Summary
After injury, residual epithelial cells coordinate contextual clues from cell–cell and cell–matrix interactions to polarize and migrate over the wound bed. Protrusion formation, cell body translocation and rear retraction is a repetitive process that allows the cell to move across the substratum. Fundamental to this process is the assembly and disassembly of focal adhesions that facilitate cell adhesion and protrusion formation. Here, we identified syndecan-1 as a regulator of focal adhesion disassembly in migrating lung epithelial cells. Syndecan-1 altered the dynamic exchange of adhesion complex proteins, which in turn regulates migration speed. Moreover, we provide evidence that syndecan-1 controls this entire process through Rap1. Thus, syndecan-1 restrains migration in lung epithelium by activating Rap1 to slow focal adhesion disassembly.
Key words: Syndecan-1, Focal adhesion, Migration
Introduction
Re-epithelialization is arguably the hallmark event characterizing successful repair following disruptive injury. Pathological conditions (e.g. tumor invasion and metastasis, chronic inflammation, fibrosis, etc.) arise when the epithelium either cannot repair or loses the contextual controls that shut down the migration process (Ridley et al., 2003; Crosby and Waters, 2010; Nathan and Ding, 2010). Cell migration involves multiple signaling and cytoskeletal changes and is modified by specific cell–matrix and cell–cell interactions (Montell, 2008). For migration to occur, cells must balance leading edge protrusion and attachment with trailing edge release and retraction (Huttenlocher and Horwitz, 2011). Cell migration would be limited if not for the traction provided by the adhesion to the matrix. Rear retraction is equally important and can limit the migration speed by restraining forward progress.
Once stimulated to migrate, cellular protrusions engage the substratum primarily through integrin receptors, which cluster and recruit adaptor proteins to form nascent adhesions (Kaverina et al., 2002; Wehrle-Haller, 2012). Nascent adhesions contain many components of classical focal adhesions and are generally short-lived and turnover while within the lamellipodia or filopodia (Parsons et al., 2010; Webb et al., 2002). However, some nascent adhesions do not turnover, but instead, mature into focal adhesions (FAs) (Broussard et al., 2008). Whereas nascent adhesions tightly bind the substratum to generate traction forces for forward migration, FAs are positioned at the termini of stress fibers and act as anchors for cell contraction (Beningo et al., 2001; Galbraith and Sheetz, 1997; Galbraith et al., 2002; Broussard et al., 2008).
Disassembly of FAs in the leading edge of migrating cells facilitate formation of new protrusions and adhesions (Webb et al., 2002; Huttenlocher and Horwitz, 2011). Leading edge FAs play an important role in regulating cell migration speed so controlling assembly and disassembly must be tightly regulated (Webb et al., 2004; Gupton and Waterman-Storer, 2006; Millon-Frémillon et al., 2008; Lawson et al., 2012; Webb et al., 2002). Assembly requires actin polymerization and is largely regulated by the Rho family GTPases (Alexandrova et al., 2008; Choi et al., 2008; Vicente-Manzanares et al., 2009; Raftopoulou and Hall, 2004). Disassembly of leading edge FAs is an equally complicated process that mechanistically converges through focal adhesion kinase (FAK) and Src signaling (Huttenlocher and Horwitz, 2011).
We recently identified syndecan-1 as a regulator of lung epithelial migration in vitro and re-epithelialization in vivo (Chen et al., 2009). Syndecan-1, a transmembrane heparan sulfate proteoglycan, mediates many effects on cellular function by coupling with integrins and regulating their allosteric state (Couchman, 2003; Morgan et al., 2007; Chen et al., 2009; Beauvais et al., 2009; Beauvais and Rapraeger, 2003). Given the importance of integrins in regulating FAs, we investigated if syndecan-1 controls cell migration through FA turnover. We determined that syndecan-1 regulates FA disassembly in lung epithelial cells to control migration speed. Additionally, syndecan-1 mediated its effects via the small GTPase, Rap1, independently of integrin activation.
Results
Syndecan-1 attenuates FA disassembly
We reported that cell migration is slower in cells expressing syndecan-1 compared to cells with shRNA-mediated downregulated expression (Chen et al., 2009). Our previous studies only found this effect on collagen matrices so all the following studies are performed on a Type I collagen matrix.
Because FA turnover is a major determinant of cell migration speed (Webb et al., 2002), we compared the presence and pattern of paxillin-positive FAs between migrating lung epithelial cells with syndecan-1 (B2bshRNA.scr) and cells lacking syndecan-1 expression (B2bshRNA.hSdc1; Fig. 1). Nascent adhesions lined the cell front in all conditions. Additionally, B2bshRNA.scr cells at the wound front had notable leading edge FAs. In contrast, B2bshRNA.hSdc1 cells had fewer FAs that were much less prominent when compared to B2bshRNA.scr cells.
Fig. 1.
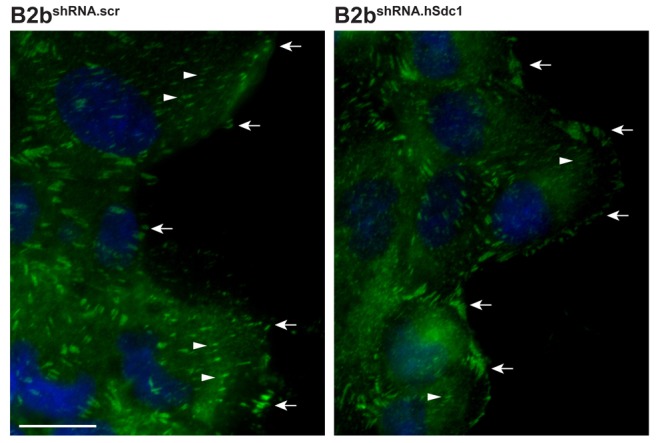
Syndecan-1 increases FAs. B2bshRNA.scr and B2bshRNA.hSdc1 cells were allowed to migrate for 2 h and immunostained for paxillin (green) and DAPI (blue). Nascent adhesions were seen at the cell front (arrows). FAs (arrowheads) are seen throughout migrating B2bshRNA.scr cells. In contrast, B2bshRNA.hSdc1 cells had fewer and less prominent FAs in migrating cells. These are representative images from reproducible experiments. Scale bar: 20 µm.
FA turnover is a dynamic process that is not well represented by static images. Therefore, we stably expressed paxillin–eGFP in B2bshRNA.scr and B2bshRNA.hSdc1 cells to follow the assembly and disassembly of FAs (Laukaitis et al., 2001). Using time-lapse total internal reflection fluorescent (TIRF) microscopy, FA assembly and disassembly was observed in migrating cells with a high spatiotemporal resolution (supplementary material Movie 1). B2bshRNA.hSdc1 cells migrated faster than B2bshRNA.scr cells, which is consistent with our previous observations (Chen et al., 2009). Similar to our findings in Fig. 1, both cell lines formed nascent adhesions at the front edge of the cell. However, FAs appeared to be more persistent in B2bshRNA.scr compared to B2bshRNA.hSdc1 cells.
