Summary
A variety of stress conditions induce mRNA and protein aggregation into mRNA silencing foci, but the signalling pathways mediating these responses are still elusive. Previously we demonstrated that PKA catalytic isoforms Tpk2 and Tpk3 localise with processing and stress bodies in Saccharomyces cerevisiae. Here, we show that Tpk2 and Tpk3 are associated with translation initiation factors Pab1 and Rps3 in exponentially growing cells. Glucose starvation promotes the loss of interaction between Tpk and initiation factors followed by their accumulation into processing bodies. Analysis of mutants of the individual PKA isoform genes has revealed that the TPK3 or TPK2 deletion affects the capacity of the cells to form granules and arrest translation properly in response to glucose starvation or stationary phase. Moreover, we demonstrate that PKA controls Rpg1 and eIF4G1 protein abundance, possibly controlling cap-dependent translation. Taken together, our data suggest that the PKA pathway coordinates multiple stages in the fate of mRNAs in association with nutritional environment and growth status of the cell.
Key words: Saccharomyces cerevisiae, Translation, PKA
Introduction
In eukaryotic translation initiation a closed loop complex forms on the mRNA (Wells et al., 1998). In this complex eIF4E interacts with the 5′ mRNA cap structure along with a molecular scaffold protein, eIF4G, and the eIF4A ATP-dependent RNA helicase. Pab1 interacts both with the mRNA 3′ poly(A) tail and with eIF4G. In addition, a 43S complex forms via the interaction of a host of translation initiation factors (eIF1, eIF1A, eIF3, eIF2 and eIF5) with the small ribosomal (40S) subunit (Pestova et al., 2007). The interaction of the closed loop mRNP complex with the 43S complex leads to the formation of the 48S pre initiation complex (Jackson et al., 2010).
In mammalian and yeast cells, PBs (processing bodies) have been defined as cytoplasmic bodies containing various components of the mRNA metabolic machinery such as Dcp1p, Dcp2p, Dhh1p, Pat1p, Lsm1p and Xrn1p (Sheth and Parker, 2003). A diverse array of functions has been ascribed to these RNA granules, including roles in mRNA localisation, degradation and storage as well as in the micro-RNA pathway (Anderson and Kedersha, 2006). PBs are dynamic structures affected by a range of cellular perturbations including glucose deprivation, osmotic stress, exposure to UV light, and stage of cell growth (Kedersha et al., 2005; Teixeira et al., 2005; Wilczynska et al., 2005). Another group of RNP granules containing mRNA, eIF4E, eIF4G, and Pab1, has been described to form in yeast during glucose deprivation and to either overlap or be distinct from PBs (Brengues and Parker, 2007; Hoyle et al., 2007). These granules, initially referred to as EGP-bodies, have been shown to be analogous to mammalian stress granules (SGs) (Buchan et al., 2008). SGs and PBs share some protein components, can dock or overlap with one another and can contain the same mRNAs (Kedersha et al., 2005), suggesting that mRNAs may move between these granules.
An unresolved issue concerning both SGs and PBs function is the mechanism that allows cells to rapidly adapt to stress and to re-establish a basal unstressed state. The involvement of signal transduction pathway enzymes into the dynamics of PB and SG formation and dissolution is an ideal mechanism for modulating mRNA function in response to changing cellular conditions. The role of post-translational modifications in translational control and mRNP granule formation has been recently discussed (Hilliker and Parker, 2008), and the possibility exists that the precise combination of modifications on proteins bound to specific mRNAs may dictate the localisation, translation and degradation rate of these individual mRNAs.
In Saccharomyces cerevisiae, cells starved for glucose or in stationary phase show a characteristic inhibition of protein synthesis and accumulation of mRNA in PBs and SGs (Ashe et al., 2000; Brengues and Parker, 2007; Hoyle et al., 2007). Recently, it has been proposed that glucose depletion inhibits translation initiation via loss of the eIF4A helicase from the preinitiation complex and a resulting temporary stabilization of the eIF3–eIF4G interaction on the 48S complex (Castelli et al., 2011).
The cAMP–PKA pathway in S. cerevisiae plays a major role in the control of metabolism, stress resistance and proliferation, particularly in connection with carbon source signalling. PKA is a hetero-tetramer composed of two regulatory subunits encoded by BCY1 gene, and two catalytic subunits encoded by three partially redundant genes, TPK1, TPK2 and TPK3 (Toda et al., 1987). The cAMP–PKA pathway is under positive control of an intracellular and extracellular glucose sensing system (Beullens et al., 1988; Rolland et al., 2000). This pathway is transiently hyperactivated upon addition of glucose to cells grown on a non-fermentable carbon source or to stationary-phase cells (Kraakman et al., 1999) and downregulated during stationary-phase or nutrient starvation (Santangelo, 2006).
PKA activity is also a requirement for the inhibition of translation when glucose becomes depleted (Ashe et al., 2000; Lui et al., 2010). Moreover, it has recently been demonstrated that PKA inhibits the aggregation of PBs by directly phosphorylating Pat1, a conserved constituent of these foci that functions as a scaffold during the assembly process (Ramachandran et al., 2011). We have recently analysed the subcellular localisation of PKA subunits from S. cerevisiae in fermentative, respiratory and stationary phases of growth and found that Tpk2 and Tpk3 isoforms, but not Bcy1, are associated with PBs and SGs during stationary phase, and that Tpk3 accumulates into PBs during glucose starvation and hyper osmotic stress (Tudisca et al., 2010).
In this work, we study the role of the PKA isoforms both in the formation of SGs and PBs as well as in translational regulation using two model conditions: glucose starvation and stationary phase. We observe that Tpk2 and Tpk3, but not Tpk1, are associated with translation initiation factors Pab1 and Rps3 in exponentially growing cells. Glucose starvation promotes the loss of interaction between Tpk2/3 and initiation factors followed by Tpk2 and Tpk3 accumulation into PBs. Tpk2 and Tpk3 show distinct mechanisms of accumulation in PBs. TPK3 deletion affects the capacity of the cells to form granules and arrest translation properly promoting as a consequence a more rapid resumption of translation under favourable conditions. However, once cells reach stationary phase, either TPK2 or TPK3 deletion reduces the ability of cells to inhibit translation and form PBs and SGs. Finally, TPK gene deletion increases the abundance of translation initiation factors such as eIF4G1 and Rpg1 (a subunit of eIF3 factor) in stationary phase. Taken together, these results show that the cAMP–PKA pathway coordinates multiple stages in the fate of mRNAs in association with the precise nutritional environment and growth status of the cell.
Results
Tpk1, Tpk2 and Tpk3 differentially associate with PBs and SGs under stationary phase or glucose starvation
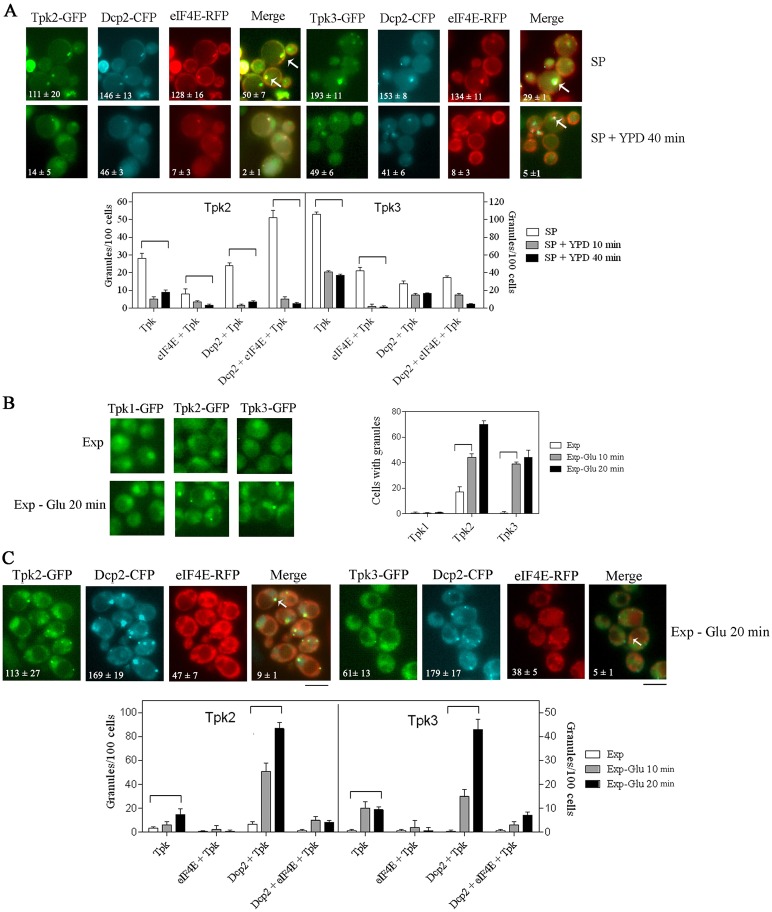
We have previously demonstrated that in S. cerevisiae cells, grown up to stationary phase, Tpk2 and Tpk3, but not Tpk1 nor Bcy1, colocalise with markers of PBs and markers of SGs (Tudisca et al., 2010). Since it has been described that SGs and PBs can dock or overlap with each other in a dynamic manner (Buchan et al., 2008; Hoyle et al., 2007) we decided to compare Tpk2 or Tpk3 distribution with markers of both PBs and SGs simultaneously in the same cell. The strategy used was to analyse chromosomally-tagged Tpk2–GFP or Tpk3–GFP colocalisation with Dcp2–CFP and eIF4E–RFP as PB or SG markers, respectively (Fig. 1).
