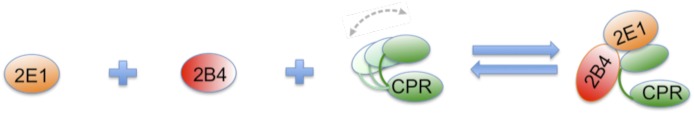
Abstract
Studies in microsomal and reconstituted systems have shown that the presence of one cytochrome P450 isoform can significantly influence the catalytic activity of another isoform. In this study, we assessed whether CYP2E1 could influence the catalytic activity of CYP2B4 under steady-state turnover conditions. The results show that CYP2E1 inhibits CYP2B4-mediated metabolism of benzphetamine (BNZ) with a Ki of 0.04 µM. However, CYP2B4 is not an inhibitor of CYP2E1-mediated p-nitrophenol hydroxylation. When these inhibition studies were performed with the artificial oxidant tert-butyl hydroperoxide, CYP2E1 did not significantly inhibit CYP2B4 activity. Determinations of the apparent KM and kcat of CYP2B4 for CPR in the presence of increasing concentrations of CYP2E1 revealed a mixed inhibition of CYP2B4 by CYP2E1. At low concentrations of CYP2E1, the apparent KM of CYP2B4 for CPR increased up to 23-fold with virtually no change in the kcat for the reaction, however, at higher concentrations of CYP2E1, the apparent KM of CYP2B4 for CPR decreased to levels similar to those observed in the absence of CYP2E1 and the kcat also decreased by 11-fold. Additionally, CYP2E1 increased the apparent KM of CYP2B4 for BNZ by 8-fold and the apparent KM did not decrease to its original value when saturating concentrations of CPR were used. While the individual apparent KM values of CYP2B4 and CYP2E1 for CPR are similar, the apparent KM of CYP2E1 for CPR in the presence of CYP2B4 decreased significantly, thus suggesting that CYP2B4 enhances the affinity of CYP2E1 for CPR and this may allow CYP2E1 to out-compete CYP2B4 for CPR.
Introduction
To interrogate the catalytic properties of the P450 under investigation, the majority of studies performed in reconstituted systems are generally conducted using saturating, or nearly saturating concentrations of CPR; however, in the endoplasmic reticulum (ER), P450s exist in vast excess (up to 20-fold) over CPR (Estabrook et al., 1971). Despite the plethora of information regarding the mechanisms by which CPR reduces P450s, very little is known about the spatial organization and interactions of the cytochromes (P450 and b5) and flavoproteins (CPR and cytochrome b5 reductase) within the membrane of the ER.
Several models have been proposed to describe the distribution of P450s and their redox partners in the lipid membrane of the ER (Peterson et al., 1976; Backes and Kelley, 2003; Brignac-Huber et al., 2011). For example, Peterson et al. proposed that the N-terminal hydrophobic tail of CPR anchors it in the membrane while its catalytic domain protrudes from the membrane into the cytosolic region of the cell. In this model, several P450s are envisioned to cluster around a central CPR molecule while a portion of the microsomal P450s are thought to be loosely associated with the CPR and may even be free-floating in the membrane matrix (Peterson et al., 1976). The disparity in the P450/CPR ratio combined with the likely organization of these microsomal proteins in the membrane suggests that at any given time only a portion of the total microsomal P450s can be in functional complexes with CPR and thus capable of catalyzing the metabolism of drugs. For that reason, the outcome of the metabolism of a drug in a given tissue may not only be a function of the particular P450 that metabolizes the drug, but also its accessibility to CPR, which may be influenced by the presence and abundance of other P450s in the ER that may compete for the CPR (Eyer and Backes, 1992).
There have been a number of reports on the interaction of P450s not only with CPR but also with each other leading to alterations in catalytic activity. For example, CYP2C19 has been shown to be more active than CYP2C9 for the metabolism of methoxychlor in the purified reconstituted system (Hazai and Kupfer, 2005). However, when the role of CYP2C19 in the metabolism of methoxychlor was assessed in human liver microsomes by monoclonal antibodies raised against CYP2C19, there was no change in methoxychlor metabolism, yet antibodies raised against CYP2C9 were efficient in inhibiting methoxychlor-O-demethylation (Hu et al., 2004). When the metabolism of methoxychlor was assessed in a reconstituted system containing both CYP2C9 and CYP2C19, the demethylation of methoxychlor by CYP2C19 was significantly inhibited, suggesting that interactions among P450 isoforms can modulate their catalytic activities. In addition to the interactions between CYP2C9 and CYP2C19, evidence also exists to support interactions between other P450s, including CYP2B4 and CYP1A2 (Cawley et al., 1995, 2001; Backes et al., 1998; Davydov et al., 2001; Reed et al., 2010), CYP1A2 and CYP2E1(Kelley et al., 2006), CYP2C9 and CYP3A4 (Subramanian et al., 2010), and CYP2C9 and CYP2D6 (Subramanian et al., 2009).
Among the many human P450s, CYP2E1 and CYP2B6 are known to play important roles in liver toxicity and drug metabolism, respectively. CYP2E1 can bioactivate low molecular-weight compounds, such as acetaminophen (Patten et al., 1993), N-nitrosodimethylamine (Levin et al., 1986), and carbon tetrachloride (Guengerich et al., 1991), into reactive metabolites that lead to chemical toxicity. Furthermore, CYP2E1 is known to be a highly uncoupled P450, and the induction of CYP2E1 by alcohol (Oneta et al., 2002) or fasting (Johansson et al., 1988) likely contributes to an increase in the generation of reactive oxygen species, which ultimately can lead to oxidative stress (Dey and Cederbaum, 2006). CYP2B6 constitutes approximately 5% of the total P450 in the liver and plays an important role in the metabolism of a number of clinically used drugs, including bupropion, cyclophosphamide, efavirenz, and propofol (Walsky et al., 2006; Wang and Tompkins, 2008). Its highly polymorphic nature is of great importance in drug development, as differences in amino acid sequence are known to affect both catalytic activity (Zhang et al., 2011) and hepatic protein levels (Lang et al., 2001). While the ultimate objective of this investigation is to better understand the catalytic activity of human CYP2B6, the studies reported here were performed initially on rabbit CYP2B4 since the final protein yields from the bacterial overexpression and purification of CYP2B6 are quite low and do not yield sufficient protein for these types of studies (Scott et al., 2001). CYP2B4, the rabbit homolog to the human CYP2B6, is significantly more highly expressed in Escherichia coli and shares 88% sequence similarity with CYP2B6, making it a reasonable model system to understand CYP2B6 (Oezguen et al., 2008).
The prediction of human in vivo pharmacokinetic data from in vitro data depends, in part, on our ability to understand the factors that influence P450-mediated drug metabolism in vivo. One of these factors that is poorly understood is the effect of P450-P450 interactions on intrinsic drug clearance. Since the importance of CYP2B6 and CYP2E1 in drug metabolism is well established and the protein levels of these P450s are highly inducible, it is critical that we understand how the presence of one P450 may alter the catalytic properties of the other if we are to make accurate in vitro to in vivo correlations of intrinsic clearance. Therefore, the objective of this study was to assess whether CYP2E1 and CYP2B4 could influence each other’s catalytic activities, to characterize the kinetic nature of such interactions, and to propose a preliminary model that explains the physical basis for the kinetic results with the ultimate goal of using this information to improve our ability to predict clearance in vivo.
Materials and Methods
Chemicals.
All chemicals used are of ACS grade unless otherwise specified and were obtained from commercial vendors. Benzphetamine (BNZ), para-nitrophenol (p-NP), NADPH, sodium dithionite, ascorbic acid, and tert-butyl hydroperoxide were purchased from Sigma. Trifluoroacetic acid (TFA) was purchased from Pierce Chemicals (Rockford, IL). Dilauroylphosphatidylcholine (DLPC) was purchased from Doosan Serdary Research Laboratory (Toronto, ON, Canada). Carbon monoxide gas (purity >99.5%) was purchased from Cryogenic Gases (Detroit, MI).
Construction of CYP2E1 Y422D Variant.
Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit according to the manufacturer’s protocol (Stratagene, La Jolla, CA). The forward and reverse mutagenic primers for Y422D were GGAAAGTTCAAGGACAGTGACTATTTCAAGCC and GGCTTGAAATAGTCACTGTCCTTGAACTTTCC, respectively. DNA sequencing performed at the University of Michigan DNA Sequencing Core confirmed the desired site-specific mutation.
Overexpression and Purification of Enzymes.
