Summary
The impact of aberrant centrosomes and/or spindles on asymmetric cell division in embryonic development indicates the tight regulation of bipolar spindle formation and positioning that is required for mitotic progression and cell fate determination. WD40-repeat protein 62 (WDR62) was recently identified as a spindle pole protein linked to the neurodevelopmental defect of microcephaly but its roles in mitosis have not been defined. We report here that the in utero electroporation of neuroprogenitor cells with WDR62 siRNAs induced their cell cycle exit and reduced their proliferative capacity. In cultured cells, we demonstrated cell-cycle-dependent accumulation of WDR62 at the spindle pole during mitotic entry that persisted until metaphase–anaphase transition. Utilizing siRNA depletion, we revealed WDR62 function in stabilizing the mitotic spindle specifically during metaphase. WDR62 loss resulted in spindle orientation defects, decreased the integrity of centrosomes displaced from the spindle pole and delayed mitotic progression. Additionally, we revealed JNK phosphorylation of WDR62 is required for maintaining metaphase spindle organization during mitosis. Our study provides the first functional characterization of WDR62 and has revealed requirements for JNK/WDR62 signaling in mitotic spindle regulation that may be involved in coordinating neurogenesis.
Key words: Centrosome, Spindle pole, WD-repeat proteins, Mitosis, Cell division, Neurogenesis
Introduction
The bipolar spindle is a highly organized microtubule-based superstructure with essential functions in faithful sister chromatid segregation during cell division (Compton, 2000). During mitosis, the minus ends of spindle microtubules in anti-parallel arrangements are focused at the poles and anchored at centrosomes. Centrosomes consist of paired centrioles surrounded by proteinaceous pericentriolar material (PCM). In the early stages of mitosis, the separation of duplicated centrosomes is accompanied by a considerable expansion of the PCM via the recruitment of γ-tubulin- and γ-tubulin-associated proteins (e.g. NEDD1, pericentrin and cdk5rap2) that facilitates astral microtubule nucleation and bipolar spindle assembly (Fong et al., 2008; Haren et al., 2009; Lüders et al., 2006; Oshimori et al., 2009; Zimmerman et al., 2004). Moreover, MTOC activity and proper positioning of centrosomes are required to specify the plane of division and dictate precise cellular segregation into daughter cells (Lesage et al., 2010; Megraw et al., 2011). Importantly, defects in centrosome-directed functions result in aberrant bipolar spindle formation, cell cycle progression and are linked to human disease (Nigg and Raff, 2009; Zyss and Gergely, 2009). Proteins with critical roles in centrosome/spindle regulation have been revealed by genetic studies linking centrosomal protein mutations with autosomal recessive primary microcephaly (MCPH) in which defects in neuronal progenitor cell division are regarded as underlying disease development (Thornton and Woods, 2009).
WD40-repeat protein 62 (WDR62) was first characterized as involved in the assembly of signaling complexes associated with cytoplasmic stress granule formation (Wasserman et al., 2010). More recently, WDR62 gene mutations were linked to MCPH and more severe brain malformations, thus implicating critical contributions by WDR62 to cortical development (Bilgüvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010). WDR62 is 170 kDa protein characterized by 13 annotated WD40 domain repeats that span the N-terminal half of the protein (Wasserman et al., 2010). WD40 repeat proteins facilitate protein–protein interactions and are involved in large protein complex formation (Stirnimann et al., 2010). WDR62 binds components of the c-Jun N-terminal kinase (JNK) pathway to potentiate stress-stimulated signal transduction (Cohen-Katsenelson et al., 2011; Wasserman et al., 2010). The observed diverse intracellular distribution of WDR62 suggests pleiotropic functions that may be dependent on cellular context (Bilgüvar et al., 2010; Nicholas et al., 2010; Wasserman et al., 2010). For example, WDR62 is localized to stress granules in response to cell stress (Wasserman et al., 2010). In post-mitotic neurons WDR62 is localized to the nucleus, whilst in neuronal progenitors undergoing mitosis, WDR62 is present at centrosomes/spindle poles (Bilgüvar et al., 2010; Nicholas et al., 2010). Global proteomic analyses also identified WDR62 as a mitotically regulated protein (Dephoure et al., 2008; Santamaria et al., 2011). Although these observations are consistent with a cell cycle regulatory function that may be indispensable for cell divisions associated with neurogenesis, the precise contributions of WDR62 in cell cycle regulation are unknown.
In this study, we have shown for the first time that WDR62 depletion with siRNA resulted in reduced cell proliferation in the developing embryonic mouse brain. Exploiting human cell cultures to define underlying biochemical mechanistic links, we revealed WDR62 to be a mitotic phosphoprotein localized to spindle poles from prophase to metaphase in a process that requires microtubule-dependent transport. Importantly, WDR62 was required for proper progression through mitosis and its depletion led to spindle orientation defects, metaphase spindle abnormalities, centrosome–spindle uncoupling and reduced centrosome integrity. Furthermore, we demonstrated that WDR62 phosphorylation by JNK in mitosis was involved in the regulation of metaphase spindle architecture. Our studies provide the first functional analyses of WDR62 in neurogenesis, centrosome/spindle organization and cell cycle regulation with important implications for centrosome-associated pathologies characterized by microcephaly.
Results
WDR62 knockdown in vivo results in reduced proliferation of neuroprogenitors
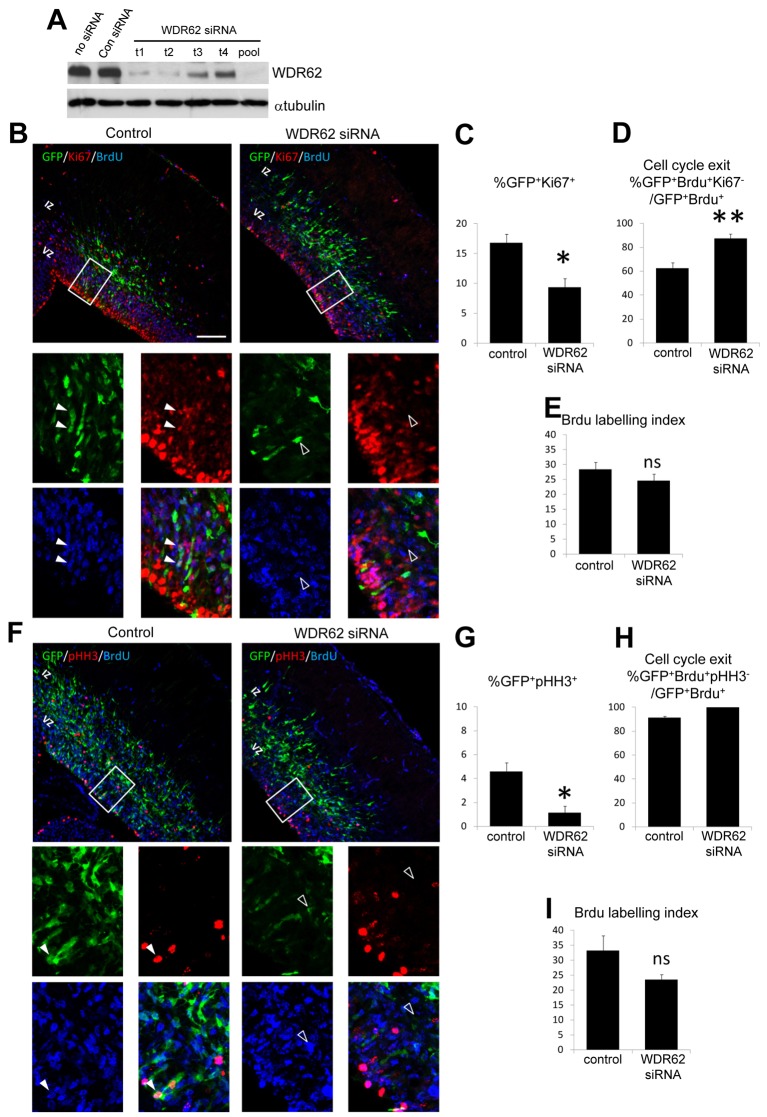
WDR62 was recently identified as the second most commonly mutated gene linked to primary microcephaly or microcephaly accompanied by severe cortical malformations (Bilgüvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010) albeit that its functions during brain development are unknown. The detection of WDR62 in neural precursors of the developing cerebral cortex (Nicholas et al., 2010) suggests its importance in regulating neuroprogenitor cell cycle progression. To investigate this, we performed in utero electroporation of embryonic mouse (E14) brain to co-introduce a GFP expression construct together with experimentally validated WDR62 siRNAs or non-targeting control siRNAs (Fig. 1A). We then examined the proliferative properties of the cortical progenitor cells. Twenty-four hours post-electroporation, a single dose of BrdU was administered to label cells undergoing S-phase DNA replication before the embryonic brains were finally harvested at 48 h (E16) to identify progenitor cells expressing Ki67 or phospho-histone H3 (pHH3) as markers of actively cycling and mitotic cells, respectively (Fig. 1).
Fig. 1.
WDR62 depletion decreased cell proliferation within the embryonic cortex. (A) Neuro2a cells were transfected with individual mouse WDR62-targeting siRNA (t1, t2, t3 and t4), a combined mouse WDR62 siRNA pool, non-targeting siRNA (Con siRNA) or not treated with siRNA (no siRNA) and immunoblotted for WDR62 and α-tubulin. (B) Images of the coronal sections of embryonic mouse cerebral cortex electroporated with control or mouse WDR62 siRNA, together with GFP to label electroporated cells. Embryos were pulse labeled with BrdU 24 h post-electroporation, and brain sections finally stained for Ki67 to identify actively proliferating cells at time of harvest (48 h post-electroporation). Magnified panels (below) of the ventricular zone (VZ) highlight proliferating (GFP+/BrdU+/Ki67+, filled arrowheads) and non-proliferating cells (GFP+/BrdU+/Ki67−, open arrowheads). (C) Quantification of cell proliferation (GFP+/Ki67+) in brain sections from control and mouse WDR62-siRNA-treated embryos. (D) The cell cycle exit index in response to WDR62 depletion was determined by counting GFP+/BrdU+/Ki67− cells and expressing this as a proportion of GFP+/BrdU+ cells. (E) BrdU labeling index in control and WDR62-siRNA-treated brain sections used in the analysis depicted in B. (F) Coronal sections of embryos electroporated with control or mouse WDR62 siRNA were stained for phospho-histone H3 (pHH3). Magnified panels (below) of the VZ highlight cells in M phase (GFP+/BrdU+/pHH3+, filled arrowheads) and non-mitotic cells (GFP+/BrdU+/pHH3−, open arrowheads). (G) The mitotic index in response to WDR62 depletion was determined by the proportion of cells double labeled for GFP and pHH3 (GFP+/pHH3+). (H) A cell cycle exit index analysis performed with pHH3 and BrdU immunostaining was determined by counting GFP+/BrdU+/pHH3− cells within the population of GFP+/BrdU+ cells (statistical analysis could not be performed because 100% of cells were GFP+BrdU+pHH3− following WDR62 siRNA treatment). (I) BrdU labeling index in control and WDR62-siRNA-treated brain sections used in the analysis depicted in (F). Scale bar: 100 µm, while all graphs plot means ± s.e.m. (*P<0.05, **P<0.01, ns denotes not statistically significant).