Fig. 1.
Analysis of Tpk accumulation and distribution between PBs and SGs during stationary phase and glucose starvation. (A) Cells coexpressing Dcp2–CFP, eIF4E–RFP and Tpk2–GFP or Tpk3–GFP were grown to stationary phase (SP), and refed with YPD medium for 10 or 40 minutes. (B) Cells expressing Tpk1–GFP, Tpk2–GFP or Tpk3–GFP or (C) coexpressing Dcp2–CFP, eIF4E–RFP and Tpk2–GFP or Tpk3–GFP grown to exponential phase and subjected to glucose starvation for 10 or 20 minutes. The numbers in each image indicate the total number of Tpk2–GFP, Tpk3–GFP, Dcp2–CFP, eIF4E–RFP granules, and triple colocalisation granules/100 cells in the merged images. The arrows indicate triple colocalisation granules. Scale bar: 5 µm. The bar chart shows the distribution of Tpk–GFP-containing granules/100 cells under each condition. Values are means ± s.d. (n = 5). Brackets denote significant differences (P<0.005).
During stationary phase (SP), we have observed that 77% of the granules could be defined as PBs (50% with Dcp2 alone and 27% containing both Dcp2 and eIF4E), while the remaining 23% were SGs (containing exclusively eIF4E–RFP; Fig. 1A, SP). Fresh media addition promoted the dissolution of these granules (Fig. 1A, SP+YPD 40 min). Expression of Tpk2–GFP or Tpk3–GFP did not alter Dcp2–CFP or eIF4E–RFP protein expression levels or the proportion of each granule population when compared with an untagged Tpk version strain (data not shown).
The majority of Tpk2–GFP was found associated with PBs (P<0.005). However, Tpk3–GFP showed a different profile, with similar levels of colocalisation with PBs and SGs. Intriguingly, a large number of Tpk3 granules did not colocalise with either Dcp2 and/or eIF4E (P<0.005). However, we have previously observed that Tpk3 mostly colocalised with Dcp1 (Tudisca et al., 2010), suggesting that this Tpk3 granule population has a composition different from canonical SGs described in Saccharomyces cerevisiae. Glucose addition to stationary phase cultures promoted a dramatic and rapid dissolution of Tpk2- and Tpk3-containing granules (Fig. 1A, SP+YPD 40 min).
Previously we have demonstrated that acute glucose depletion from exponentially growing cells also causes Tpk3 accumulation into PBs (Tudisca et al., 2010). Here we have extended the analysis to investigate Tpk2 and Tpk1 localisation (Fig. 1B). When exponentially growing Tpk1–GFP-, Tpk2–GFP- or Tpk3–GFP-expressing yeasts were starved for glucose, Tpk2–GFP and Tpk3–GFP accumulated into cytoplasmic granules as soon as 10 minutes after glucose starvation. In contrast, the localisation of Tpk1–GFP was diffusely nucleo-cytoplasmic without a significant accumulation of granules (Fig. 1B). To characterize Tpk2–GFP and Tpk3–GFP containing cytoplasmic granules after glucose starvation, the colocalisation with coexpressing Dcp2–CFP and eIF4E–RFP was assessed (Fig. 1C). As in previous studies (Hoyle et al., 2007), at early time points after glucose starvation (20 minutes), Dcp2 and eIF4E total granule population was predominantly comprised by PBs (84%), with only 16% of very newly formed SGs. The distribution analysis of Tpk2–GFP and Tpk3–GFP granules indicates that both isoforms accumulate mainly in PBs (P<0.005).
Overall, it seems that each Tpk isoform shows a differential subcellular distribution following glucose starvation and subsequently, after entry into stationary phase. The cytoplasmic localisation of Tpk1 seems not to be affected by the nutritional status of the cell, whereas Tpk3 and Tpk2 accumulate into RNP-granules under the same nutrient limitation conditions that trigger translational arrest (Lui et al., 2010).
Tpk2 and Tpk3 show a differential dynamics and mechanism of association with PBs
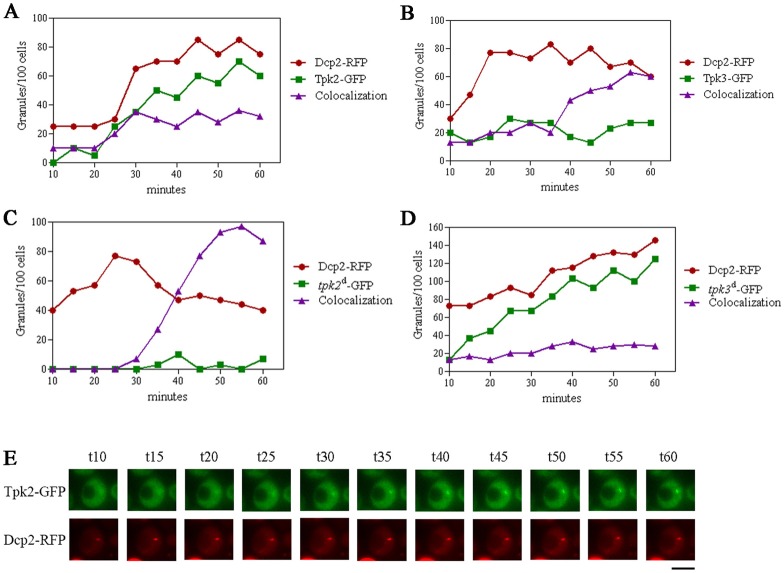
The above results suggest that Tpk2 and Tpk3 predominantly localise to PBs early after glucose starvation. To investigate the mechanism of Tpk2 and Tpk3 accumulation into PBs, we analysed Tpk–GFP granule formation in strains expressing Dcp2–RFP (PBs) following individual cells after glucose starvation. As shown in Fig. 2A, when cells are deprived of glucose, there is an increase in the amount of Dcp2, Tpk2 and colocalisation granules. Since exclusively Tpk2–GFP or Dcp2–RFP containing granules increase all along the stimulus (and there is not a decrease in the accumulation of one protein at the expense of another), we can conclude that Tpk2 localises to PBs at the same time as PBs are formed, with Dcp2 marking this event. However, even though both proteins likely assemble simultaneously into PBs, detailed inspection showed that the colocalising granules mature during the starvation (Fig. 2E). At early stages, Tpk2–GFP exhibits a diffuse pattern, with weak fluorescence intensity in the granules. As glucose starvation progresses, Tpk2–GFP granule intensity increases and thus the mature granules exhibit a well defined and delimited structure. This suggests that even though Tpk2 enters the granules when they form, new molecules of the kinase are added continuously during PB maturation.
Fig. 2.
Tpk2 and Tpk3 show different dynamics and mechanisms for their association with PBs. Strains DCP2–RFP TPK2–GFP (A), DCP2–RFP TPK3–GFP (B), DCP2–RFP tpk2d–GFP (C), DCP2–RFP tpk3d–GFP (D) grown to exponential phase on SCD were washed with SC and visualized by fluorescence microscopy at 5-minute intervals, over a period of 60 minutes. The experimental setup dictates that the earliest possible time for image acquisition is 10 minutes after glucose starvation. The graphs show the number of granules/100 cells. Granules were categorized as those containing only Dcp2–RFP, only Tpk–GFP, or both proteins (colocalisation). (E) A representative cell from the experiment shown in A. Scale bar: 5 µm.
In order to assess whether PKA activity is required for the localisation, an inactive kinase mutant of Tpk2 (Fig. 2C, tpk2d) is used. Kinase dead Tpk2–GFP accumulates almost exclusively in PBs with few granules containing exclusively Tpk2–GFP, indicating that Tpk2 activity is not required for the association with PBs but is required for the formation of Tpk2-only containing granules.
