholar]
- Fueshko S. M., Key S., Wray S. (1998a). GABA inhibits migration of luteinizing hormone-releasing hormone neurons in embryonic olfactory explants. J. Neurosci. 18, 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fueshko S. M., Key S., Wray S. (1998b). Luteinizing hormone releasing hormone (LHRH) neurons maintained in nasal explants decrease LHRH messenger ribonucleic acid levels after activation of GABA(A) receptors. Endocrinology 139, 2734–2740 10.1210/en.139.6.2734 [DOI] [PubMed] [Google Scholar]
- Gao C., Noden D. M., Norgren R. B., Jr (2000). LHRH neuronal migration: heterotypic transplantation analysis of guidance cues. J. Neurobiol. 42, 95–103 [DOI] [PubMed] [Google Scholar]
- Giacobini P., Messina A., Wray S., Giampietro C., Crepaldi T., Carmeliet P., Fasolo A. (2007). Hepatocyte growth factor acts as a motogen and guidance signal for gonadotropin hormone-releasing hormone-1 neuronal migration. J. Neurosci. 27, 431–445 10.1523/JNEUROSCI.4979-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greciano P. G., Moyano J. V., Buschmann M. M., Tang J., Lu Y., Rudnicki J., Manninen A., Matlin K. S. (2012). Laminin 511 partners with laminin 332 to mediate directional migration of Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell 23, 121–136 10.1091/mbc.E11-08-0718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon A., Nahon J. L. (2007). Multiple actions of the chemokine stromal cell-derived factor-1alpha on neuronal activity. J. Mol. Endocrinol. 38, 365–376 10.1677/JME-06-0013 [DOI] [PubMed] [Google Scholar]
- Guyon A., Banisadr G., Rovère C., Cervantes A., Kitabgi P., Melik–Parsadaniantz S., Nahon J. L. (2005). Complex effects of stromal cell-derived factor-1 alpha on melanin-concentrating hormone neuron excitability. Eur. J. Neurosci. 21, 701–710 10.1111/j.1460-9568.2005.03890.x [DOI] [PubMed] [Google Scholar]
- Hatten M. E. (2002). New directions in neuronal migration. Science 297, 1660–1663 10.1126/science.1074572 [DOI] [PubMed] [Google Scholar]
- Hu L., Wada K., Mores N., Krsmanovic L. Z., Catt K. J. (2006). Essential role of G protein-gated inwardly rectifying potassium channels in gonadotropin-induced regulation of GnRH neuronal firing and pulsatile neurosecretion. J. Biol. Chem. 281, 25231–25240 10.1074/jbc.M603768200 [DOI] [PubMed] [Google Scholar]
- Hutchins B. I., Li L. (2009). EphrinA and TrkB interact to promote axon branching. J. Neurosci. 29, 4329–4331 10.1523/JNEUROSCI.0238-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovane A., Aumas C., de Roux N. (2004). New insights in the genetics of isolated hypogonadotropic hypogonadism. Eur. J. Endocrinol. 151 Suppl. 3, U83–U88 10.1530/eje.0.151U083 [DOI] [PubMed] [Google Scholar]
- Klenke U., Taylor–Burds C. (2012) Culturing embryonic nasal explants for developmental and physiological study. Curr. Protoc. Neurosci. 3, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenke U., Constantin S., Wray S. (2010). Neuropeptide Y directly inhibits neuronal activity in a subpopulation of gonadotropin-releasing hormone-1 neurons via Y1 receptors. Endocrinology 151, 2736–2746 10.1210/en.2009-1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro H., Rakic P. (1995). Dynamics of granule cell migration: a confocal microscopic study in acute cerebellar slice preparations. J. Neurosci. 15, 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer P. R., Wray S. (2000). Midline nasal tissue influences nestin expression in nasal-placode-derived luteinizing hormone-releasing hormone neurons during development. Dev. Biol. 227, 343–357 10.1006/dbio.2000.9896 [DOI] [PubMed] [Google Scholar]