The plasmids for the N-terminal truncated and C-terminal His-tagged CYP2B4dH (hereon referred to as CYP2B4) and human CYP2E1 were a generous gift from Dr. James Halpert. These two P450s and the CYP2E1 Y422D variant were overexpressed in E. coli C41 (DE3) cells separately and purified using a Ni-NTA affinity column as described previously (Scott et al., 2003; Pratt-Hyatt et al., 2010). The concentrations of CYP2B4 and CYP2E1 were determined using an extinction coefficient of ∆ε450–490 nm of 91 mM−1 cm−1 as described by Omura and Sato, 1964. NADPH-dependent cytochrome P450 reductase (CPR) was expressed and purified from E. coli as described previously (Zhang et al., 2007). The concentration of CPR was determined using an extinction coefficient of 21 mM−1 cm−1 at 456 nm for the oxidized enzyme (Vermilion and Coon, 1978).
Determination of the Inhibition of CYP2B4-Mediated N-Demethylation of BNZ by CYP2E1 WT and the CYP2E1 Y422D Variant.
The extent to which varying concentrations of either CYP2E1 WT or CYP2E1 Y422D inhibited the rate of formaldehyde formation by CYP2B4 (0.25 μM) was assessed at 37°C using a fixed concentration of CPR and BNZ. CYP2B4 was reconstituted in triplicate with increasing concentrations of CYP2E1 WT or CYP2E1 Y422D (0.25, 0.5, 0.75, 1.0, 1.25, 1.50 μM), CPR (0.5 μM) and 0.03 mg/ml DLPC on ice for 1 hour. The reconstituted mixtures were then added to 50 mM potassium phosphate buffer, pH 7.4, and BNZ (1.2 mM). After the samples were equilibrated at 37°C for 15 min, the reactions were initiated by adding 7.5 μl of 20 mM NADPH to give a final reaction volume of 500 μl. The reactions were incubated for 5 minutes at 37°C and then quenched by the addition of 25 μl of 50% TFA. The proteins were precipitated by centrifugation at 13.2k rpm in an Eppendorf 5415D microcentrifuge for 5 minutes and a 500-μl aliquot of the supernatant was assayed for formaldehyde using the Nash reaction (Nash, 1953).
Difference Spectra of the Carbon-Monoxy Ferrous WT CYP2E1 and Y422D Variant.
Wild type (WT) CYP2E1 and the Y422D mutant (0.5 nmol) were reconstituted with CPR (1 nmol) at 22°C for 30 minutes in 0.5 ml of suspension buffer (100 mM potassium phosphate buffer, 20% glycerol, and 0.1 mM EDTA, pH 7.4). After adding 100 μM 4-methylpyrazole and 0.5 mM NADPH for the baseline, the samples were bubbled with CO, and the visible absorbance spectra were determined by scanning from 400 to 500 nm on a UV-2501PC spectrophotometer (Shimadzu Corporation, Kyoto, Japan) until a steady state was attained. A trace amount of sodium dithionite was added and additional scans were performed until no further changes were observed.
Determination of the Apparent KM and kcat Values for CYP2B4 for CPR Using the N-Demethylation of BNZ in the Presence and Absence of CYP2E1.
The apparent KM and kcat values for CYP2B4 WT for CPR were determined at 37°C by measuring the rate of formaldehyde formation as a result of the N-demethylation of BNZ at a constant P450 concentration with increasing concentrations of CPR, as previously described (Kenaan et al., 2011). In brief, CYP2B4 WT (0.25 μM) was reconstituted in triplicate with varying concentrations of CPR (0.1, 0.2, 0.3, 0.6, 0.8, 1.2, and 1.4 μM) and 0.03 mg/ml DLPC on ice for 1 hour. The reconstituted mixtures were then added to 50 mM potassium phosphate buffer, pH 7.4, containing 1.2 mM BNZ. After the samples were equilibrated at 37°C for 15 min, the reactions were initiated by adding 7.5 μl of 20 mM NADPH to give a final reaction volume of 500 μl. The reactions were incubated for 5 minutes at 37°C and then quenched by the addition of 25 μl of 50% TFA. The proteins were precipitated by centrifugation at 13.2k rpm in an Eppendorf 5415D microcentrifuge for 5 minutes, and a 500-μl aliquot of the supernatant was assayed for formaldehyde using the Nash reaction. The kinetic parameters were determined by fitting the data to the Michaelis-Menten equation using GraphPad Prism 5.0 from GraphPad Software (La Jolla, CA). The free concentrations of CPR in the plot were calculated from the initial concentration of CPR and the apparent KM. To determine the effect of CYP2E1 on the apparent KM and kcat of the CYP2B4-CPR complex the procedure outlined in this paragraph was repeated in the presence of 0.125, 0.25, 0.75, 1.25, and 1.50 μM CYP2E1.
Graphical Analysis of Steady-State Activity Data.
By use of GraphPad Prism, the inverse of the reaction velocities obtained for the determination of the apparent KM and kcat values for CYP2B4 and CPR was plotted against the inverse of the concentrations of CPR used under varying concentrations of CYP2E1 to obtain a Lineweaver-Burk plot. To obtain a Ki for CYP2E1 as an inhibitor of the CYP2B4-CPR complex, two methods were used. The first one used a nonlinear global fitting analysis of the data to a mixed inhibition model using GraphPad Prism, and the second involved plotting KM obs against varying concentrations of CYP2E1 (0–0.75 μM) for which competitive inhibition was observed. The Ki was obtained from the x-intercept of a linear regression analysis of this data (Kakkar et al., 1999).
Characterization of the tert-Butyl Hydroperoxide-Supported Metabolism of BNZ by CYP2B4 WT in the Presence of Increasing Concentrations of CYP2E1 WT.
To determine the rates for the tert-butyl hydroperoxide–supported metabolism of BNZ, a final concentration of 0.25 μM CYP2B4 WT was reconstituted with varying concentrations of CYP2E1 (0.25, 0.5, 0.75, 1.0, 1.25, 1.50 μM) and 0.03 mg/ml DLPC on ice for 1 hour. The samples were then aliquoted to solutions containing 50 mM potassium phosphate buffer, pH 7.4, and 1.2 mM BNZ, and incubated at 37°C for 15 min. tert-Butyl hydroperoxide (52.5 μl of 1 M solution) was added to give a final volume of 500 μl, and the reactions were incubated for 5 minutes and then terminated by the addition of 25 μl of 50% TFA. The samples were centrifuged at 13.2k rpm in an Eppendorf 5415D microcentrifuge for 5 minutes, and 500-μl aliquots of the supernatants were assayed for formaldehyde using the Nash reaction. This procedure was then performed in the presence of 0.25, 0.5, 0.75, 1.0, 1.25, 1.50 μM CYP2E1 WT.
Determination of the KM and kcat Values for the Metabolism of BNZ by CYP2B4 in the Presence and Absence of CYP2E1 WT.
The KM and kcat values for BNZ metabolism by CYP2B4 in the presence or absence of CYP2E1 were determined at 37°C at constant P450 and CPR concentrations with increasing concentrations of BNZ. CYP2B4 WT (0.50 μM) was reconstituted in triplicate with an equal concentration of CPR and 0.03 mg/ml DLPC with or without 2 μM CYP2E1 WT on ice for 1 hour. The reconstituted mixtures were then added to 50 mM potassium phosphate buffer, pH 7.4, containing varying concentrations of BNZ (0.05, 0.1, 0.2, 0.4, 0.6, 0.8 mM). After the samples were equilibrated at 37°C for 15 min, the reactions were initiated by adding 7.5 μl of 20 mM NADPH to give a final reaction volume of 500 μl. The reactions were incubated for 5 minutes at 37°C and then quenched by the addition of 25 μl of 50% TFA. The protein was precipitated by centrifugation at 13.2k rpm in an Eppendorf 5415D microcentrifuge for 5 minutes, and a 500-μl aliquot of the supernatant was assayed for formaldehyde using the Nash reaction. To determine whether the original KM value in the absence of CYP2E1 could be observed by using saturating concentrations of CPR, this experiment was repeated in the presence of 2.5 μM CPR. The kinetic parameters were determined by fitting the data to the Michaelis-Menten equation using GraphPad Prism 5.0.
Spectral Dissociation Constant (Ks) for the Binding of BNZ to CYP2B4 in the Presence and Absence of CYP2E1.