Tpk3–GFP does not colocalise with PBs at their formation (10-20 minutes) but much later; colocalising with Dcp2–RFP after 35 minutes of glucose starvation and then continuing to increase until the end of the experiment (Fig. 2B). This result indicates that in contrast to Tpk2, Tpk3 accumulates on pre-formed PBs. Using a strain carrying a Tpk3 kinase dead version (Fig. 2D, tpk3d), we observed significant aggregation of Tpk3 into granules that do not correspond to PBs. Indeed little or no colocalisation of the kinase dead form of Tpk3 was observed with Dcp2. Therefore, the kinase activity is required for the accumulation of Tpk3 into pre-formed PBs. The differences in the kinetics, and the kinase activity requirement, suggests that each Tpk isoform might have a fundamentally different mode of interaction with PBs.
Glucose dependent interaction between Tpk2 and Tpk3 with translation initiation factors in vivo
Translation initiation components eIF4E, eIF4G1 and Pab1 have been shown to accumulate in PBs after approximately 25 minutes of glucose starvation, concomitant with re-sedimentation of these factors away from ribosomal fractions of a sucrose density gradient (Hoyle et al., 2007). We have found Tpk2 and Tpk3, but not Tpk1 associated with PBs and SGs during translational inhibition caused by glucose starvation and stationary phase. To investigate if the PKA catalytic subunits, Tpk1, Tpk2 or Tpk3 are associated with translated mRNAs and ribosomes we performed cosedimentation analysis across sucrose gradients using extracts prepared before and after 20 minutes of glucose deprivation (Fig. 3A). As previously reported, following glucose depletion there is an accumulation of 80S monosomes via polysome run-off (Ashe et al., 2000), accompanied by a movement of the ribosomal proteins Rps3 and Rpl35A, among others, towards the 80S fractions (Hoyle et al., 2007).
Fig. 3.

Tpk2 and Tpk3 are associated in vivo with translation initiation complexes during exponential growth. (A) Polysomal profile analysis and immunoblots of 15–50% sucrose gradient fractions from cells expressing Tpk1-TAP (top), Tpk2-TAP (middle) or Tpk3-TAP (bottom) grown to exponential phase in YPD and subjected to glucose starvation for 20 minutes (YP). (B) Expression levels of each Tpk under exponential growth (YPD) or after glucose starvation (YP) were determined by immunoblot. The abundance of each Tpk represents 5% of the input fraction used in A. (C) Tpk2-TAP and untagged strains were purified from 20-minute glucose-starved (YP) or unstarved (YPD) cultures. Immunoprecipitated samples were subjected to western blot analysis with anti-TAP, anti-Pab1, anti-Rps3 and anti-Rpl35A antibodies. The input represents 1% of total protein used in the immunoprecipitation assay.
As shown in Fig. 3A, Tpk1 does not sediment with any ribosome associated fraction both during exponential growth and after glucose starvation (fractions 1–2, YPD and YP). In contrast, Tpk2 cofractioned with the free 40S ribosomal subunit in the presence of glucose (Fig. 3A, middle graph, YPD fractions 4–5). Improved 40S particle fractionation using shallower sucrose gradients confirms this result (supplementary material Fig. S3A). Moreover, the Tpk2 kinase dead version also cofractioned with the 40S ribosomal subunit (supplementary material Fig. S3B) suggesting that Tpk2 activity is not required for its co sedimentation with the small ribosomal subunit.
After glucose starvation, Tpk2 was found in the sub-monosomal fractions (fraction 4), probably reflecting changes in the association of Tpk2 isoforms with ribosomes. Tpk3 sedimentation pattern was similar to that observed for Tpk2 (Fig. 3A, lower panel, YPD versus YP), even though the results are not so clear due to the low expression levels of this isoform (Fig. 3B). These results highlight the possibility that both Tpk2 and Tpk3 associate with the small ribosomal subunit.
To further investigate this possibility, we undertook a tandem affinity purification (TAP) strategy. As shown in Fig. 3C, Tpk2 was found associated with Pab1 and with the small ribosomal subunit Rps3 when glucose is available. In contrast, Tpk2 was not found associated with the Rpl35A (60S ribosomal subunit). During glucose starvation, Pab1 and Rps3 were not immunoprecipitated with Tpk2. These results suggest that Tpk2, and possibly Tpk3, are associated with translation initiation components, such as Pab1 and Rps3 in exponentially growing cells and that following glucose starvation this interaction is compromised allowing the accumulation of Tpk2 and Tpk3 in RNP granules.
Tpk3 activity is required for proper granule accumulation in response to glucose starvation
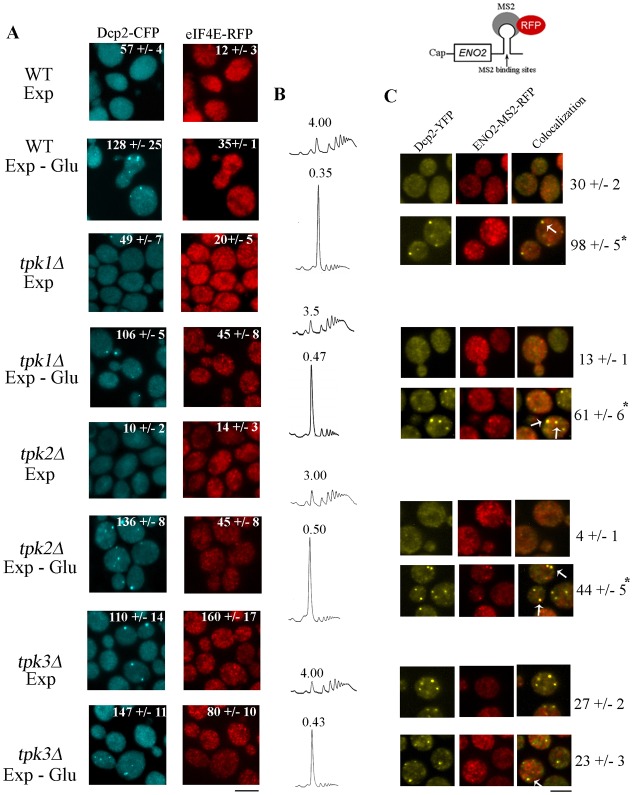
To analyse the role of PKA catalytic subunits in translational arrest and RNP granule accumulation following glucose starvation, we constructed strains deleted for each TPK gene and coexpressing Dcp2–CFP and eIF4E–RFP. In order to evaluate mRNA localisation to PBs, we constructed TPK deletion strains coexpressing Dcp2–YFP and ENO2-mRNA-MS2–RFP from their chromosomal locus. Deletion of the TPK genes individually did not affect Dcp2 or eIF4E protein levels (supplementary material Fig. S1B).
During exponential phase the number of Dcp2–CFP and eIF4E–RFP granules was significantly reduced in wild type (WT) cells when compared to cells in stationary phase (Fig. 1). In response to glucose starvation, the number of both Dcp2 and eIF4E-containing granules increased in the wild type strain (Fig. 4A, WT EXP versus WT EXP-Glu), consistent with the reduction in translational activity, as measured by the polysome/monosome ratio (Fig. 4B).
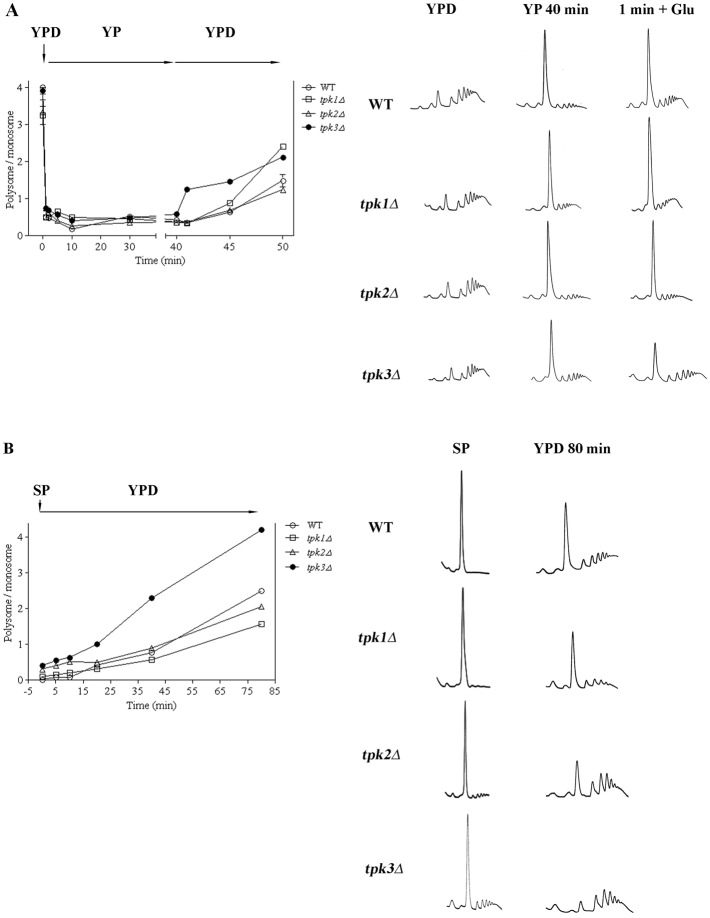
Fig. 4.