We revealed that the treatment of cortical progenitors with WDR62 siRNA in vivo caused a significant decrease in Ki67 marker expression compared with control siRNA treatment (Fig. 1B,C). Moreover, WDR62 knockdown reduced the proliferative capacity of the progenitor compartment, as judged by a significant increase in the cell cycle exit index [defined as the fraction of GFP+BrdU+ cells which do not immunostain for Ki67 versus all GFP+BrdU+ cells (GFP+BrdU+Ki67−/GFP+BrdU+)] compared with control treatment (Fig. 1D). Similarly, we found a significant decrease in the fraction of WDR62-siRNA-treated cells that express the mitotic marker pHH3 (Fig. 1F,G), as well as a concomitant increase in their cell cycle exit (Fig. 1H). Importantly, the incorporation of BrdU by cortical cells was not significantly affected by siRNA treatment (Fig. 1E,I), and we excluded apoptosis as a contributing factor in our analysis by showing no differences in cleaved caspase 3 staining in the different cell populations (data not shown). Thus, the depletion of WDR62 in vivo resulted in reduced proliferation and enhanced cell cycle exit indicative of altered neuroprogenitor self-renewal during cortical development.
Cell-cycle-dependent WDR62 localization as a spindle pole protein
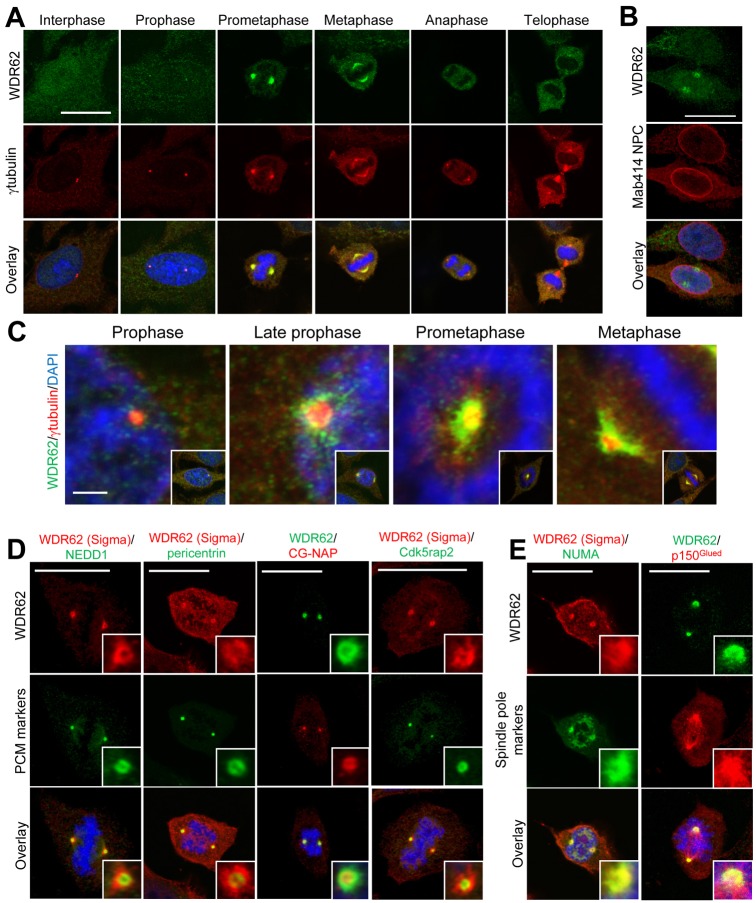
A requirement for WDR62 in sustaining the proliferative capacity of neuroprogenitors suggests important cell cycle regulatory functions and is consistent with the shared centrosome association observed for the different MCPH proteins. In addition, the multi-compartment localization of WDR62 (Bilgüvar et al., 2010; Nicholas et al., 2010; Wasserman et al., 2010; Yu et al., 2010) prompted our analysis of the intracellular distribution of endogenous WDR62 throughout the cell cycle. As three commercially available antibodies detected human but not murine WDR62 protein by immunofluorescence, we analyzed human cell cultures. Immunofluorescence analysis in HeLa cells using antibodies recognizing distinct WDR62 epitopes consistently revealed a cytosolic and nuclear distribution of WDR62 during interphase (Fig. 2A; supplementary material Fig. S1A,B). During interphase, WDR62 was only weakly associated with centrosomes marked by γ-tubulin co-staining (supplementary material Fig. S1A). In contrast, we observed prominent WDR62 colocalization with γ-tubulin in mitotic cells indicating centrosome/spindle accumulation of WDR62 under these conditions (Fig. 2A). All three WDR62 antibodies consistently revealed dynamic WDR62 association with centrosomes in mitotic cells emphasizing the specificity of this localization (Fig. 2A; supplementary material Fig. S1A,B). With staining for nuclear pore complex proteins, we demonstrated that the initial stages of WDR62 centrosomal accumulation during prophase preceded nuclear envelope breakdown (Fig. 2B). Further higher magnification imaging and detailed dissection of the kinetics of WDR62 centrosomal association revealed WDR62 accumulation around γ-tubulin-stained centrosomes beginning at prophase (Fig. 2C). This culminated in prominent WDR62 localization at the spindle pole that enveloped centrosomes during metaphase (Fig. 2C). In contrast, during anaphase and telophase, WDR62 no longer showed obvious centrosomal association (Fig. 2A). These results highlight a cell-cycle-dependent distribution of WDR62 to the spindle pole.
Fig. 2.
Accumulation of WDR62 at spindle poles during mitosis. (A) WDR62 subcellular localization in HeLa cells at different mitotic stages. DAPI-stained condensed chromatin and γ-tubulin immunostaining revealed centrosomes and spindle poles. Scale bar: 20 µm. (B) WDR62 association with centrosomes occurred prior to nuclear envelope breakdown as demonstrated by co-staining with nuclear pore complex proteins (Mab414 NPC). Scale bar: 20 µm. (C) Higher magnification images of WDR62 localization around γ-tubulin-stained centrosomes from prophase to metaphase. Scale bar: 2 µm. Insets are lower magnification images showing entire cell and mitotic stage. (D) Co-staining of WDR62 with PCM markers (NEDD1, pericentrin, CG-NAP and Cdk5rap2). (E) WDR62 co-localization with spindle pole markers (NUMA and p150Glued). Insets in D and E are high magnification images of centrosomes. Scale bars: 20 µm.
To extend this analysis of mitotic WDR62 distribution, we evaluated WDR62 colocalization with other key centrosomal proteins. During prometaphase/metaphase, WDR62 surrounded but did not show strict colocalization with the PCM proteins NEDD-1, pericentrin, centrosome and Golgi-localized PKN-associated protein (CG-NAP) or cdk5 regulatory subunit associated protein 2 (cdk5rap2; Fig. 2D). In contrast, WDR62 colocalized with the microtubule cross-linker, nuclear mitotic apparatus (NUMA) protein, and the dynactin complex component, p150Glued, at spindle poles (Fig. 2E). Taken together, these results identify WDR62 as a prominent spindle pole protein with dynamic localization that peaks during prometaphase/metaphase.
WDR62 depletion delays completion of mitosis
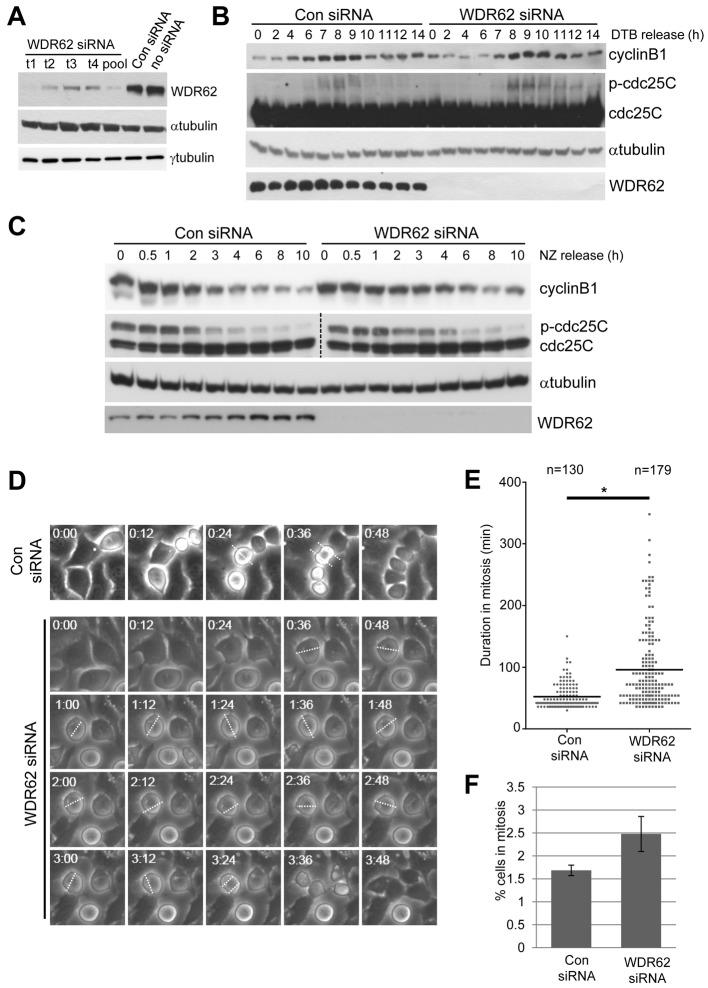
To define the potential roles of WDR62 in cell division, as suggested by its spatial and temporal regulation during mitosis, we depleted WDR62 in HeLa cells with siRNA. Four individual siRNAs all substantially silenced WDR62 when compared to a non-targeting Con siRNA whereas levels of α- and γ-tubulin were unchanged (Fig. 3A), and depletion was sustained for at least 72 h post-transfection (supplementary material Fig. S2A). All subsequent findings utilized WDR62 depletion by WDR62 target 1 siRNA but were re-confirmed with at least two other independent siRNAs (typically WDR62 target 2 and target 3 siRNA) to ensure specificity of the knockdown phenotype. Interestingly, proliferation and cell viability were not markedly altered despite substantially reduced WDR62 levels (supplementary material Fig. S2B,C). Thus, WDR62 loss did not induce cell cycle arrest or prevent division in HeLa cells. In contrast, we found that WDR62 depletion altered the duration of mitosis.