BNZ binding to ferric CYP2B4 was monitored spectrophotometrically, as previously described, by measuring the type I spectral changes (Zhang et al., 2009). In brief, 1 μM CYP2B4 and 0.1 mg/ml DLPC were reconstituted in the presence or absence of 4 μM CYP2E1 on ice for 1 hour. Equal volumes of the reconstitutions were then aliquoted to solutions containing 0.1 M potassium phosphate, pH 7.4, and 0.1 mg/ml DLPC and were added to the sample and reference cuvettes in a UV-2501PC spectrophotometer (Shimadzu Corporation). A baseline was recorded after thermal equilibration at 30°C for 5 min. CYP2B4 in the sample cuvette was titrated with aliquots of 20 mM BNZ, while an equal volume of water was added to the reference cuvette. The difference spectra were recorded from 350 to 500 nm, and the difference in absorbance (A) between the wavelength maximum (386 nm) and minimum (421 nm) was plotted as a function of varying BNZ concentration (5–1200 μM). The data were then fit to eq. 1 to obtain the Ks.
| (1) |
Determination of the Apparent KM and kcat Values of CYP2E1 for CPR Using the Hydroxylation of p-NP in the Presence and Absence of CYP2B4.
Saturating concentrations of p-NP, a fixed concentration of CYP2E1 and varying concentrations of CPR were used to determine the apparent KM and kcat of CYP2E1 for CPR. CYP2E1 (0.1 μM) was reconstituted with varying concentrations of CPR (0.08, 0.12, 0.24, 0.72, 1.44, 2.00 μM) and 0.03 mg/ml DLPC in the presence or absence of 0.1 μM CYP2B4 on ice for 1 hour. The reconstituted mixtures were then added to 50 mM potassium phosphate buffer, pH 7.4, containing 0.3 mM p-NP and 2 mM ascorbic acid. After the samples were equilibrated at 37°C for 15 min, the reactions were initiated by adding 20 μl of 20 mM NADPH to give a final reaction volume of 1000 μl. The reactions were incubated for 20 minutes at 37°C before they were quenched with 300 μl of 20% TCA. The samples were then incubated on ice for 10 minutes and centrifuged at 13.2k rpm in an Eppendorf 5415D microcentrifuge for 5 min. The absorbance at 510 nm was then measured by transferring a 1000-μl aliquot of the reaction mixture to a cuvette containing 100 μl of 10 M NaOH. The kinetic parameters were determined by fitting the data to the Michaelis-Menten equation using GraphPad Prism 5.0 from GraphPad Software. The free concentrations of CPR in the plot were calculated from the initial concentration of CPR and the apparent KM.
Results
Effect of Increasing Concentrations of CYP2E1 on BNZ Metabolism by CYP2B4.
To preclude the possibility that CYP2E1 metabolizes BNZ to form formaldehyde, or any other detectable product, the metabolism of BNZ by CYP2E1 was assessed by electrospray ionization liquid chromatography mass spectrometry using previously established methods (Kent et al., 2004). Our results showed that CYP2B4 metabolizes BNZ primarily to nor-benzphetamine and that metabolism by CYP2E1 is negligible compared with CYP2B4, even at high concentrations of CYP2E1 (data not shown). The converse was also shown to be true for the metabolism of p-NP by CYP2E1 and CYP2B4. That is, CYP2E1 metabolizes p-NP relatively well, whereas CYP2B4 shows very little activity toward p-NP, even at high concentrations (data not shown). Therefore, these control experiments demonstrate that the N-demethylation of BNZ activity to form formaldehyde and the hydroxylation of p-NP to form 4-nitrocatechol (4-NC) are specific reporters of CYP2B4 and CYP2E1 activity, respectively, in this system.
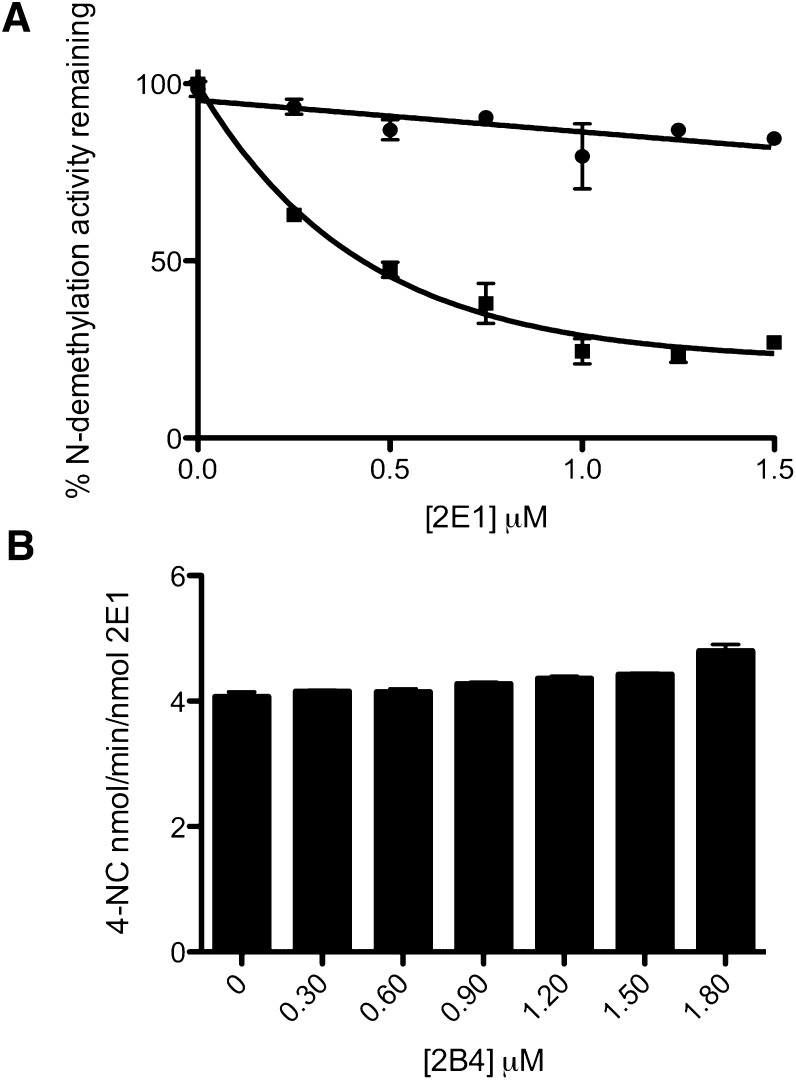
To assess the extent to which CYP2E1 could inhibit the catalytic activity of CYP2B4 toward its probe substrate (BNZ) under conditions of constant CPR, increasing concentrations of CYP2E1 were reconstituted with CYP2B4, CPR, and DLPC. The results shown in Fig. 1A illustrate that increasing concentrations of CYP2E1 can inhibit the activity of CYP2B4 up to approximately 80% of the activity observed in the absence of CYP2E1. The data also show that the inhibition can be fit to a rectangular hyperbolic function and follows saturation kinetics. To determine whether the decrease in the CYP2B4 catalytic activity is due to a disruption of the interaction between CYP2B4 and CPR or to direct interaction between CYP2B4 and CYP2E1, the metabolism of BNZ by CYP2B4 supported by tBHP was determined in the presence of increasing concentrations of CYP2E1. Although the presence of CYP2E1 significantly decreased the metabolism of BNZ by CYP2B4 when supported by CPR, CYP2E1 had virtually no effect on BNZ metabolism by CYP2B4 when supported by tBHP. Because hydroperoxides and other artificial oxygen donors are able to support substrate metabolism by P450s in the absence of redox partners such as CPR, the decrease in the CPR-supported BNZ activity, with no change in the tBHP-supported activity, suggests that inhibition is not attributed to direct CYP2B4-CYP2E1 interactions but rather to an effect on CPR-CYP2B4 interaction. The ability of CYP2B4 to inhibit or stimulate the metabolism of p-NP by CYP2E1 was also investigated, and the results shown in Fig. 1B demonstrate that CYP2B4 does not have any significant effect on the catalytic activity of CYP2E1. Therefore, the inhibition observed in the reconstituted system is solely an effect of CYP2E1 on CYP2B4, which is not reciprocated.
Fig. 1.