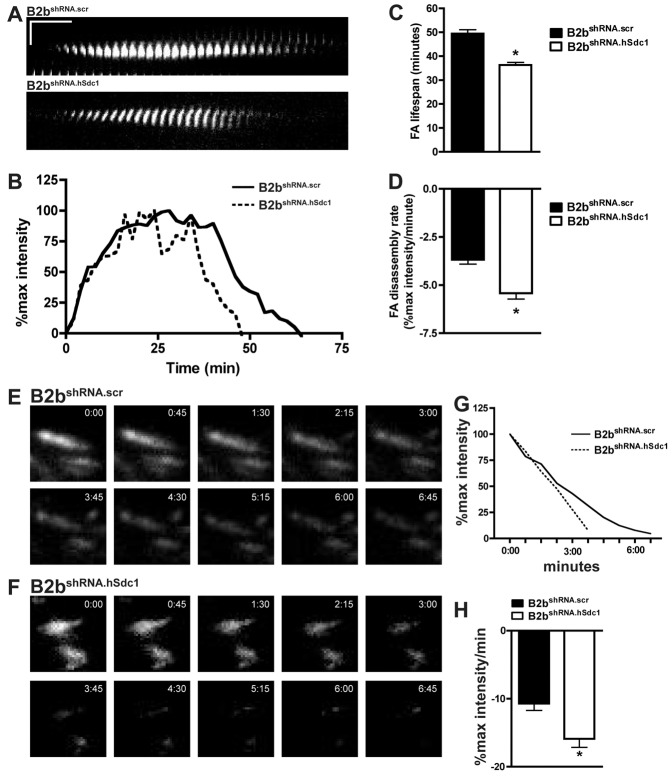
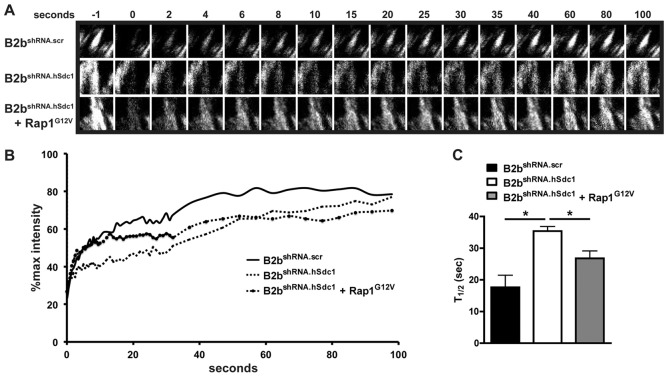
The difference in the leading edge FA lifespan in migrating cells was better illustrated by examining kymographs of individual FAs (Fig. 2A). To quantify the FA dynamics, the intensity of FAs of migrating cells was measured over time to follow adhesion complex assembly and disassembly (Fig. 2B). The FA lifespan was longer in B2bshRNA.scr compared to B2bshRNA.hSdc1 cells (49.5±1.6 versus 36.3±1.1 min, respectively; P<0.0001; Fig. 2C). Additionally, the disassembly rate was slower in migrating B2bshRNA.scr cells compared to B2bshRNA.hSdc1 cells (−3.68±0.23 versus −5.43±0.30% max intensity/min, respectively; P<0.0001; Fig. 2D). The FA assembly rate and FA size did not differ between cell lines (supplementary material Figs S1, S2). Thus, the shorter FA lifespan was largely driven by a faster disassembly of FAs in conditions lacking syndecan-1.
Fig. 2.
Syndecan-1 slows FA disassembly in lung epithelial cells. (A–D) Migrating B2bshRNA.scr and B2bshRNA.hSdc1 cells stably expressing paxillin–eGFP were observed by time-lapse TIRF microscopy (supplementary material Movie 1). (A) Kymographs revealed a longer FA lifespan in B2bshRNA.scr compared to B2bshRNA.hSdc1 cells. Horizontal line: 10 min; vertical line: 5 µm. (B) Normalized fluorescent intensity (% of max intensity) of the FAs in panel A over time. (C) The total FA lifespan and (D) the rate of the FA disassembly were determined for migrating lung epithelial cells. *P<0.0001 by Student's t-test; n = 4 independent experiments. (E–H) B2bshRNA.scr and B2bshRNA.hSdc1 stably expressing paxillin–eGFP were treated with nocodazole (10 µM; 4 h). After washing the cells with fresh medium, FA disassembly was observed with time-lapse TIRF microscopy in (E) B2bshRNA.scr and (F) B2bshRNA.hSdc1 cells. (G) The intensity curves of the FAs in panels E and F. (H) The FA disassembly rate was determined in B2bshRNA.scr and B2bshRNA.hSdc1 cells. *P<0.05 by Student's t-test; n = 3 independent experiments.
To further study FA dynamics, we evaluated FA disassembly with a nocodazole washout assay (Millon-Frémillon et al., 2008; Kaverina et al., 1999). Microtubule polymerization after nocodazole washout induced a rapid disassembly of FAs in lung epithelial cells. Congruous with the data from migrating cells, syndecan-1 also attenuated FA disassembly after nocodazole washout (Fig. 2E–H). The FA disassembly rate was more rapid in B2bshRNA.hSdc1 cells compared to B2bshRNA.scr cells (−15.90±1.25 versus −10.69±1.02% max intensity/min, respectively; P<0.05; Fig. 2H).
Syndecan-1 facilitates paxillin exchange within the FA
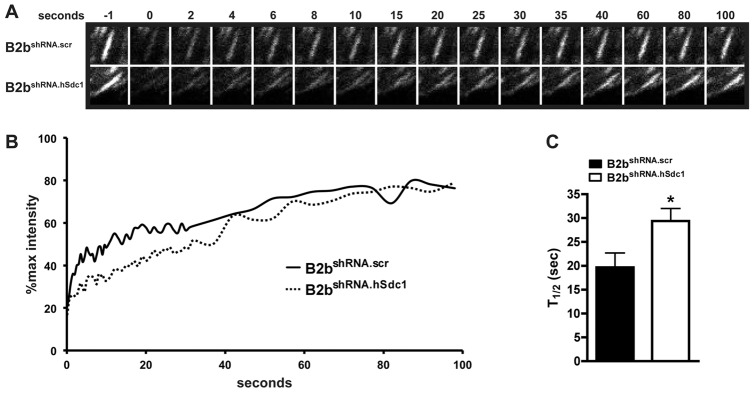
Differences in the exchange of adhesion complex proteins (e.g. paxillin, FAK, zyxin) between cytoplasmic pools and the stable FA controls cell migration by altering FA stability (von Wichert et al., 2003; Webb et al., 2004). FAs within migrating cells at the wound front were selectively photobleached, and the recovery of fluorescence was evaluated to quantify this process (Fig. 3A,B). The amount of the immobilized fraction was similar between the cell lines (unpublished data). However, the dynamics of paxillin cycling within the FA was different with B2bshRNA.scr cells having a shorter half-time to recovery of fluorescent signal in comparison to B2bshRNA.hSdc1 cells (19.68±3.03 versus 29.41±2.63 sec; P<0.05; Fig. 3C). These findings indicate that syndecan-1 alters the dynamic exchange of adhesion complex proteins in the FA thus regulating FA disassembly, which ultimately governs cell migration.
