Abstract
This is a report on a symposium sponsored by the American Society for Pharmacology and Experimental Therapeutics and held at the Experimental Biology 2012 meeting in San Diego, California, on April 25, 2012. The symposium speakers summarized and critically evaluated our current understanding of the physiologic, pharmacological, and toxicological roles of NADPH–cytochrome P450 oxidoreductase (POR), a flavoprotein involved in electron transfer to microsomal cytochromes P450 (P450), cytochrome b5, squalene mono-oxygenase, and heme oxygenase. Considerable insight has been derived from the development and characterization of mouse models with conditional Por deletion in particular tissues or partial suppression of POR expression in all tissues. Additional mouse models with global or conditional hepatic deletion of cytochrome b5 are helping to clarify the P450 isoform- and substrate-specific influences of cytochrome b5 on P450 electron transfer and catalytic function. This symposium also considered studies using siRNA to suppress POR expression in a hepatoma cell–culture model to explore the basis of the hepatic lipidosis phenotype observed in mice with conditional deletion of Por in liver. The symposium concluded with a strong translational perspective, relating the basic science of human POR structure and function to the impacts of POR genetic variation on human drug and steroid metabolism.
Introduction
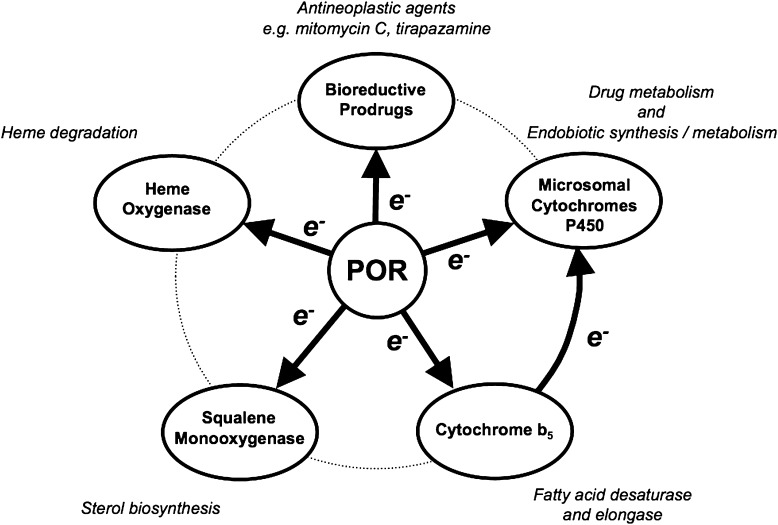
Cytochrome P450 (P450) enzymes located in the endoplasmic reticulum of hepatocytes and a wide range of additional cell types play important roles in xenobiotic biotransformation and in both the biosynthesis and metabolism of numerous endogenous substances (e.g., vitamins, cholesterol, bile acids, and steroid hormones). These microsomal P450s function in partnership with their obligate electron donor enzyme known as NADPH–cytochrome P450 oxidoreductase (CPR or POR). The physiologic functions of POR are quite diverse (Hart and Zhong, 2008), a reflection of the ability of POR to donate electrons to multiple acceptors in addition to the microsomal P450s (Fig. 1). Electron transfer from POR to cytochrome b5, a small microsomal hemoprotein, is important in fatty acid metabolism since it supports fatty acid desaturase and elongase activities. Cytochrome b5 also plays a role in electron transfer to the microsomal P450s. Squalene mono-oxygenase utilizes electrons donated by POR to support sterol biosynthesis. An additional electron acceptor is heme oxygenase, the enzyme that degrades heme to biliverdin, iron, and carbon monoxide. As well, POR can directly catalyze the one-electron reductive bioactivation of prodrugs such as the antineoplastic agents mitomycin C and tirapazamine.
Fig. 1.
The acceptors for electron transfer from POR and their roles in physiology, pharmacology, and toxicology.
As a “partner” enzyme, POR is often viewed as functioning in the background, with the electron acceptors such as the P450s taking center stage in pharmacology and toxicology investigations. This symposium placed the POR enzyme squarely in the spotlight to scrutinize critically its biologic roles in animal and cellular models and in human drug and steroid metabolism. Germ-line deletion of Por in mice results in multiple developmental defects and embryonic lethality (Shen et al., 2002; Otto et al., 2003), confirming the importance of POR in normal physiology. Recent years have witnessed the generation of a panel of mouse models with conditional knockout of Por in particular tissues/cell types, e.g., hepatic reductase null (HRN) mice (Henderson et al., 2003) or liver-Cpr-null (LCN) mice (Gu et al., 2003). This symposium considered the insights that such genetically modified mouse models have provided into the physiologic functions of the P450/POR system in various organs, as well as the roles of hepatic versus extrahepatic P450/POR systems in drug metabolism and xenobiotic toxicity. Data from additional mouse models with global or conditional hepatic knockout of cytochrome b5 highlighted in this symposium helped to clarify the P450 isoform- and substrate-specific influences of cytochrome b5 on P450 electron transfer and catalytic function (Finn et al., 2008; Finn et al., 2011).
Conditional hepatic Por deletion in HRN and LCN mice results in a profound decrease in all hepatic microsomal P450 functions, a compensatory increased expression of several P450 proteins, reduced circulating levels of cholesterol and triglycerides, and hepatic lipidosis (Gu et al., 2003; Henderson et al., 2003). This symposium considered studies using small interfering RNA (siRNA) to suppress POR expression in a hepatoma cell–culture model to explore the basis of the hepatic lipidosis phenotype (Porter et al., 2011).
The symposium concluded with a consideration of the impact of POR genetic variation on human drug and steroid metabolism. Human POR deficiency is associated with disordered steroidogenesis and the Antley-Bixler skeletal malformation syndrome (ABS). In addition to the human POR mutations that cause disease, relatively common POR allelic variants have been found to influence P450 activity in ways that are P450 isoform- and substrate-specific (Flück et al., 2007; Flück and Pandey, 2011; Miller et al., 2011), and the structural, functional, and clinical implications of this were a focus of this symposium.
Engineered Mouse Models Harboring Null or Hypomorphic Alleles for NADPH–Cytochrome P450 Oxidoreductase
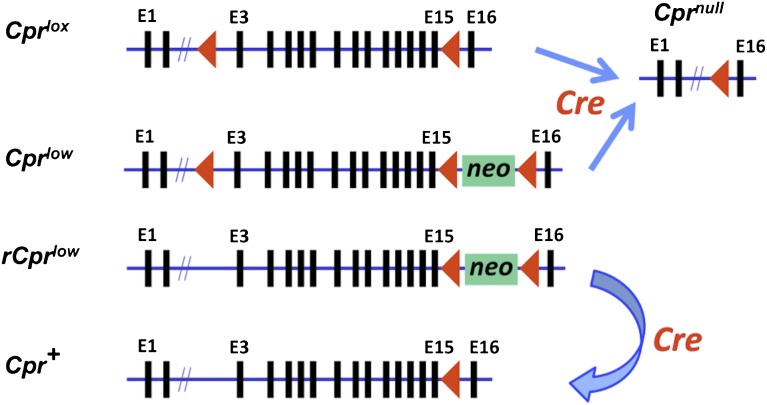
The Cpr (also know as Por) gene contains 16 exons. We have used two strategies to modify the function of CPR in mice: either through a deletion of exons 2–15, leading to a null allele, or through insertion of a neomycin-resistance gene (neo) in intron 15, leading to a hypomorphic allele (Fig. 2). Conditional deletion of Cpr required the generation of a “floxed” Cpr (Cpr lox) allele, via insertion of loxP sites in introns 2 and 15. Cpr-lox (Cpr lox/lox) mice are indistinguishable from wild-type mice, except for the gene modification at the Cpr locus (Wu et al., 2003).
Fig. 2.
Structures of the various modified Cpr alleles and strategy for conditional deletion or rescue of the Cpr gene. Arrowheads represent LoxP sequences and indicate orientation. Selected exons are numbered at the top of the gene structure schematic. Insertion of the floxed neomycin-resistance gene (neo) in the last Cpr intron leads to the generation of a hypomorphic (Cpr low) or a reversible hypomorphic (rCpr low) allele, with global decreases in Cpr expression. Cre-mediated deletion of the Cpr exons 3–15 from the floxed Cpr (Cpr lox) allele or the Cpr low allele leads to conditional Cpr deletion in cells expressing Cre (Cpr null), whereas Cre-mediated deletion of the floxed neo from the rCpr low allele leads to conditional Cpr rescue in cells expressing Cre, regenerating the wild-type Cpr (Cpr +) allele.
The Cpr lox allele can be efficiently deleted, yielding the Cpr null allele (Fig. 2) in a variety of cells and organs via Cre-mediated recombination, following intercrosses between the Cpr-lox mouse and a Cre-transgenic mouse. Mice with tissue-specific Cpr deletion have been generated, with Cpr-lox as a parental strain, for many organs, including the liver (Gu et al., 2003), lung (Weng et al., 2007), heart (Fang et al., 2008), intestine (Zhang et al., 2009), brain (Conroy et al., 2010), mammary gland (Lin et al., 2012), and bone (Panda et al., 2012).
The Cpr low allele contains a floxed neo gene in the last Cpr intron (Fig. 2), leading to global suppression of CPR expression in all cells (Wu et al., 2005; Wei et al., 2010). Upon Cre-mediated recombination, the Cpr low allele can also be converted to the Cpr null allele. Elimination of the loxP site in intron 2 of the Cpr low allele led to the generation of a reversible version of the Cpr-low mice (named rCpr-low or r-CL) (Wei et al., 2010), in which the floxed neo in intron 15 could be removed through Cre-mediated recombination, thus allowing a full recovery of CPR expression and function in Cre-expressing cells (Fig. 2). The r-CL model was used to generate an extrahepatic Cpr-low (xh-CL) mouse (Wei et al., 2010), in which CPR expression in hepatocytes was restored to normal level.