(A) effect of increasing concentrations of CYP2E1 on CYP2B4-catalyzed N-demethylation of BNZ. N-demethylation activity supported by t-BHP (●) and CPR/NADPH (▪) was determined as described under Materials and Methods in attempts to differentiate the effect of direct CYP2E1-CYP2B4 interactions on CYP2B4 activity from competition for CPR. Excess BNZ (1.2 mM) was used in the incubations to compensate for possible perturbations in substrate binding affinity due to direct P450-P450 interactions. The concentrations of CYP2B4 and CPR were fixed at 0.25 and 0.5 μM respectively, while the concentration of CYP2E1 varied from 0 to 1.50 μM). (B) effect of Increasing Concentrations of CYP2B4 on CYP2E1-catalyzed p-NP hydroxylation. To determine whether inhibition of CYP2E1 was observed with CYP2B4, the CYP2E1 catalyzed hydroxylation of p-NP was assessed in the presence of increasing concentrations of CYP2B4 with concentrations of p-NP well above saturation. The concentration of CYP2E1 and CPR used was 0.30 μM while CPR was varied from 0 to 1.80 μM. Data points for BNZ and p-NP metabolism represent the mean of three experiments, done at least in duplicate, while error bars represent the standard deviations.
Inhibition of CYP2B4 Activity by the CYP2E1 Y422D Variant.
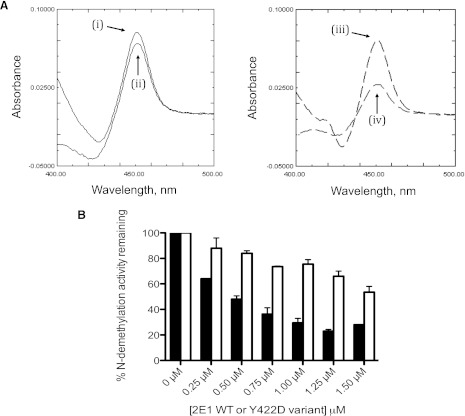
Previous studies by Lin et al. (Lin et al., 2007) led us to express and purify the CYP2E1 Y422D mutant to dissect out whether the inhibitory phenomenon observed in Fig. 1A was due to the CYP2E1-CPR interaction. The Tyr 422 residue of CYP2E1 can be nitrated by peroxynitrite and its modification results in a loss in the CPR-supported CYP2E1 activity when compared with tBHP-supported activity (Lin et al., 2007). Since the FMN domain of CPR, which is highly negatively charged, is believed to interact with the proximal side of CYP2E1, we hypothesized that replacing Tyr 422 with Asp using site-directed mutagenesis would create a charge- repulsive interaction between the CYP2E1 Y422D and its residue counterpart in CPR that would perturb the apparent affinity of the two proteins for each other. By use of stopped-flow studies, we have previously shown that the extent of reduced CO-complex formation upon addition of NADPH is an indicator of the extent of P450-CPR complex, and thus, it reflects the affinity of the P450 under investigation for CPR (Kenaan et al., 2011). The extent of reduction of the CYP2E1 Y422D mutant by CPR and NADPH relative to the reduction of WT CYP2E1 by CPR and NADPH can be determined by measuring the absorbance of the Y422D-CO complex at 450 nm and comparing this to the absorbance of the WT-CO complex at the same wavelength. As shown in Fig. 2A, the interaction of CYP2E1 Y422D with CPR decreases by 50% compared with WT. Furthermore, the apparent KM value of this variant for CPR increased significantly as a result of mutagenesis (data not shown). Interestingly, when the inhibitory potential of this mutant for CYP2B4 was investigated, the results showed that it inhibited CYP2B4 activity to a significantly lesser extent than the WT CYP2E1 (Fig. 2B).
Fig. 2.
Effect of the mutation of Tyr 422 to Asp in CYP2E1 on its ability to form the reduced–carbon monoxide (CO) complex when reduced with NADPH and for the CYP2E1 WT or the Y422D variant to inhibit CYP2B4 activity. (A) reduced CO spectrum of 0.5 nmol WT CYP2E1 (left) and the 0.5 nmol CYP2E1 Y422D variant (right). The reduced CO difference spectra were measured in the following reconstitutions, as described under Materials and Methods. Samples (i) and (iii) were chemically reduced with dithionite, whereas samples (ii) and (iv) were reconstituted with 1 nmol CPR and reduced by the addition of NADPH. (B) inhibition of CYP2B4 BNZ activity by CYP2E1 WT and the Y422D variant. The ability of the Y422D variant to inhibit CYP2B4 activity was assessed (open bars) and compared with WT CYP2E1 (filled bars), as described under Materials and Methods.
Determination of the Apparent KM and kcat Values for the Interactions of CYP2B4 WT with CPR.
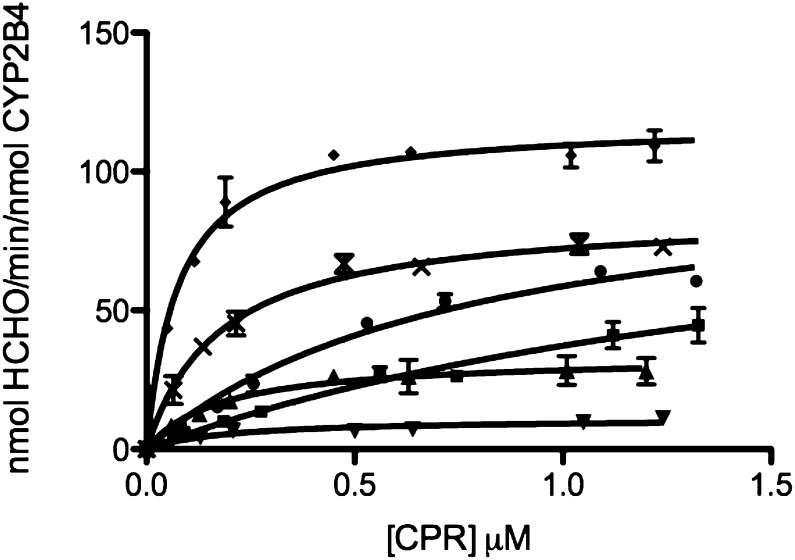
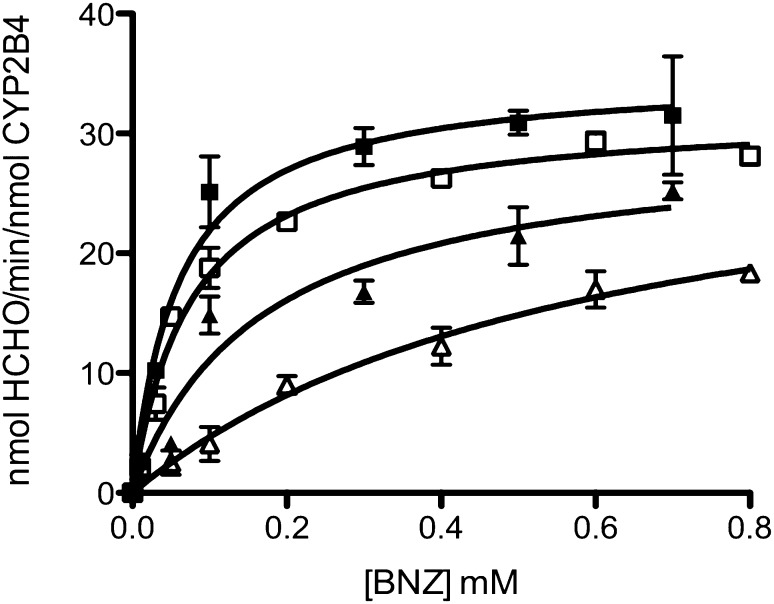
Formation of a CPR-P450 complex is essential for the transfer of electrons to the heme that is required for substrate oxidation. The rate of substrate oxidation is believed to be directly proportional to the concentration of the CPR-CYP2B4 complex (Miwa et al., 1979; Bridges et al., 1998). Thus, by measuring the rate of BNZ oxidation in the presence of a fixed concentration of CYP2B4 and increasing concentrations of CPR, apparent KM and kcat values for CPR binding to CYP2B4 can be determined across a range of CYP2E1 concentrations. By comparing these kinetic parameters at increasing concentrations of CYP2E1, we can investigate the nature of the inhibitory interactions observed in Fig. 1A. The competition between CYP2E1 and CYP2B4 for CPR should disrupt CYP2B4-CPR complex formation and decrease the concentration of CPR-complexed CYP2B4 that is able to oxidize substrate. As shown in Fig. 3, the rate of BNZ oxidation as a function of varying CPR concentrations follows a rectangular hyperbolic relationship, which can be fit to the Michaelis-Menten equation. For CYP2E1 concentrations ranging from 0 to 0.75 μM, the apparent KM of CYP2B4 for CPR increased by 23-fold while the kcat remained virtually unchanged (Fig. 3; Table 1), suggesting a competitive inhibition. For concentrations higher than 0.75 μM CYP2E1, an unusual noncompetitive kinetic behavior was observed with a 11-fold decrease in the kcat that was accompanied by a decrease in the apparent KM value to 0.20 μM in the presence of 1.50 μM CYP2E1.