Fig. 3.
Syndecan-1 facilitates the recovery of fluorescence after photobleaching in migrating lung epithelial cells. A scratch wound was created on monolayers of B2bshRNA.scr and B2bshRNA.hSdc1 cells stably expressing paxillin–eGFP, and FAs at the wound front were identified for photobleaching. (A) Representative FA intensities prior to (−1 second) and after photobleaching. (B) The normalized intensity of the representative FA in A was plotted over time. (C) The FRAP recovery half-time was determined for B2bshRNA.scr and B2bshRNA.hSdc1 cells. More than 15 FAs were analyzed across three independent experiments. *P<0.05 by Student's t-test.
Syndecan-1 augments Rap1 activation to restrain cell migration
Small GTPases are vital for cell migration to occur (Raftopoulou and Hall, 2004; Ridley et al., 2003). Rap1 is a small GTPase that has pleotropic effects on cell migration and has been specifically implicated in modulating integrin affinity states (Boettner and Van Aelst, 2009). Because our previous work showed syndecan-1 regulates α2β1 integrin activation in lung epithelial cells (Chen et al., 2009), we evaluated if syndecan-1 regulates Rap1 activation.
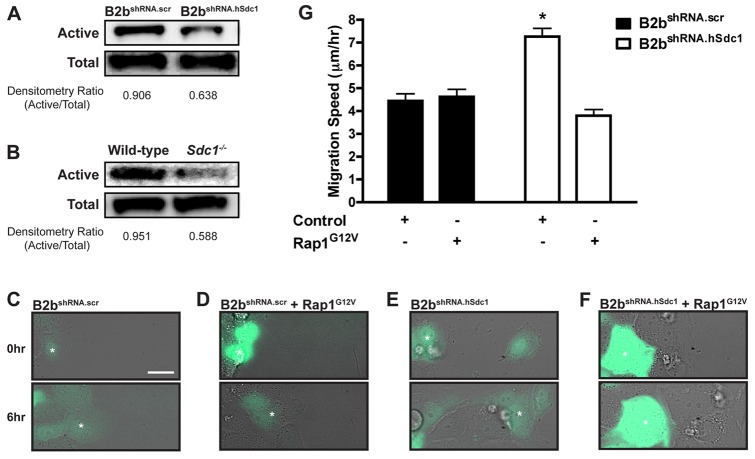
Rap1 was more activated in cells with syndecan-1 (B2bshRNA.scr cells and wild-type cultures) compared to cells that lack syndecan-1 (B2bshRNA.hSdc1 cells and Sdc1−/− cultures; Fig. 4A,B). To determine if syndecan-1 augmentation of Rap1 activity affected the α2β1 integrin affinity state, we overexpressed a dominant-active Rap1 (Rap1G12V) in lung epithelial cells. Surprisingly, overexpression of Rap1G12V had no effect on α2β1 integrin activation (supplementary material Fig. S3) indicating that syndecan-1 does not modulate integrin affinity via Rap1 activation.
Fig. 4.
Syndecan-1 restrains lung epithelial cell migration by activating Rap1. Western blot for active and total Rap1 in (A) B2bshRNA.scr and B2bshRNA.hSdc1 cells and (B) wild-type and Sdc1−/− primary ALI cultures. (C–F) Migration of (C) B2bshRNA.scr transduced with eGFP, (D) B2bshRNA.scr co-transduced with eGFP and Rap1G12V, (E) B2bshRNA.hSdc1 cells transduced with eGFP, or (F) B2bshRNA.hSdc1 cells co-transduced with eGFP and Rap1G12V (supplementary material Movie 2). The white asterisk identifies the same cell at 0 h and 6 h after migration. (G) Cell migration speed was measured for all conditions. *P<0.001 by one-way ANOVA and post-test Bonferroni analysis; n = 3 independent experiments.
Rap1 can affect cell migration independent of effects on the integrin affinity state (Takahashi et al., 2008; Jossin and Cooper, 2011). Therefore, we evaluated if syndecan-1 regulation of Rap1 modulates cell migration. Control conditions (eGFP expression in B2bshRNA.scr and B2bshRNA.hSdc1 cells) reproduced the phenotype where B2bshRNA.hSdc1 cells migrated faster than B2bshRNA.scr cells (supplementary material Movie 2; Fig. 4C,E). Co-expression of Rap1G12V and eGFP in B2bshRNA.hSdc1 cells slowed the migration of eGFP positive cells only and had no effect on B2bshRNA.scr cells (supplementary material Movie 2; Fig. 4D,F).
We quantified the migration speed in all conditions and found in comparison to B2bshRNA.scr and B2bshRNA.hSdc1 cells expressing eGFP only, Rap1G12V expression in B2bshRNA.hSdc1 cells slowed migration speed (4.45±0.30 versus 7.26±0.36 versus 3.80±0.27 µm/h, respectively; P<0.001; Fig. 4G). Rap1G12V transduction into B2bshRNA.scr cells did not change migration speed (4.62±0.33 µm/h) from control B2bshRNA.scr cells (4.45±0.30 µm/h) suggesting maximal activation of Rap1 in B2bshRNA.scr cells (Fig. 4G). Importantly, expression of Rap1G12V in B2bshRNA.scr cells did not slow migration whereas Rap1G12V expression in B2bshRNA.hSdc1 cells did reduce migration speed to the rate of B2bshRNA.scr control conditions, which in essence rescued the migration phenotype and indicated the effect of Rap1 is directly associated with syndecan-1.