Notably, several conditional Cpr-knockout models have also been generated by the Wolf/Henderson laboratory (University of Dundee, Dundee, United Kingdom). These were developed from Cpr alleles that contained loxP sites in introns 4 and 15.
LCN Mouse.
The LCN mice are fertile and display normal growth; however, they have decreased plasma cholesterol and triglyceride levels, substantial changes in hepatic fatty acid profiles, and enlarged and fatty livers (Gu et al., 2003; Weng et al., 2005; Gonzalez et al., 2011). The fatty liver phenotypes presenting in the LCN mouse appear to resemble those typically found in nonalcoholic fatty liver disease (Gonzalez et al., 2011). In addition to displaying a large number of genomic changes (Weng et al., 2005), the LCN mouse also has decreased (∼30%) hepatic nonprotein sulfhydryl levels (Gu et al., 2005), and decreased hepatic (∼40%) and renal (∼20%) F2-isoprostane levels (Dostalek et al., 2008), changes that are likely related to the altered lipid homeostasis. Interestingly, the fatty liver phenotype was partially relieved by dietary interventions with highly unsaturated fatty acids in the LCN mice (Gonzalez et al., 2011).
The LCN mouse has been used to study the role of hepatic P450 enzymes in the systemic clearance of numerous drugs or toxicants, such as acetaminophen (Gu et al., 2005), nifedipine (Zhang et al., 2007), and benzo[a]pyrene (Fang and Zhang, 2010). The LCN mouse has also been used in several studies to determine whether hepatic P450-mediated metabolism is important for tissue toxicity in extrahepatic target organs induced by drugs or other xenobiotics, such as the lung procarcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (Weng et al., 2007).
Intestinal Epithelium-Cpr-Null Mouse.
The intestinal epithelium-Cpr-null (IECN) mice, in which Cpr deletion occurs specifically in enterocytes in the small intestine and colon, are viable, fertile, normal in size, and do not display any histologic abnormalities in the structure of the various intestinal segments (Zhang et al., 2009). The utility of the IECN model for in vivo drug metabolism studies has been demonstrated (Zhang et al., 2009; Zhu et al., 2011). Clearance of orally (but not intravenously or intraperitoneally) administered nifedipine and lovastatin was much slower in the IECN mice than in wild-type mice, thus confirming the substantial role of intestinal P450 enzymes in the first-pass clearance of oral nifedipine and lovastatin. The IECN mouse has also been used to test the impact of intestinal P450-mediated metabolism on systemic exposure of other orally administered xenobiotics and dietary P450 inducers (Zhang et al., 2009; Fang and Zhang, 2010). In addition, the IECN mouse was valuable for demonstrating the essential role of intestinal P450 enzymes in the bioactivation of diclofenac, and in diclofenac-induced intestinal toxicity (Zhu and Zhang, 2012).
The IECN mouse was used in a recent study to identify potential biologic roles of CPR-dependent enzymes in the intestine (D'Agostino et al., 2012). Cholesterol biosynthetic activity was greatly reduced, whereas the levels of the cholesterol precursor farnesyl pyrophosphate and its derivative geranylgeranyl pyrophosphate were increased in the enterocytes of the IECN mice. These metabolic changes were associated with changes in gene expression in cholesterol biosynthesis and antigen presentation/processing pathways; the latter included genes of the major histocompatibility complex class II. Thus, enterocyte CPR-dependent enzymes modulate expression of genes important for intestinal immunity, possibly through intermediates in cholesterol biosynthesis.
Other Tissue-Specific Cpr-Null Mouse Models.
A lung-specific Cpr-null mouse was developed, and used for the investigation of the roles of pulmonary P450-mediated metabolic activation in 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone–induced lung tumorigenesis (Weng et al., 2007); the sites of Cpr deletion were restricted to subsets of airway epithelial cells and alveolar type II cells. A heart Cpr-null mouse was generated and used for studying the roles of CPR/P450 in the acute cardiotoxicity induced by doxorubicin (Fang et al., 2008); the sites of Cpr deletion were restricted to cardiomyocytes of both the ventricle and atrium.
A brain Cpr-null mouse was generated for studies of the in vivo functions of CPR and CPR-dependent enzymes in the brain (Conroy et al., 2010). The sites of Cpr deletion were restricted to forebrain neurons. No obvious abnormities were observed in brain morphology; however, although the null mice responded normally to painful stimuli, they were highly resistant to the antinociceptive properties of morphine. That finding led to the proposal that brain P450 epoxygenases play essential roles in the pain-relieving circuits of μ opioids.
Two other models have been described recently. A mammary epithelium-specific Cpr-null mouse was generated and used for studying the roles of mammary epithelial P450 enzymes in the bioactivation and disposition of 7,12-dimethylbenz[a]anthracene, a breast carcinogen (Lin et al., 2012). A bone-specific Cpr-null mouse was generated (by the laboratory of B.S. Masters, University of Texas Health Science Center, San Antonio, TX), which displayed ABS-like craniofacial and bone mass defects (Panda et al., 2012).
Cpr-Low Mouse Models.
CPR expression decreased by 70–90% in various tissues of the Cpr-low (CL) mice. Adult male CL mice had decreased body weight and organ weights, whereas females were infertile, presumably resulting from increased serum and tissue levels of testosterone and progesterone (Wu et al., 2005; Wei et al., 2010). CL mice also exhibited lowered plasma cholesterol levels and mild hepatic lipidosis. The CL mice serve as important disease models for human patients harboring mutations that affect CPR expression. Indeed, more studies are needed to identify additional biologic, pharmacological, or pathologic phenotypes in the CL mice, which may also occur in people with substantially reduced CPR expression. In that regard, the r-CL mouse (Wei et al., 2010) is valuable for further mechanistic studies of the roles of CPR in disease-related phenotypes seen in the CL mice.
For studies on the role of P450 enzymes in xenobiotic metabolism and bioactivation in extrahepatic target tissues, the CL mouse models, particularly the xh-CL mouse, can be alternative or complementary models to conditional Cpr-knockout models with tissue-specific Cpr deletion in an extrahepatic organ. The xh-CL mouse is a valuable screening tool for testing the collective functions of extrahepatic CPR-dependent enzymes. The “extra-hepatic Cpr-low” status is established around birth in the xh-CL model, making it useful for investigating the roles of CPR-dependent enzymes in xenobiotic metabolism in neonatal, as well as adult, animals.
In summary, we have described the generation and utility of a panel of engineered mouse models harboring null or hypomorphic Cpr alleles. The various mouse models with conditional modification of the Cpr gene are valuable for mechanistic studies on the roles of CPR-dependent enzymes in the in vivo metabolism and toxicity of xenobiotic compounds. These novel mouse models are also useful for exploration of the biologic functions of CPR-dependent enzymes in diverse organs and cell types, and for the involvement of these enzymes in the various known or still-to-be-identified disease phenotypes associated with human CPR genetic deficiencies.
NADPH–Cytochrome P450 Oxidoreductase and Cytochrome b5 in Physiology, Drug Metabolism, and Cancer
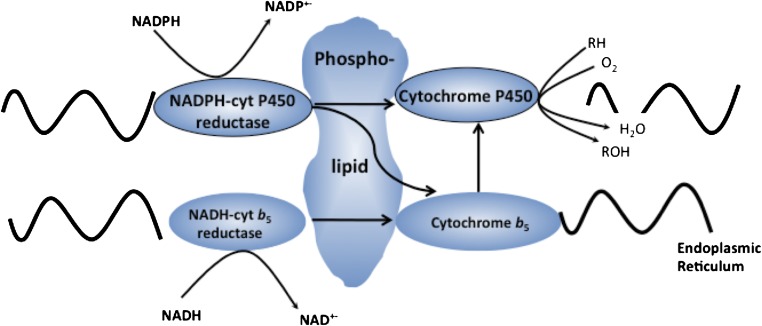
The P450-dependent mono-oxygenase system, as proposed by Estabrook and colleagues in 1970, involves two electron-transport chains, the primary electron source being via POR. A secondary electron-transport system involving cytochrome b5 and cytochrome b5 reductase using NADH as cofactor has also been proposed (Fig. 3).
Fig. 3.
Electron-transport pathways in the cytochrome P450 system.
To understand the relative roles of these two electron transfer pathways as determinants of P450-mediated oxidation reactions, and also to develop models that allow the role of P450s in drug and endogenous compound metabolism to be established, we have generated transgenic mouse lines in which either Por or cytochrome b5 has been deleted from the mouse genome. Both complete and conditional deletion models have been created; in the latter case this allows either of these two proteins to be deleted in any mouse tissue by expressing Cre recombinase from a tissue-specific constitutive or inducible promoter system.