Fig. 3.
WDR62 depletion perturbs mitotic progression. (A) HeLa cells were transiently transfected with individual human WDR62-targeting siRNA (t1, t2, t3 and t4), a WDR62 siRNA pool, non-targeting siRNA (Con siRNA) or not treated with siRNA (no siRNA) and immunoblotted for WDR62. α- and γ-tubulin were blotted as controls. (B) HeLa cells transfected with WDR62 or Con siRNA were synchronized at G1/S (DTB). Cell cycle progression following thymidine release (2–14 h as indicated) was determined by immunoblotting for cyclin B1 and cdc25C phosphorylation. (C) HeLa cells transfected with WDR62 or Con siRNA were synchronized at M-phase [nocodazole (NZ) treated; 350 nM, 16 h]. Mitotic progression following NZ release (0.5–10 h as indicated) was determined by immunoblotting for cyclin B1 and phospho-cdc25C. α-tubulin was blotted for protein loading and WDR62 levels determined to confirm knockdown. (D) HeLa cells, treated with Con siRNA or WDR62 siRNA, were synchronized (DTB) and imaged under phase-contrast optics at 6-min intervals from 7 h post thymidine release. Representative images of mitotic progression are shown. White dotted lines indicate the position of the metaphase plate. (E) Duration of mitotis in WDR62-depleted and control HeLa cells, measured from nuclear envelope breakdown until two daughter cells were observed, are shown on a vertical scatter plot. Horizontal lines indicate mean values and n values are the total cells counted from three independent experiments (*P<0.01). (F) WDR62 was depleted in HeLa cells for 48 h and the proportion of cells in M phase identified by DAPI staining. Values are means ± s.e.m. from three independent experiments.
When the effect of WDR62 loss was evaluated in synchronized HeLa cells, we observed delayed mitotic progression following double thymidine block (DTB) release. Levels of cyclin B1 and phosphorylated cdc25C are typically elevated during mitotic entry and persist until the metaphase–anaphase transition (Hutchins and Clarke, 2004; Takizawa and Morgan, 2000), and so we used these as indicators of cell cycle progression. In WDR62-siRNA-treated cells cyclin B1 degradation and dephosphorylation of cdc25C were delayed, extending to 11–12 h post-release compared to the return of these mitotic markers to interphase levels by 10 h post-release in Con-siRNA-treated cells (Fig. 3B). Similarly, in cells synchronized by nocodazole treatment, elevated cyclinB1 and phospho-cdc25C levels were extended to 6 h post-release from nocodazole compared to the loss of these mitotic markers by 4 h post-nocodazole release in Con-siRNA-treated cells (Fig. 3C). Time-lapse imaging of WDR62-siRNA-treated cells further showed that the duration of mitosis was prolonged (Fig. 3D,E) with WDR62-siRNA-treated HeLa cells taking on average nearly twice as long (96 min) to complete mitosis when compared with Con-siRNA-treated cells (54 min; Fig. 3E). Some WDR62-depleted cells took >200 min (approximately four times average duration of control cells) to complete mitosis (Fig. 3E) without cell cycle arrest. Furthermore, substantial movement and rotation of the metaphase plate was observed suggestive of spindle orientation defects during periods of prolonged mitosis (Fig. 3D, broken white lines). Reflecting this mitotic delay, a modest increase in mitotic index in WDR62-siRNA-treated cells compared to controls was observed (Fig. 3F). Thus, knockdown of WDR62 delays the completion of mitosis indicating a requirement for WDR62 in regulating timely mitotic progression.
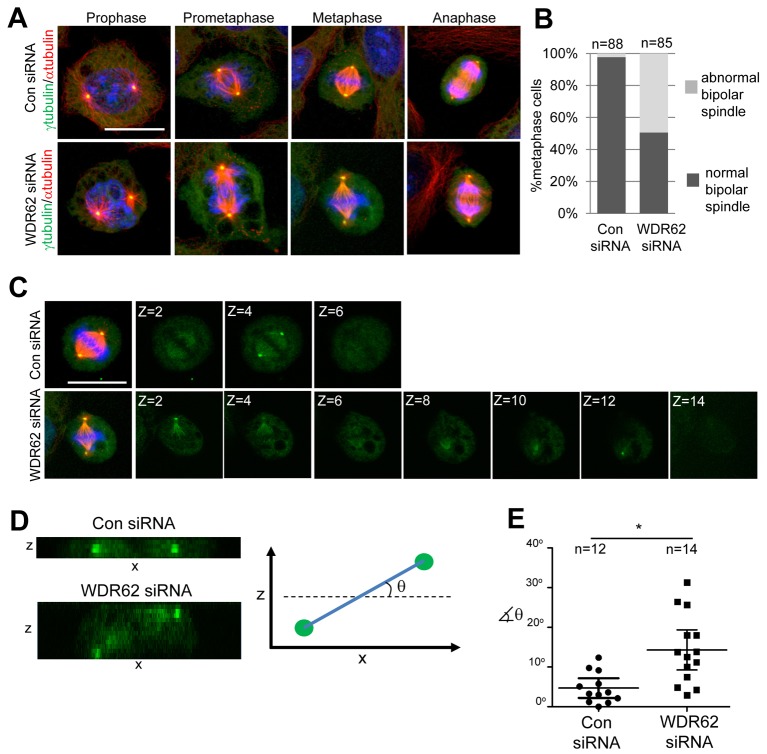
Metaphase spindle defects with WDR62 knockdown
To interrogate the features underlying delayed mitotic progression and spindle orientation defects, we examined the changes in mitotic spindle organization as a result of WDR62 loss. On closer examination of spindle assembly in prophase and prometaphase, intact centrosomes anchoring astral microtubules (as noted by α- and γ-tubulin localizations) were observed for both control and WDR62-depleted cells (Fig. 4A). Strikingly, WDR62 depletion resulted in abnormal metaphase spindles characterized by a displacement of centrosomes from the spindle (Fig. 4A). Quantitation of the metaphase cell population indicated that approximately half of WDR62-depleted cells exhibited an abnormal bipolar spindle (Fig. 4B). In addition, while metaphase spindles in cultured cells commonly attain a planar orientation, with the division plane parallel to surface of the culture dish, the metaphase spindle polarity axis in WDR62-depleted cells exhibited substantial rotation in the z-axis (Fig. 4C). The rotation of the spindle in the z-axis was more clearly seen through orthogonal sectioning through the z-stacks (Fig. 4D) and our measurements of the angle of rotation from planar orientation showed a greater degree of spindle rotation in the z-axis in WDR62-depleted cells compared to controls (Fig. 4E). The localization of NUMA and dynein/dynactin motors at spindle poles and the cell cortex are critical for pole focusing activities and the cortical capture of astral microtubules for spindle positioning, respectively (Radulescu and Cleveland, 2010). However, the cortical and spindle pole localization of NUMA and p150Glued was unaltered by WDR62 loss (supplementary material Fig. S3). Thus, mechanisms involved in spindle pole focusing and the cortical capture of astral microtubules for spindle positioning appeared largely intact. Rather, our results highlight a metaphase specific role for spindle localized WDR62 in stabilizing or maintaining spindle pole integrity to establish a polarity axis for cell division.
Fig. 4.
WDR62 is required for metaphase spindle maintenance and positioning. (A) HeLa cells were transfected with WDR62 or Con siRNA and synchronized (DTB). Following thymidine release (8–10 h), cells were fixed and stained for α-tubulin/γ-tubulin. Representative cells at different mitotic stages are shown. (B) The proportion of metaphase cells with abnormal spindles characterized by displaced centrosomes was quantified. (C) Individual z-stacks of γ-tubulin staining in WDR62-depleted or control HeLa cells. Overlay images show maximum intensity projections of z-stacks. All scale bars: 20 µm. (D) Orthogonal slice along spindle polarity axis viewed from a side-on perspective of γ-tubulin immunostained z-sections reveals spindle rotation in z-axis. ‘θ’ is the angle of rotation from planar orientation. (E) The angle of spindle rotation was measured in WDR62- or Con-siRNA-treated HeLa cells and plotted on a vertical scatter plot. Horizontal lines depict means ± s.e.m. and n values are total number of cells counted from two independent experiments (*P<0.05).
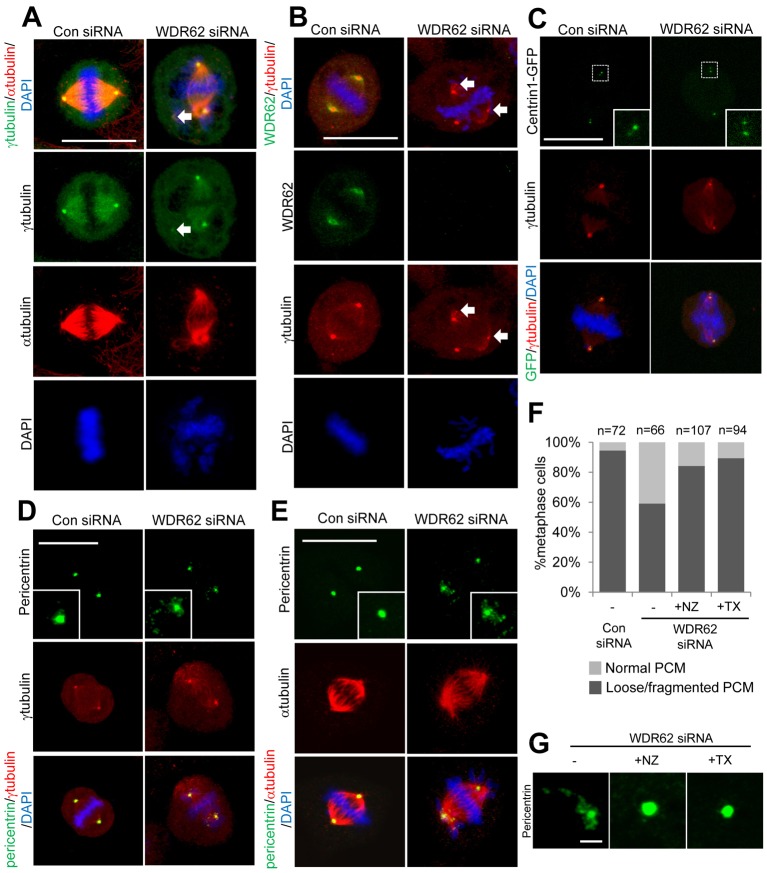
Centrosome integrity and coupling to spindle poles are compromised with WDR62 loss
Our closer interrogation of abnormal metaphase spindles in WDR62-depleted cells revealed that apolar centrosomes remained juxtaposed to the spindle pole and retained microtubule attachment to a spindle pole (Fig. 5A). Strikingly, we observed multiple γ-tubulin puncta in WDR62-depleted cells (Fig. 5A,B, arrows). When we investigated centriole pairs, visualized by GFP-tagged centrin1 expression, we found these to be intact in displaced centrosomes (Fig. 5C). Thus, there did not appear to be a loss of centriole cohesion with WDR62 loss. Staining for pericentrin in WDR62-depleted cells revealed loose and disrupted PCM of displaced centrosomes that co-localized with γ-tubulin puncta and which was suggestive of reduced centrosome integrity (Fig. 5D,E). Furthermore, the extent of PCM fragmentation was reduced in the presence of low-dose nocodazole or Taxol (Fig. 5F,G) indicating microtubule dynamics as a contributing factor to the reduced centrosome integrity in WDR62-depleted cells. Furthermore, with DAPI staining we noted the presence of unaligned chromosomal DNA in WDR62-siRNA-treated cells during metaphase (Fig. 5A–E). Our quantitative analysis, demonstrating a substantial increase in the proportion of cells with chromosomes not aligned at the metaphase plate following WDR62 knockdown (supplementary material Fig. S4A), was consistent with delayed mitotic progression and disrupted centrosome/bipolar spindle organization.