Rap1 slows FA disassembly in syndecan-1-deficient lung epithelial cells
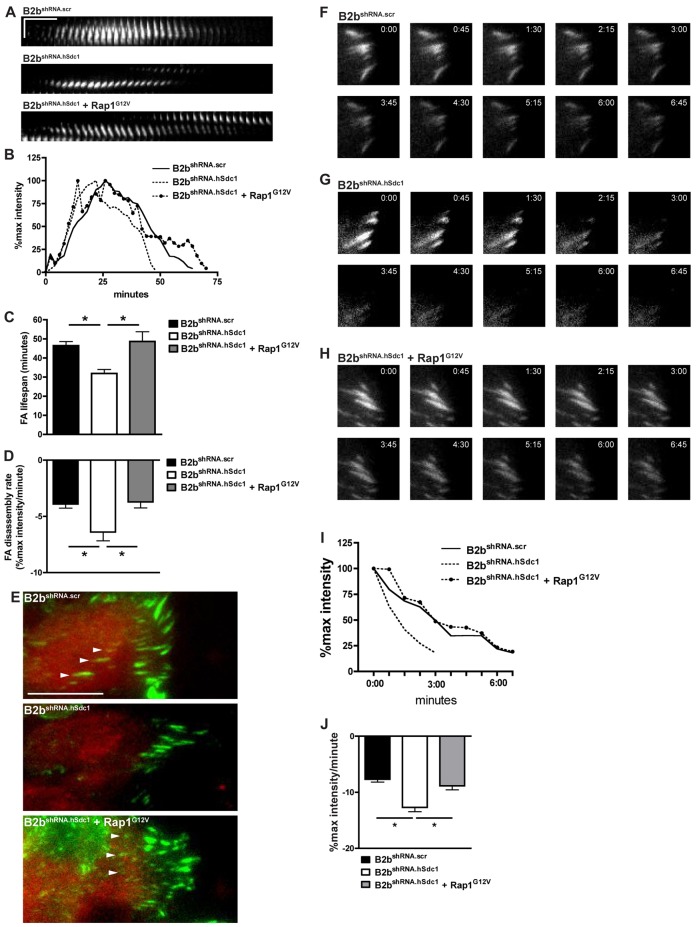
Our data indicate that syndecan-1 regulates cell migration by slowing FA disassembly (Fig. 2). Using B2bshRNA.scr and B2bshRNA.hSdc1 cells that stably express paxillin–eGFP, we transduced Rap1G12V and identified cells by co-expression of mCherry to determine if syndecan-1 regulates FA disassembly via Rap1. FA disassembly occurred faster in control (mCherry expression only) migrating B2bshRNA.hSdc1 cells compared to B2bshRNA.scr cells (Fig. 5). However, when B2bshRNA.hSdc1 cells expressed Rap1G12V, FA dynamics changed to resemble that of B2bshRNA.scr cells (Fig. 5A,B). Indeed, when compared to B2bshRNA.scr and B2bshRNA.hSdc1 cells, FA lifespan was prolonged in B2bshRNA.hSdc1 cells expressing Rap1G12V (46.46±2.18 versus 31.91±2.05 versus 48.67±5.13 min, respectively; P<0.05; Fig. 5C), and the FA disassembly rate was slower (−3.90±0.36 versus −6.40±0.77 versus −3.71±0.53% max intensity/min, respectively; P<0.001; Fig. 5D). Accordingly, B2bshRNA.scr cells and B2bshRNA.hSdc1 cells expressing Rap1G12V had FAs identified near the interior of the cell in contrast to B2bshRNA.hSdc1 cells, which is consistent with the data presented in Fig. 1 and congruous with the longer FA lifespan.
Fig. 5.
Rap1 slows FA disassembly in lung epithelial cells lacking syndecan-1 expression. (A–E) A scratch wound was created on monolayers of B2bshRNA.scr and B2bshRNA.hSdc1 cells stably expressing paxillin–eGFP, and FA assembly and disassembly was observed by time-lapse TIRF microscopy. All cells were identified by expression of mCherry. In control conditions (B2bshRNA.scr and B2bshRNA.hSdc1), cells were transduced with mCherry only, whereas B2bshRNA.hSdc1 + Rap1G12V cells were transduced with both mCherry and Rap1G12V. (A) Kymographs (horizontal line: 10 min; vertical line: 5 µm) and (B) intensity curves of FAs of the conditions indicated. (C) The total FA lifespan and (D) the rate of the FA disassembly were determined. *P<0.05 by one-way ANOVA and post-test Bonferroni analysis; n = 3 independent experiments. (E) FAs (arrowheads) are present in the center of B2bshRNA.scr cells and B2bshRNA.hSdc1 cells + Rap1G12V, and lacking in B2bshRNA.hSdc1 cells. Green, paxillin; Red, mCherry; Scale bar: 20 µm. (F–J) Cells stably expressing paxillin–eGFP were treated with nocodazole (10 µM; 4 h). In control conditions (B2bshRNA.scr and B2bshRNA.hSdc1), cells were transduced with mCherry only, whereas B2bshRNA.hSdc1 + Rap1G12V cells were transduced with both mCherry and Rap1G12V. After washing with fresh medium, FA disassembly was observed by time-lapse TIRF microscopy in (F) B2bshRNA.scr, (G) B2bshRNA.hSdc1 and (H) B2bshRNA.hSdc1 + Rap1G12V cells. (I) The intensity curves of the FAs in panels F–H. (J) The FA disassembly rate was determined in all conditions. *P<0.001 by one-way ANOVA and post-test Bonferroni analysis; n = 3 independent experiments.
We also evaluated FA disassembly with the nocodazole washout assay (Fig. 5F–H). After washout, FA disassembly was slower in B2bshRNA.scr cells (−7.67±0.50% max intensity/min) compared to B2bshRNA.hSdc1 cells (−12.70±0.75% max intensity/min; P<0.001; Fig. 5I,J) and was consistent with the data presented in Fig. 2H. In comparison to B2bshRNA.hSdc1 cells, Rap1G12V expression in B2bshRNA.hSdc1 cells again significantly slowed the FA disassembly rate after nocodazole washout (−8.85±0.69% max intensity/min; P<0.001).
Rap1 accelerates the exchange of adhesion complex proteins in syndecan-1-deficient lung epithelial cells
Migrating B2bshRNA.scr and B2bshRNA.hSdc1 cells stably expressing paxillin–eGFP had FAs photobleached to determine if the dynamic cycling of adhesion complex proteins is regulated by Rap1 activation. B2bshRNA.hSdc1 cells expressing Rap1G12V changed the dynamic exchange of paxillin within the stable FA to behave similarly to B2bshRNA.scr cells (Fig. 6A,B). Compared to the half-time of B2bshRNA.hSdc1 cells (35.40±1.45 sec), Rap1G12V expressed in B2bshRNA.hSdc1 cells significantly shortened the half-time of fluorescent recovery (26.77±2.37 sec; P<0.01; Fig. 6C).
Fig. 6.
Rap1 accelerates the recovery of fluorescent after photobleaching in migrating lung epithelial cells lacking syndecan-1 expression. A scratch wound was created on monolayers of B2bshRNA.scr and B2bshRNA.hSdc1 cells stably expressing paxillin–eGFP, and FAs at the wound front were identified for photobleaching. In control conditions (B2bshRNA.scr and B2bshRNA.hSdc1), cells were transduced with mCherry only whereas B2bshRNA.hSdc1 + Rap1G12V cells were transduced with both mCherry and Rap1G12V. (A) Representative FA intensities prior to (−1 sec) and after photobleaching. (B) The normalized intensity of the representative FA in A was plotted over time. (C) The FRAP recovery half-time was determined for B2bshRNA.scr and B2bshRNA.hSdc1 cells. More than 15 FAs were analyzed across three independent experiments. *P<0.01 by one-way ANOVA and post-test Bonferroni analysis.