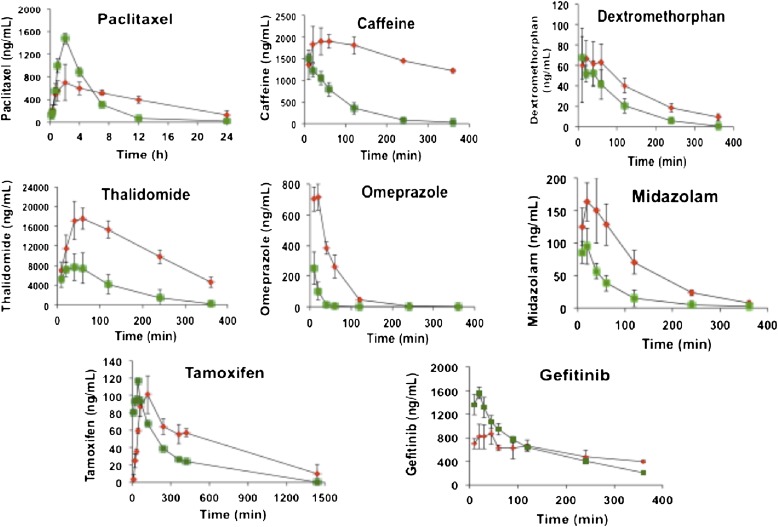
By crossing floxed Por mice with mice expressing Cre recombinase under control of the albumin promoter, the HRN mouse line has been generated, in which POR expression in the liver is essentially absent (> 99% of hepatocytes) (Henderson et al., 2003). The hepatic deletion of POR did not result in an overt phenotype; the mice grew and developed normally and were fertile. However, livers were enlarged and steatotic, the latter a consequence of triglyceride accumulation. In addition, there was a profound increase in the constitutive expression of P450 isozymes. HRN mice also exhibited a profound change in the in vivo pharmacokinetics of a wide range of drugs and foreign compounds that are substrates for the P450 system (Fig. 4). In addition, we found that compounds such as acetaminophen, which are metabolically activated by these enzymes to cytotoxic products, no longer caused hepatotoxicity. This model therefore provides a powerful approach in understanding whether the P450-dependent mono-oxygenase system plays an important role in drug disposition and also, in cases where hepatotoxicity is observed, whether it is the parent or metabolites that are responsible for the cytotoxic effects.
Fig. 4.
Altered in vivo drug pharmacokinetics in HRN mice. Adult male HRN mice (n = 3) were administered the following drugs intraperitoneally (except tamoxifen, which was given orally): paclitaxel 10 mg/kg, caffeine 1 mg/kg, dextromethorphan 1 mg/kg, thalidomide 20 mg/kg, omeprazole 1 mg/kg, midazolam 1 mg/kg, tamoxifen 10 mg/kg, gefitinib 5 mg/kg. Serial blood samples were taken from the tail vein at the time points shown and analyzed by liquid chromatography–tandem mass spectrometry for levels of parent drug. Green symbols represent wild-type mice (Por lox/lox), and red symbols represent HRN mice (Por lox/lox + Cre ALB). Data shown are mean ± SD.
We have extended the utility of this model by crossing the HRN mice with immunocompromised nude mice to generate a system that allows the potential antitumor effects of drugs in development to be established. This model has some significant advantages over currently-used nude models in that it allows the assessment of antitumor effects of compounds that are rapidly metabolized or may induce their own metabolism over the period of the xenograft experiment. In drug development, this model also provides a means of establishing the efficacy of a potential anticancer drug prior to any chemical modification targeted at improving phase I metabolic properties. In addition, an increasing number of anticancer drugs are pro-drugs that require metabolism to exert their therapeutic effects, for example, cyclophosphamide, tegafur, and tamoxifen. The HRN nude model allows the determination of whether the parent compound or its metabolites, or both, are antitumorigenic.
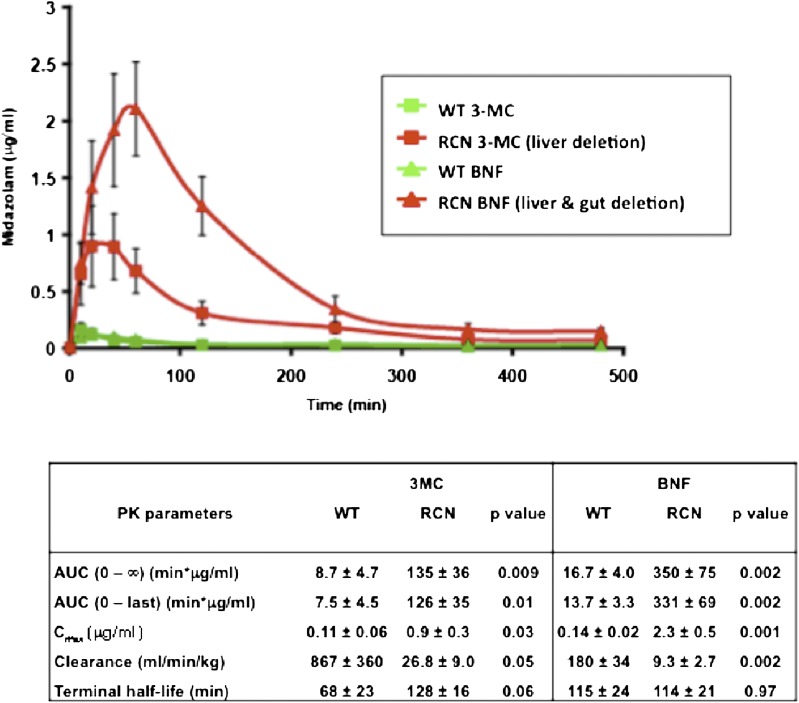
In a further development we have generated a mouse line in which Cre-recombinase expression is driven by the CYP1A1 promoter, and crossed these mice with floxed Por mice to create reductase conditional null (RCN) mice (Finn et al., 2007). In this model, the mice are phenotypically wild-type until an aryl hydrocarbon receptor (AHR) activator such as 3-methylcholanthrene (3-MC) is administered to the mice. This should have resulted in the deletion of Por in all tissues containing a functional AHR (Campbell et al., 1996). Intriguingly, however, depending on the AHR activator and the dose used, Por can be deleted in the liver or in the liver and the gastrointestinal tract (Finn et al., 2007). Two versions of the CYP1A1-Cre system have been developed: a random transgenic model initially generated by Ireland et al. (2004), in which Cre-recombinase expression is driven by the rat CYP1A1 promoter, and more recently where Cre recombinase has been cloned into the murine Cyp1a1 gene locus. The advantage of the latter model, referred to as endogenous reductase locus, is that much lower doses of AHR ligands are required to generate the deleted POR phenotypes. These models provide a powerful approach for establishing the impact on the oral bioavailability of drugs by metabolism either in the gastrointestinal tract or the liver (Fig. 5). This is illustrated by studies on the pharmacokinetics of midazolam, which has low oral bioavailability in wild-type mice but following Por deletion in the liver or gut and liver a highly significant increase in exposure is observed (Fig. 5). A further advantage of these models is that the hepatic lipid accumulation observed in HRN mice can be attenuated by placing the animals on a low-fat diet; as a result, the profound overexpression of hepatic P450 isozymes, including Cyp2b10 and Cyp3a11 [regulated by the transcription factors constitutive androstane receptor (CAR) and pregnane X receptor (PXR)], characteristic of the HRN phenotype, is abolished. To establish the role of CAR and PXR in the overexpression of hepatic P450s in HRN mice, we crossed RCN mice with animals in which either Car or Pxr, or both, had been deleted. In such mice lacking CAR, the overexpression of the P450s was lost; furthermore, the primary endogenous activator of CAR was found to be unsaturated fatty acids, and specifically linoleic acid, indicating that marked changes in drug metabolism can occur depending on the dietary status (Finn et al., 2009). The mechanism(s) underlying the accumulation of unsaturated fatty acids in HRN mice remain to be elucidated; however, these experiments demonstrate that potent endogenously controlled signaling mechanisms regulate the hepatic P450 system. Animal models whose P450 system can be attenuated or inactivated in a tissue-specific manner have a number of powerful applications, not only in defining pathways important in the development and use of drugs but also in elucidating pathways of chemical toxicity and in improving models to determine the efficacy of drugs in development.
Fig. 5.
Use of the RCN model to dissect the role of liver versus gut in drug metabolism. Adult male RCN or wild-type (WT) mice (n = 3) were treated with either 3-MC (40 mg/kg in corn-oil, intraperitoneally, single dose) or β-naphthoflavone (BNF, 80 mg/kg in corn-oil, intraperitoneally, daily for 4 days). After 10 days the mice were administered midazolam, orally at 2.5 mg/kg, and a pharmacokinetic study carried out. Serial blood samples were taken from the tail vein at the time points shown and analyzed by liquid chromatography–tandem mass spectrometry for levels of parent drug. Green symbols represent wild-type mice (Por lox/lox) treated with 3-MC (squares) or BNF (triangles), and red symbols represent RCN mice (Por lox/lox + Cre CYP1A1) treated with 3-MC (squares) or BNF (triangles). Data shown are mean ± SD. The table lists the pharmacokinetic parameters and associated statistical significance.
Since the early 1970s it has been postulated that cytochrome b5 is an important electron donor to P450s, although the evidence for this has been based entirely on in vitro studies. To establish the role of cytochrome b5 in P450 catalysis in vivo we carried out a complete and conditional (liver) deletion of this hemoprotein in mice (Finn et al., 2008; McLaughlin et al., 2010). As a consequence of the reported housekeeping functions of cytochrome b5—for example, in the reduction of methemoglobin to hemoglobin and in the biosynthesis of steroid hormones—we anticipated that the complete constitutive deletion of this enzyme would result in embryonic or neonatal lethality. However, cytochrome b5 complete null (BCN) mice were viable but exhibited some significant and intriguing phenotypes. BCN mice were smaller than their wild-type counterparts, developed more slowly in the post-weaning period and had a severe skin phenotype very similar to the inherited human condition, ichthyosis. When BCN mice were nurtured on surrogate wild-type dams there was a significant improvement in growth rate and a reversal of many of the developmental phenotypes, and we could associate the nurturing and skin phenotypes to the role of cytochrome b5 in fatty acid desaturation (Finn et al., 2011).