Fig. 5.
Centrosome integrity is reduced with WDR62 depletion. WDR62-depleted or control HeLa cells in metaphase were stained with (A) α- and γ-tubulin or (B) WDR62 and γ-tubulin. Arrows indicate γ-tubulin puncta. DAPI staining of DNA is shown in overlay images. (C) HeLa cells were transfected with WDR62 or Con siRNA together with Centrin1–GFP to label centrioles in addition to staining for γ-tubulin. (D,E) WDR62-depleted or control HeLa cells in metaphase were stained with pericentrin and (D) γ-tubulin or (E) α-tubulin. Insets in D,E are higher magnification images of the centrosomal region. All scale bars: 20 µm. (F) WDR62-depleted cells were synchronized (DTB) and treated with nocodazole (NZ, 5 nM) or Taxol (TX, 5 nM) at 7 h post-thymidine release. The number of cells in metaphase with loose/fragmented PCM, as revealed by pericentrin staining, was quantified and expressed as a proportion of total metaphase cells. Con siRNA treatment was included for comparison. n values are the total number of cells counted from three independent experiments. (G) Representative images of pericentrin staining in WDR62-depleted cells treated with nocodazole or Taxol.
With our observations of defects in centrosome–spindle-pole coupling, we also measured spindle length in WDR62-depleted cells and found this to be significantly increased relative to control cells (supplementary material Fig. S4B). This reinforces the recent appreciation of the relationship between proper centrosome attachment to the spindle pole and spindle length determination (Dumont and Mitchison, 2009; Silk et al., 2009). WDR62 depletion also increased the frequency of multipolar spindles (15% versus 3%, supplementary material Fig. S4C). However, we did not observe differences in interphase centrosome numbers in WDR62-depleted cells synchronized at late G2 when compared to controls (supplementary material Fig. S4D). Thus, centrosome duplication and cohesion appeared to not be overtly perturbed. Our studies show spindle defects following WDR62 loss were typified during metaphase by displaced centrosomes with reduced integrity. This was also associated with inefficient chromosome congression and elongated spindle length. Thus, WDR62 is required to maintain metaphase spindle architecture.
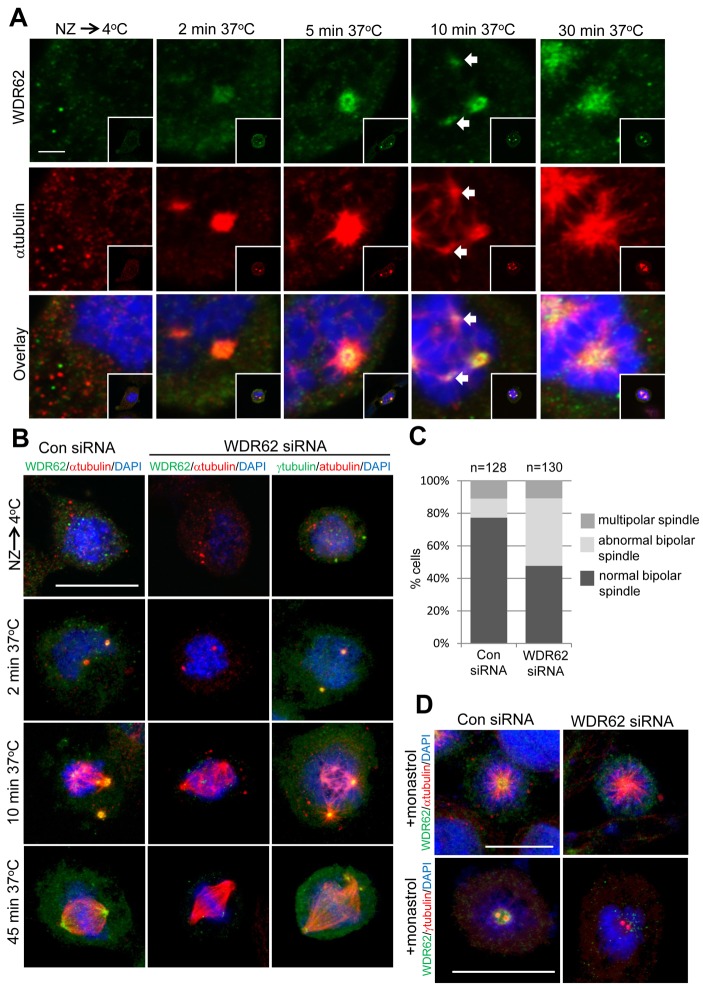
WDR62 is required for metaphase spindle maintenance but not bipolar spindle formation
WDR62 spindle pole localization during early mitotic stages parallels centrosome maturation and the enhanced nucleation of astral microtubules that facilitate formation of a bipolar spindle. To identify WDR62 functions in microtubule nucleation and bipolar spindle formation, we employed microtubule regrowth assays. WDR62 centrosomal localization was lost following microtubule depolymerization by cold treatment and was rapidly returned (<2 min) during astral microtubule regrowth (Fig. 6A). This further reinforced WDR62 as a microtubule-associated spindle pole protein. Surprisingly, at 10 min regrowth, WDR62 was also localized to regions of acentrosomal microtubule formation originating from mitotic chromatin. We confirmed WDR62 localization to calcium-resistant kinetochore microtubules (supplementary material Fig. S5A). WDR62 localization then coalesced at the poles as spindle bipolarity was re-established at 30 min (Fig. 6A). WDR62 was therefore associated with sites of microtubule nucleation at both the centrosomes and chromosomes.
Fig. 6.
WDR62 is required for spindle/centrosome maintenance after spindle bipolarity is established. (A) G1/S synchronized HeLa cells (DTB) were released into nocodazole (350 nM) before microtubules were depolymerized at 4°C (30 min). Following nocodazole washout, cells were incubated at 37°C for various time intervals before staining for WDR62 and microtubule regrowth with α-tubulin. Scale bar: 2 µm. (B) Microtubule regrowth in WDR62-depleted HeLa cells was analyzed as in A and compared to control (Con siRNA) cells. In addition, centrosomes were revealed by γ-tubulin co-staining. Scale bar: 20 µm. (C) Following MT regrowth (45 mins, 37°C) the proportion of cells with normal, abnormal or multi-polar spindles was determined. n values are total number of cells counted from three independent experiments. (D) HeLa cells synchronized to G1/S (DTB) were released into monastrol (100 µM) before fixing and immunostaining for WDR62 and α- or γ-tubulin. Scale bars: 20 µm.
We next established WDR62 requirements for microtubule regrowth. In interphase cells, no overt defects in centrosome-nucleated microtubule formation in WDR62-siRNA-treated cells were observed (supplementary material Fig. S5B). This was not surprising as WDR62 was not principally located at the MTOC during this cell cycle stage. Intriguingly, in mitotic cells, the extent of microtubule nucleation and regrowth at both centrosomal and non-centrosomal sites did not differ when WDR62-depleted and control HeLa cells were compared (Fig. 6B). However, as the bipolar spindle was re-established (45 min), defects in centrosome integrity and positioning were observed in WDR62-siRNA-treated cells (Fig. 6B). The proportion of WDR62-depleted cells with spindle abnormalities observed in our microtubule regrowth assays were consistent with our measurements during metaphase (Fig. 6C compared to Fig. 4B).
To determine whether spindle defects arose due to microtubule forces associated with bipolar spindle formation, we examined centrosome integrity in DTB-synchronized HeLa cells subsequently released in the presence of monastrol, an inhibitor of kinesin-5 motor proteins required for establishing spindle tension and bipolarity (Mayer et al., 1999). Monastrol treatment arrested both control and WDR62-siRNA-treated cells in mitosis with monopolar spindles (Fig. 6D). Detection of γ-tubulin showed that centrosome integrity was unperturbed in WDR62-depleted cells arrested in monastrol (Fig. 6D). Taken together, our study indicates that microtubule nucleation and spindle assembly does not require WDR62. In contrast, reduced centrosome integrity and spindle attachment defects occur subsequent to bipolar spindle formation and this is consistent with the previously defined requirement for microtubule dynamics (Fig. 5F).