TPK3 deletion promotes aberrant granule accumulation during exponential phase. (A) WT, tpk1Δ, tpk2Δ or tpk3Δ cells coexpressing Dcp2–CFP and eIF4E–RFP, grown to exponential phase (Exp) and subjected to glucose starvation (Exp-Glu) for 20 minutes were visualized by fluorescence microscopy. The number in each image indicates the total number of Dcp2–CFP- or eIF4E–RFP-containing granules per 100 cells. Scale bar: 5 µm. (B) Polysomal profiles of the strains shown in A. The numbers represent the polysome/monosome area. (C) WT, tpk1Δ, tpk2Δ or tpk3Δ cells coexpressing Dcp2–YFP and ENO2-MS2–RFP mRNA were incubated as described in A. The numbers are the number of Dcp2–YFP granules containing ENO2-MS2–RFP mRNA/100 cells. The arrows indicate granules containing both Dcp2–YFP and ENO2-MS2–RFP. Scale bar: 5 µm. Values are mean ± s.d. (n = 5). *P<0.001 Exp versus Exp-Glu.
In response to starvation, mRNAs are stored in PBs and SGs (Lui et al., 2010). In order to analyse mRNA movement from cytoplasm to PBs under glucose starvation, we examined the number of Dcp2–YFP granules colocalising with a reporter ENO2 mRNA-MS2. ENO2 encodes the enolase enzyme that is required for both glycolysis and gluconeogenesis (McAlister and Holland, 1982). As shown Fig. 4C, when WT cells are starved for glucose, there is an increase in ENO2 mRNA/Dcp2–YFP colocalising granules indicating that ENO2 mRNA is stored in PBs probably as a result of the significant reduction in translation that occurs when cells shutdown fermentative metabolism. Control experiments indicate that ENO2 mRNA levels were not affected by MS2 tagging or by deletion of each Tpk (supplementary material Fig. S1A).
tpk1Δ and tpk2Δ strains were indistinguishable from WT cells in that translation initiation was inhibited and the cells accumulated both Dcp2 and eIF4E into granules in response to glucose starvation (Fig. 4A). The deletion of these isoforms does not impair ENO2-mRNA accumulation into PBs in response to glucose starvation (Fig. 4C). In contrast, PB and SG accumulation seems to be drastically affected in the absence of the Tpk3 isoform. Even though the translational inhibition caused by glucose withdrawal is still normal (Fig. 4B), this strain showed a high number of Dcp2–CFP and eIF4E–RFP containing granules even in exponentially growing cells that remain almost unchanged in number following glucose starvation (Fig. 4A). The abnormal granule accumulation observed in tpk3Δ cells is therefore not due to a defect in translation initiation. This result indicates that the inhibition of translation initiation and granule formation, which are normally tightly coupled, have become uncoupled in this strain. Moreover, the number of Dcp2–YFP granules that contain the ENO2 mRNA observed after glucose withdrawal in the tpk3Δ strain was drastically reduced when compared to either the WT or the other tpk mutants (Fig. 4C). Therefore, we can conclude that tpk3Δ strain fails to efficiently accumulate ENO2-mRNA into Dcp2 containing granules. By analogy with mRNA decay mutants such as xrn1Δ and dcp1Δ which accumulate granules constitutively (Sheth and Parker, 2003), these results suggest that TPK3 mutant has some rate limiting step in either mRNA decay or granule formation. If the TPK3 mutant had a problem in mRNA decay then mRNA should accumulate in the granules, however, as observed in Fig. 4C this is not the case since the majority of the Dcp2 containing granules are devoid of mRNA. This result is in agreement with a recent report that shows that PKA activity does not influence global mRNA decay (Ramachandran et al., 2011). We therefore favour the hypothesis that tpk3Δ mutant has some deficiency in granule formation which leads to the accumulation of partially formed or functionally compromised PBs.
To further characterize the Dcp2 and eIF4E containing granules that accumulate in vivo in the tpk3Δ strain, we performed an immunoblot analysis of granule enriched fractions (Teixeira et al., 2005) obtained from WT and tpk3Δ exponentially growing cells as well as from cells submitted to 20 minutes of glucose starvation. The pellet from glucose-starved WT cells was enriched for Dcp2, eIF4E, eIF4G1 and Pab1 (Fig. 5A) and these interactions were dependent on RNA (Fig. 5C). The absence of Pyk1 from the pellet fraction indicates that the granule preparation was free from soluble cytoplasmic contaminants. The presence of Dcp2 and eIF4E confirms our microscopy data (Fig. 4). In agreement with recent data (Castelli et al., 2011), Rps3 and eIF4A were not present in the granule fraction. Unlike WT cells, granule-enriched fractions obtained from tpk3Δ cells growing on glucose showed the presence of Rpg1 and Rps3 as well as Dcp2, eIF4G and Pab1. Protein expression levels remained similar before and after glucose starvation in WT and tpk3Δ cells (supplementary material Fig. S1B). Even though we have microscopically observed the formation of granules containing eIF4E in tpk3Δ strain (Fig. 4), this protein was not found in the granular fraction after biochemical analysis. These observations suggest that eIF4E granules formed in the presence of glucose in tpk3Δ strain are unlikely to correspond to SGs. A possibility is that these granules are more fragile and therefore are sensitive to biochemical fractionation.
Fig. 5.
Biochemical characterization of granules from Tpk mutant cells. WT or tpk3Δ strains were (A) grown to exponential phase (Exp) and then subjected to glucose starvation for 20 minutes (Exp-Glu 20 min) or (B) grown to stationary phase (SP) and re-fed with YPD for 10 minutes (SP+YPD). Immunoblots from granule-enriched fractions (P) and their corresponding supernatants (S). The graphs represent the relative abundance for each protein between the P and S fractions, determined by densitometric quantification of the bands. (C) P fractions from a WT strain after glucose starvation or tpk3Δ strain grown to exponential phase in the absence (−) or presence (+) of RNase A.
Except for Dcp2, the abundance of each protein in the pellet decreased after glucose starvation, suggesting that the granules formed in tpk3Δ strain are unstable and dynamic in response to glucose availability.
Overall, these results indicate that the granules that accumulate in cells lacking Tpk3 activity do not correspond to typical PBs or SGs (see Discussion).
PKA activity is required for proper translational arrest in stationary phase
The post-diauxic-shift growth phase is characterized by one to three doublings over a period of one week, after which cells enter stationary phase, and the yeast genome remains unreplicated (Werner-Washburne et al., 1993; Werner-Washburne et al., 1996; Zakrajšek et al., 2011). While overall rates of transcription and translation are diminished in stationary phase cells, the abundance of transcripts of some stress-responsive genes is increased (Davidson et al., 2011; Werner-Washburne et al., 1989). The Ras/PKA signalling pathway negatively controls the entry into stationary phase (Werner-Washburne et al., 1989).
To evaluate the role of the Tpk isoforms on the translational status of the cell during stationary phase, we analysed polysomal profiles, and followed mRNA accumulation in PBs and SGs. Using the same strains as in Fig. 4, stationary phase cells were resuspended in fresh YPD medium. Deletion of each TPK did not affect the expression levels of ENO2 mRNA during stationary phase or after YPD addition (supplementary material Fig. S1A). The expression levels of several translation initiation factors were similar in WT and tpkΔ strains with the exception of Rpg1 and eIF4G1 (see below, supplementary material Fig. S1D).
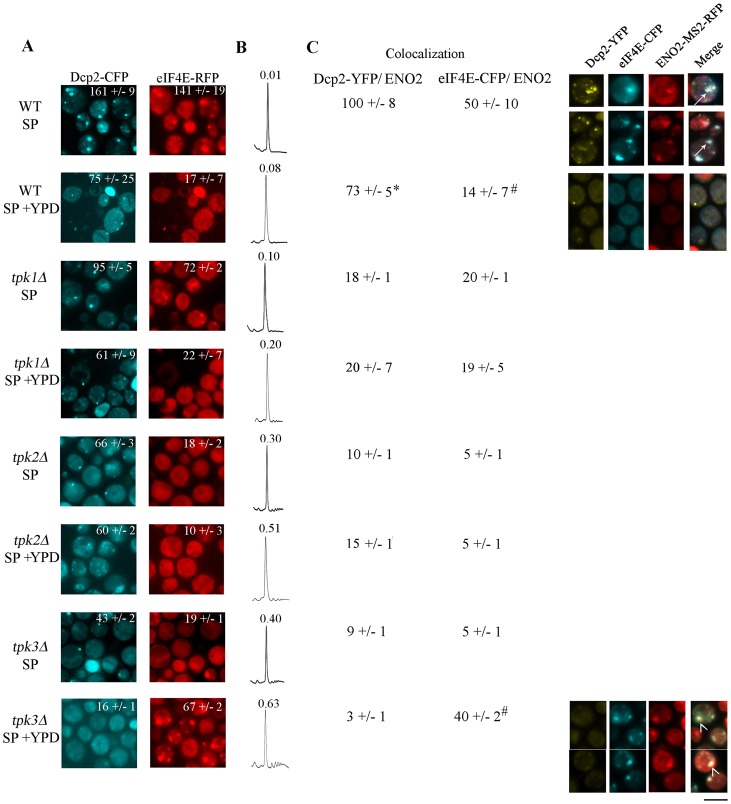
As shown Fig. 6A, seven-day-old cultures of WT cells showed an accumulation of Dcp2–CFP and eIF4E–RFP containing granules. The accumulation of these granules correlates with an extreme translational inhibition as shown by a very low polysome/monosome area ratio (Fig. 6B). As expected, PBs and SGs in stationary phase cells contain ENO2-mRNA, indicating that like the vast majority of mRNAs, ENO2 is translationally repressed. Colocalisation analysis shows that ENO2-mRNA is associated preferentially with Dcp2. A resumption of translation initiation in the first minutes after the addition of glucose to stationary phase WT cells can be observed in the slight recovery of the polysome/monosome ratio, a decrease in the number of both Dcp2 and eIF4E-containing granules, and a significant reduction in the level of ENO2-mRNA associated with PBs or SGs (Fig. 6A,C; WT SP versus WT SP+YPD).