Conditional hepatic cytochrome b5 null (HBN) mice were generated by crossing mice carrying the floxed cytochrome b5 gene with those in which Cre expression was driven by the liver-specific albumin promotor; HBN mice were phenotypically normal. In both BCN and HBN mice the metabolism of a wide range of drugs was profoundly altered (Finn et al., 2008; McLaughlin et al., 2010). Intriguingly, this effect was not restricted to metabolism by specific P450 enzymes but was dependent on the substrate used. These data demonstrate that electron transfer from cytochrome b5 to the P450 system is a key determinant of in vivo rates of drug metabolism, and that variability in cytochrome b5 concentration may significantly affect rates of drug disposition in humans. In vitro drug metabolism assays in microsomal fractions derived from a number of extrahepatic tissues from the BCN mice also demonstrated that cytochrome b5 can play a major role in extrahepatic drug metabolism and disposition, particularly in tissues such as the testes and lungs (McLaughlin et al., 2010).
In conclusion, the application of transgenic animal models has allowed the detailed dissection of the relative importance of different electron transfer pathways in defining P450-dependent mono-oxygenase activities in hepatic and extrahepatic tissues. As a consequence, the capacity to modulate phase I metabolism in these tissues provides a powerful approach to understanding the role of metabolism both in drug development and defining the therapeutic or adverse effects of drugs.
Replication of the Hepatic Lipidosis Seen in Hepatic Por-Null Mice in a Hepatoma Cell–Culture Model: A Role for Farnesoid X Receptor?
In 2003 the laboratories of Roland Wolf (University of Dundee, Dundee, United Kingdom) and Xinxin Ding (New York State Department of Health, Albany, NY) independently published their studies on the conditional deletion of POR expression in mouse liver (Gu et al., 2003; Henderson et al., 2003). While these studies focused on the role of POR in hepatic drug metabolism, these mice exhibited a remarkable reduction in blood lipids while accumulating triglycerides in the liver, leading to a hepatic lipidosis resembling nonalcoholic fatty liver disease. The basis for this lipidosis was not readily explained given the lack of a known role for POR in triglyceride catabolism or secretion. POR is essential to the synthesis of cholesterol (Porter, 2012) and bile acids; in addition, and as confirmed by Gu et al. (2003), POR is necessary for heme catabolism via heme oxygenase and is essential to steroid hormone synthesis and vitamin D activation. POR is both directly involved in fatty acid metabolism by P450s in the CYP4 family, resulting in ω-oxidation, and indirectly through electron transfer to cytochrome b5 in the fatty acid desaturation (Enoch and Strittmatter, 1979) and elongation pathways (Ilan et al., 1981). Thus, there are several pathways by which POR expression might indirectly impact triglyceride metabolism and secretion. To elucidate the mechanism by which loss of POR expression leads to hepatic lipid accumulation, we developed an in vitro, hepatoma cell–culture model in which POR expression was suppressed by siRNA (Porter et al., 2011). This model faithfully replicated the lipidosis seen in hepatic Por-null mice and allowed us to explore the role of extracellular (serum) lipids in this lipidosis. Here we review our findings and present some of our more recent data on the role of serum lipids, and suggest how the farnesoid X receptor (FXR) and peroxisome proliferator–activated receptors (PPAR) may be involved in this lipidosis.
An oligonucleotide predicted to form a “hairpin loop” from the 3′-nontranslated segment of the rat POR mRNA sequence proved effective at decreasing POR mRNA levels by 80% in McA-RH7777 rat hepatoma cells by 5 days post-transfection of the siRNA-expressing plasmid, corresponding to greater than 70% suppression of POR protein. Protein suppression was maintained through 20 days, and lipid accumulation was evident as early as day 12, and was fully developed and statistically significant by day 15. Cells remained viable through 20 days of culture.
We hypothesized that a decrease in cellular cholesterol levels in Por-null cells, secondary to the loss of cholesterol synthesis, might prevent very-low-density lipoprotein formation and secretion, given that cholesterol is a required component of this lipoprotein complex. To test this hypothesis, cholesterol was added to the medium of POR-suppressed cells at 50 mg/l beginning on day 2. Cholesterol supplementation did not prevent the lipid accumulation in these cells, indicating that cholesterol was not rate-limiting for very-low-density lipoprotein secretion in this model. Consistent with this conclusion, siRNA-mediated suppression of CYP51 (lanosterol demethylase), an enzyme unique to the cholesterol synthesis pathway, did not engender triglyceride accumulation, and in fact lowered triglyceride levels significantly at 10 and 15 days post-transfection. Thus it appears unlikely that a lack of cholesterol in these cells is responsible for the triglyceride accumulation, which is in line with our studies (Li and Porter, 2007) and those of others (Finn et al., 2009) that showed that cholesterol levels remain normal in Por-null liver despite the loss of hepatic cholesterol synthesis.
The inability of Por-null cells to catabolize cholesterol to bile acids raised the possibility that oxysterols might be forming and accumulating in these cells; oxysterols activate the liver X receptor (LXR), which stimulates sterol regulatory element–binding protein-1c (SREBP-1c) transcription, leading to excessive fatty acid and triglyceride synthesis and hepatic lipidosis (Kalaany and Mangelsdorf, 2006). However, gas chromatography–mass spectrometric analysis of POR-suppressed cells did not reveal the presence of oxysterols in levels greater than those seen in normal hepatoma cells, indicating that LXR activation was not a likely cause of the triglyceride accumulation. This finding is consistent with the microarray studies carried out by the Ding and Wolf laboratories on Por-null liver (Wang et al., 2005; Weng et al., 2005), where a gene expression pattern consistent with LXR activation was not evident.
We then tested the hypothesis that the loss of bile acid synthesis might be responsible for an increase in triglyceride synthesis through a loss of FXR signaling. FXR activation suppresses SREBP-1c transcription, and so in the absence of bile acids (the natural activators of FXR) this suppression would be lessened. Indeed, the addition of chenodeoxycholate (50 μM) to POR-suppressed cells prevented lipid accumulation, lowering triglyceride levels to 40% of normal. This presumed suppression of SREBP-1c expression by bile acid–mediated FXR activation suggested that the triglycerides that accumulate in POR-suppressed and Por-null cells are newly synthesized, rather than taken up from the medium. To confirm this hypothesis we removed all lipids from the culture medium by diethyl ether extraction. Surprisingly, removal of lipids from the medium prevented the triglyceride accumulation in POR-suppressed cells, indicating that uptake, rather than synthesis, was largely responsible for the lipid accumulation. This finding was again in accord with the microarray studies from the Ding and Wolf laboratories, where the expression of genes involved in fatty acid and triglyceride synthesis was not markedly elevated in Por-null mouse liver (Wang et al., 2005; Weng et al., 2005). Moreover, Finn et al. (2009) found that placing Por-null mice on a fat-deficient diet prevented the hepatic lipidosis, providing strong evidence that the hepatic lipids are largely and perhaps entirely derived from the diet through the circulation.
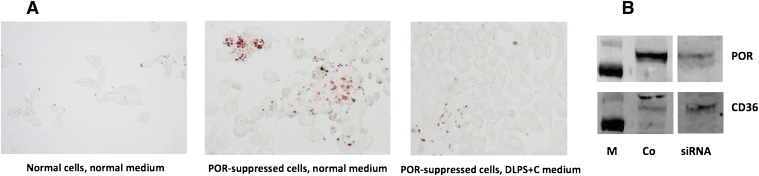
Our POR-suppressed cells could not survive past day 15 in delipidated medium, which we suspected was due to the lack of cholesterol. The poor viability of these cells compromised the studies already noted here on the role of extracellular lipids in the lipid accumulation in these cells, and so we added cholesterol back to the medium as a cyclodextrin complex at 10 μg/ml. As shown in Fig. 6A, POR-suppressed cells grown in this cholesterol-supplemented, delipidated medium retained good viability through 15 days but did not accumulate lipids, providing convincing evidence that the lipid accumulation otherwise seen in these cells is largely derived from the medium. Why the presumed loss of FXR signaling does not markedly increase fatty acid and triglyceride synthesis remains to be determined, but it should be noted that neither activity has yet been measured in POR-suppressed cells or Por-null liver tissue.
Fig. 6.
(A) McA-RH7777 rat hepatoma cells were cultured in medium containing 10% fetal bovine serum (normal medium) or in medium from which the lipids had been extracted and cholesterol added back at 10 μg/ml as a cyclodextrin complex (DLPS+C). Cells were stained for lipid with oil red O at 10 days post-transfection. (B) Immunoblot of POR and CD36 expression in control (Co) and POR-suppressed cells (siRNA) after 5 days. M, mass markers at 100 and 70 kDa.