Fig. 3.
Determination of the kinetics for the N-demethylation of BNZ by CYP2B4 in the presence of increasing concentrations of CYP2E1 WT. The N-demethylation of BNZ to produce formaldehyde was measured at a constant concentration of CYP2B4 (0.25 μM) with increasing concentrations of CPR (0.1, 0.2, 0.3, 0.6, 0.8, 1.2 μM) and the following concentrations of CYP2E1 WT: 0 (♦), 0.125 (×), 0.25 (●), 0.75 (▪), 1.25 (▴), 1.50 μM (▾), as described under Materials and Methods. The plots were corrected to account for the free concentration of CPR at each titration. Error bars are the standard deviations from three measurements done at least in triplicate.
TABLE 1.
Apparent KM and kcat values of CYP2B4 for CPR in the presence of increasing concentrations of CYP2E1 WT
The apparent KM and kcat values of CYP2B4 for CPR were determined as described under Materials and Methods. Since CYP2E1 does not metabolize BNZ, the kinetic parameters for CPR binding to CYP2B4 could be determined by measuring the rate of formaldehyde formation using fixed concentrations of CYP2B4 and BNZ with increasing concentrations of CPR and CYP2E1. The kinetic values given here were derived from data in Fig. 3.
| [2E1] | KM | kcat |
|---|---|---|
| μM | μM | min–1 |
| 0 | 0.08 ± 0.01 | 118 ± 2.2 |
| 0.125 | 0.18 ± 0.014 | 85 ± 1.8 |
| 0.25 | 0.77 ± 0.13 | 103 ± 8.7 |
| 0.75 | 1.86 ± 0.61 | 106 ± 23 |
| 1.25 | 0.18 ± 0.04 | 33 ± 2.2 |
| 1.50 | 0.20 ± 0.06 | 11 ± 1.0 |
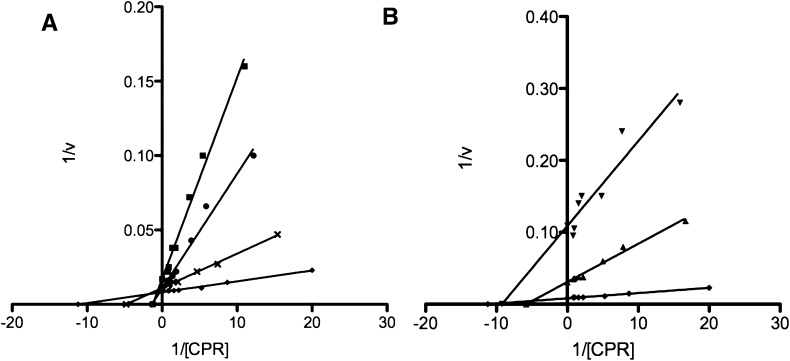
Linear Regression Analysis of the Steady-State Activity Data.
By plotting the inverse of the BNZ activity against the inverse of CPR concentrations as a function of varying CYP2E1 concentrations, it was observed that this data could be divided into two sets (Fig. 4). These data sets were grouped based on whether the linear regression analysis resulted in an intersection at a common value on the y-axis but diverged on the x-axis or whether the lines diverged on the y-axis and intersected on the x-axis. The data shown in Fig. 4A clearly demonstrate that between 0 and 0.75 μM CYP2E1 the apparent KM values change significantly while the kcat values remain almost unchanged. This is indicative of a competitive interaction. Figure 4B illustrates the noncompetitive nature of CYP2E1 inhibition, as the apparent KM values for 1.25 and 1.50 μM CYP2E1 are relatively similar to the apparent KM value observed in the absence of CYP2E1, yet the kcat decreases significantly as proven by an increase in the y-intercept value compared with 0 μM CYP2E1. Therefore, under our experimental conditions, it appears that CYP2E1 inhibits the formation of the CYP2B4-CPR complex by acting as a competitive inhibitor at low concentrations and as a noncompetitive inhibitor at higher concentrations.
Fig. 4.
Lineweaver-Burk plots of the steady-state activity data presented in Fig. 3. The inverse of the rate of formaldehyde formation was plotted against the inverse of CPR concentrations (from Fig. 3) as a function of varying CYP2E1 concentrations. These data were divided into two sets based on whether the linear regressions intersected at a common intercept or not. For the following concentrations of CYP2E1 WT: 0 (♦), 0.125 (×), 0.25 (●), 0.75 μM (▪), CYP2E1 displayed competitive inhibition kinetics (A) while at higher concentrations, 1.25 (▴) and 1.50 μM (▾), CYP2E1 behaved as a noncompetitive (B) inhibitor.
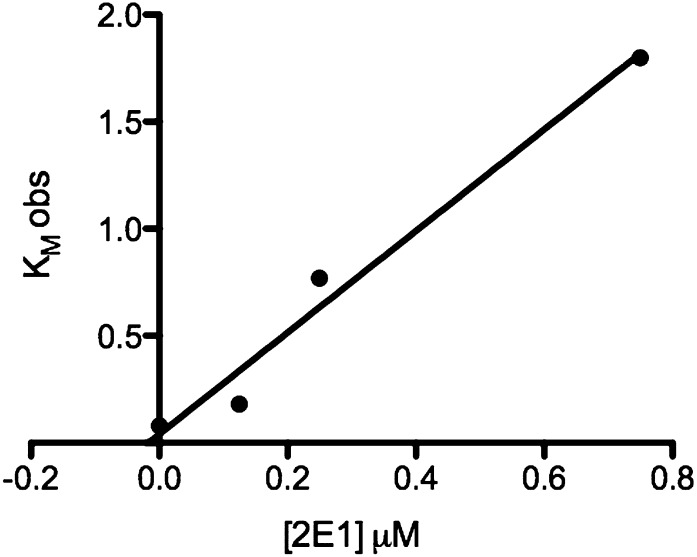
To estimate a Ki for CYP2E1 as a competitive inhibitor of the CYP2B4-CPR complex we plotted the KM obs as a function of CYP2E1 concentration (Kakkar et al., 1999). These data were fit to a straight line that intersected the x-axis at an absolute value of 0.02 μM (Fig. 5). Therefore, CYP2E1 is a potent CYP2B4 inhibitor with a nanomolar affinity. Global nonlinear analysis of CYP2E1 concentrations 0–1.50 μM by GraphPad Prism revealed a Ki of 0.038 μM.
Fig. 5.
Plot of KM obs versus CYP2E1 concentration used to estimate the inhibition constant Ki. Estimates of KM obs were obtained from analysis of the activity data shown in Fig. 3. The solid line is the linear regression analysis of data for CYP2E1 concentrations from 0 to 0.75 μM. The absolute value of the x-intercept of the line represents Ki (approximately 20 nM).
Steady-State Activity of CYP2B4 for the Metabolism of BNZ in the Presence and Absence of CYP2E1.
Previous reports have suggested an interplay between redox partner affinity and substrate affinity for P450s (French et al., 1980; Bonfils et al., 1981). Because the binding affinity of BNZ to CYP2B4 may be enhanced by the interaction between CYP2B4 and CPR (and vice versa), we hypothesized that a perturbation in the formation of the CYP2B4-CPR complex by CYP2E1 might lead to changes in the kinetic parameters of CYP2B4 for BNZ. The KM and kcat of CYP2B4 for BNZ were determined initially by varying the concentrations of BNZ in the reaction mixture while keeping the concentrations of CYP2B4 and CPR constant and equal. This was then repeated in the presence of a 4-fold excess in the concentration of CYP2E1 over CYP2B4. The data in Fig. 6 and Table 2 show that the apparent KM increases 8-fold while the kcat remains essentially unchanged. If this increase in the apparent KM for BNZ arises from a competition between CYP2E1 and CYP2B4 for reduction by CPR, then supplying saturating concentrations of CPR should shift the apparent KM back to favor the formation of the catalytically active CYP2B4-CPR complex, as demonstrated by a decrease in the apparent KM of CYP2B4 for BNZ activity at high concentrations of CPR (Fig. 3; Table 2). Therefore, the determination of KM and kcat of CYP2B4 for BNZ in the presence and absence of CYP2E1 was performed under high concentrations of CPR (5-fold over the concentration of CYP2B4). The results show that the presence of high concentrations of CPR decreased the apparent KM of CYP2B4 for BNZ by 20 and 72% in the absence and presence of CYP2E1, respectively, compared with lower concentrations of CPR (Table 2). These data are consistent with the results shown in Fig. 3, which taken together demonstrate that the presence of increased concentrations of CPR reduce the inhibitory effect of CYP2E1 on CYP2B4. Alleviation of this inhibition leads to a significant decrease of the apparent KM of CYP2B4 for BNZ back toward that observed in the absence of CYP2E1.