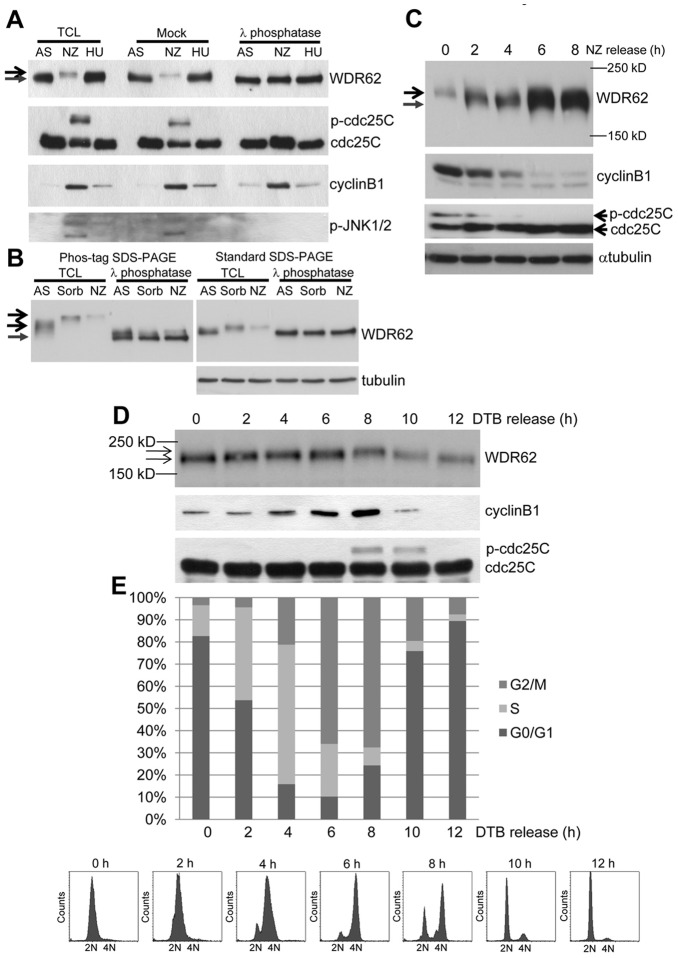
WDR62 is phosphorylated in mitosis
In considering regulatory mechanisms that control WDR62 function or localization, the phosphorylation of centrosomal and spindle proteins is critical for mitotic regulation (Lüders et al., 2006; Oshimori et al., 2006). WDR62 is likely targeted for phosphorylation as previous studies have identified WDR62 as a substrate of JNK and polo-like kinase 1 (Plk1) in the context of cell stress and mitosis, respectively (Santamaria et al., 2011; Wasserman et al., 2010). Indeed, M-phase arrest with nocodazole decreased the mobility of WDR62 in SDS-PAGE (Fig. 7A). Furthermore, WDR62 band detection in nocodazole-arrested cells was weaker but λ-phosphatase treatment rendered the migration and detection of the WDR62 band in lysates from the M-phase-arrested cells indistinguishable from asynchronous (AS) or S-phase-arrested (using hydroxyurea; HU) cells consistent with a phosphorylation modification (Fig. 7A). We obtained similar results regardless of the specific WDR62 antibody used (data not shown). As identical cell lysates were used for the λ-phosphatase or mock treatments, the reduced detection of WDR62 during nocodazole arrest indicated reduced antibody affinity for phosphorylated WDR62 on SDS-PAGE, rather than changes in WDR62 protein levels. However, λ-phosphatase treatment did not alter our detection of WDR62 subcellular localization indicating that phosphorylation did not impact on our immunofluorescence analyses in fixed cells (data not shown). The mobility shift of WDR62 in response to nocodazole-induced mitotic arrest or hyperosmolarity was more easily visualized in SDS-PAGE resolved with polyacrylamide-bound Mn2+-Phos-tag (Fig. 7B) (Kinoshita et al., 2006). In addition, while not evident on standard SDS-PAGE, the phosphorylation of WDR62 in untreated AS cells was revealed by λ-phosphatase treatment and electrophoresis on a Phos-tag gel (Fig. 7B). These results confirm WDR62 phosphorylation under basal conditions which was further increased with cell stress or M phase synchronization.
Fig. 7.
WDR62 is phosphorylated during mitosis. (A) Protein lysates from asynchronous (AS), S-phase-arrested (HU, 2 mM, 16 h) or mitotically arrested (NZ, 350 nM, 16 h) HeLa cells were treated with λ-phosphatase, mock treated without enzyme or left untreated (TCL) before blotting with the indicated antibodies. Arrows indicate decreased WDR62 band migration on SDS-PAGE. (B) Protein lysates from AS-, NZ- (350 nM, 16 h) or sorbitol-treated (Sorb, 0.5 M, 60 min) HeLa cells were treated with λ-phosphatase or left untreated (TCL) before resolving on Phos-tag gels or standard SDS-PAGE followed by immunoblotting. Arrows indicated reduced mobility of phosphorylated WDR62. (C) HeLa cells were arrested at M phase (NZ, 350 nM, 16 h) before release into normal serum medium for the indicated times. WDR62 band mobility, cyclin B1, α-tubulin and phospho-cdc25C were determined by immunoblot analysis. Arrows indicate increased WDR62 gel migration and carets (<) antibody detection with time following NZ release. (D) HeLa cells were synchronized at G1/S (DTB). WDR62, cyclin B1 and cdc25C levels were then analyzed by immunoblotting. (E) Cell cycle distribution was determined by FACS at time 0 min before release and at regular time intervals following thymidine release. Representative histograms are shown.
The M-phase arrest of cells treated with nocodazole was verified by increased cyclin B1 and phospho-cdc25C levels (Fig. 7A). Further, nocodazole washout increased WDR62 band mobility and detection in parallel with reduced cyclinB1 and phospho-cdc25C (Fig. 7C). Finally, as G1/S synchronized cells by DTB were tracked following thymidine block release, the WDR62 band shift closely correlated with mitotic progression as determined by biochemical markers (cyclin B1 and phospho-cdc25C) and FACS analysis of the cell cycle (Fig. 7D,E). These results revealed phosphorylation of WDR62 during mitotic progression that coincided with spindle pole localization.
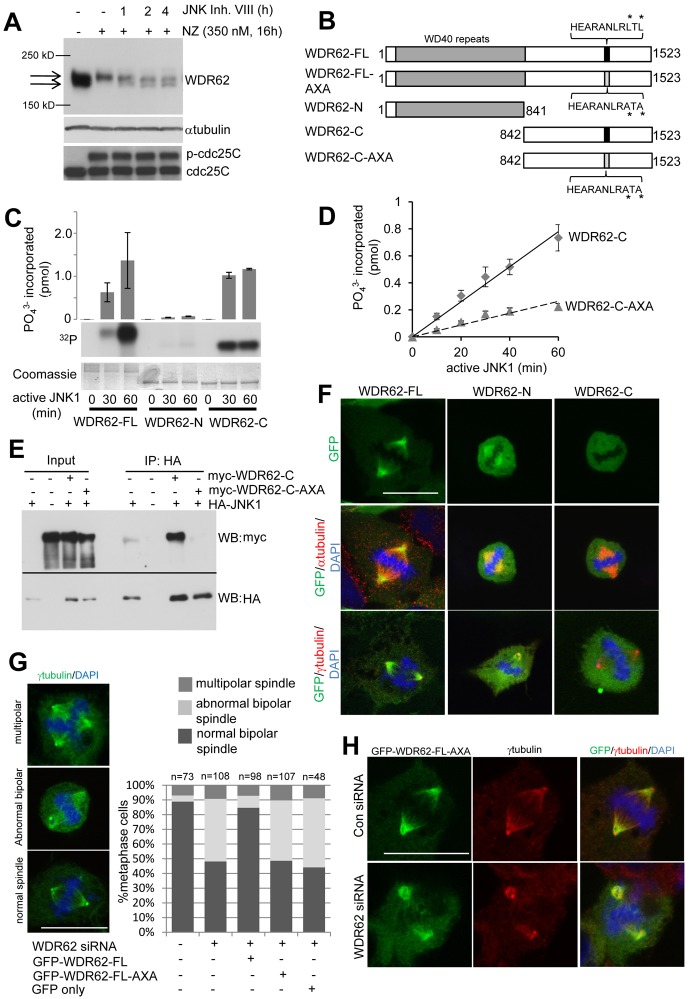
JNK mediated WDR62 phosphorylation is required for spindle regulation
As WDR62 was previously shown to be a JNK-interacting protein (Wasserman et al., 2010), we investigated JNK regulation of WDR62 and its contributions to mitosis. WDR62 phosphorylation in M-phase-arrested cells was partially reversed by JNK inhibition whereas cdc25C phosphorylation status was not altered indicating that cells remained arrested in M-phase following JNK inhibitor treatment (Fig. 8A). Thus, our results show that JNK activity is directly involved in mitotic WDR62 phosphorylation. In contrast, Aurora A inhibition or Plk1 knockdown did not prevent mitotic WDR62 phosphorylation as observed by reduced SDS-PAGE mobility and decreased detection of WDR62 in synchronized cells that was reversed by phosphatase treatment (supplementary material Fig. S6A,C). As Aurora A and Plk1 have well defined functions in bipolar spindle formation (Barr and Gergely, 2007; Barr et al., 2004), their inhibition resulted in a prometaphase arrest with spindle morphologies consistent with previous reports (supplementary material Fig. S6B,D) (Sumara et al., 2004). In this situation, we observed WDR62 phosphorylation without additional chemical synchronization (supplementary material Fig. S6A,C). In contrast, treatment of M-phase-arrested cells with a Cdk1 inhibitor reversed WDR62 phosphorylation that coincided with mitotic exit (supplementary material Fig. S6E). We also observed that WDR62 movement to centrosomes was not perturbed in cells arrested in prophase with the Cdk1 inhibitor (supplementary material Fig. S6F). These results indicate that mitotic WDR62 phosphorylation occurred subsequent to mitotic entry during prometaphase and that this phosphorylation was not required for WDR62 movement to spindle poles.
Fig. 8.
JNK-mediated WDR62 phosphorylation is involved in spindle regulation. (A) M-phase-arrested HeLa cells (NZ, 350 nM, 16 h) were treated with JNK inhibitor VIII (20 µM) and samples immunoblotted for WDR62 and α-tubulin. (B) Schematic representation of WDR62 truncation and JNK-binding (JBD) mutants. Asterisks indicate replacement of leucines with alanines in the JBD motif. (C) JNK1-mediated phosphorylation of full-length WDR62 (WDR62-FL) and truncation mutants containing WDR62 N- (WDR62-N) or C-terminal (WDR62-C) regions as measured by in vitro kinase activity assays. Values are means ± s.d. 32P (pmol) incorporated into substrates from three independent experiments. A representative autoradiograph and Coomassie stain for protein loading are shown. (D) JNK1-mediated phosphorylation of WDR62-C-AXA was determined by in vitro kinase activity assays and compared to the WDR62-C truncation mutant. (E) Myc-tagged WDR62-C and WDR62-C-AXA were transiently expressed with HA–JNK1 in HeLa cells. HA–JNK1 was immunoprecipitated and WDR62 co-immunoprecipitation determined by immunoblotting for the myc tag. Protein expression (input) in total cell lysates was also determined. (F) GFP-labeled WDR62-FL and truncation mutants were expressed in Ad293 cells. Their subcellular localization were determined during metaphase and colocalization with α- or γ-tubulin and DAPI was examined. (G) Ad293 cells were co-transfected with WDR62 siRNA and either GFP–WDR62-FL, GFP–WDR62-FL-AXA or GFP alone. The proportion of GFP-positive cells with normal bipolar, abnormal bipolar or multipolar spindles in metaphase was determined. n values are the total number of cells counted from three independent experiments. Representative images of multipolar, abnormal or normal bipolar spindles in Ad293 cells stained for γ-tubulin and DAPI are shown. (H) Representative images of GFP–WDR62-FL-AXA spindle localization in Ad293 cells treated with WDR62 or Con siRNA and stained for γ-tubulin and DAPI. Images in F and H are single z-sections.