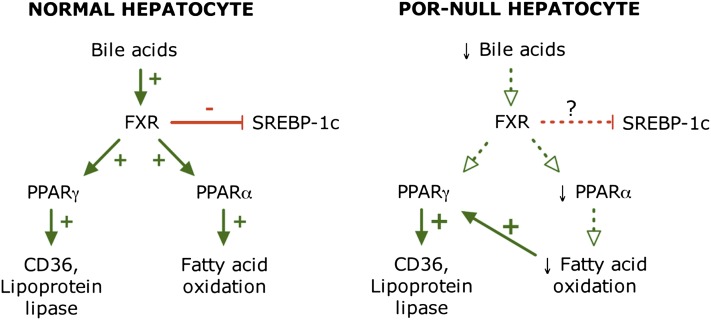
A loss of FXR signaling, as seen in Fxr-null mice, readily leads to hepatic triglyceride accumulation resembling nonalcoholic fatty liver disease (Sinal et al., 2000; Kong et al., 2009). This is generally assumed to result from derepression of SREBP-1c signaling, as already noted. However, in our POR-suppressed hepatoma cells and in hepatic Por-null mice (where FXR signaling is presumably lost due to the loss of bile acids), increased lipid uptake appears to predominate over fatty acid and triglyceride synthesis. The connection between FXR signaling and lipid uptake is not well defined. Hepatic Por-null mice have elevated expression of two genes involved in lipid uptake: CD36, a fatty acid uptake transporter, and lipoprotein lipase, which releases fatty acids from triglycerides in lipoproteins (Weng et al., 2005). We also see an increase in CD36 expression in POR-suppressed hepatoma cells (Fig. 6B). Neither of these genes is known to be regulated by FXR, but they are upregulated by PPARγ, whose expression is increased 1.6-fold in Por-null liver. Because FXR has been reported to positively regulate PPARγ expression, the presumed loss of FXR signaling in POR-suppressed cells and Por-null liver could not contribute to this increase in PPARγ expression in Por-null cells. However, a second factor proposed to contribute to the lipidosis of hepatic Por-null mice is a decrease in fatty acid catabolism, a metabolic pathway positively regulated by PPARα. Importantly, PPARα expression was significantly decreased in Por-null liver (Weng et al., 2005), which would support a decrease in fatty acid oxidation. Because PPARα expression is regulated by FXR (Pineda Torra et al., 2003), the absence of FXR signaling might well contribute to the decrease in PPARα expression and thereby decrease fatty acid catabolism in these mice. As proposed by Weng et al. (2005), the resulting increase in fatty acid content and changes in composition may induce PPARγ expression, leading to increased expression of genes involved in fatty acid uptake (CD36 and lipoprotein lipase) and augmenting the hepatic lipidosis. These putative signaling pathways in normal and Por-null or suppressed cells are diagrammed in Fig. 7. The ability of FXR to promote fatty acid catabolism by increasing PPARα expression would thus explain our finding that adding chenodeoxycholate to POR-suppressed hepatoma cells prevents the lipidosis otherwise seen in these cells and in Por-null mouse liver. Further studies will be needed to fully elucidate the interaction between FXR and the PPAR isoforms and to dissect their role in lipid metabolism in the hepatocyte.
Fig. 7.
Proposed FXR, PPARα, and PPARγ signaling pathways in normal and Por-null hepatocytes. The absence of bile acids in Por-null cells suppresses FXR signaling, leading to decreased PPARα and PPARγ signaling. A subsequent decrease in fatty acid oxidation may be responsible for the activation of PPARγ and enhanced lipid uptake from the medium (or circulation).
Clinical, Structural, and Functional Implications of NADPH–Cytochrome P450 Oxidoreductase Deficiency
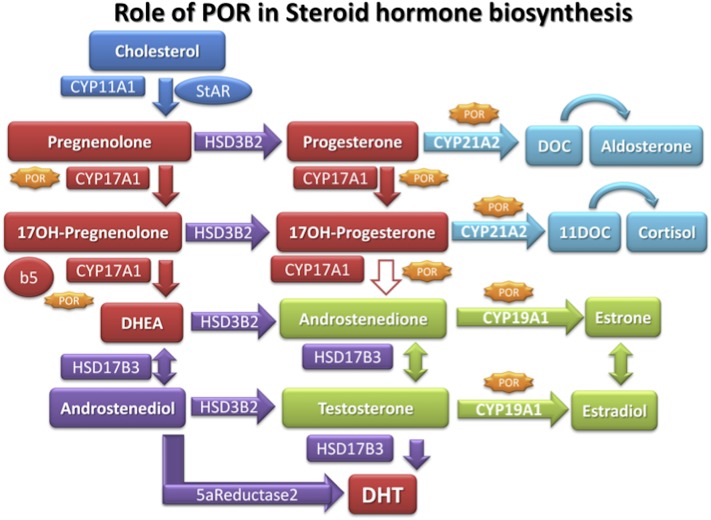
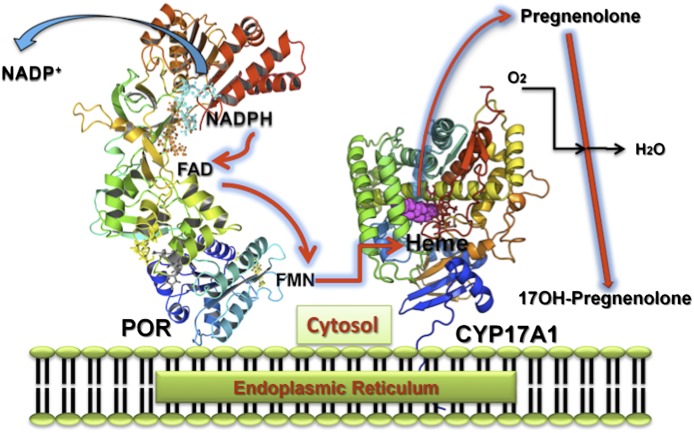
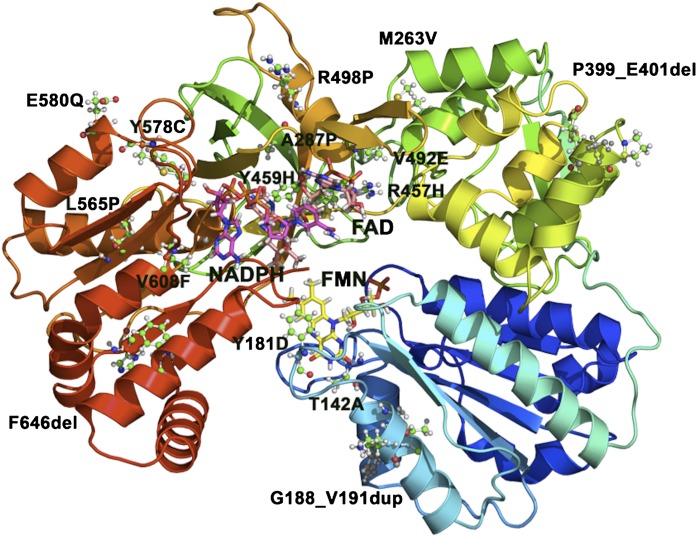
Patients with POR deficiency have disordered steroidogenesis due to malfunctioning of several key enzymes in the steroidogenesis pathway (Flück et al., 2007; Flück and Pandey, 2011; Miller et al., 2011; Miller and Auchus, 2011) (Fig. 8). In 1985, a 46,XY patient with genital abnormalities and an abnormal urinary steroid profile that suggested combined defects in CYP17A1 (17α-hydroxylase/17,20 lyase) and CYP21A2 (21-hydroxylase) was reported by Peterson et al. (1985). That led to suggestions of a potentially defective POR (Miller, 1986), since both CYP17A1 and CYP21A2 require POR for supply of electrons from NADPH for their metabolic reactions (Fig. 9); however, when the Por knockout was found to be embryonically lethal in mice (Shen et al., 2002; Otto et al., 2003), POR was excluded as a candidate gene. Almost two decades later in a 2004 report, Flück et al. (2004) described the first four patients with mutations in POR. Three of these patients had ambiguous genitalia and ABS [Online Mendelian Inheritance in Man (OMIM): 207410, 201750]; one patient was an adult with steroid abnormalities, primary amenorrhea, and polycystic ovaries (Flück et al., 2004). ABS is a skeletal malformation syndrome that was first described in 1975 (Antley and Bixler, 1975). ABS is characterized by craniosynostosis, midface hypoplasia, radiohumeral synostosis, joint contractures, and bowing of the femora (Crisponi et al., 1997). About 50% of patients with ABS also have genital anomalies (Crisponi et al., 1997; Reardon et al., 2000). Subsequent studies confirmed the presence of POR mutations in patients with similar patterns of steroid abnormalities with and without ABS (Huang et al., 2005; Flück and Pandey, 2011). In the patients of European descent A287P is most common; while Japanese patients often have the R457H mutation (Huang et al., 2005) (Fig. 10). A larger study of 35 Japanese patients found POR mutations on all alleles (Fukami et al., 2009), ruling out manifesting heterozygosity for POR. A study of 842 healthy unrelated individuals identified 140 single nucleotide polymorphisms, 13 novel missense variations, and 8 indels (Huang et al., 2008). The duplication mutation G188_V191dup, R498P, Y578C, and E580Q have recently been reported but had not been tested in enzymatic assays (Krone et al., 2012) (Fig. 10).
Fig. 8.
Steroid biosynthetic pathways. Enzymes requiring POR as electron donor are shown with gene names. The 17,20-lyase activity of human CYP17A1 converts 17α-hydroxypregnenolone to dehydroepiandrosterone (DHEA) but is less effective in converting 17α-hydroxyprogesterone to androstenedione. Abbreviations used in this figure but not appearing in text: DHT, dihydrotestosterone; DOC, 11-deoxycorticosterone; 11DOC, 11-deoxycortisol; HSD, hydroxysteroid dehydrogenase; StAR, steroidogenic acute regulatory protein.
Fig. 9.
Electron transfer by POR to microsomal P450 enzymes such as CYP17A1, CYP21A2, and CYP19A1. NADPH interacts with POR, which is bound to the endoplasmic reticulum, and donates a pair of electrons, which are transferred to flavin adenine dinucleotide (FAD) eliciting a conformational change causing the isoalloxazine rings of the FAD and flavin mononucleotide (FMN) moieties to move closer for electron transfer from the FAD to the FMN. Another conformational change returns the POR to its open position, the FMN domain interacts with the P450, and the P450 heme iron accepts electrons in two discrete single electron transfers to support P450 catalytic activities.