Fig. 6.
Effect of CYP2E1 and CPR on the Kinetics of BNZ Metabolism by CYP2B4. To investigate the effect of CYP2E1 on BNZ metabolism by CYP2B4, the apparent KM and kcat values for BNZ were determined. The reaction mixtures contained 0.50 μM CYP2B4 as shown. They also contained increasing concentrations of BNZ: 0.50 μM CPR (□), 0.50 μM CPR + 2 μM CYP2E1 (▵), 2.50 μM CPR + 2 μM CYP2E1 (▴), and 2.50 μM CPR (▪). Incubations and determinations of formaldehyde product formed were performed as described under Materials and Methods.
TABLE 2.
The effect of CYP2E1 and CPR on the apparent KM and kcat of CYP2B4 for the metabolism of BNZ
The rate of BNZ N-demethylation by CYP2B4 was determined as described under Materials and Methods. All samples contained 0.50 μM CYP2B4 and increasing concentrations of BNZ. The “control” sample contained equal concentrations of CPR and CYP2B4, “plus CYP2E1” contained 2 μM CYP2E1 in addition to the components in the “control” sample, “plus CYP2E1 and excess CPR” contained 2.5 μM CPR in addition to the components in the “plus CYP2E1” sample and, “plus saturating CPR” contained the same components as the “control” sample except for the addition of 2.5 μM CPR. The kinetic values given here were derived from data plotted in Fig. 6.
| CYP2B4 |
||||
|---|---|---|---|---|
| Control | Plus CYP2E1 | Plus Saturating CPR | Plus CYP2E1 and Saturating CPR | |
| KM (mM) | 0.075 ± 0.010 | 0.60 ± 0.13 | 0.060 ± 0.012 | 0.17 ± 0.051 |
| kcat (min–1) | 32 ± 0.78 | 33 ± 3.7 | 35 ± 1.6 | 30 ± 3.0 |
Effect of CYP2E1 on the Spectral Binding Constant of BNZ to CYP2B4.
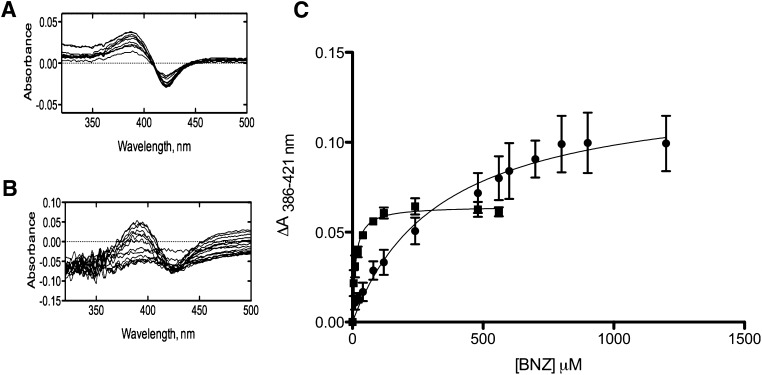
Attempts to completely reduce the apparent KM of BNZ for CYP2B4 in the presence of CYP2E1 to that in the absence of CYP2E1 by incubation with excess CPR proved only partially successful (Fig. 6; Table 2). Therefore, we hypothesized that an alternative mechanism of inhibition may account for this difference. Since the previous studies on the inhibition of CYP2B4 by CYP2E1 were performed in the presence of excess BNZ (1.2 mM), a perturbation in the affinity of CYP2B4 for BNZ by CYP2E1 may have eluded our detection in these studies. BNZ induces a type I spectral shift upon binding to CYP2B4 but not CYP2E1 (data not shown), and this can be exploited to determine the effect of CYP2E1 on the affinity of CYP2B4 for its substrate. The results in Fig. 7 show that BNZ binds to CYP2B4 with a Ks of 11.4 μM and that, in the presence of CYP2E1, this increases by approximately 30-fold to 339 μM. Thus, CYP2E1 not only inhibits complex formation between CYP2B4 and CPR but also affects complex formation between CYP2B4 and BNZ (Fig. 7).
Fig. 7.
Spectral binding titrations of BNZ binding to CYP2B4. Changes in the UV-visible spectrum of 1 μM CYP2B4 induced by the addition of increasing concentrations of BNZ to CYP2B4 were recorded in the absence (A) and presence of 4 μM CYP2E1 (B), as described under Materials and Methods. Absorbance changes as measured by absorbance at 386 nm minus the absorbance at 421 nm were plotted as a function of BNZ concentration (C) to determine the Ks value for BNZ binding in the absence CYP2E1 (▪) and the presence of CYP2E1 (●), as described under Materials and Methods. The values were 11.4 ± 1.10 μM and 339 ± 60.2 μM, respectively.
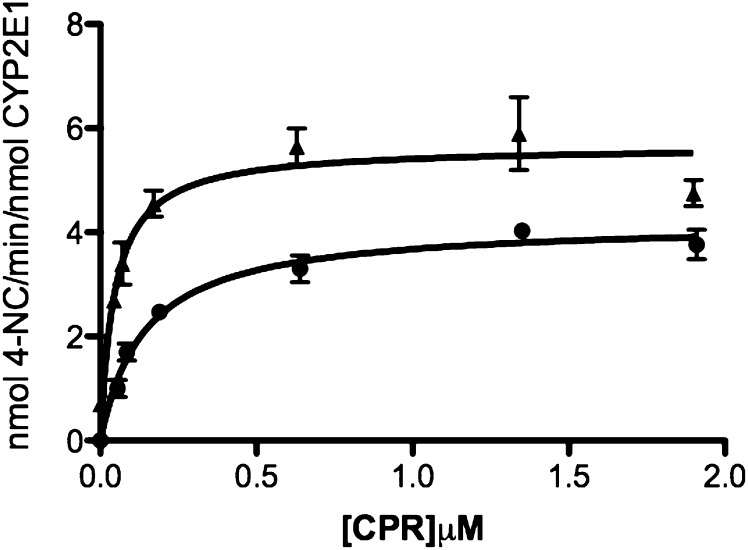
Determination of the Apparent KM and kcat Values for the Interactions of CYP2E1 WT with CPR.
The ability of CYP2E1 to out-compete CYP2B4 for CPR could be attributed to a higher affinity (lower KM) of CYP2E1 for CPR compared with CYP2B4 for CPR. However, our results show that CYP2E1 and CYP2B4 have similar affinities for CPR with apparent KM values of 0.14 and 0.08 μM, respectively (Tables 1 and 3). Interestingly, when the apparent KM of CYP2E1 for CPR was measured in the presence of CYP2B4, we observed a 68% decrease in the apparent KM and an increase in the kcat (Fig. 8; Table 3). The final apparent KM value, 0.04 μM, is close to the Ki of CYP2E1 for the CYP2B4-CPR complex, which is 0.02 μM determined in Fig. 5 and 0.038 μM as determined by global nonlinear fitting. This suggests that the presence of CYP2B4 leads to the formation of a CYP2E1-CPR complex with a higher affinity than the CYP2B4-CPR complex.
TABLE 3.
Apparent KM and kcat values of CYP2E1 WT for CPR as measured by p-NP hydroxylation in the absence and presence of CYP2B4
CYP2E1 (0.1 μM) was reconstituted with varying concentrations of CPR in the presence or absence of 0.1 μM CYP2B4. The reconstituted mixtures were then added to 50 mM potassium phosphate buffer, pH 7.4, containing 0.3 mM p-NP, and 2 mM ascorbic acid. The reactions were initiated by adding 0.4 mM NADPH, and the incubations and determination of 4-NC formation were performed as described under Materials and Methods. The kinetic values given here were derived from data as plotted in Fig. 8.
| CYP2E1 WT |
||
|---|---|---|
| Control | Plus CYP2B4 | |
| KM (μM) | 0.14 ± 0.020 | 0.045 ± 0.01 |
| kcat (min–1) | 4.2 ± 0.015 | 5.65 ± 0.29 |
Fig. 8.