The C-terminal region of WDR62 was phosphorylated by JNK and the JNK-binding domain (JBD) of WDR62 required for kinase docking interactions have been identified (Cohen-Katsenelson et al., 2011; Wasserman et al., 2010). Despite these advances, the biological function of WDR62 phosphorylation is undetermined. We generated truncation and JBD mutants of WDR62 (Fig. 8B) and confirmed that JNK phosphorylated the C-terminal half (WDR62-C, amino acids 842–1523) of WDR62 but not the N-terminal region containing the WD40 domains (WDR62-N, amino acids 1–841; Fig. 8C). Furthermore, replacement of critical leucines with alanine in the WDR62 JBD (AXA, L1299A/L1301A) inhibited JNK phosphorylation of WDR62-C in vitro and completely abrogated interaction with JNK when ectopically expressed in HeLa cells (Fig. 8D,E). We then compared the spindle localization properties of WDR62 mutants expressed as GFP fusion proteins. However, due to difficulties with aggregation of GFP-labeled full-length WDR62 expressed in HeLa cells, we analyzed Ad293 cells which displayed a greater proportion of cells that retained diffuse cytoplasmic distribution (supplementary material Fig. S7A). Interestingly, expression of the GFP-labeled WDR62-N truncation mutant led to a greatly enhanced proportion of cells with aggregates while GFP–WDR62-C expression was predominantly cytoplasmic despite higher expression levels (supplementary material Fig. S7A,B). Thus, the WDR62 N-terminal region containing WD40 repeats was responsible for the propensity of the ectopically expressed protein to aggregate. However, in cells with non-aggregate GFP–WDR62-N, we observed GFP–WDR62-N localization to the mitotic spindle albeit with reduced efficiency compared to full-length GFP–WDR62 (GFP–WDR62-FL; Fig. 8F). In contrast, GFP–WDR62-C failed to localize to the mitotic spindle (Fig. 8F). These observations indicate a requirement for the N-terminal domains of WDR62 in its mitotic localization.
We lastly sought to identify the contribution of JNK-mediated WDR62 phosphorylation to spindle regulation. Chemical inhibition of JNK during mitosis caused spindle abnormalities and centrosome attachment defects reminiscent of WDR62 depletion (supplementary material Fig. S7C). To determine the contribution of JNK signaling to WDR62 specifically, we performed rescue experiments. WDR62 depletion in Ad293 cells resulted in abnormal metaphase spindles as similarly observed in HeLa cells (Fig. 8G). Importantly, the concomitant expression of siRNA-resistant GFP–WDR62-FL reversed the frequency of the spindle defects in WDR62-depleted cells confirming siRNA effects were specific to WDR62 depletion. In contrast, expression of the JBD mutant (WDR62-FL-AXA) in WDR62-depleted cells was without effect (Fig. 8G) despite localization to the mitotic spindle (Fig. 8H) and comparable expression levels (supplementary material Fig. S7B). Thus, our studies show JNK-mediated phosphorylation of the WDR62 C-terminus is dispensable for its spindle localization but required for WDR62 functions in maintaining metaphase spindle architecture.
Discussion
Our combined in vitro and in vivo studies are consistent in identifying a mitotic role for WDR62 that is critical for neurogenesis. Specifically, we have provided the first functional characterization of WDR62 in mitosis and revealed a key role in metaphase spindle organization required for normal mitotic progression. WDR62 has a role in stabilizing the metaphase spindle for timely transition to anaphase but is not required for proliferation per se in cultured cells. In contrast, we demonstrate that WDR62 depletion impacts on the pools of self-renewing neural progenitors leading to their reduced proliferation in embryonic brains consistent with previously reported expression of WDR62 in mitotic neuronal precursor cells (Nicholas et al., 2010). One possibility is that defects in the timing of mitosis and/or spindle orientation following WDR62-depeletion impacts specifically on cortical neurogenesis. Indeed, centrosome-directed spindle orientation has been suggested as a critical determinant for asymmetric division of neuroprogenitors, a necessary property for their self-renewal within the germinal compartment of the embryonic nervous system (Lesage et al., 2010). This is compatible with the notion that centrosome-associated mitotic defects during neurogenesis may underlie the clinical manifestation of WDR62 mutations in MCPH (Nicholas et al., 2010; Yu et al., 2010).
Our analysis of WDR62 intracellular distribution and cellular functions reinforces the association of MCPH with centrosome-associated proteins and highlights the role of spindle pole integrity in neurogenesis (Thornton and Woods, 2009). The prominent spindle pole localization of WDR62 is shared by another MCPH protein, abnormal spindle microcephaly related gene (ASPM) (Higgins et al., 2010; Pulvers et al., 2010). In contrast, other MCPH proteins, microcephalin, cdk5rap2, CENPJ, CEP152, and STIL are principally centrosomal in their localization (Delattre et al., 2004; Fong et al., 2008; Varmark et al., 2007; Zhong et al., 2006). Interestingly, ASPM and WDR62 are most frequently mutated in MCPH accounting for 50% and 10% of cases, respectively (Thornton and Woods, 2009). We have demonstrated spindle orientation defects as a consequence of WDR62 depletion in cultured human cells. The depletion of ASPM in cultured cells has revealed similar requirements for spindle organization and spindle positioning (Higgins et al., 2010). Moreover, the loss of ASPM in embryonic mice resulted in premature cell cycle exit due to altered cleavage plane orientation in dividing neuroprogenitors (Fish et al., 2006). The shared depletion phenotypes of WDR62 and ASPM suggest that mitotic spindle and cleavage plane positioning may represent a mechanism that links these proteins that are most commonly mutated in MCPH.
The molecular basis that underlies the development of MCPH may also involve the disruption and/or dysfunction of centrosomes (Thornton and Woods, 2009). The depletion of PCM proteins cdk5rap2 or pericentrin, with the latter also genetically linked to primordial dwarfism with associated microcephaly, similarly altered neuroprogenitor division in embryonic mouse brains with reduced cell proliferation and enhanced cell cycle exit (Buchman et al., 2010; Lizarraga et al. 2010). However, unlike MCPH proteins (cdk5rap2, CENPJ, STIL, Cep152, Cep135 and Cep63) with well characterized functions in centriole duplication (Barrera et al. 2010; Delattre et al., 2004; Hatch et al., 2010; Hussain et al., 2012; Sir et al., 2011; Vulprecht et al., 2012), we have demonstrated that WDR62 is not principally a centrosome protein and its depletion did not obviously impact on centrosome number or structure during interphase. We cannot completely exclude WDR62 functions as an integral centrosomal protein. However, the WDR62 depletion phenotype was observed specifically during metaphase and coincided with its most prominent spindle localization. Thus, centrosome/spindle defects in WDR62 knockdown cells were likely the result of the loss of spindle localized WDR62 during mitosis. Specifically, we revealed defects with centrosome–spindle-pole coupling in cells lacking WDR62.
Centrosome–spindle-pole attachment defects were previously observed with the disruption of centrosomal cdk5rap2, NUMA or dynein/dynactin subunits (Barr et al., 2010; Morales-Mulia and Scholey, 2005; Silk et al., 2009). Interestingly, we noted that centrosome displacement occurred subsequent to bipolar spindle formation suggesting that microtubule forces may underlie centrosome uncoupling. However, WDR62 depletion did not result in unfocused spindle poles. In addition, the spindle pole and cortical localization of NUMA and dynein were unaltered by WDR62 loss. Our study therefore suggests that the disruption of centrosome–spindle coupling is linked to microtubule forces associated with establishing spindle bipolarity and spindle orientation but not readily explained by the mislocalization of previously identified factors critical for spindle–centrosome attachment.
We have additionally revealed that the PCM of uncoupled centrosomes appeared diffuse with multiple puncta positively stained for pericentrin. It is possible that PCM recruitment to MT minus ends on the uncoupled spindle pole may also lead to such a phenotype. However, disrupted centrosomes were smaller in size and as we did not find defects in centrosome maturation or astral microtubule formation, our findings suggest reduced centrosome integrity in response to WDR62 knockdown. As with centrosome/spindle pole uncoupling, defects in centrosome integrity were observed subsequent to bipolar spindle formation and required microtubule dynamics, which suggests that centrosome uncoupling and fragmentation events may be related. However, previous studies that have inactivated cdk5rap2, NUMA or dynein have shown that loss of spindle pole attachment does not necessarily lead to reduced centrosome integrity (Barr et al., 2010; Morales-Mulia and Scholey, 2005; Silk et al., 2009). Notably, NUMA and dynein/dynactin motors perform multiple functions at several mitotic locations (Radulescu and Cleveland, 2010). Thus, the differences in spindle/centrosome defects may reflect different mechanisms underlying WDR62 function in metaphase spindle maintenance compared to previously characterized spindle pole proteins. We propose that the spindle localization of WDR62 is required to maintain tight centrosome–spindle-pole coupling, centrosome integrity and the maintenance of spindle orientation that is involved in determining cleavage plane position and division symmetry. Disruption of these processes with WDR62 loss or mutation may impact on self-renewing divisions in the embryonic brain that ultimately manifests as microcephaly.
As WDR62 is a multi-functional protein with dynamic intracellular distribution dependent on cellular context, this raises the significant question of the range of regulatory mechanisms for its actions. Our study has now provided first insights into the mechanisms that regulate mitotic WDR62 activity. WDR62 is mitotically phosphorylated. However, WDR62 movement to centrosomes, prior to mitotic entry, preceded its phosphorylation. This indicates that mitotic phosphorylation is not likely involved in regulating WDR62 movement. In support of this, the N-terminal region of WDR62, lacking C-terminal phosphorylation sites, is sufficient for spindle localization. Rather, WDR62 phosphorylation appears more likely to be involved in functional regulation.
We revealed JNK contribution to mitotic WDR62 phosphorylation. WDR62 was initially characterized as a component of the JNK signaling pathway (Wasserman et al., 2010) and our findings demonstrate that this is maintained during mitosis. JNK expression and activity regulates various cell cycle stages (Kennedy and Davis, 2003; MacCorkle and Tan, 2005). Specifically, JNK is activated during early mitotic stages and targets substrates such as cdc25C, histone H3 and Cdh1 to facilitate mitotic progression (Gutierrez et al., 2010a; Gutierrez et al., 2010b; Lee and Song, 2008). JNK has also previously been reported to be centrosome/spindle pole associated during cell division (Huang et al., 2011; MacCorkle-Chosnek et al., 2001). Importantly, we have now shown that JNK phosphorylation of WDR62 is involved in spindle regulation.
There are multiple serine/threonine residues at the C-terminal region of WDR62 that are likely JNK target sites (Wasserman et al., 2010). Future detailed analyses should reveal those sites critical for the mitotic activity of WDR62. However, we have taken advantage of the previously characterized WDR62 JBD (Cohen-Katsenelson et al., 2011) and utilized mutations to this motif (AXA) as a strategy to abrogate JNK signaling to WDR62. This leaves unchanged the proline-directed serine/threonine residues that may be additionally targeted by other mitotically regulated kinases (e.g. cdks). WDR62-FL-AXA mutants failed to rescue spindle abnormalities caused by endogenous WDR62 depletion highlighting JNK contribution to spindle regulation through the targeting of WDR62. It is likely that WDR62 is targeted by multiple kinases, particularly as JNK inhibition only partially reverses WDR62 phosphorylation in response to M-phase arrest. Although Plk1 and Aurora A inhibition did not attenuate mitotic WDR62 phosphorylation, at least on residues responsible for gel shift, this does not exclude roles for these mitotic kinases in WDR62 regulation. However, investigations into the precise contributions of Plk1, Aurora A and Cdk1 are complicated by their critical requirements for bipolar spindle formation and mitotic entry.