Discussion
Syndecan-1 modulates re-epithelialization of the wounded lung epithelium (Chen et al., 2009). Here, we show that syndecan-1 controls the turnover of the leading edge FAs in lung epithelial cells to regulate cell migration. Our data demonstrate that syndecan-1 governs FA dynamics by regulating the exchange of adhesion complex proteins, which in turn slows FA disassembly and restrains migration. Moreover, we show that syndecan-1 controls this process by activating Rap1.
FA turnover in migrating cells is integral for generating traction forces as well as maintaining cell–matrix contacts (Broussard et al., 2008). Our data reveal that the syndecan-1 prolongs the leading edge FA lifespan and is consistent with other reports that demonstrate decreased FA turnover slows cell migration (Webb et al., 2004; Gupton and Waterman-Storer, 2006; Millon-Frémillon et al., 2008; Lawson et al., 2012). Maximal cell migration involves a complicated interplay between the cell and extracellular matrix. Indeed, intermediate levels of cell adhesion are ideal for maximal migration velocity (Palecek et al., 1997; DiMilla et al., 1993; Gupton and Waterman-Storer, 2006). However, the optimal substrate concentration can be shifted by altering the FA turnover dynamics (Gupton and Waterman-Storer, 2006; Millon-Frémillon et al., 2008).
Syndecan-1 had no effect on the assembly rate and primarily regulates the turnover of leading edge FAs in migrating cells by controlling FA disassembly. Furthermore, our data indicate that syndecan-1 restricts FA disassembly by increasing the trafficking of adhesion complex protein within the stable FA. This finding is consistent with others that have showed faster exchange of FA proteins stabilizes the FA and slows cell migration (Webb et al., 2004; von Wichert et al., 2003; Hamadi et al., 2005; Deramaudt et al., 2011). Phosphorylation of adapter proteins such as paxillin and FAK alter the kinetics of cycling and work in concert to stabilize the FA (Webb et al., 2004; Zaidel-Bar et al., 2007). FAs can also be destabilized by external factors such as targeting by microtubules to induce disassembly (Kaverina et al., 1998; Kaverina et al., 1999; Ezratty et al., 2005; Bhatt et al., 2002). Syndecan-1 could be modulating one or more of these factors in stabilizing the FA in migrating cells.
Our findings indicate that syndecan-1 mediates its effects on FA disassembly through Rap1. Rap1 is a Ras-family GTPase that associates with various effectors to change the way the cell interacts with the environment (Boettner and Van Aelst, 2009; Kooistra et al., 2007). By associating with either RapL or RIAM, Rap1 switches integrins to a high affinity state to govern cell adhesion (Han et al., 2006; Carmona et al., 2009; Katagiri et al., 2003). Rap1 did not affect α2β1 integrin activation in our studies. However, syndecan-1 can also regulate the allosteric state of the αvβ3 and αvβ5 integrins (Beauvais et al., 2009; Beauvais et al., 2004). Our previous work demonstrated that syndecan-1 primarily regulates cell migration via effects on the collagen binding integrin, α2β1, and was not dependent on the αvβ3 and αvβ5 integrins (Chen et al., 2009). Plus, our experiments were all conducted on collagen matrices corroborating the idea that Rap1 is facilitating FA disassembly independent of direct effects on integrin activation. However, we cannot rule out Rap1 effects on other integrins in the lung epithelium independent of syndecan-1. Re-organization of adherens junctions, which can be mediated by Rap1, is a large determinant of collective cell migration (Montell, 2008; Knox and Brown, 2002; Price et al., 2004; Hogan et al., 2004). In fact, cadherin and integrin mediated signals are highly interconnected so changes to the cell–cell junctions indirectly alters cell–matrix dynamics (Weber et al., 2011). Syndecan-1 has been functionally linked to E-cadherin so it is plausible that syndecan-1 modifies cell–cell contacts through Rap1 to circuitously control FA turnover (Leppä et al., 1996; Kato et al., 1995).
Syndecan-1 forms multimeric complexes to transduce intracellular signaling and alter cellular function (Beauvais and Rapraeger, 2010; Beauvais and Rapraeger, 2003; Beauvais et al., 2009; McQuade et al., 2006; Hayashida et al., 2008). Therefore, syndecan-1 could be directly interacting with Rap1 to facilitate its activation. Indeed, Rab5, a small GTPases, has been shown to associated with syndecan-1 through the syndecan-1 cytoplasmic domain to control syndecan-1 shedding (Hayashida et al., 2008). Alternatively, syndecan-1 could be associating with guanine nucleotide exchange factors (GEFs) and/or GTPase-activating proteins (GAPs). Rap1 is controlled by cycling between a GTP-bound (on) state and a GDP-bound (off) state, which is governed by GEFs and GAPs (Gloerich and Bos, 2011). Several Rap-1 GEFs and GAPs have PDZ domains that could associate with the PDZ binding domain on syndecan-1 to spatiotemporally control Rap1 (Nourry et al., 2003; García-Mata and Burridge, 2007).
Syndecan-1 shedding from the cell surface is an important process that regulates multiple cellular functions including stimulating cell migration (Bass et al., 2009; Endo et al., 2003; Chen et al., 2009). Therefore, we propose that syndecan-1 shedding from injured lung epithelium induces a migratory phenotype that is mediated by effects on Rap1. Our data indicate that the loss of syndecan-1 on lung epithelial cells causes Rap1 to assume an inactive state. In turn, the loss of Rap1 activity slows adhesion complex proteins exchange, which accelerates FA disassembly causing faster cell migration and facilitating wound closure.
Materials and Methods
Cloning
AAV vectors were used for transient transduction of exogenous genes in cultured epithelial cells. The AAV expression vector (AAV-DJ Helper Free Expression System; Cell Biolabs; San Diego, CA) was modified to include an internal ribosomal entry site (IRES)-eGFP immediately 3′ to the multiple cloning site by subcloning from the pBM-IRES-eGFP plasmid (Addgene; Cambridge, MA; deposited by Garry Nolan) with BamHI (5′) and SalI (3′) to create an AAV-IRES-eGFP vector. To create an AAV-IRES-mCherry vector, we subcloned into the AAV-IRES-eGFP vector used a KpnI restriction site that is located within the IRES and the SalI site 3′ to eGFP. The cDNA sequence for a 5′ KpnI restriction site, intervening IRES sequence, mCherry cDNA (GenBank: AB512478.1), and a 3′ SalI restriction site was synthesized (Genscript; Piscataway, NJ) and subcloned into the AAV-IRES-eGFP.
The mRNA sequence for human Rap1 (NM_001010935) was modified with a single point mutation (G→T) at position 307 resulting in a glycine (G) to valine (V) mutation of the 12th amino acid to create a constitutively active Rap1 (Rap1G12V) (Han et al., 2006). The cDNA sequence for Rap1G12V flanked by BamHI (5′) and XhoI (3′) restriction sites was commercially synthesized (Genscript) and subcloned into the AAV-IRES-eGFP and AAV-IRES-mCherry plasmids. The plasmid sequence of the final expression vectors were confirmed by DNA sequencing.