Fig. 6.
PKA catalytic subunits are required for proper accumulation of PBs and SGs during stationary phase. (A) WT, tpk1Δ, tpk2Δ or tpk3Δ cells coexpressing Dcp2–CFP and eIF4E–RFP were grown to stationary phase (SP), refed with YPD (SP+YPD) for 10 minutes and visualized by fluorescence microscopy. The inset number in each picture indicates the total number of Dcp2–CFP or eIF4E–RFP/100 cells. (B) Polysomal profiles of the strains shown in A. Numbers represent the polysome/monosome area. (C) Cells coexpressing Dcp2–YFP, eIF4E–CFP and ENO2-MS2–RFP mRNA were incubated as described in A. Data are the number of Dcp2–YFP or eIF4E–CFP granules containing ENO2-MS2–RFP mRNA/100 cells. Values are mean ± s.d. n = 5). *P<0.0001 and #P<0.0001, SP versus SP+YPD. Scale bar: 5 µm.
Attainment of stationary phase for the TPK mutants is not different from the WT strain as judged by growth rate, budding capacity, and cell viability followed for 10 days (supplementary material Fig. S2; and data not shown). However deletion of TPK1, TPK2 or TPK3 seems to have different effects on the translational arrest in stationary phase. Deletion of the TPK1 partially impairs Dcp2–CFP and eIF4E–RFP accumulation into granules (Fig. 6A) and translational activity is slightly higher than in WT cells (Fig. 6B). Finally, colocalisation analysis of ENO2-mRNA in both Dcp2 and eIF4E containing granules suggest that tpk1Δ strains failed to accumulate mRNA into PBs and SGs. Glucose addition to the tpk1Δ strain promoted a reduction in the number of both PBs and SGs, in correlation with the higher translational activity (Fig. 6A–C; tpk1Δ SP versus tpk1Δ SP+YPD) and with a decrease in the number of mRNA containing granules.
In the TPK2 and TPK3 mutant strains, there was a drastic reduction in the level of Dcp2–CFP and eIF4E–RFP granules in stationary phase cells, and as consequence a very low level of ENO2-mRNA aggregation. These reduced levels of PBs and SGs correlated with the slightly improved levels of translation observed in the tpk2Δ and tpk3Δ (Fig. 6A–C; tpk2Δ SP, tpk3Δ SP versus WT SP). Glucose addition to stationary phase tpk2Δ cells promoted translational activation without great effect on the PBs and SGs pattern. Surprisingly, glucose addition to tpk3Δ cells promoted eIF4E–RFP to accumulate in granules with the ENO2 mRNA (Fig. 6A–C; tpk3Δ SP+YPD). Overall, the results shown in Fig. 6 indicate that deletion of each TPK promotes slightly higher translational levels in stationary phase; with TPK2 and TPK3 deletion being more severe than TPK1. The increased translational activity in stationary phase for the TPK mutants presumably explains their higher translational activities following re-addition of glucose. The TPK3 mutant in particular, promotes an aberrant eIF4E-ENO2 mRNA aggregation in response to addition of nutrients. Moreover, mRNAs trapped in polysomes by cycloheximide addition prior to YPD addition to tpk3Δ stationary cells did not prevent this eIF4E granule aggregation (data not shown), suggesting that these granules do not correspond to RNP granules produced by translational arrest (Parker and Sheth, 2007; Sheth and Parker, 2003). Recently, eIF4E has been postulated to function as a granule nucleation factor (Ferrero et al., 2012), therefore the eIF4E-ENO2 mRNA granules observed in tpk3Δ cells could be a reflection of a non canonical function of eIF4E.
In order to characterize the RNP granules observed during refeeding of stationary phase tpk3Δ cells, granule enriched fractions were prepared from stationary phase samples with and without nutrient resupplementation in tpk3Δ and WT cells. Stationary phase granules from the WT strain were enriched for several components of PBs and SGs, such as Dcp2, eIF4E, eIF4G1 and Pab1 (Fig. 5B). Dcp2 and eIF4E enrichment in the granular pellet was consistent with the microscopic results (Fig. 6). In addition, Rpg1, eIF4A, Rps3 and Rpl35A ribosomal subunit proteins were also found in the granular fraction. These observations differ from previous results in which Rpg1 was not found in granules (Brengues and Parker, 2007), however, the precise timing of stationary phase are different; here observations were made after 7 days of culture, whereas in Brengues and Parker (Brengues and Parker, 2007), microscopy was performed after only 3 days of stationary phase. It is entirely possible that in stationary phase there is a greater propensity for many dormant proteins to exist in aggregates as already described (Narayanaswamy et al., 2009). We think that the aggregation of the 80S particle into a cytoplasmic punctuate distribution could represent an economic mechanism to allow a quick translational reactivation once favourable nutritional conditions are restored.
When cells were refed with YPD, there was a redistribution of all the components found in the pellet towards the supernatant. This is consistent with the disassembly of granules observed in WT cells after fresh medium addition and with translation reactivation (Fig. 6, WT cells).
Even though the microscopic results show an increase in the amount of granules containing eIF4E after glucose readdition (Fig. 6B, tpk3Δ SP+YP), we did not observe enrichment of this protein in the granule fraction (Fig. 5). This could be due to the protein composition of these eIF4E granules affecting their sedimentation properties. However, this difference in the biochemical properties of eIF4E within granules in the tpk3Δ mutant does suggest that these eIF4E containing granules do not correspond to canonical PBs or SGs.
PKA affects protein abundance of Rpg1 and eIF4G1 translation factors during stationary phase
Expression level analysis of translation factors from stationary phase cells showed that specific factors, such as Rpg1 and eIF4G1 are more abundant in tpk1Δ, tpk2Δ and tpk3Δ strains than in a WT strain. This difference was not evident in exponentially growing cells (Fig. 7, lanes YPD). Semi-quantitative RT-PCR demonstrates that the differences observed in the abundance of these proteins is not caused by an increase of RPG1 and eIF4G1 (TIF4631) mRNA levels (Fig. 7B). These results indicate PKA is involved in the regulation of Rpg1 and eIF4G1 protein levels via a post-transcriptional mechanism during stationary phase. As Rpg1 is a subunit of eIF3, a factor involved in mRNA recruitment to the 43S translation complex via its interaction with the 40S ribosomal subunit (Chiu et al., 2010), and eIF4G1 is a limiting factor in the Cap-dependent mediated translation initiation (Berset et al., 1998), we suggest that a high accumulation of Rpg1 and eIF4G1 could explain the decrease in the shut-down of translational activity for the tpkΔ strains during stationary phase.
Fig. 7.