Fig. 10.
Mutations in POR. The structure of human POR, with missense mutations identified in patients with POR deficiency, is shown as a ribbon model. Mutations are shown as ball-and-stick models, whereas FMN (yellow), FAD (orange), and NADPH (magenta) are shown as stick models. Mutations that result in a stop codon and/or cause truncations in POR are not shown.
In the initial report, the POR mutants A287P, R457H, V492E, C569Y, and V608F were tested with 17α-hydroxylase and 17,20 lyase activities of CYP17A1 (Flück et al., 2004). A good correlation between the clinical features and the CYP17A1 activities was observed. Later on we tested the effects of some POR variants on CYP19A1 (aromatase) activity (Pandey et al., 2007). Mutations R457H and V492E located at the flavin adenine dinucleotide (FAD) binding site of POR (Fig. 10) caused a complete loss of CYP19A1 activity (Table 1), confirming that POR mutations disrupting the FAD binding and electron transfer will severely affect all P450s. By contrast, POR mutants A287P, C569Y, and V608F had variable effects on CYP19A1 and CYP17A1 activities (Table 1). POR mutations C569Y and V608F, located in the NADPH-binding site (Fig. 10), were in a compound heterozygote state in a female patient with a polycystic ovary syndrome–like condition, and had greater than half of the wild-type CYP17A1 activities but less than 50% of wild-type CYP19A1 activity (Pandey et al., 2007). The mutants C569Y and V608F were more sensitive to the variation in NADPH concentration (Pandey et al., 2007). The A287P mutant did not affect CYP19A1 activity but considerably lowered the CYP17A1 activities (Table 1). Mutation Y181D, which is involved in binding flavin mononucleotide (FMN) (Fig. 10), impaired both CYP21A2 and CYP17A1 activities. We have recently reported a novel POR mutation P399_E401del found in two unrelated Turkish families (Flück et al., 2011a). The novel POR mutation P399_E401del had a clinical phenotype of ABS and disordered sex development but only subclinical cortisol deficiency. In vitro functional testing of POR mutant P399_E401del on single enzymes showed an activity loss of 68–85% for different P450s. The severity of aromatase inhibition by P399_E401del (∼85%) and in utero virilization may be linked as we have suggested earlier for other mutations in the NADPH-binding domain of POR (Pandey et al., 2007). Computational studies suggest that P399_E401del causes structural instability and may impair electron transfer from NADPH to FAD. We found that POR mutations in the FAD- and FMN-binding domains (Fig. 10) also inhibit cytochrome b5 reduction (Nicolo et al., 2010). Supplementing flavin in our in vitro POR assays, we were able to restore activities of cytochrome b5 reduction not only for POR Y181D but also for A287P, which is not near the cofactor binding sites (Nicolo et al., 2010). However, which POR variant with what partner P450 may respond to external FMN, and whether flavin can be used as a treatment of affected patients, remains to be tested. We have found that POR mutants Y181D, A457H, Y459H, V492E, and R616X lost more than 99% of drug metabolizing CYP3A4 activity, while 60–85% activity loss was observed for POR mutations A287P, C569Y, and V608F (Flück et al., 2010) (Table 1). Loss of CYP3A4 activity may result in increased risk of drug toxicities and adverse drug reactions in patients with severe POR mutations. Among non-P450 partner proteins, we have observed that POR mutants Y181D, A457H, Y459H, V492E, and R616X result in total loss of heme oxygenase-1 (HO-1) activity, while POR mutations A287P, C569Y, and V608F lost 50–70% of HO-1 activity (Pandey et al., 2010) (Table 1). The POR variants P228L, R316W, G413S, A503V, and G504R identified as polymorphs had close to wild-type activity in HO-1 assays. The structural and molecular selectivity of POR variants toward specific P450s or other electron acceptors will become clearer as studies with more target proteins are carried out. We believe that the impact of specific POR variants that do not disrupt the structure, such as loss of FAD or FMN or a truncated protein (Flück et al., 2009; Xia et al., 2011), could not be predicted from a single assay but need to be tested with multiple P450s and other interaction partners.
TABLE 1.
Wild-type or mutant POR activities toward P450s and heme oxygenase-1. Enzyme activities are shown as a percentage of the wild-type (WT) control activities. Data were compiled from previous studies on effects of POR variants (Flück et al., 2004; Huang et al., 2005; Pandey et al., 2007; Flück et al., 2010; Pandey et al., 2010; Flück and Pandey, 2011). —, not determined or not reported.
| POR | 3A4 | Cyt c Reduction | NADPH Oxidation | 17OH-ase | 17,20 Lyase | Aromatase | HO-1 |
|---|---|---|---|---|---|---|---|
| WT | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| A115V | 85 | 63 | 41 | 80 | 71 | — | 95 |
| T142A | 85 | 49 | 52 | 60 | 54 | — | 81 |
| Q153R | 119 | 9 | 11 | 31 | 27 | — | 72 |
| Y181D | — | — | — | — | — | <1% | |
| P228L | 101 | 75 | 72 | 100 | 41 | — | 106 |
| A287P | 26 | 93 | 104 | 40 | 21 | 104 | 49 |
| R316W | 110 | 61 | 77 | 94 | 141 | — | 96 |
| G413S | 100 | 76 | 100 | 83 | 110 | — | 104 |
| R457H | — | 1 | — | 3 | — | 1 | — |
| Y459H | — | — | — | 11 | — | — | — |
| V492E | — | — | — | 3 | — | <1 | — |
| A503V | 107 | 67 | 56 | 68 | 58 | — | 97 |
| G504R | 93 | 53 | 47 | 91 | 103 | — | 107 |
| C569Y | 32 | 18 | 7 | 28 | 13 | 51 | 33 |
| V608F | 16 | 8 | 3 | 80 | 57 | 24 | 32 |
| R616X | - | — | 6 | — | — | — | — |
| V631I | 89 | 74 | 23 | 51 | 40 | — | 96 |
| delF646 | 88 | 36 | 94 | 97 | 46 | — | 95 |
Patients with POR deficiency were earlier diagnosed as ABS with genital anomalies. In a study of 32 ABS patients with and without hormonal abnormalities, 18 patients had recessive POR mutations while 10 had dominant fibroblast growth factor receptor FGFR2 or FGFR3 mutations but no mutations in POR, and one patient had compound heterozygote POR mutations and a FGFR1 mutation; in three patients no mutations in POR or FGFR were found (Huang et al., 2005). This demonstrated that mutations in POR and FGFR2 segregate, and patients with an ABS-like phenotype and anomalies in both genital development and steroid hormone production have a different disorder, namely “POR deficiency” (OMIM: 613571). Loss of 17α-hydroxylase and 17,20 lyase activities of CYP17A1 may cause elevated plasma concentrations of deoxycorticosterone and corticosterone but decreased levels of cortisol and C19 steroids (dehydroepiandrosterone, dehydroepiandrosterone sulfate, and androstenedione) (Fig. 8). This impairment of CYP17A1 activities by POR deficiency in the gonads may lead to disordered development of the external genitalia in the male fetus and failure of pubertal development and fertility in both sexes, similar to mutations in CYP17A1. Reduction or loss of CYP19A1 (aromatase) activity in POR deficiency will cause diminished production of estrogens from androgen precursors in the ovaries of affected individuals (Fig. 8). Impairment of CYP19A1 activity will affect the conversion of fetal androgen precursors to estriol and estrone and may cause virilization of the fetus and the mother (manifested by acne, voice changes, and hirsutism). Loss of CYP21A2 (21-hydroxylase) activity may lead to reduced plasma cortisol (and in rare cases aldosterone) and elevated 21-deoxycortisol concentrations. However, compared with CYP21A2 mutations, POR deficiency patients usually have only moderately elevated plasma 17α-hydroxyprogesterone, and decreased rather than increased C19 steroid concentrations due to combined deficiencies of CYP17A1 and CYP21A2. The pathogenesis of skeletal malformations in patients with severe POR deficiency is not clear. Recent mouse studies suggest a potential role for cholesterol biosynthesis and POR in bone development (Keber et al., 2011). Disorders of cholesterol biosynthesis such as the Smith-Lemli-Opitz syndrome, which is caused by mutations in 7-dehydrocholesterol reductase, may cause skeletal malformations.
Variations in effects of POR deficiency may result from its role in the alternate pathway of androgen production in which dihydrotestosterone is formed from 17α-hydroxyprogesterone or progesterone via 5α- and 3α-reductases, bypassing the usual intermediates, androstenedione and testosterone (Auchus, 2004). A role of the backdoor pathway in human androgen production has recently been established from patients with mutations in the aldo-keto reductases AKR1C2/C4, which display 3α-reductase activity (Flück et al., 2011b). The clinical spectrum of POR deficiency is quite broad, and ranges from severely handicapped ABS patients to mildly affected normal-looking adults with impaired fertility. The global prevalence of POR deficiency will become clearer after more studies from different regions of the world are completed. Several patients who were previously misdiagnosed with either 21-hydroxylase (CYP21A2) or 17α-hydroxylase (CYP17A1) deficiency have now been identified with defects in POR. The diagnosis of POR deficiency may be suggested from clinical and hormonal analysis, but it requires molecular genetic analysis for confirmation of the specific defect as treatment may depend on the severity of the effect on different POR interaction partners. Treatment may include replacement of glucocorticoids and sex steroids (and rarely mineralocorticoids) as assessed by low serum hormone levels. The skeletal malformations caused by POR deficiency require orthopedic management, and mortality is often due to skeletal abnormalities leading to respiratory problems (e.g., choanal obstruction). Computational docking studies and functional assays suggest that the activity loss of some POR variants may be rescued by flavin supplementation (Nicolo et al., 2010).