AAV vectors were created by transfecting HEK cells with calcium phosphate precipitation using 10 µg each of the expression, capsule and helper plasmids. HEK cells were collected 2 days after transfection, and AAV vectors were released by three freeze-thaw cycles. The viral supernate was added to cells for 24 hours two days prior to the desired experiment.
Cell culture
Subclones of BEAS-2b cells, a non-malignant immortalized bronchial epithelial cell line, were created by stably expressing shRNA toward syndecan-1 (B2bshRNA.hSdc1) or a scramble control (B2bshRNA.scr) as previously described (Chen et al., 2009). Cultures were maintained in bronchial epithelial growth medium (BEGM) supplemented with growth factors and retinoic acid (Lonza; Walkersville, MD). Selection of B2bshRNA.scr and B2bshRNA.hSdc1 cells were ensured by supplementing the culture medium with puromycin (5 µg/ml; Thermo Fisher Scientific; Waltham, MA) and periodic characterization of cell surface human syndecan-1 levels (clone BA38-RPE; Serotec; Raleigh, NC) with a Guava bench top flow cytometer (Millipore; Billerica, MA).
Paxillin tagged with a C-terminal eGFP in the pEGFP-N3 vector was purchased from Addgene (deposited by Rick Horwitz). B2bshRNA.scr and B2bshRNA.hSdc1 cells were transfected with the paxillin-eGFP plasmid using Lipofectamine 2000 (Invitrogen; Carlsbad, CA), and stable clones were sorted by FACS for GFP-positive cells (Aria II; BD Biosciences; San Jose, CA). Additionally, B2bshRNA.scr and B2bshRNA.hSdc1 cells that stably express paxillin–eGFP were confirmed to maintain the respective high and low expression of human syndecan-1 by flow cytometry (clone BA38-Alexa 647; Serotec; supplementary material Fig. S4).
Primary cultures of airway epithelial cells grown at an air-liquid interface (ALI) were created from wild-type and Sdc1-null (Sdc1−/−) C57BL/6 mice as previously described (Chen et al., 2009). Cultures were maintained in mouse tracheal epithelial cell (MTEC) growth medium supplemented with 2% Nuserum (BD Biosciences) (You et al., 2002).
Immunofluorescence assay
For immunofluorescence and live-cell assays, B2bshRNA.scr and B2bshRNA.hSdc1 cells were plated into chambered no. 1.5 glass coverglass (Nunc International; Rochester, NY) coated with rat tail Type I collagen (BD Biosciences) at 2 µg/cm2.
Cultures were fixed with 4% paraformaldehyde for 15 min at 37°C before being processed for immunofluorescence. Focal adhesions were identified by immunostaining for paxillin (clone 5H11; Millipore) followed by an anti-mouse Alexa Fluor 488 secondary antibody (Invitrogen). F-actin was labeled with phalloidin–Alexa Fluor 568 (Invitrogen). Immunofluorescence images were obtained with a DeltaVision Olympus IX71 inverted microscope using a 1.35 40×/1.35 NA U Plan Apo oil immersion objective.
Focal adhesion turnover assays
Live cell time-lapse microscopy was performed using a Nikon TiE inverted widefield fluorescence microscope that has a chamber to maintain cells at 37°C and 5% CO2. Images were obtained by total internal reflection fluorescent (TIRF) microscopy using a CFI 60×/1.49 NA Apo TIRF oil immersion objective.
Monolayers of B2bshRNA.scr and B2bshRNA.hSdc1 cells that stably express paxillin–eGFP were scratched with a p-100 pipette tip to simulate a wound. Cells were allowed to initiate migration for at least 2 h before starting time-lapse TIRF microscopy. Images were captured every 2 min for up to 6 hours. Each experiment was repeated at least three times. For each independent experiment, FAs at the leading edge of migrating cells were evaluated in at least 10 different randomly chosen cells. Within each cell, we analyzed between one and three FAs over time. Therefore, we evaluated ∼100 focal adhesions per condition.
FA disassembly was induced with a nocodazole washout assay (Millon-Frémillon et al., 2008; Kaverina et al., 1999). Sub-confluent B2bshRNA.scr and B2bshRNA.hSdc1 cells that stably express paxillin–eGFP were incubated with 10 µM of nocodazole in BEGM for 4 h. Cells were then thoroughly washed with warm BEGM immediately before the start of time-lapse TIRF microscopy. Images were captured every 45 sec for 30 min. Each experiment was repeated a minimum of three times. In each independent experiment, at least five different cells and two FAs per cell was evaluated. In all, we evaluated at least 30 FAs per condition.
FA assembly and disassembly rates were determined as previously described (Webb et al., 2004; Millon-Frémillon et al., 2008). Fluorescence intensity of FAs in the cells was measured in raw images with ImageJ (NIH). The background intensity was subtracted from the intensity values and was then normalized so that all values are a percent of the maximum intensity of the FA. The rate of the FA disassembly was determined by linear regression analysis of the linear portion of the descending fluorescent intensity. The slope of the declining FA fluorescent intensity represents the rate of FA disassembly with a more negative slope equating to faster disassembly.
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) experiments were performed on a Nikon A1R scanning laser confocal microscope using a CFI 60×/1.40 NA Plan Apo VC oil immersion objective. A Tokai Hit stage-top incubator was installed to maintain cells in a humidified chamber at 37°C and 5% CO2.
Monolayers of B2bshRNA.scr and B2bshRNA.hSdc1 cells that stably express paxillin–eGFP were scratched with a p-100 pipet tip to simulate a wound. Cells were allowed to initiate migration for at least 2 h before starting FRAP experiments. Leading edge FAs in cells at the wound front were photobleached with both the 405 and 488 laser at 50% intensity for 1 sec, which consistently achieved ∼80% photobleaching. After photobleaching, sequential images were obtained with a low laser intensity (i.e. <3%) to minimize any further photobleaching. The frequency of image acquisition was variable to optimize the resolution during the recovery period. Images were acquired every 0.5 sec for 10 sec, 1.0 sec for 20 sec, 5 sec for 1 min, and 10 sec for 1 min. Raw images were evaluated with the Nikon Elements AR software. The intensity of the FA was normalized so that all values are a percent of the initial intensity of the FA immediately prior to photobleaching.
The recovery half-time (T1/2) of the FRAP intensity curve was determined for each experiment. The fluorescent recovery curve is best described by the equation: I(t) = IE[1−e∧(−τt)], where IE is the maximum fluorescent intensity recovered after photobleaching, t is time, and τ is a constant that is determined after fitting the fluorescent intensity recovery curve to the exponential equation (Prism; Graphpad Software; La Jolla, CA). The half-time is calculated by the following equation: T1/2 = ln0.5/−τ. Experiments were repeated a minimum of three times with at least five FAs evaluated per experiment.