Effect of CYP2B4 on the apparent KM and kcat of CYP2E1 for CPR. The hydroxylation of p-NP to produce 4-NC was measured at a constant concentration of CYP2E1 (0.1 μM) with increasing concentrations of CPR in the absence (●) or presence of 0.1 μM CYP2B4 (▴), as described under Materials and Methods. The plots were corrected to account for the free concentration of CPR at each titration. Error bars are the standard deviations from three measurements performed at least in duplicate.
Discussion
For almost four decades, it has been known that P450s exist in significant excess (∼20-fold) over CPR, yet very little is known about their spatial distribution within the membrane (Estabrook et al., 1971). Even less is known about how the disparity in the molar ratio of P450s to CPR influences the catalytic activities of the various isoforms of the P450s and whether this affects the pharmacokinetic profile of drugs that mainly undergo metabolism by specific P450s. The objective of this study was to assess the effect of varying amounts of CYP2E1, CYP2B4, and CPR on P450 catalytic activity with the ultimate goal of using this information to improve our understanding of the effect on P450-P450 interaction on drug metabolism in humans.
The ability of peroxides and hydroperoxides to bypass the need for the delivery of two electrons from CPR makes tBHP a useful experimental tool to study the catalytic properties of the P450 heme in the absence of CPR. To differentiate direct P450-P450 interactions from P450-CPR interactions that lead to inhibition, we measured CYP2B4 activity supported by CPR and tBHP. The data show that CYP2B4 was resistant to inhibition by CYP2E1 when BNZ metabolism was supported by tBHP but lost up to 80% of its BNZ demethylase activity when supported by NADPH and CPR. This suggests that competition for CPR is implicated in the inhibitory phenomenon observed in Fig. 1A. To further interrogate the contribution of the CYP2E1-CPR interaction to the inhibition of CYP2B4, we investigated whether a CYP2E1 variant (Y422D), which has a mutation in the proximal side that reduces its binding to CPR by approximately 50%, could alter the extent of inhibition of CYP2B4 activity by CYP2E1. Our results show that the extent of CYP2B4 inhibition by CYP2E1 Y422D is significantly less than that observed with the WT CYP2E1, suggesting a role for the CYP2E1-CPR interaction in the inhibition of CYP2B4 activity supported by CPR.
The notion that CYP2E1 perturbs the formation of the CYP2B4-CPR complex is further supported by the data on the effect of CYP2E1 on the apparent KM of CYP2B4 for CPR, which showed that low concentrations of CYP2E1 raised the apparent KM for CPR by 23-fold with a negligible effect on kcat compared with samples that did not contain CYP2E1. Higher CYP2E1 concentrations decreased the apparent KM of CYP2B4 for CPR but reduced the kcat by 11-fold. Inverse plots of velocities as a function of CPR concentration revealed that CYP2E1 behaved both as a competitive (at low concentrations) and noncompetitive (at higher concentrations) inhibitor of CYP2B4-CPR. To estimate a kinetic constant for the observed competitive inhibition component of CYP2E1, we plotted KM obs as a function of the CYP2E1 concentration that exhibited competitive inhibition kinetics. Linear regression analysis of these data produced a straight line, which intersected the x-axis at –0.02 μM, therefore indicating that inhibition of CYP2B4 by CYP2E1 is very potent. Global nonlinear analysis of the same data by GraphPad Prism revealed a Ki of 0.038 μM.
It is interesting to consider these results in light of the individual apparent KM values of these two P450s for CPR. If two P450s compete for the same functional binding site in CPR, the specificity, in the sense of discrimination between two P450s competing for CPR, should be determined by the respective apparent KM values for CPR. However, the measured apparent KM values of CYP2B4 and CYP2E1 for CPR are similar: 0.08 and 0.14 μM, respectively (Tables 1 and 3). Therefore, one would expect that both P450s could compete with each other for CPR with almost equal inhibitory potency, yet the data show that CYP2E1 can compete with CYP2B4 for CPR but CYP2B4 does not compete with CYP2E1 for reduction by CPR. This discrepancy led us to hypothesize that an alternative mechanism for the inhibition of CYP2B4-CPR complex formation by CYP2E1 likely confers a higher binding affinity of CYP2E1 for CPR.
Backes and co-workers have previously suggested that interactions between P450s and CPR may lead to changes in the affinity of P450s for CPR (Backes and Kelley, 2003; Reed and Backes, 2012). Determination of the apparent KM and kcat of CYP2E1 for CPR in the presence and absence of CYP2B4 revealed that the apparent KM decreased by 68% to 0.045 μM while the kcat increased by 1.3-fold to 5.7 min−1 in the presence of CYP2B4. It is noteworthy that the value of this enhanced apparent KM is similar to the Ki of CYP2E1 for the CYP2B4-CPR complex, thus supporting the notion that a decrease in the apparent KM of CYP2E1 for CPR, due to CYP2B4, contributes to CYP2E1’s inhibitory properties.
To propose a tentative model to explain our kinetic data, we took into account the following findings: 1) direct CYP2B4-CYP2E1 interactions alone do not lead to inhibition of CYP2B4 activity in the presence of saturating concentrations of BNZ (Fig. 1A), 2) CYP2B4 and CYP2E1 interact directly to reduce the affinity of CYP2B4 for BNZ (Fig. 7), and 3) CYP2E1 has a higher affinity for CPR in the presence of CYP2B4 (Fig. 8). In light of these findings, one possible mechanism that may explain the inhibitory behavior of CYP2E1 toward CYP2B4 is depicted in Fig. 9. In this model, CYP2E1 and CYP2B4 interact to form a CYP2E1-CYP2B4 complex. This complex may then interact with CPR in such a way that CYP2E1, in the CYP2E1-CYP2B4 complex, interacts with the functional site of CPR with a higher affinity than CYP2E1 alone. A second, although unlikely scenario, this model raises is the possibility that CYP2E1 lowers CYP2B4’s affinity for CPR by binding directly to CPR’s docking site on CYP2B4. However, our studies with the CYP2E1 Y422D mutant point us to consider the alternate possibility for the following reason. If the interaction of CYP2E1 with CYP2B4, and not with CPR, was responsible for the observed inhibition, one should not see any change in the extent of CYP2B4 inhibition by the Y422D variant. Thus, although our data show that CYP2E1 interacts directly with CYP2B4 to perturb the affinity of CYP2B4 for BNZ, this interaction does not explain the modulation of CYP2B4’s affinity for CPR by CYP2E1.
Fig. 9.
A tentative model for the CYP2B4-CYP2E1 interaction. Inhibition of CYP2B4 activity may arise from the formation of a CYP2E1-CYP2B4 complex. CPR is believed to be highly flexible (Hamdane et al., 2009; Xia et al., 2011) in solution and a CYP2E1-CYP2B4 complex could trap CPR in a particular conformation that makes it more favorable for CYP2E1, instead of CYP2B4, to bind at a functional site on CPR with a higher affinity than CYP2E1 or CYP2B4 alone.
Despite earlier studies to investigate interactions between P450s, the inhibitory nature of CYP2E1 toward CYP2B4-mediated metabolism of BNZ and the enhancement of CYP2E1’s apparent KM for CPR by CYP2B4 (using p-NP as a probe substrate for CYP2E1) have not previously been reported. For example, studies by (Kelley et al., 2006) demonstrated that interactions between CYP1A2 and CYP2E1 lead to significantly enhanced rates of 7-ethoxyresorufin (7-ER) and 7-pentoxyresorufin (7-PR). However, when interactions between CYP2E1 and CYP2B4 were investigated using 7-ER, 7-PR, and aniline as substrates, functional interactions between these two P450s were not observed, suggesting that they did not form P450-P450 complexes. Alternatively, it is possible that complex formation had no effect on any of the activities measured. Therefore, the differences between our results and those of Kelley et al. (2006) may be attributed to differences in the substrates used, as previous work has shown that interactions between P450s are highly dependent on the substrates under investigation (Cawley et al., 1995). Since Cawley et al. (1995) used full-length CYP2B4 and CYP2E1, it is also possible that N-terminal truncation of CYP2B4 and CYP2E1 used in the present study may account for the observed differences.