Acknowledgments
The expert technical assistance of Catherine Meakin and Susanne van Schelven with animal work is gratefully acknowledged.
Abbreviations
- ABS
Antley-Bixler syndrome
- AHR
aryl hydrocarbon receptor
- BCN
cytochrome b5 complete null
- CAR
constitutive androstane receptor
- CL
Cpr-low
- CPR or POR
NADPH–cytochrome P450 oxidoreductase
- FAD
flavin adenine dinucleotide
- FGFR
fibroblast growth factor receptor
- FMN
flavin mononucleotide
- FXR
farnesoid X receptor
- HBN
hepatic cytochrome b5 null
- HO-1
heme oxygenase-1
- HRN
hepatic reductase null
- IECN
intestinal epithelium-Cpr-null
- LCN
liver-Cpr-null
- LXR
liver X receptor
- 3-MC
3-methylcholanthrene
- P450
cytochrome P450
- PPAR
peroxisome proliferator–activated receptor
- PXR
pregnane X receptor
- r-CL
reversible-Cpr-low
- RCN
reductase conditional null
- siRNA
small interfering RNA
- SREBP
sterol regulatory element–binding protein
- xh-CL
extrahepatic-Cpr-low
Authorship Contributions
Participated in research design: Riddick, Ding, Wolf, Porter, Pandey, Zhang, Gu, Finn, Ronseaux, Henderson, Zou, Flück.
Conducted experiments: Pandey, Finn, Ronseaux, McLaughlin, Henderson, Zou, Flück.
Contributed new reagents or analytic tools: not applicable.
Performed data analysis: Porter, Pandey, Finn, Ronseaux, McLaughlin, Zou, Flück.
Wrote or contributed to the writing of the manuscript: Riddick, Ding, Wolf, Porter, Pandey, Henderson, Flück.
Footnotes
This work was supported by the Canadian Institutes of Health Research [Grant MOP-93759]; the National Institutes of Health National Cancer Institute [Grant CA092596]; the National Institutes of Health National Institute of Environmental Health Sciences [Grants ES007462, ES018884]; the National Institutes of Health National Institute of General Medical Sciences [Grant GM082978]; the National Institutes of Health National Center for Complementary and Alternative Medicine [Grant AT005235]; Cancer Research UK [Programme Grant C4639/A5661]; and the Swiss National Science Foundation [Grants 3100A0-113719, 31003A-134926].
D.S.R., X.D., C.R.W., T.D.P., and A.V.P. contributed equally to this work.
References
- Antley R, Bixler D. (1975) Trapezoidocephaly, midfacial hypoplasia and cartilage abnormalities with multiple synostoses and skeletal fractures. Birth Defects Orig Artic Ser 11:397–401 [PubMed] [Google Scholar]
- Auchus RJ. (2004) The backdoor pathway to dihydrotestosterone. Trends Endocrinol Metab 15:432–438 [DOI] [PubMed] [Google Scholar]
- Campbell SJ, Carlotti F, Hall PA, Clark AJ, Wolf CR. (1996) Regulation of the CYP1A1 promoter in transgenic mice: an exquisitely sensitive on-off system for cell specific gene regulation. J Cell Sci 109:2619–2625 [DOI] [PubMed] [Google Scholar]
- Conroy JL, Fang C, Gu J, Zeitlin SO, Yang W, Yang J, VanAlstine MA, Nalwalk JW, Albrecht PJ, Mazurkiewicz JE, et al. (2010) Opioids activate brain analgesic circuits through cytochrome P450/epoxygenase signaling. Nat Neurosci 13:284–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisponi G, Porcu C, Piu ME. (1997) Antley-Bixler syndrome: case report and review of the literature. Clin Dysmorphol 6:61–68 [PubMed] [Google Scholar]
- D’Agostino J, Ding X, Zhang P, Jia K, Fang C, Zhu Y, Spink DC, Zhang QY. (2012) Potential biological functions of cytochrome P450 reductase-dependent enzymes in small intestine: novel link to expression of major histocompatibility complex class II genes. J Biol Chem 287:17777–17788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostalek M, Hardy KD, Milne GL, Morrow JD, Chen C, Gonzalez FJ, Gu J, Ding X, Johnson DA, Johnson JA, et al. (2008) Development of oxidative stress by cytochrome P450 induction in rodents is selective for barbiturates and related to loss of pyridine nucleotide-dependent protective systems. J Biol Chem 283:17147–17157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch HG, Strittmatter P. (1979) Cytochrome b5 reduction by NADPH-cytochrome P-450 reductase. J Biol Chem 254:8976–8981 [PubMed] [Google Scholar]
- Fang C, Gu J, Xie F, Behr M, Yang W, Abel ED, Ding X. (2008) Deletion of the NADPH-cytochrome P450 reductase gene in cardiomyocytes does not protect mice against doxorubicin-mediated acute cardiac toxicity. Drug Metab Dispos 36:1722–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Zhang QY. (2010) The role of small-intestinal P450 enzymes in protection against systemic exposure of orally administered benzo[a]pyrene. J Pharmacol Exp Ther 334:156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, McLaren AW, Carrie D, Henderson CJ, Wolf CR. (2007) Conditional deletion of cytochrome P450 oxidoreductase in the liver and gastrointestinal tract: a new model for studying the functions of the P450 system. J Pharmacol Exp Ther 322:40–47 [DOI] [PubMed] [Google Scholar]
- Finn RD, McLaughlin LA, Ronseaux S, Rosewell I, Houston JB, Henderson CJ, Wolf CR. (2008) Defining the in vivo role for cytochrome b5 in cytochrome P450 function through the conditional hepatic deletion of microsomal cytochrome b5. J Biol Chem 283:31385–31393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Henderson CJ, Scott CL, Wolf CR. (2009) Unsaturated fatty acid regulation of cytochrome P450 expression via a CAR-dependent pathway. Biochem J 417:43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, McLaughlin LA, Hughes C, Song C, Henderson CJ, Roland Wolf C. (2011) Cytochrome b5 null mouse: a new model for studying inherited skin disorders and the role of unsaturated fatty acids in normal homeostasis. Transgenic Res 20:491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flück CE, Tajima T, Pandey AV, Arlt W, Okuhara K, Verge CF, Jabs EW, Mendonça BB, Fujieda K, Miller WL. (2004) Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat Genet 36:228–230 [DOI] [PubMed] [Google Scholar]
- Flück CE, Nicolo C, Pandey AV. (2007) Clinical, structural and functional implications of mutations and polymorphisms in human NADPH P450 oxidoreductase. Fundam Clin Pharmacol 21:399–410 [DOI] [PubMed] [Google Scholar]
- Flück CE, Mullis PE, Pandey AV. (2009) Modeling of human P450 oxidoreductase structure by in silico mutagenesis and MD simulation. Mol Cell Endocrinol 313:17–22 [DOI] [PubMed] [Google Scholar]
- Flück CE, Mullis PE, Pandey AV. (2010) Reduction in hepatic drug metabolizing CYP3A4 activities caused by P450 oxidoreductase mutations identified in patients with disordered steroid metabolism. Biochem Biophys Res Commun 401:149–153 [DOI] [PubMed] [Google Scholar]
- Flück CE, Mallet D, Hofer G, Samara-Boustani D, Leger J, Polak M, Morel Y, Pandey AV. (2011a) Deletion of P399_E401 in NADPH cytochrome P450 oxidoreductase results in partial mixed oxidase deficiency. Biochem Biophys Res Commun 412:572–577 [DOI] [PubMed] [Google Scholar]
- Flück CE, Meyer-Böni M, Pandey AV, Kempná P, Miller WL, Schoenle EJ, Biason-Lauber A. (2011b) Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual differentiation. Am J Hum Genet 89:201–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flück CE, Pandey AV. (2011) Clinical and biochemical consequences of P450 oxidoreductase deficiency. Endocr Dev 20:63–79 [DOI] [PubMed] [Google Scholar]
- Fukami M, Nishimura G, Homma K, Nagai T, Hanaki K, Uematsu A, Ishii T, Numakura C, Sawada H, Nakacho M, et al. (2009) Cytochrome P450 oxidoreductase deficiency: identification and characterization of biallelic mutations and genotype-phenotype correlations in 35 Japanese patients. J Clin Endocrinol Metab 94:1723–1731 [DOI] [PubMed] [Google Scholar]
- Gonzalez M, Sealls W, Jesch ED, Brosnan MJ, Ladunga I, Ding X, Black PN, DiRusso CC. (2011) Defining a relationship between dietary fatty acids and the cytochrome P450 system in a mouse model of fatty liver disease. Physiol Genomics 43:121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Cui HD, Behr M, Zhang L, Zhang QY, Yang WZ, Hinson JA, Ding X. (2005) In vivo mechanisms of tissue-selective drug toxicity: effects of liver-specific knockout of the NADPH-cytochrome P450 reductase gene on acetaminophen toxicity in kidney, lung, and nasal mucosa. Mol Pharmacol 67:623–630 [DOI] [PubMed] [Google Scholar]
- Gu J, Weng Y, Zhang QY, Cui HD, Behr M, Wu L, Yang WZ, Zhang L, Ding X. (2003) Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem 278:25895–25901 [DOI] [PubMed] [Google Scholar]
- Hart SN, Zhong XB. (2008) P450 oxidoreductase: genetic polymorphisms and implications for drug metabolism and toxicity. Expert Opin Drug Metab Toxicol 4:439–452 [DOI] [PubMed] [Google Scholar]
- Henderson CJ, Otto DME, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR. (2003) Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem 278:13480–13486 [DOI] [PubMed] [Google Scholar]