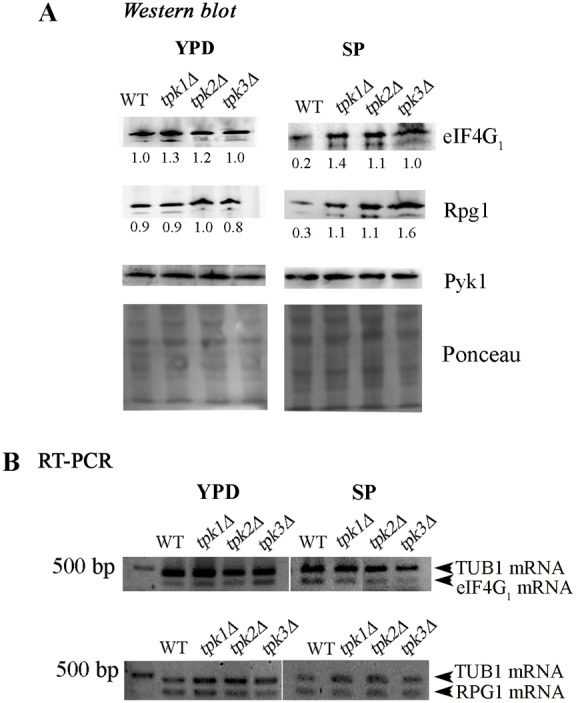
PKA regulates Rpg1 and eIF4G1 expression levels during stationary phase. (A) WT, tpk1Δ, tpk2Δ or tpk3Δ strains were grown to exponential (left panel) or stationary (right panel) phase in YPD. Endogenous expression levels of eIF4G1, Rpg1 and Pyk1 (control) were analysed by western blotting. The numbers under blots are the densitometric quantification of Rpg1 or eIF4G1 bands in relation with Pyk bands. (B) RT-PCR of eIF4G1 (TIF4631), RPG1 and TUB1 (control) mRNA during exponential (YPD) and stationary (SP) phase.
Even though in exponentially growing cells deletion of the individual TPK genes has little impact on Rpg1 and eIF4G1 levels (Fig. 7A), no changes were observed in Rpg1 and eIF4G1 levels after 20 minutes of glucose starvation or after 10 minutes of YPD re-supplementation to stationary phase cells (supplementary material Fig. S1), suggesting that the protein abundance of these factors are not immediately regulated by nutrient availability but strongly support that the PKA activity controls eIF4G1 and Rpg1 protein abundance during establishment of stationary phase.
PKA activity is required for the translational response to glucose availability and growth phase
PKA isoforms are functionally redundant for translational arrest during glucose starvation; however each TPK deletion differentially affects the translation inhibition capacity of the cell (Figs 4, 6). To further investigate the role of each Tpk on translational arrest and recovery dynamics in response to glucose availability we analysed in tpk1Δ, tpk2Δ and tpk3Δ strains the polysome profiles at various time points during the starvation-refeeding regime (Fig. 8A). The polysome profiles observed during exponential growth were similar in all the strains, indicating that the translational status of the cells is not grossly altered by the deletion of one Tpk gene. The kinetics of translational inhibition after glucose removal was indistinguishable between Tpk mutants and WT cells (Fig. 8A); in every case the level of polysomes decreased rapidly, reaching a minimum 10 minutes after glucose starvation. After glucose addition to starved cells, translational arrest of the three tpk mutant strains was recovered, but the kinetics of translational recuperation was different for each mutant. Surprisingly, 1–5 minutes after glucose readdition, the translational status observed for tpk3Δ strain was higher as a consequence of a faster recovery. In contrast, tpk1Δ and tpk2Δ did not show differences with the WT cells during the first 5 minutes of translational recuperation, but tpk1Δ reached similar values of translational recovery to tpk3Δ after 10 minutes of glucose addition; these values were significantly higher than those attained by WT and tpk2Δ strains.
Fig. 8.
Tpk3 controls translation activation dynamics. WT, tpk1Δ, tpk2Δ and tpk3Δ polysome profiles were analysed by sucrose gradient sedimentation at the different times points indicated in the figure. (A) Cells were grown to exponential phase in YPD, subjected to glucose starvation (YP) and re-fed with YPD. (B) Cells were grown to stationary phase and re-fed with YPD. The graphs show the polysome/monosome area ratio along the time course. Values are means ± s.d., n = 2. Representative polysome profiles of the times indicated are shown.
In stationary phase, the inhibition of translation for the tpk1Δ, tpk2Δ and tpk3Δ strains is not as severe as that in WT cells (Fig. 6B; Fig. 8B, SP samples). Even though all the strains recovered after glucose addition, the rate and level of recovery for the tpk3Δ were higher than for the WT, tpk1Δ and tpk2Δ cells. These results indicate that the PKA isoforms are functionally redundant in terms of their effects on translational inhibition during glucose starvation but not during the recovery phase. Specifically, the Tpk3 isoform seems to limit the translational recovery once glucose is added back to translational arrested cells.
Discussion
PKA role in translational arrest evoked by glucose starvation
During exponential growth on glucose, Tpk2 is located mainly in the nucleus, while Tpk1 and Tpk3 are homogeneously distributed between nucleus and cytoplasm (Tudisca et al., 2010). In response to glucose limitation translation is repressed and both PBs and SGs are formed (Hoyle et al., 2007). The reduction of translation initiation rates caused by glucose starvation involves alterations that take place at the level of protein-protein interactions within the 48S pre-initiation complex (Castelli et al., 2011).
In exponentially growing cells, we have observed that Tpk2 and Tpk3, but not Tpk1, cofractionate with the 48S preinitiation complex via their association with the 40S ribosomal subunit and/or Pab1 protein (Fig. 3). Similar to closed loop complex translation factors, glucose starvation promotes the dissociation of Tpk2 and – presumably – Tpk3 from the 48S pre-initiation complex allowing their accumulation in PBs and SGs (Fig. 1).
Protein kinase activity was necessary for both the accumulation of Tpk2 into granules and the association of Tpk3 with Dcp2 (Fig. 2). Recently it has been determined that specific glutamine- and/or asparagine-rich (Q/N-rich) regions in several PB components play key roles in the aggregation and assembly of PBs (Reijns et al., 2008). Protein sequence analysis of Tpk1, Tpk2 and Tpk3 showed that only Tpk2 has a Q/N rich region (19 Q, 2 N, 2P residues) positioned between Q9 and Q56 in the N-terminal region of Tpk2. Therefore, it is possible that the presence of the Q/N rich domain in Tpk2 allows or favours its interaction with translation factors and its accumulation in PBs independently of protein kinase activity.
The analysis of PB and SG formation in TPK mutants showed that the TPK3 deletion causes major alterations in both granule composition and accumulation since they are formed even in the presence of glucose. Close inspection of granule composition showed the presence of Rpg1 and Rps3 suggesting that deletion of TPK3 could lead to accumulation and aggregation of stalled 48S pre-initiation complexes, which differs from the situation in glucose-deprived cells where it has been suggested that the stalled 48S complexes breakdown to the closed loop complex prior to relocalisation to SGs (Castelli et al., 2011). The translation factors present in granules in the tpk3Δ strain resemble in part those present in stress granules induced by sodium azide, heat shock or ethanol stress (Buchan et al., 2011; Grousl et al., 2009; Kato et al., 2011). Interestingly, it has been reported that deletion of TPK3 promotes damage to the mitochondrial enzymatic content and an increase in the production of ROS (Chevtzoff et al., 2005). These results highlight the possibility that the absence of Tpk3 activity promotes RNP granule accumulation in response to defects in mitochondrial functionality. In the tpk3Δ mutant, we have observed no defects in the translational activity of cells growing on glucose or in the capacity of these cells to inhibit translation in response to glucose starvation. However, the kinetics of translational reactivation after glucose addition to arrested cells in the tpk3Δ strain was faster than in WT cells (Fig. 8A). It seems possible that the formation of stress granules even under glucose replete conditions would diminish the cells capacity to arrest translation correctly upon glucose starvation, thus favouring an immediate translational reactivation once glucose is readded.
While this work was being prepared for publication, a study has been published showing that PKA specifically inhibits the formation of the larger PB aggregates by direct phosphorylation of Pat1, a conserved constituent of PBs that functions as a scaffold during the assembly process (Ramachandran et al., 2011). In the light of our results, it seems likely that Tpk1, Tpk2 and Tpk3 are functionally redundant for Pat1 phosphorylation since all of the Tpk-deleted strains were able to accumulate PBs. Intriguingly Tpk2 and Tpk3 associate with PBs; where unphosphorylated Pat1 is expected. PKA targets residues in the C-terminus of Pat1, which is essential for the interaction with mRNA decapping factors, i.e. Dcp2, Edc4 and Lsm1-7 (Braun et al., 2010). It seems possible that the association of Tpk2 and Tpk3 with PBs occurs after the establishment of protein-protein interactions required for core PB formation (Decker et al., 2007).
A role for PKA in translational inhibition during stationary phase
The Ras/PKA signalling pathway in S. cerevisiae is thought to regulate the entry into stationary phase and thus the survival of cells within this resting state (Dechant and Peter, 2008; Herman, 2002).
Our results strongly suggest that Tpk2 and Tpk3 have a role in the translation inhibition that occurs during entry into stationary phase (Fig. 6). First, the deletion of either TPK2 or TPK3 produces a decrease in the cells capacity to accumulate Dcp2 and eIF4E containing granules that correlate with reduced translational inhibition even during the stationary phase of growth. Second, the TPK3 deletion, in particular, induced the accumulation of eIF4E granules associated with the ENO2 mRNA after glucose addition to stationary phase cells. Biochemical analysis of these granules (Fig. 5B) and their persistence after cycloheximide addition (data not shown) suggests that these eIF4E granules do not correspond to SGs (stress granules induced by glucose depletion). Third, the tpk3Δ strain showed a faster translational recovery from stationary phase (Fig. 8B) than the WT and other tpk mutants, which may be due to this aberrant granule formation.