- Kusano K., Fueshko S., Gainer H., Wray S. (1995). Electrical and synaptic properties of embryonic luteinizing hormone-releasing hormone neurons in explant cultures. Proc. Natl. Acad. Sci. USA 92, 3918–3922 10.1073/pnas.92.9.3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M., Tiong J., Maddox D. M., Condie B. G., Wray S. (2008). Temporal migration of gonadotrophin-releasing hormone-1 neurones is modified in GAD67 knockout mice. J. Neuroendocrinol. 20, 93–103 10.1111/j.1365-2826.2007.01623.x [DOI] [PubMed] [Google Scholar]
- Lim Y. S., McLaughlin T., Sung T. C., Santiago A., Lee K. F., O’Leary D. D. (2008). p75(NTR) mediates ephrin-A reverse signaling required for axon repulsion and mapping. Neuron 59, 746–758 10.1016/j.neuron.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López–Bendito G., Sánchez–Alcañiz J. A., Pla R., Borrell V., Picó E., Valdeolmillos M., Marín O. (2008). Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 28, 1613–1624 10.1523/JNEUROSCI.4651-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C., Slesinger P. A. (2010). Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat. Rev. Neurosci. 11, 301–315 10.1038/nrn2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysko D. E., Putt M., Golden J. A. (2011). SDF1 regulates leading process branching and speed of migrating interneurons. J. Neurosci. 31, 1739–1745 10.1523/JNEUROSCI.3118-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacColl G., Bouloux P., Quinton R. (2002). Kallmann syndrome: adhesion, afferents, and anosmia. Neuron 34, 675–678 10.1016/S0896-6273(02)00720-1 [DOI] [PubMed] [Google Scholar]
- Marín O., Rubenstein J. L. (2003). Cell migration in the forebrain. Annu. Rev. Neurosci. 26, 441–483 [DOI] [PubMed] [Google Scholar]
- Marler K. J., Becker–Barroso E., Martínez A., Llovera M., Wentzel C., Poopalasundaram S., Hindges R., Soriano E., Comella J., Drescher U. (2008). A TrkB/EphrinA interaction controls retinal axon branching and synaptogenesis. J. Neurosci. 28, 12700–12712 10.1523/JNEUROSCI.1915-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin T., O’Leary D. D. (2005). Molecular gradients and development of retinotopic maps. Annu. Rev. Neurosci. 28, 327–355 10.1146/annurev.neuro.28.061604.135714 [DOI] [PubMed] [Google Scholar]
- Murakami S., Arai Y. (1994). Transient expression of somatostatin immunoreactivity in the olfactory-forebrain region in the chick embryo. Brain Res. Dev. Brain Res. 82, 277–285 10.1016/0165-3806(94)90169-4 [DOI] [PubMed] [Google Scholar]
- Nadarajah B., Brunstrom J. E., Grutzendler J., Wong R. O., Pearlman A. L. (2001). Two modes of radial migration in early development of the cerebral cortex. Nat. Neurosci. 4, 143–150 10.1038/83967 [DOI] [PubMed] [Google Scholar]
- O’Rourke N. A., Dailey M. E., Smith S. J., McConnell S. K. (1992). Diverse migratory pathways in the developing cerebral cortex. Science 258, 299–302 10.1126/science.1411527 [DOI] [PubMed] [Google Scholar]
- Odemis V., Boosmann K., Heinen A., Küry P., Engele J. (2010). CXCR7 is an active component of SDF-1 signalling in astrocytes and Schwann cells. J. Cell Sci. 123, 1081–1088 10.1242/jcs.062810 [DOI] [PubMed] [Google Scholar]
- Oh S. B., Endoh T., Simen A. A., Ren D., Miller R. J. (2002). Regulation of calcium currents by chemokines and their receptors. J. Neuroimmunol. 123, 66–75 10.1016/S0165-5728(01)00485-4 [DOI] [PubMed] [Google Scholar]