- Huang NW, Pandey AV, Agrawal V, Reardon W, Lapunzina PD, Mowat D, Jabs EW, Van Vliet G, Sack J, Flück CE, Miller WL. (2005) Diversity and function of mutations in P450 oxidoreductase in patients with Antley-Bixler syndrome and disordered steroidogenesis. Am J Hum Genet 76:729–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Agrawal V, Giacomini KM, Miller WL. (2008) Genetics of P450 oxidoreductase: sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc Natl Acad Sci USA 105:1733–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan Z, Ilan R, Cinti DL. (1981) Evidence for a new physiological role of hepatic NADPH:ferricytochrome (P-450) oxidoreductase. Direct electron input to the microsomal fatty acid chain elongation system. J Biol Chem 256:10066–10072 [PubMed] [Google Scholar]
- Ireland H, Kemp R, Houghton C, Howard L, Clarke AR, Sansom OJ, Winton DJ. (2004) Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of β-catenin. Gastroenterology 126:1236–1246 [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. (2006) LXRs and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol 68:159–191 [DOI] [PubMed] [Google Scholar]
- Keber R, Motaln H, Wagner KD, Debeljak N, Rassoulzadegan M, Ačimovič J, Rozman D, Horvat S. (2011) Mouse knockout of the cholesterogenic cytochrome P450 lanosterol 14α-demethylase (Cyp51) resembles Antley-Bixler syndrome. J Biol Chem 286:29086–29097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B, Luyendyk JP, Tawfik O, Guo GL. (2009) Farnesoid X receptor deficiency induces nonalcoholic steatohepatitis in low-density lipoprotein receptor-knockout mice fed a high-fat diet. J Pharmacol Exp Ther 328:116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone N, Reisch N, Idkowiak J, Dhir V, Ivison HE, Hughes BA, Rose IT, O'Neil DM, Vijzelaar R, Smith MJ, et al. (2012) Genotype-phenotype analysis in congenital adrenal hyperplasia due to P450 oxidoreductase deficiency. J Clin Endocrinol Metab 97:E257–E267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Porter TD. (2007) Hepatic cytochrome P450 reductase-null mice reveal a second microsomal reductase for squalene monooxygenase. Arch Biochem Biophys 461:76–84 [DOI] [PubMed] [Google Scholar]
- Lin Y, Yao Y, Liu S, Wang L, Moorthy B, Xiong D, Cheng T, Ding X, Gu J. (2012) Role of mammary epithelial and stromal P450 enzymes in the clearance and metabolic activation of 7,12-dimethylbenz[a]anthracene in mice. Toxicol Lett 212:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin LA, Ronseaux S, Finn RD, Henderson CJ, Roland Wolf C. (2010) Deletion of microsomal cytochrome b5 profoundly affects hepatic and extrahepatic drug metabolism. Mol Pharmacol 78:269–278 [DOI] [PubMed] [Google Scholar]
- Miller WL. (1986) Congenital adrenal hyperplasia. N Engl J Med 314:1321–1322 [PubMed] [Google Scholar]
- Miller WL, Agrawal V, Sandee D, Tee MK, Huang N, Choi JH, Morrissey K, Giacomini KM. (2011) Consequences of POR mutations and polymorphisms. Mol Cell Endocrinol 336:174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL, Auchus RJ. (2011) The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev 32:81–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolo C, Flück CE, Mullis PE, Pandey AV. (2010) Restoration of mutant cytochrome P450 reductase activity by external flavin. Mol Cell Endocrinol 321:245–252 [DOI] [PubMed] [Google Scholar]
- Otto DME, Henderson CJ, Carrie D, Davey M, Gundersen TE, Blomhoff R, Adams RH, Tickle C, Wolf CR. (2003) Identification of novel roles of the cytochrome P450 system in early embryogenesis: effects on vasculogenesis and retinoic acid homeostasis. Mol Cell Biol 23:6103–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda SP, Buntur AR, Kar R, Tang K, Masters BS. (2012) Conditional deletion of cytochrome P450 reductase in mouse bone results in Antley-Bixler syndrome-like craniofacial and bone mass defects. FASEB J 26:784.8 (abstract). [Google Scholar]
- Pandey AV, Kempná P, Hofer G, Mullis PE, Flück CE. (2007) Modulation of human CYP19A1 activity by mutant NADPH P450 oxidoreductase. Mol Endocrinol 21:2579–2595 [DOI] [PubMed] [Google Scholar]
- Pandey AV, Flück CE, Mullis PE. (2010) Altered heme catabolism by heme oxygenase-1 caused by mutations in human NADPH cytochrome P450 reductase. Biochem Biophys Res Commun 400:374–378 [DOI] [PubMed] [Google Scholar]
- Peterson RE, Imperato-McGinley J, Gautier T, Shackleton C. (1985) Male pseudohermaphroditism due to multiple defects in steroid-biosynthetic microsomal mixed-function oxidases. A new variant of congenital adrenal hyperplasia. N Engl J Med 313:1182–1191 [DOI] [PubMed] [Google Scholar]
- Pineda Torra I, Claudel T, Duval C, Kosykh V, Fruchart JC, Staels B. (2003) Bile acids induce the expression of the human peroxisome proliferator-activated receptor α gene via activation of the farnesoid X receptor. Mol Endocrinol 17:259–272 [DOI] [PubMed] [Google Scholar]
- Porter TD, Banerjee S, Stolarczyk EI, Zou L. (2011) Suppression of cytochrome P450 reductase (POR) expression in hepatoma cells replicates the hepatic lipidosis observed in hepatic Por-null mice. Drug Metab Dispos 39:966–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter TD. (2012) New insights into the role of cytochrome P450 reductase (POR) in microsomal redox biology. Acta Pharmaceutica Sinica B 2:100–104 [Google Scholar]
- Reardon W, Smith A, Honour JW, et al. (2000) Evidence for digenic inheritance in some cases of Antley-Bixler syndrome? J Med Genet 37:26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen AL, O’Leary KA, Kasper CB. (2002) Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J Biol Chem 277:6536–6541 [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. (2000) Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102:731–744 [DOI] [PubMed] [Google Scholar]
- Wang XJ, Chamberlain M, Vassieva O, Henderson CJ, Wolf CR. (2005) Relationship between hepatic phenotype and changes in gene expression in cytochrome P450 reductase (Por) null mice. Biochem J 388:857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zhou X, Fang C, Li L, Kluetzman K, Yang W, Zhang QY, Ding X. (2010) Generation of a mouse model with a reversible hypomorphic cytochrome P450 reductase gene: utility for tissue-specific rescue of the reductase expression, and insights from a resultant mouse model with global suppression of P450 reductase expression in extrahepatic tissues. J Pharmacol Exp Ther 334:69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, DiRusso CC, Reilly AA, Black PN, Ding X. (2005) Hepatic gene expression changes in mouse models with liver-specific deletion or global suppression of the NADPH-cytochrome P450 reductase gene. Mechanistic implications for the regulation of microsomal cytochrome P450 and the fatty liver phenotype. J Biol Chem 280:31686–31698 [DOI] [PubMed] [Google Scholar]
- Weng Y, Fang C, Turesky RJ, Behr M, Kaminsky LS, Ding X. (2007) Determination of the role of target tissue metabolism in lung carcinogenesis using conditional cytochrome P450 reductase-null mice. Cancer Res 67:7825–7832 [DOI] [PubMed] [Google Scholar]
- Wu L, Gu J, Weng Y, Kluetzman K, Swiatek P, Behr M, Zhang QY, Zhuo XL, Xie Q, Ding X. (2003) Conditional knockout of the mouse NADPH-cytochrome P450 reductase gene. Genesis 36:177–181 [DOI] [PubMed] [Google Scholar]
- Wu L, Gu J, Cui HD, et al. (2005) Transgenic mice with a hypomorphic NADPH-cytochrome P450 reductase gene: effects on development, reproduction, and microsomal cytochrome P450. J Pharmacol Exp Ther 312:35–43 [DOI] [PubMed] [Google Scholar]
- Xia C, Panda SP, Marohnic CC, Martásek P, Masters BS, Kim JJ. (2011) Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency. Proc Natl Acad Sci USA 108:13486–13491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QY, Kaminsky LS, Dunbar D, Zhang J, Ding X. (2007) Role of small intestinal cytochromes P450 in the bioavailability of oral nifedipine. Drug Metab Dispos 35:1617–1623 [DOI] [PubMed] [Google Scholar]
- Zhang QY, Fang C, Zhang J, Dunbar D, Kaminsky L, Ding X. (2009) An intestinal epithelium-specific cytochrome P450 (P450) reductase-knockout mouse model: direct evidence for a role of intestinal P450s in first-pass clearance of oral nifedipine. Drug Metab Dispos 37:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, D’Agostino J, Zhang QY. (2011) Role of intestinal cytochrome P450 (P450) in modulating the bioavailability of oral lovastatin: insights from studies on the intestinal epithelium-specific P450 reductase knockout mouse. Drug Metab Dispos 39:939–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Zhang QY. (2012) Role of intestinal cytochrome P450 enzymes in diclofenac-induced toxicity in the small intestine. J Pharmacol Exp Ther 343:362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]