Rap1 assays
The activated and total Rap1 levels in cell lysates were determined by western blot using the Rap1 Activation Assay Kit (Millipore). In time-lapse microscopy experiments, control or Rap1G12V-transduced cells were identified by either eGFP or mCherry expression.
Statistics
Student's t-test was used to determine statistical significance between two conditions. One-way ANOVA with post-test Bonferroni analysis was used for multiple conditions. P<0.05 was considered statistically significant. All data are means ± standard error of the mean unless otherwise stated.
Supplementary Material
Acknowledgments
We thank Ron Seifert, PhD for his help with the imaging studies. The imaging studies were supported in part by the Mike and Lynn Garvey Cell Imaging Lab at the UW Institute for Stem Cell and Regenerative Medicine.
Footnotes
Funding
This work was supported by grants from the National Institutes of Health: [grant numbers HL084396 to P.C., HL103868 to P.C., HL086883 to W.A.A., and HL029594 to W.C.P.]; the American Heart Association Beginning Grant-in-Aid (to P.C.); the American Lung Association (to P.C.); and the Cystic Fibrosis Foundation Research Development Program (to W.C.P.). Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.109884/-/DC1
References
- Alexandrova A. Y., Arnold K., Schaub S., Vasiliev J. M., Meister J-J., Bershadsky A. D., Verkhovsky A. B. (2008). Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PLoS ONE 3, e3234 10.1371/journal.pone.0003234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass M. D., Morgan M. R., Humphries M. J. (2009). Syndecans shed their reputation as inert molecules. Sci. Signal. 2, pe18 10.1126/scisignal.264pe18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais D. M., Rapraeger A. C. (2003). Syndecan-1-mediated cell spreading requires signaling by alphavbeta3 integrins in human breast carcinoma cells. Exp. Cell Res. 286, 219–232 10.1016/S0014-4827(03)00126-5 [DOI] [PubMed] [Google Scholar]
- Beauvais D. M., Rapraeger A. C. (2010). Syndecan-1 couples the insulin-like growth factor-1 receptor to inside-out integrin activation. J. Cell Sci. 123, 3796–3807 10.1242/jcs.067645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais D. M., Burbach B. J., Rapraeger A. C. (2004). The syndecan-1 ectodomain regulates alphavbeta3 integrin activity in human mammary carcinoma cells. J. Cell Biol. 167, 171–181 10.1083/jcb.200404171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais D. M., Ell B. J., McWhorter A. R., Rapraeger A. C. (2009). Syndecan-1 regulates alphavbeta3 and alphavbeta5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J. Exp. Med. 206, 691–705 10.1084/jem.20081278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo K. A., Dembo M., Kaverina I., Small J. V., Wang Y. L. (2001). Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153, 881–888 10.1083/jcb.153.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A., Kaverina I., Otey C., Huttenlocher A. (2002). Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain. J. Cell Sci. 115, 3415–3425 [DOI] [PubMed] [Google Scholar]
- Boettner B., Van Aelst L. (2009). Control of cell adhesion dynamics by Rap1 signaling. Curr. Opin. Cell Biol. 21, 684–693 10.1016/j.ceb.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard J. A., Webb D. J., Kaverina I. (2008). Asymmetric focal adhesion disassembly in motile cells. Curr. Opin. Cell Biol. 20, 85–90 10.1016/j.ceb.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Carmona G., Göttig S., Orlandi A., Scheele J., Bäuerle T., Jugold M., Kiessling F., Henschler R., Zeiher A. M., Dimmeler S.et al. (2009). Role of the small GTPase Rap1 for integrin activity regulation in endothelial cells and angiogenesis. Blood 113, 488–497 10.1182/blood-2008-02-138438 [DOI] [PubMed] [Google Scholar]
- Chen P., Abacherli L. E., Nadler S. T., Wang Y., Li Q., Parks W. C. (2009). MMP7 shedding of syndecan-1 facilitates re-epithelialization by affecting alpha(2)beta(1) integrin activation. PLoS ONE 4, e6565 10.1371/journal.pone.0006565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. K., Vicente–Manzanares M., Zareno J., Whitmore L. A., Mogilner A., Horwitz A. R. (2008). Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couchman J. R. (2003). Syndecans: proteoglycan regulators of cell-surface microdomains? Nat. Rev. Mol. Cell Biol. 4, 926–938 10.1038/nrm1257 [DOI] [PubMed] [Google Scholar]
- Crosby L. M., Waters C. M. (2010). Epithelial repair mechanisms in the lung. Am. J. Physiol. Lung Cell Mol. Physiol. 298, L715–L731 10.1152/ajplung.00361.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deramaudt T. B., Dujardin D., Hamadi A., Noulet F., Kolli K., De Mey J., Takeda K., Rondé P. (2011). FAK phosphorylation at Tyr-925 regulates cross-talk between focal adhesion turnover and cell protrusion. Mol. Biol. Cell 22, 964–975 10.1091/mbc.E10-08-0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMilla P. A., Stone J. A., Quinn J. A., Albelda S. M., Lauffenburger D. A. (1993). Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J. Cell Biol. 122, 729–737 10.1083/jcb.122.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K., Takino T., Miyamori H., Kinsen H., Yoshizaki T., Furukawa M., Sato H. (2003). Cleavage of syndecan-1 by membrane type matrix metalloproteinase-1 stimulates cell migration. J. Biol. Chem. 278, 40764–40770 10.1074/jbc.M306736200 [DOI] [PubMed] [Google Scholar]
- Ezratty E. J., Partridge M. A., Gundersen G. G. (2005). Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7, 581–590 10.1038/ncb1262 [DOI] [PubMed] [Google Scholar]
- Galbraith C. G., Sheetz M. P. (1997). A micromachined device provides a new bend on fibroblast traction forces. Proc. Natl. Acad. Sci. USA 94, 9114–9118 10.1073/pnas.94.17.9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith C. G., Yamada K. M., Sheetz M. P. (2002). The relationship between force and focal complex development. J. Cell Biol. 159, 695–705 10.1083/jcb.200204153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García–Mata R., Burridge K. (2007). Catching a GEF by its tail. Trends Cell Biol. 17, 36–43 10.1016/j.tcb.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Gloerich M., Bos J. L. (2011). Regulating Rap small G-proteins in time and space. Trends Cell Biol. 21, 615–623 10.1016/j.tcb.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Gupton S. L., Waterman–Storer C. M. (2006). Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 125, 1361–1374 10.1016/j.cell.2006.05.029 [DOI] [PubMed] [Google Scholar]
- Hamadi A., Bouali M., Dontenwill M., Stoeckel H., Takeda K., Rondé P. (2005). Regulation of focal adhesion dynamics and disassembly by phosphorylation of FAK at tyrosine 397. J. Cell Sci. 118, 4415–4425 10.1242/jcs.02565 [DOI] [PubMed] [Google Scholar]