- Oh W. J., Westmoreland J. J., Summers R., Condie B. G. (2010). Cleft palate is caused by CNS dysfunction in Gad1 and Viaat knockout mice. PLoS ONE 5, e9758 10.1371/journal.pone.0009758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled A., Wald O., Burger J. (2012). Development of novel CXCR4-based therapeutics. Expert Opin. Investig. Drugs 21, 341–353 10.1517/13543784.2012.656197 [DOI] [PubMed] [Google Scholar]
- Poluch S., Jablonska B., Juliano S. L. (2008). Alteration of interneuron migration in a ferret model of cortical dysplasia. Cereb. Cortex 18, 78–92 10.1093/cercor/bhm032 [DOI] [PubMed] [Google Scholar]
- Pujol F., Kitabgi P., Boudin H. (2005). The chemokine SDF-1 differentially regulates axonal elongation and branching in hippocampal neurons. J. Cell Sci. 118, 1071–1080 10.1242/jcs.01694 [DOI] [PubMed] [Google Scholar]
- Rakic P. (1990). Principles of neural cell migration. Experientia 46, 882–891 10.1007/BF01939380 [DOI] [PubMed] [Google Scholar]
- Rashid T., Upton A. L., Blentic A., Ciossek T., Knöll B., Thompson I. D., Drescher U. (2005). Opposing gradients of ephrin-As and EphA7 in the superior colliculus are essential for topographic mapping in the mammalian visual system. Neuron 47, 57–69 10.1016/j.neuron.2005.05.030 [DOI] [PubMed] [Google Scholar]
- Represa A., Ben–Ari Y. (2005). Trophic actions of GABA on neuronal development. Trends Neurosci. 28, 278–283 10.1016/j.tins.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Rugarli E. I. (1999). Kallmann syndrome and the link between olfactory and reproductive development. Am. J. Hum. Genet. 65, 943–948 10.1086/302600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez–Alcañiz J. A., Haege S., Mueller W., Pla R., Mackay F., Schulz S., López–Bendito G., Stumm R., Marín O. (2011). Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron 69, 77–90 10.1016/j.neuron.2010.12.006 [DOI] [PubMed] [Google Scholar]
- Schwanzel–Fukuda M., Pfaff D. W. (1989). Origin of luteinizing hormone-releasing hormone neurons. Nature 338, 161–164 10.1038/338161a0 [DOI] [PubMed] [Google Scholar]
- Schwarting G. A., Raitcheva D., Bless E. P., Ackerman S. L., Tobet S. (2004). Netrin 1-mediated chemoattraction regulates the migratory pathway of LHRH neurons. Eur. J. Neurosci. 19, 11–20 10.1111/j.1460-9568.2004.03094.x [DOI] [PubMed] [Google Scholar]
- Schwarting G. A., Henion T. R., Nugent J. D., Caplan B., Tobet S. (2006). Stromal cell-derived factor-1 (chemokine C-X-C motif ligand 12) and chemokine C-X-C motif receptor 4 are required for migration of gonadotropin-releasing hormone neurons to the forebrain. J. Neurosci. 26, 6834–6840 10.1523/JNEUROSCI.1728-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel J., Niggemann B., Punzel M., Fischer J., Zänker K. S., Dittmar T. (2007). The neurotransmitter GABA is a potent inhibitor of the stromal cell-derived factor-1alpha induced migration of adult CD133+ hematopoietic stem and progenitor cells. Stem Cells Dev. 16, 827–836 10.1089/scd.2007.0004 [DOI] [PubMed] [Google Scholar]
- Simen A. A., Lee C. C., Simen B. B., Bindokas V. P., Miller R. J. (2001). The C terminus of the Ca channel alpha1B subunit mediates selective inhibition by G-protein-coupled receptors. J. Neurosci. 21, 7587–7597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiveron M. C., Cremer H. (2008). CXCL12/CXCR4 signalling in neuronal cell migration. Curr. Opin. Neurobiol. 18, 237–244 10.1016/j.conb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Tiveron M. C., Boutin C., Daou P., Moepps B., Cremer H. (2010). Expression and function of CXCR7 in the mouse forebrain. J. Neuroimmunol. 224, 72–79 10.1016/j.jneuroim.2010.05.011 [DOI] [PubMed] [Google Scholar]