In S. cerevisiae, it has been observed that nutritional deprivation or interruption of the TOR signal transduction pathway induces eIF4G1 degradation which would negatively regulate translation, allowing growth arrest in G1 phase. Degradation of this factor could be required for proper entry of the cells into the stationary phase (Berset et al., 1998). Here, for the first time, we have determined that Rpg1 protein abundance drastically decreases during stationary phase. On the contrary, tpk1Δ, tpk2Δ and tpk3Δ strains showed high abundance of eIF4G1 and Rpg1 proteins, with levels similar to those observed in cells growing in exponential phase (Fig. 7). This observation could justify, at least partly, the higher translational activity observed in TPK-deleted strains when compared with WT cells. Moreover, we observed that TPK-deleted strains grew quicker than WT cells during the diauxic shift as a consequence of a delay in cell cycle arrest (supplementary material Fig. S2). Our results suggest that PKA signalling would control the Rpg1 and eIF4G1 protein abundance allowing the entry into stationary phase (supplementary material Fig. S2; Fig. 7). The characterization of translation factors present in the PBs and SGs in stationary phase (Figs 5, 6) shows that the Rpl35A ribosomal protein is present. The possibility that the large ribosomal subunit is in granules during stationary phase highlights that a proportion of ribosomes may exist as 80S ribosomes. If these 80S ribosomes are mRNA associated, their presence in protein aggregates could facilitate a rapid resumption of translation in appropriate conditions.
Conclusion
Our results suggest that PKA may connect glucose availability with the cell cycle and with the robustness of cellular translation. PKA seems to be involved in multiple steps of translation regulation including interaction with the translational machinery, localisation in RNP granules, control of mRNA storage and granule composition, as well as the regulation of Rpg1 and eIF4G1 protein abundance. This work highlights the intricate and multitudinous network of inputs that signalling pathways, such as the Ras/PKA pathway, can have on fundamental processes such as the synthesis of proteins especially under changing cellular conditions.
Materials and Methods
Yeast strains and plasmids
The strains and plasmids used in this study are listed in supplementary material Tables S1 and S2.
Media, growth conditions and drug treatment
Strains were grown on rich medium containing 2% bactopeptone, 1% yeast extract and 2% glucose (YPD). Synthetic medium containing 0.67% yeast nitrogen base without amino acids, 2% glucose, plus the necessary additions to fulfil auxotrophic requirements (SCD) was used to maintain the selectable plasmid. Solid media contained 2% agar. Cells in stationary phase were obtained by growth on YPD for 7 days or in SCD for 3 days at 30°C. The re-feeding experiments were performed by transferring the cells to fresh YPD medium. Glucose-depleted cells were obtained from cells in exponential phase in YPD or SCD, washed twice with YP or SC medium, resuspension in the same medium lacking glucose for different times. For the cycloheximide treatment, cells were grown to exponential phase in YPD, treated with 100 µg/ml cycloheximide for 10 minutes before glucose addition in the presence of cycloheximide.
Crude extract western blotting
Strains were grown in the appropriate medium until exponential or stationary phase, lysed by disruption with glass beads at 4°C in an appropriate buffer (Portela et al., 2002), and centrifuged at 5000 g for 5 minutes. The crude extract was resolved by SDS-PAGE and analysed by western blotting. Blots were probed using the relevant primary antibody. Samples containing SG components were prepared as described previously (Teixeira et al., 2005). Pellet fraction was treated with 1 µg/µl RNase A for 30 minutes at room temperature. Pellets and supernatants were analysed by western blotting for the presence of translation factors and PB and SG markers. The immunoblots shown are representative of two independent experiments.
TAP protein purification
Yeast cultures were grown to an OD600 of 0.6; cells were pelleted, resuspended in YPD or YP, incubated for 20 minutes at 30°C, pelleted, snap frozen and ground under liquid nitrogen. TAP-affinity purification was carried out on the cell lysates as described previously (Castelli et al., 2011). TAP tagged proteins were detected with a horseradish peroxidase (HRP) conjugated primary antibody to Protein A (Abcam).
Sucrose density gradient sedimentation analysis
Sucrose density gradients were performed as described previously (Ashe et al., 2000). Briefly, cells were grown to stationary or exponential phase, resuspended in medium either with or without glucose and incubated at different times (as indicated in each figure). 10 µg/ml cycloheximide was added and cells were lysed with glass beads. Nine A260 units of pre-cleared lysate were loaded onto 15–50% linear sucrose gradients. After centrifugation for 2.5 h at 40,000 rpm using a SW41Ti rotor (Beckman), the gradients were fractionated from the top. The A254 was measured continuously using an ISCO UA6 gradient collection apparatus. Lower sucrose gradients (7.5–30%) (Nielsen et al., 2004) were centrifuged for 4.5 h at 35,000 rpm using a SW55Ti rotor (Beckman). Individual gradient fractions were collected and precipitated with 10% trichloroacetic acid, washed twice with acetone and resuspended in Laemmli buffer. Proteins were analysed by SDS-PAGE and western blotting.
Fluorescence microscopy
For fixed-time epifluorescence microscopy, cells were grown to stationary or exponential phase (OD600 of 0.6), incubated for 10 minutes with medium containing or not glucose and fixed with 7.4% formaldehyde. Confocal images were taken at room temperature by a confocal microscope (Eclipse E600; Nikon) using a 100×/0.5–1.3 NA plan APO oil objective and camera (Axiocam MRm). Images were acquired using Axiovision 4.5 software (Carl Zeiss MicroImaging, Inc.). Representative cells are shown from experiments repeated at least five times. Granules of approximately 100–200 nm diameter were counted in >100 cells. For clarity the images shown are single planes. The numbers on the images represent the mean ± s.d. of five independent experiments.
For time-course epifluorescence microscopy, cells were grown to stationary or exponential phase, washed twice with SC medium either with or without glucose and 2 µl of cell suspension was applied to poly-lysine-coated glass slides. Real-time 2D deconvoluted projections generated from continuous Z-sweep acquisition were captured at room temperature using a microscope system (Delta Vision RT; Applied Precision) with a 100×/1.40NA differential interference contrast oil plan Apo objective (Olympus) and camera (CoolSNAP HQ; Roper Scientific) using Softworx 1.1 software (Applied Precision) and 2×2 binning. Images were taken over the course of 1 h every 5 minutes, allowing 10 minutes initially for experimental set-up. A representative cell is shown from an experiment repeated at least five times. For colocalisation scoring per cell, replicates were analysed using ImageJ (National Institutes of Health).
RNA extraction and semi-quantitative RT-PCR
Total RNA extraction was performed using hot phenol protocol as described previously (Ocampo et al., 2009). Semi-quantitative reverse transcription-PCR (RT-PCR) of each mRNA was performed using the TUB1 gene as an internal standard. ENO2 for+963: 5′-TGACTTGACTGTCACCAACCCAGCTAGAATTG-3′, ENO2rev+1257: 5′-GGAAGTTTTCACCGGCGTAG-3′, ENO2for+166: 5′-AAGTGGATGGGTAAGGGTGTTATGAAC-3′, ENO2rev+294: 5′-CTTGGACTTGTTGGCGGTACCA-3′, TIF4631for+183: 5′-GAACTAAAGAAAGGTAAGCTGC-3′, TIF4631rev+466: 5′-CGATATGATTCCCAAGTAAATC-3′, eIF4G1for+203: 5′-GATATAACAACCGTGGCAAC-3′, eIF4G1rev+518: 5′-GAAGTAGAAGTAGAATCAGAAG-3′, TUB1for: 5′-CAAGGGTTCTTGTTTACCCATTC-3′, TUB1rev: 5′-GGATAAGACTGGAGAATATGAAAC-3′. The PCR products were electophoresed in a 2% agarose gel.
Statistical analysis of the data
All the experiments were repeated several times with independent cultures and enzymatic preparations. The data shown in figures were analysed using ANOVA and Tukey's HSD test, α: 0.05.
Supplementary Material
Acknowledgments
We are grateful to M. Pool (The University of Manchester) for the kind gift of α-Rps3 and α-Rpl35A antibodies, J. Gerst (Weizmann Institute for Science) for MS2 tagging reagents and R. Parker for the kind gift of pRP115. We are grateful to the Journal for Cell Science for a Travelling Fellowship.