- Han J., Lim C. J., Watanabe N., Soriani A., Ratnikov B., Calderwood D. A., Puzon–McLaughlin W., Lafuente E. M., Boussiotis V. A., Shattil S. J.et al. (2006). Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 16, 1796–1806 10.1016/j.cub.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Hayashida K., Stahl P. D., Park P. W. (2008). Syndecan-1 ectodomain shedding is regulated by the small GTPase Rab5. J. Biol. Chem. 283, 35435–35444 10.1074/jbc.M804172200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C., Serpente N., Cogram P., Hosking C. R., Bialucha C. U., Feller S. M., Braga V. M. M., Birchmeier W., Fujita Y. (2004). Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 24, 6690–6700 10.1128/MCB.24.15.6690-6700.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A., Horwitz A. R. (2011). Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 3, a005074 10.1101/cshperspect.a005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y., Cooper J. A. (2011). Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat. Neurosci. 14, 697–703 10.1038/nn.2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K., Maeda A., Shimonaka M., Kinashi T. (2003). RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 4, 741–748 10.1038/ni950 [DOI] [PubMed] [Google Scholar]
- Kato M., Saunders S., Nguyen H., Bernfield M. (1995). Loss of cell surface syndecan-1 causes epithelia to transform into anchorage-independent mesenchyme-like cells. Mol. Biol. Cell 6, 559–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I., Rottner K., Small J. V. (1998). Targeting, capture, and stabilization of microtubules at early focal adhesions. J. Cell Biol. 142, 181–190 10.1083/jcb.142.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J. V. (1999). Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J. Cell Biol. 146, 1033–1044 10.1083/jcb.146.5.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J. V. (2002). Regulation of substrate adhesion dynamics during cell motility. Int. J. Biochem. Cell Biol. 34, 746–761 10.1016/S1357-2725(01)00171-6 [DOI] [PubMed] [Google Scholar]
- Knox A. L., Brown N. H. (2002). Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295, 1285–1288 10.1126/science.1067549 [DOI] [PubMed] [Google Scholar]
- Kooistra M. R. H., Dubé N., Bos J. L. (2007). Rap1: a key regulator in cell-cell junction formation. J. Cell Sci. 120, 17–22 10.1242/jcs.03306 [DOI] [PubMed] [Google Scholar]
- Laukaitis C. M., Webb D. J., Donais K., Horwitz A. F. (2001). Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153, 1427–1440 10.1083/jcb.153.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C., Lim S. T., Uryu S., Chen X. L., Calderwood D. A., Schlaepfer D. D. (2012). FAK promotes recruitment of talin to nascent adhesions to control cell motility. J. Cell Biol. 196, 223–232 10.1083/jcb.201108078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppä S., Vleminckx K., Van Roy F., Jalkanen M. (1996). Syndecan-1 expression in mammary epithelial tumor cells is E-cadherin-dependent. J. Cell Sci. 109, 1393–1403 [DOI] [PubMed] [Google Scholar]
- McQuade K. J., Beauvais D. M., Burbach B. J., Rapraeger A. C. (2006). Syndecan-1 regulates alphavbeta5 integrin activity in B82L fibroblasts. J. Cell Sci. 119, 2445–2456 10.1242/jcs.02970 [DOI] [PubMed] [Google Scholar]
- Millon–Frémillon A., Bouvard D., Grichine A., Manet–Dupé S., Block M. R., Albiges–Rizo C. (2008). Cell adaptive response to extracellular matrix density is controlled by ICAP-1-dependent beta1-integrin affinity. J. Cell Biol. 180, 427–441 10.1083/jcb.200707142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell D. J. (2008). Morphogenetic cell movements: diversity from modular mechanical properties. Science 322, 1502–1505 10.1126/science.1164073 [DOI] [PubMed] [Google Scholar]
- Morgan M. R., Humphries M. J., Bass M. D. (2007). Synergistic control of cell adhesion by integrins and syndecans. Nat. Rev. Mol. Cell Biol. 8, 957–969 10.1038/nrm2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C., Ding A. (2010). Nonresolving inflammation. Cell 140, 871–882 10.1016/j.cell.2010.02.029 [DOI] [PubMed] [Google Scholar]
- Nourry C., Grant S. G. N., Borg J-P. (2003). PDZ domain proteins: plug and play! Sci. STKE 2003, RE7 10.1126/stke.2003.179.re7 [DOI] [PubMed] [Google Scholar]
- Palecek S. P., Loftus J. C., Ginsberg M. H., Lauffenburger D. A., Horwitz A. F. (1997). Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature 385, 537–540 10.1038/385537a0 [DOI] [PubMed] [Google Scholar]
- Parsons J. T., Horwitz A. R., Schwartz M. A. (2010). Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price L. S., Hajdo–Milasinovic A., Zhao J., Zwartkruis F. J. T., Collard J. G., Bos J. L. (2004). Rap1 regulates E-cadherin-mediated cell-cell adhesion. J. Biol. Chem. 279, 35127–35132 10.1074/jbc.M404917200 [DOI] [PubMed] [Google Scholar]
- Raftopoulou M., Hall A. (2004). Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23–32 10.1016/j.ydbio.2003.06.003 [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003). Cell migration: integrating signals from front to back. Science 302, 1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Rikitake Y., Nagamatsu Y., Hara T., Ikeda W., Hirata K-I., Takai Y. (2008). Sequential activation of Rap1 and Rac1 small G proteins by PDGF locally at leading edges of NIH3T3 cells. Genes Cells 13, 549–569 10.1111/j.1365-2443.2008.01187.x [DOI] [PubMed] [Google Scholar]
- Vicente–Manzanares M., Choi C. K., Horwitz A. R. (2009). Integrins in cell migration – the actin connection. J. Cell Sci. 122, 199–206 10.1242/jcs.018564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wichert G., Haimovich B., Feng G-S., Sheetz M. P. (2003). Force-dependent integrin-cytoskeleton linkage formation requires downregulation of focal complex dynamics by Shp2. EMBO J. 22, 5023–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb D. J., Parsons J. T., Horwitz A. F. (2002). Adhesion assembly, disassembly and turnover in migrating cells – over and over and over again. Nat. Cell Biol. 4, E97–E100 10.1038/ncb0402-e97 [DOI] [PubMed] [Google Scholar]
- Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. (2004). FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6, 154–161 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- Weber G. F., Bjerke M. A., DeSimone D. W. (2011). Integrins and cadherins join forces to form adhesive networks. J. Cell Sci. 124, 1183–1193 10.1242/jcs.064618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrle–Haller B. (2012). Structure and function of focal adhesions. Curr. Opin. Cell Biol. 24, 116–124 10.1016/j.ceb.2011.11.001 [DOI] [PubMed] [Google Scholar]
- You Y., Richer E. J., Huang T., Brody S. L. (2002). Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell Mol. Physiol. 283, L1315–L1321 [DOI] [PubMed] [Google Scholar]
- Zaidel–Bar R., Milo R., Kam Z., Geiger B. (2007). A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120, 137–148 10.1242/jcs.03314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.