In targeting mitotically regulated WDR62, we have now defined functions in centrosome/spindle organization, mitotic regulation and neurogenesis. Our study has provided the first in vitro and in vivo functional analysis of WDR62 and revealed novel contributions of a JNK/WDR62 signaling mechanism towards mitotic spindle and cell cycle regulation. Moreover, studies in transgenic mice have demonstrated that JNK was required for early brain development (Kuan et al., 1999). Thus, we have revealed a novel signaling mechanism linking JNK activity and neurogenesis that warrants further investigation.
Materials and Methods
Antibodies and reagents
Polyclonal WDR62 antibodies were from Bethyl Laboratories (A301-560A) or Novus Biologicals (NB100-77302). CG-NAP and p150Glued antibodies were from BD Biosciences. NUMA, dynein intermediate chain (DIC), and mouse monoclonal (Mab414) antibodies to Nuclear Pore Complex proteins were from Abcam. α-Tubulin antibody was obtained from Santa Cruz Biotechnology. Plk1, phospho-TCTP (Ser46), cyclin B1 and cdc25C antibodies were from Cell Signaling Technology. Antibodies to Ki67 and phosphorylated histone H3 were from Leica and Merck-Millipore, respectively. Cdk1 inhibitor I and Aurora kinase inhibitor III were from Calbiochem. Phos-tag reagent was from Wako Chemicals. Cell culture and transfection reagents including DMEM and fetal bovine serum were from Invitrogen-GibcoBRL. All other reagents, including thymidine, nocodazole, hydroxyurea, monoclonal and polyclonal γ-tubulin and monoclonal WDR62 (W3269) antibodies were obtained from Sigma-Aldrich. Unless specified otherwise, immunoblot and immunofluorescence analysis were performed with anti-WDR62 from Bethyl Laboratories.
In utero electroporation and brain slice analysis
Mice were maintained within the animal facilities at Monash University, with all animal procedures approved by the relevant Animal Ethics Committee. In utero electroporation was performed on pregnant, time-mated female C57/B6 mice as previously described (Heng et al., 2008). Mouse embryos (E14) were electroporated with WDR62 siRNA or a non-targeting control siRNA (10 µM of siRNA in solution) together with a GFP expression plasmid (1 µg/µl). BrdU pulse labeling (100 mg/kg) was performed by injecting pregnant dams 24 h post-electroporation. Following an additional 24 h, the embryonic brains were dissected and fixed in 4% paraformaldehyde/PBS solution overnight, before washing with PBS followed by incubation in 20% sucrose/PBS solution. Finally, the brains were embedded in OCT and sectioned at 16 µm thickness using a cryostat (Leica) and immunostained using previously defined protocols (Heng et al., 2008). The ventricular zone/subventricular zone and intermediate zones were identified by nuclei density and images were acquired on a confocal microscope. Cell counts were performed on representative fields followed by statistical analysis (GraphPad). Data presented for the in utero electroporation experiments is plotted from three or four brains per condition.
Immunofluorescence and live-cell microscopy
Cell culture and treatments were performed on uncoated glass coverslips. Cells were washed in PBS before fixation either with 4% (w/v) paraformaldehyde (20 min, room temperature) or cold methanol (5 min, −20°C). Sample preparation was as previously described (Ng et al., 2011). Images were acquired on a Leica TCS SP2 or SP5 confocal microscope using 100×1.35 NA objectives. Unless specified otherwise, images are maximum intensity projections of z-stacks of 8–14 sections of 0.4 µm thickness that encompasses the entire spindle. Z-stack spindle rotation analysis was performed with ImageJ using the Z-function plug-in. Spindle length measurements were performed on single stack images using Metamorph software by measuring the linear distance between the apexes of bipolar spindles poles.
Plasmids and protein purification
SiRNA-resistant full-length WDR62 (WDR62-FL) was codon optimized and synthesized by GeneArt (Invitrogen). WDR62 truncation mutants (WDR62-N, WDR62-C) were generated by PCR and, along with full-length WDR62, labeled with GFP or Myc tag by cloning into pEGFP-N3 or pXJ40-Myc vectors, respectively. WDR62-FL-AXA and WDR62-C-AXA (L1299A/L1301A) mutations were generated by site-directed mutagenesis. For recombinant protein expression, WDR62 cDNAs were cloned into pGEX-6P-1 to generate GST fusions. Where specified, the GST tag was removed with Precision Protease cleavage. HA–JNK1 was made by subcloning PCR amplified Jnk1α1 into pXJ41-HA. Active JNK1 was generated using baculoviral expression in insect cells as described previously (Ngoei et al., 2011). All constructs were subjected to restriction digestion and full sequencing analysis prior to use. GFP–centrin1 encoded in pCMV6 vector was obtained from Origene.
RNAi
ON-TARGETPlus human WDR62 siRNA, mouse WDR62 siRNA pool, human Plk1 siRNA pool, non-targeting control siRNA (D-001810-01-20) and non-targeting siRNA pool (D-001810-10-20) were obtained from Dharmacon and resuspended in RNAase free water at 100 µM. siRNA were transiently transfected using Lipofectamine™ 2000 according to the manufacturer’s protocol.
Cell culture and transient transfection
HeLa and Ad293 were maintained in DMEM and genetically modified U2OS-GFP–TUB1A (Sigma-Aldrich) was grown in McCoy’s 5A medium. All growth media were supplemented with 10% fetal calf serum and 100 U/ml penicillin/streptomycin and cells cultured in a humidified 5% CO2 environment. Liposomal-mediated transfection was performed with Lipofectamine™ 2000 and antibiotic-free Opti-MEM medium according to manufacturer’s instructions.
Cell synchronization and cell cycle arrest
HeLa cells were synchronized by DTB or nocodazole arrest. Briefly, sub-confluent HeLa cells were cultured in the presence of thymidine (2 mM) for 20 h before washing with PBS (three washes) and release into normal growth medium for 6 h. Cells were then cultured in the presence of thymidine (2 mM) for a further 20 h before final release from late G1 arrest into normal growth medium. Cells were then collected at specified times regular post-release for subsequent analysis. M-phase or S-phase arrests were with nocodazole (350 nM) or hydroxyurea (2 mM) treatment respectively for 16 h. Kinase inhibitors were applied at the time or subsequent to cell cycle arrest as specified.
Cell lysates and immunoblots
Total cell lysates were prepared in RIPA buffer [50 mM Tris-HCl, pH 7.3, 150 mM NaCl, 0.1 mM EDTA, 1% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100, 0.2% (w/v) NaF and 100 µM Na3VO4] supplemented with protease inhibitors and immunoblotted as previously described (Ng et al., 2010) with the exception that protein lysates were resolved on SDS-PAGE without sample boiling due to the heat sensitivity of WDR62 (Wasserman et al. 2010). For phosphatase treatment, lysates were prepared in RIPA buffer lacking EDTA, NaF and Na3VO4. Lysates (100 µg) were treated with λ-phosphatase (400 U, New England Biolabs) for 20 min at 30°C. For Phos-tag SDS-PAGE, gels were supplemented with a dinuclear manganese complex of acrylamide-pendant Phos-tag ligand (Mn2+-Phos-tag; 50 µM).
In vitro kinase assay
Purified recombinant protein substrate (10 µg) was incubated with active kinase (10 ng), and 32P-radiolabeled ATP ([γ32-P]ATP, 1 µCi/reaction) in a kinase reaction buffer (20 mM HEPES pH 7.6, 20 mM MgCl2.6H2O, 75 µM ATP, 20 mM β-glycerophosphate and supplemented with 25 µM Na3VO4 and 100 µM DTT) over a 120 min time course at 30°C. Reactions were stopped with the addition of Laemmli sample buffer. Samples were then resolved by SDS-PAGE, stained with Gelcode Blue Stain reagent and analyzed by autoradiography and Cerenkov counting.
Microtubule regrowth assays, Eg5 inhibition and CaCl2 extraction
HeLa cells were synchronized (DTB) and released into nocodazole (350 nM, 8–10 h). Nocodazole was removed by washing (3×) with cold PBS then cold growth medium added. Mitotic microtubules were completely depolymerized by cold treatment (4°C, 30 min). Microtubule regrowth was initiated by incubating cells at 37°C for between 2 and 60 min before fixation. For kinesin Eg5 inhibition, HeLa cells were synchronized (DTB) and released into monastrol (100 µM, 8 h). To reveal kinetochore microtubules, synchronized cells were extracted with Calcium Extraction Buffer (1 mM CaCl2, 100 mM PIPES pH 6.8, 0.1 mM MgCl2 and 0.1% v/v Triton X-100) for 3 min before fixing and immunofluorescence analysis.
Cell viability and cell proliferation
Cell viability was determined by labeling metabolically active cells with the yellow tetrazolium salt, XTT {sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulfonic acid hydrate}, using the XTT Kit (Roche) according to manufacturer specifications. WDR62 protein levels were blotted in parallel to confirm WDR62 depletion. Cell numbers were counted using a hemocytometer and Trypan Blue exclusion.
Supplementary Material
Acknowledgments
We thank Prof. Sharad Kumar (Centre for Cancer Biology/South Australia Pathology) for the NEDD1 antibody and Prof. Leann Tilley (University of Melbourne) for critical reading of this manuscript.