Footnotes
Funding
This work was supported by a PhD fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas to V.T.; a European Molecular Biology Organization Short Term Fellowship to V.T.; the Agencia Nacional de Promoción Científica y Tecnológica PICT 2195 [grant number 2008-2195 to P.P.]; the University of Buenos Aires [grant number UBAX-528 to P.P.]; Consejo Nacional de Investigaciones Científicas y Técnicas [grant number PIP0519 to S.M.]; C.S. was supported by a Wellcome Trust project grant [grant number 088141/Z/09/Z to M.P.A.]; L.C. was supported by a Biotechnology and Biological Sciences Research Council [grant numbers Lola BB/G012571/1 to M.P.A.]; both N.H. and J.L. were supported by BBSRC funded studentships. Deposited in PMC for immediate release.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.111534/-/DC1
References
- Anderson P., Kedersha N. (2006). RNA granules. J. Cell Biol. 172, 803–808 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M. P., De Long S. K., Sachs A. B. (2000). Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11, 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset C., Trachsel H., Altmann M. (1998). The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95, 4264–4269 10.1073/pnas.95.8.4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beullens M., Mbonyi K., Geerts L., Gladines D., Detremerie K., Jans A. W., Thevelein J. M. (1988). Studies on the mechanism of the glucose-induced cAMP signal in glycolysis and glucose repression mutants of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 172, 227–231 10.1111/j.1432-1033.1988.tb13877.x [DOI] [PubMed] [Google Scholar]
- Braun J. E., Tritschler F., Haas G., Igreja C., Truffault V., Weichenrieder O., Izaurralde E. (2010). The C-terminal alpha-alpha superhelix of Pat is required for mRNA decapping in metazoa. EMBO J. 29, 2368–2380 10.1038/emboj.2010.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Parker R. (2007). Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2592–2602 10.1091/mbc.E06-12-1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Muhlrad D., Parker R. (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441–455 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Yoon J. H., Parker R. (2011). Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124, 228–239 10.1242/jcs.078444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli L. M., Lui J., Campbell S. G., Rowe W., Zeef L. A., Holmes L. E., Hoyle N. P., Bone J., Selley J. N., Sims P. F.et al. (2011). Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up-regulated. Mol. Biol. Cell 22, 3379–3393 10.1091/mbc.E11-02-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevtzoff C., Vallortigara J., Avéret N., Rigoulet M., Devin A. (2005). The yeast cAMP protein kinase Tpk3p is involved in the regulation of mitochondrial enzymatic content during growth. Biochim. Biophys. Acta 1706, 117–125 10.1016/j.bbabio.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Chiu W. L., Wagner S., Herrmannová A., Burela L., Zhang F., Saini A. K., Valásek L., Hinnebusch A. G. (2010). The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 30, 4415–4434 10.1128/MCB.00280-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G. S., Joe R. M., Roy S., Meirelles O., Allen C. P., Wilson M. R., Tapia P. H., Manzanilla E. E., Dodson A. E., Chakraborty S.et al. (2011). The proteomics of quiescent and nonquiescent cell differentiation in yeast stationary-phase cultures. Mol. Biol. Cell 22, 988–998 10.1091/mbc.E10-06-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R., Peter M. (2008). Nutrient signals driving cell growth. Curr. Opin. Cell Biol. 20, 678–687 10.1016/j.ceb.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Decker C. J., Teixeira D., Parker R. (2007). Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437–449 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero P. V., Layana C., Paulucci E., Gutiérrez P., Hernández G., Rivera–Pomar R. V. (2012). Cap binding-independent recruitment of eIF4E to cytoplasmic foci. Biochim. Biophys. Acta 1823, 1217–1224 10.1016/j.bbamcr.2012.03.013 [DOI] [PubMed] [Google Scholar]
- Grousl T., Ivanov P., Frýdlová I., Vasicová P., Janda F., Vojtová J., Malínská K., Malcová I., Nováková L., Janosková D.et al. (2009). Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122, 2078–2088 10.1242/jcs.045104 [DOI] [PubMed] [Google Scholar]
- Haim L., Zipor G., Aronov S., Gerst J. E. (2007). A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat. Methods 4, 409–412 [DOI] [PubMed] [Google Scholar]
- Herman P. K. (2002). Stationary phase in yeast. Curr. Opin. Microbiol. 5, 602–607 10.1016/S1369-5274(02)00377-6 [DOI] [PubMed] [Google Scholar]
- Hilliker A., Parker R. (2008). Stressed out? Make some modifications! Nat. Cell Biol. 10, 1129–1130 10.1038/ncb1008-1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle N. P., Castelli L. M., Campbell S. G., Holmes L. E., Ashe M. P. (2007). Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179, 65–74 10.1083/jcb.200707010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Hellen C. U., Pestova T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Yamamoto Y., Izawa S. (2011). Severe ethanol stress induces assembly of stress granules in Saccharomyces cerevisiae. Yeast 28, 339–347 10.1002/yea.1842 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke–Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman L., Lemaire K., Ma P., Teunissen A. W., Donaton M. C., Van Dijck P., Winderickx J., de Winde J. H., Thevelein J. M. (1999). A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32, 1002–1012 10.1046/j.1365-2958.1999.01413.x [DOI] [PubMed] [Google Scholar]
- Lui J., Campbell S. G., Ashe M. P. (2010). Inhibition of translation initiation following glucose depletion in yeast facilitates a rationalization of mRNA content. Biochem. Soc. Trans. 38, 1131–1136 10.1042/BST0381131 [DOI] [PubMed] [Google Scholar]
- McAlister L., Holland M. J. (1982). Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J. Biol. Chem. 257, 7181–7188 [PubMed] [Google Scholar]
- Narayanaswamy R., Levy M., Tsechansky M., Stovall G. M., O'Connell J. D., Mirrielees J., Ellington A. D., Marcotte E. M. (2009). Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA 106, 10147–10152 10.1073/pnas.0812771106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K. H., Szamecz B., Valásek L., Jivotovskaya A., Shin B. S., Hinnebusch A. G. (2004). Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 23, 1166–1177 10.1038/sj.emboj.7600116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo J., Fernandez Nuñez L., Silva F., Pereyra E., Moreno S., Garre V., Rossi S. (2009). A subunit of protein kinase a regulates growth and differentiation in the fungus Mucor circinelloides. Eukaryot. Cell 8, 933–944 10.1128/EC.00026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T., Takano I. (1980). Mutants Showing Heterothallism from a Homothallic Strain of SACCHAROMYCES CEREVISIAE. Genetics 94, 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Pestova T. V., Lorsch J. R., Hellen C. U. T. (2007). Mechanisms of translation initiation in eukaryotes. Translational Control in Biology and Medicine (ed. Mathews M B, Sonenberg N, Hershey J W B.), pp. 87–128 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Portela P., Howell S., Moreno S., Rossi S. K. (2002). In vivo and in vitro phosphorylation of two isoforms of yeast pyruvate kinase by protein kinase A. J. Biol. Chem. 277, 30477–30487 [DOI] [PubMed] [Google Scholar]
- Ramachandran V., Shah K. H., Herman P. K. (2011). The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43, 973–981 10.1016/j.molcel.2011.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M. A., Alexander R. D., Spiller M. P., Beggs J. D. (2008). A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121, 2463–2472 10.1242/jcs.024976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., De Winde J. H., Lemaire K., Boles E., Thevelein J. M., Winderickx J. (2000). Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38, 348–358 10.1046/j.1365-2958.2000.02125.x [DOI] [PubMed] [Google Scholar]
- Rothstein R. (1991). Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194, 281–301 10.1016/0076-6879(91)94022-5 [DOI] [PubMed] [Google Scholar]
- Santangelo G. M. (2006). Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 253–282 10.1128/MMBR.70.1.253-282.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia–Sanchez M. A., Brengues M., Parker R. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. (1987). Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudisca V., Recouvreux V., Moreno S., Boy–Marcotte E., Jacquet M., Portela P. (2010). Differential localization to cytoplasm, nucleus or P-bodies of yeast PKA subunits under different growth conditions. Eur. J. Cell Biol. 89, 339–348 10.1016/j.ejcb.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140 10.1016/S1097-2765(00)80122-7 [DOI] [PubMed] [Google Scholar]
- Werner–Washburne M., Becker J., Kosic–Smithers J., Craig E. A. (1989). Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J. Bacteriol. 171, 2680–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner–Washburne M., Braun E., Johnston G. C., Singer R. A. (1993). Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57, 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner–Washburne M., Braun E. L., Crawford M. E., Peck V. M. (1996). Stationary phase in Saccharomyces cerevisiae. Mol. Microbiol. 19, 1159–1166 10.1111/j.1365-2958.1996.tb02461.x [DOI] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. (2005). The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 118, 981–992 10.1242/jcs.01692 [DOI] [PubMed] [Google Scholar]
- Zakrajšek T., Raspor P., Jamnik P. (2011). Saccharomyces cerevisiae in the stationary phase as a model organism–characterization at cellular and proteome level. J. Proteomics 74, 2837–2845 10.1016/j.jprot.2011.06.026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.