- Toba Y., Pakiam J. G., Wray S. (2005). Voltage-gated calcium channels in developing GnRH-1 neuronal system in the mouse. Eur. J. Neurosci. 22, 79–92 10.1111/j.1460-9568.2005.04194.x [DOI] [PubMed] [Google Scholar]
- Toba Y., Tiong J. D., Ma Q., Wray S. (2008). CXCR4/SDF-1 system modulates development of GnRH-1 neurons and the olfactory system. Dev. Neurobiol. 68, 487–503 10.1002/dneu.20594 [DOI] [PubMed] [Google Scholar]
- Tobet S. A., Chickering T. W., King J. C., Stopa E. G., Kim K., Kuo–Leblank V., Schwarting G. A. (1996). Expression of gamma-aminobutyric acid and gonadotropin-releasing hormone during neuronal migration through the olfactory system. Endocrinology 137, 5415–5420 10.1210/en.137.12.5415 [DOI] [PubMed] [Google Scholar]
- Tran P. B., Miller R. J. (2003). Chemokine receptors: signposts to brain development and disease. Nat. Rev. Neurosci. 4, 444–455 10.1038/nrn1116 [DOI] [PubMed] [Google Scholar]
- Wang H., Olsen R. W. (2000). Binding of the GABA(A) receptor-associated protein (GABARAP) to microtubules and microfilaments suggests involvement of the cytoskeleton in GABARAPGABA(A) receptor interaction. J. Neurochem. 75, 644–655 10.1046/j.1471-4159.2000.0750644.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Li G., Stanco A., Long J. E., Crawford D., Potter G. B., Pleasure S. J., Behrens T., Rubenstein J. L. (2011). CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron 69, 61–76 10.1016/j.neuron.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciak–Stothard B., Ridley A. J. (2003). Shear stress-induced endothelial cell polarization is mediated by Rho and Rac but not Cdc42 or PI 3-kinases. J. Cell Biol. 161, 429–439 10.1083/jcb.200210135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S. (2002). Molecular mechanisms for migration of placodally derived GnRH neurons. Chem. Senses 27, 569–572 10.1093/chemse/27.6.569 [DOI] [PubMed] [Google Scholar]
- Wray S., Gähwiler B. H., Gainer H. (1988). Slice cultures of LHRH neurons in the presence and absence of brainstem and pituitary. Peptides 9, 1151–1175 10.1016/0196-9781(88)90103-9 [DOI] [PubMed] [Google Scholar]
- Wray S., Nieburgs A., Elkabes S. (1989). Spatiotemporal cell expression of luteinizing hormone-releasing hormone in the prenatal mouse: evidence for an embryonic origin in the olfactory placode. Brain Res. Dev. Brain Res. 46, 309–318 10.1016/0165-3806(89)90295-2 [DOI] [PubMed] [Google Scholar]
- Wray S., Key S., Qualls R., Fueshko S. M. (1994). A subset of peripherin positive olfactory axons delineates the luteinizing hormone releasing hormone neuronal migratory pathway in developing mouse. Dev. Biol. 166, 349–354 10.1006/dbio.1994.1320 [DOI] [PubMed] [Google Scholar]
- Wray S., Fueshko S. M., Kusano K., Gainer H.1996). GABAergic neurons in the embryonic olfactory pit/vomeronasal organ: maintenance of functional GABAergic synapses in olfactory explants. Dev. Biol. 180631–645 10.1006/dbio.1996.0334 [DOI] [PubMed] [Google Scholar]
- Yamada M., Inanobe A., Kurachi Y. (1998). G protein regulation of potassium ion channels. Pharmacol. Rev. 50, 723–760 [PubMed] [Google Scholar]
- Zhang C., Atasoy D., Araç D., Yang X., Fucillo M. V., Robison A. J., Ko J., Brunger A. T., Südhof T. C. (2010). Neurexins physically and functionally interact with GABA(A) receptors. Neuron 66, 403–416 10.1016/j.neuron.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]