Footnotes
Funding
This work was supported by a Faculty of Medicine, Dentistry and Health Science (MDHS) CR Roper Fellowship [to D.C.H.N.]; an National Health and Medical Research Council Career Development Fellowship [grant number APP1011505 to J.I.H.]; and National Health and Medical Research Council Project Grants [grant numbers 628335 and 566804 to D.C.H.N and M.A.B., respectively.]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.107326/-/DC1
References
- Barr A. R., Gergely F. (2007). Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120, 2987–2996 10.1242/jcs.013136 [DOI] [PubMed] [Google Scholar]
- Barr F. A., Silljé H. H., Nigg E. A. (2004). Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–441 10.1038/nrm1401 [DOI] [PubMed] [Google Scholar]
- Barr A. R., Kilmartin J. V., Gergely F. (2010). CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J. Cell Biol. 189, 23–39 10.1083/jcb.200912163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera J. A., Kao L. R., Hammer R. E., Seemann J., Fuchs J. L., Megraw T. L. (2010). CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev. Cell 18, 913–926 10.1016/j.devcel.2010.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgüvar K., Oztürk A. K., Louvi A., Kwan K. Y., Choi M., Tatli B., Yalnizoğlu D., Tüysüz B., Cağlayan A. O., Gökben S.et al. (2010). Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467, 207–210 10.1038/nature09327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman J. J., Tseng H. C., Zhou Y., Frank C. L., Xie Z., Tsai L. H. (2010). Cdk5rap2 interacts with pericentrin to maintain the neural progenitor pool in the developing neocortex. Neuron 66, 386–402 10.1016/j.neuron.2010.03.036 [DOI] [PubMed] [Google Scholar]
- Cohen–Katsenelson K., Wasserman T., Khateb S., Whitmarsh A. J., Aronheim A. (2011). Docking interactions of the JNK scaffold protein WDR62. Biochem. J. 439, 381–390 10.1042/BJ20110284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D. A. (2000). Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95–114 10.1146/annurev.biochem.69.1.95 [DOI] [PubMed] [Google Scholar]
- Delattre M., Leidel S., Wani K., Baumer K., Bamat J., Schnabel H., Feichtinger R., Schnabel R., Gönczy P. (2004). Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6, 656–664 10.1038/ncb1146 [DOI] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008). A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105, 10762–10767 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S., Mitchison T. J. (2009). Compression regulates mitotic spindle length by a mechanochemical switch at the poles. Curr. Biol. 19, 1086–1095 10.1016/j.cub.2009.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish J. L., Kosodo Y., Enard W., Pääbo S., Huttner W. B. (2006). Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA 103, 10438–10443 10.1073/pnas.0604066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K. W., Choi Y. K., Rattner J. B., Qi R. Z. (2008). CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell 19, 115–125 10.1091/mbc.E07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez G. J., Tsuji T., Chen M., Jiang W., Ronai Z. A. (2010a). Interplay between Cdh1 and JNK activity during the cell cycle. Nat. Cell Biol. 12, 686–695 10.1038/ncb2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez G. J., Tsuji T., Cross J. V., Davis R. J., Templeton D. J., Jiang W., Ronai Z. A. (2010b). JNK-mediated phosphorylation of Cdc25C regulates cell cycle entry and G(2)/M DNA damage checkpoint. J. Biol. Chem. 285, 14217–14228 10.1074/jbc.M110.121848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Stearns T., Lüders J. (2009). Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE 4, e5976 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E. M., Kulukian A., Holland A. J., Cleveland D. W., Stearns T. (2010). Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191, 721–729 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J. I., Nguyen L., Castro D. S., Zimmer C., Wildner H., Armant O., Skowronska–Krawczyk D., Bedogni F., Matter J. M., Hevner R.et al. (2008). Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455, 114–118 10.1038/nature07198 [DOI] [PubMed] [Google Scholar]
- Higgins J., Midgley C., Bergh A. M., Bell S. M., Askham J. M., Roberts E., Binns R. K., Sharif S. M., Bennett C., Glover D. M.et al. (2010). Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol. 11, 85 10.1186/1471-2121-11-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Tong J. S., Wang Z. B., Yang C. R., Qi S. T., Guo L., Ouyang Y. C., Quan S., Sun Q. Y., Qi Z. Q.et al. (2011). JNK2 participates in spindle assembly during mouse oocyte meiotic maturation. Microsc. Microanal. 17, 197–205 10.1017/S1431927610094456 [DOI] [PubMed] [Google Scholar]
- Hussain M. S., Baig S. M., Neumann S., Nürnberg G., Farooq M., Ahmad I., Alef T., Hennies H. C., Technau M., Altmüller J.et al. (2012). A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am. J. Hum. Genet. 90, 871–878 10.1016/j.ajhg.2012.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins J. R., Clarke P. R. (2004). Many fingers on the mitotic trigger: post-translational regulation of the Cdc25C phosphatase. Cell Cycle 3, 40–44 10.4161/cc.3.1.595 [DOI] [PubMed] [Google Scholar]
- Kennedy N. J., Davis R. J. (2003). Role of JNK in tumor development. Cell Cycle 2, 199–201 [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita–Kikuta E., Takiyama K., Koike T. (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5, 749–757 10.1074/mcp.T500024-MCP200 [DOI] [PubMed] [Google Scholar]
- Kuan C. Y., Yang D. D., Samanta Roy D. R., Davis R. J., Rakic P., Flavell R. A. (1999). The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22, 667–676 10.1016/S0896-6273(00)80727-8 [DOI] [PubMed] [Google Scholar]
- Lee K., Song K. (2008). Basal c-Jun N-terminal kinases promote mitotic progression through histone H3 phosphorylation. Cell Cycle 7, 216–221 10.4161/cc.7.2.5155 [DOI] [PubMed] [Google Scholar]
- Lesage B., Gutierrez I., Martí E., Gonzalez C. (2010). Neural stem cells: the need for a proper orientation. Curr. Opin. Genet. Dev. 20, 438–442 10.1016/j.gde.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Lizarraga S. B., Margossian S. P., Harris M. H., Campagna D. R., Han A. P., Blevins S., Mudbhary R., Barker J. E., Walsh C. A., Fleming M. D. (2010). Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development 137, 1907–1917 10.1242/dev.040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Patel U. K., Stearns T. (2006). GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- MacCorkle R. A., Tan T. H. (2005). Mitogen-activated protein kinases in cell-cycle control. Cell Biochem. Biophys. 43, 451–462 10.1385/CBB:43:3:451 [DOI] [PubMed] [Google Scholar]
- MacCorkle–Chosnek R. A., VanHooser A., Goodrich D. W., Brinkley B. R., Tan T. H. (2001). Cell cycle regulation of c-Jun N-terminal kinase activity at the centrosomes. Biochem. Biophys. Res. Commun. 289, 173–180 10.1006/bbrc.2001.5948 [DOI] [PubMed] [Google Scholar]
- Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. (1999). Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Sharkey J. T., Nowakowski R. S. (2011). Cdk5rap2 exposes the centrosomal root of microcephaly syndromes. Trends Cell Biol. 21, 470–480 10.1016/j.tcb.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales–Mulia S., Scholey J. M. (2005). Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell 16, 3176–3186 10.1091/mbc.E04-12-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. C., Zhao T. T., Yeap Y. Y., Ngoei K. R., Bogoyevitch M. A. (2010). c-Jun N-terminal kinase phosphorylation of stathmin confers protection against cellular stress. J. Biol. Chem. 285, 29001–29013 10.1074/jbc.M110.128454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. C., Ng I. H., Yeap Y. Y., Badrian B., Tsoutsman T., McMullen J. R., Semsarian C., Bogoyevitch M. A. (2011). Opposing actions of extracellular signal-regulated kinase (ERK) and signal transducer and activator of transcription 3 (STAT3) in regulating microtubule stabilization during cardiac hypertrophy. J. Biol. Chem. 286, 1576–1587 10.1074/jbc.M110.128157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoei K. R., Catimel B., Church N., Lio D. S., Dogovski C., Perugini M. A., Watt P. M., Cheng H. C., Ng D. C., Bogoyevitch M. A. (2011). Characterization of a novel JNK (c-Jun N-terminal kinase) inhibitory peptide. Biochem. J. 434, 399–413 10.1042/BJ20101244 [DOI] [PubMed] [Google Scholar]
- Nicholas A. K., Khurshid M., Désir J., Carvalho O. P., Cox J. J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A.et al. (2010). WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 42, 1010–1014 10.1038/ng.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E. A., Raff J. W. (2009). Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Oshimori N., Ohsugi M., Yamamoto T. (2006). The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat. Cell Biol. 8, 1095–1101 10.1038/ncb1474 [DOI] [PubMed] [Google Scholar]
- Oshimori N., Li X., Ohsugi M., Yamamoto T. (2009). Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J. 28, 2066–2076 10.1038/emboj.2009.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvers J. N., Bryk J., Fish J. L., Wilsch–Bräuninger M., Arai Y., Schreier D., Naumann R., Helppi J., Habermann B., Vogt J.et al. (2010). Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc. Natl. Acad. Sci. USA 107, 16595–16600 10.1073/pnas.1010494107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu A. E., Cleveland D. W. (2010). NuMA after 30 years: the matrix revisited. Trends Cell Biol. 20, 214–222 10.1016/j.tcb.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A., Wang B., Elowe S., Malik R., Zhang F., Bauer M., Schmidt A., Sillje H. H., Korner R., Nigg E. A. (2011). The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell Proteomics 10, M110 004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk A. D., Holland A. J., Cleveland D. W. (2009). Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184, 677–690 10.1083/jcb.200810091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir J. H., Barr A. R., Nicholas A. K., Carvalho O. P., Khurshid M., Sossick A., Reichelt S., D’Santos C., Woods C. G., Gergely F. (2011). A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 43, 1147–1153 10.1038/ng.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann C. U., Petsalaki E., Russell R. B., Müller C. W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574 10.1016/j.tibs.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Sumara I., Giménez–Abián J. F., Gerlich D., Hirota T., Kraft C., de la Torre C., Ellenberg J., Peters J. M. (2004). Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 14, 1712–1722 10.1016/j.cub.2004.09.049 [DOI] [PubMed] [Google Scholar]
- Takizawa C. G., Morgan D. O. (2000). Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr. Opin. Cell Biol. 12, 658–665 10.1016/S0955-0674(00)00149-6 [DOI] [PubMed] [Google Scholar]
- Thornton G. K., Woods C. G. (2009). Primary microcephaly: do all roads lead to Rome? Trends Genet. 25, 501–510 10.1016/j.tig.2009.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmark H., Llamazares S., Rebollo E., Lange B., Reina J., Schwarz H., Gonzalez C. (2007). Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17, 1735–1745 10.1016/j.cub.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M. S., Zentgraf H., Izraeli S.et al. (2012). STIL is required for centriole duplication in human cells. J. Cell Sci. 125, 1353–1362 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wasserman T., Katsenelson K., Daniliuc S., Hasin T., Choder M., Aronheim A. (2010). A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell 21, 117–130 10.1091/mbc.E09-06-0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T. W., Mochida G. H., Tischfield D. J., Sgaier S. K., Flores–Sarnat L., Sergi C. M., Topçu M., McDonald M. T., Barry B. J., Felie J. M.et al. (2010). Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 42, 1015–1020 10.1038/ng.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Pfeifer G. P., Xu X. (2006). Microcephalin encodes a centrosomal protein. Cell Cycle 5, 457–458 10.4161/cc.5.4.2481 [DOI] [PubMed] [Google Scholar]
- Zimmerman W. C., Sillibourne J., Rosa J., Doxsey S. J. (2004). Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642–3657 10.1091/mbc.E03-11-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyss D., Gergely F. (2009). Centrosome function in cancer: guilty or innocent? Trends Cell Biol. 19, 334–346 10.1016/j.tcb.2009.04.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.