A significant amount of effort is expended by pharmaceutical and pharmacokinetic modeling companies to predict in vivo pharmacokinetic profiles of drugs from in vitro data (Rostami-Hodjegan and Tucker, 2007). Since protein-protein interactions in the P450 system may confound in vitro to in vivo drug metabolism extrapolations, it is important that in-depth kinetic studies be conducted to determine the kinetic basis and associated kinetic constants that govern these interactions. The information obtained from these types of studies is invaluable as it can play an important role in improving our ability to predict in vivo drug clearance and drug-drug interactions from in vitro data.
To assess the biological relevance of the findings reported in the present study, it would be of interest to investigate the possibility of in vivo interactions between CYP2E1 and CYP2B4 in rabbit. For example, if either concomitant or chronic administration of ethanol (a CYP2E1 inducer and substrate) alters the metabolism of benzphetamine by CYP2B4, this would suggest that these interactions may confound traditional models used to explain metabolism of compounds and support biological relevance for CYP2E1-CYP2B4 interactions.
In conclusion, we have shown that the presence of CYP2E1 significantly reduces the N-demethylase activity of CYP2B4 for BNZ. This is not mediated through a simple competition for CPR since CYP2B4 does not decrease the catalytic activity of CYP2E1. It can be concluded from our studies that CYP2B4 and CYP2E1 interact to lower CYP2E1’s apparent KM for CPR, which may allow CYP2E1 to out-compete CYP2B4 for CPR. These findings warrant further physical and kinetic investigation to determine the precise structural basis for these interactions, and the impact these types of interactions may have on drug-drug interactions.
Acknowledgments
We thank Drs. David Ballou and Jorge Iniguez-Lluhi from the Departments of Biological Chemistry and Pharmacology, respectively, at the University of Michigan (Ann Arbor, MI) for insightful discussions during the preparation of this manuscript.
Abbreviations
- 7-ER
7-Ethoxyresorufin
- 7-PR
7-pentoxyresorufin
- 4-NC
4-nitrocatechol
- BNZ
benzphetamine
- CO
carbon monoxide
- CPR
cytochrome P450 reductase
- CYP or P450
cytochrome P450
- DLPC
dilauroylphosphatidylcholine
- ER
endoplasmic reticulum
- FMN
flavin mononucleotide
- p-NP
para-nitrophenol
- tBHP
tert-butyl hydroperoxide
- TCA
trichloroacetic acid
- TFA
trifluoroacetic acid
- WT
wild-type
- Y422D
Tyr 422 to Asp mutation in CYP2E1
Authorship Contributions
Participated in research design: Kenaan, Zhang, Hollenberg.
Conducted experiments: Kenaan, Shea, Lin.
Contributed new reagents or analytic tools: Kenaan, Lin, Pratt-Hyatt.
Performed data analysis: Kenaan, Hollenberg.
Wrote or contributed to the writing of the manuscript: Kenaan, Hollenberg, Lin.
Footnotes
This work was supported, in part, by National Institutes of Health National Cancer Institute [Grant CA16954] and an ASPET Institutional Summer Undergraduate Research Fellowship.
References
- Backes WL, Batie CJ, Cawley GF. (1998) Interactions among P450 enzymes when combined in reconstituted systems: formation of a 2B4-1A2 complex with a high affinity for NADPH-cytochrome P450 reductase. Biochemistry 37:12852–12859 [DOI] [PubMed] [Google Scholar]
- Backes WL, Kelley RW. (2003) Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol Ther 98:221–233 [DOI] [PubMed] [Google Scholar]
- Bonfils C, Balny C, Maurel P. (1981) Direct evidence for electron transfer from ferrous cytochrome b5 to the oxyferrous intermediate of liver microsomal cytochrome P-450 LM2. J Biol Chem 256:9457–9465 [PubMed] [Google Scholar]
- Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. (1998) Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem 273:17036–17049 [DOI] [PubMed] [Google Scholar]
- Brignac-Huber L, Reed JR, Backes WL. (2011) Organization of NADPH-cytochrome P450 reductase and CYP1A2 in the endoplasmic reticulum—microdomain localization affects monooxygenase function. Mol Pharmacol 79:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley GF, Batie CJ, Backes WL. (1995) Substrate-dependent competition of different P450 isozymes for limiting NADPH-cytochrome P450 reductase. Biochemistry 34:1244–1247 [DOI] [PubMed] [Google Scholar]
- Cawley GF, Zhang S, Kelley RW, Backes WL. (2001) Evidence supporting the interaction of CYP2B4 and CYP1A2 in microsomal preparations. Drug Metab Dispos 29:1529–1534 [PubMed] [Google Scholar]
- Davydov DR, Petushkova NA, Bobrovnikova EV, Knyushko TV, Dansette P. (2001) Association of cytochromes P450 1A2 and 2B4: are the interactions between different P450 species involved in the control of the monooxygenase activity and coupling? Adv Exp Med Biol 500:335–338 [DOI] [PubMed] [Google Scholar]
- Dey A, Cederbaum AI. (2006) Alcohol and oxidative liver injury. Hepatology 43(2, Suppl 1)S63–S74 [DOI] [PubMed] [Google Scholar]
- Estabrook RW, Franklin MR, Cohen B, Shigamatzu A, Hildebrandt AG. (1971) Biochemical and genetic factors influencing drug metabolism. Influence of hepatic microsomal mixed function oxidation reactions on cellular metabolic control. Metabolism 20:187–199 [DOI] [PubMed] [Google Scholar]
- Eyer CS, Backes WL. (1992) Relationship between the rate of reductase-cytochrome P450 complex formation and the rate of first electron transfer. Arch Biochem Biophys 293:231–240 [DOI] [PubMed] [Google Scholar]
- French JS, Guengerich FP, Coon MJ. (1980) Interactions of cytochrome P-450, NADPH-cytochrome P-450 reductase, phospholipid, and substrate in the reconstituted liver microsomal enzyme system. J Biol Chem 255:4112–4119 [PubMed] [Google Scholar]
- Guengerich FP, Kim DH, Iwasaki M. (1991) Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem Res Toxicol 4:168–179 [DOI] [PubMed] [Google Scholar]
- Hamdane D, Xia C, Im SC, Zhang H, Kim JJ, Waskell L. (2009) Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J Biol Chem 284:11374–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazai E, Kupfer D. (2005) Interactions between CYP2C9 and CYP2C19 in reconstituted binary systems influence their catalytic activity: possible rationale for the inability of CYP2C19 to catalyze methoxychlor demethylation in human liver microsomes. Drug Metab Dispos 33:157–164 [DOI] [PubMed] [Google Scholar]
- Hu Y, Krausz K, Gelboin HV, Kupfer D. (2004) CYP2C subfamily, primarily CYP2C9, catalyses the enantioselective demethylation of the endocrine disruptor pesticide methoxychlor in human liver microsomes: use of inhibitory monoclonal antibodies in P450 identification. Xenobiotica 34:117–132 [DOI] [PubMed] [Google Scholar]
- Johansson I, Ekström G, Scholte B, Puzycki D, Jörnvall H, Ingelman-Sundberg M. (1988) Ethanol-, fasting-, and acetone-inducible cytochromes P-450 in rat liver: regulation and characteristics of enzymes belonging to the IIB and IIE gene subfamilies. Biochemistry 27:1925–1934 [DOI] [PubMed] [Google Scholar]
- Kakkar T, Boxenbaum H, Mayersohn M. (1999) Estimation of Ki in a competitive enzyme-inhibition model: comparisons among three methods of data analysis. Drug Metab Dispos 27:756–762 [PubMed] [Google Scholar]
- Kelley RW, Cheng D, Backes WL. (2006) Heteromeric complex formation between CYP2E1 and CYP1A2: evidence for the involvement of electrostatic interactions. Biochemistry 45:15807–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenaan C, Zhang H, Shea EV, Hollenberg PF. (2011) Uncovering the role of hydrophobic residues in cytochrome P450-cytochrome P450 reductase interactions. Biochemistry 50:3957–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent UM, Pascual L, Roof RA, Ballou DP, Hollenberg PF. (2004) Mechanistic studies with N-benzyl-1-aminobenzotriazole-inactivated CYP2B1: differential effects on the metabolism of 7-ethoxy-4-(trifluoromethyl)coumarin, testosterone, and benzphetamine. Arch Biochem Biophys 423:277–287 [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Fischer J, Nüssler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, Zanger UM. (2001) Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11:399–415 [DOI] [PubMed] [Google Scholar]
- Levin W, Thomas PE, Oldfield N, Ryan DE. (1986) N-demethylation of N-nitrosodimethylamine catalyzed by purified rat hepatic microsomal cytochrome P-450: isozyme specificity and role of cytochrome b5. Arch Biochem Biophys 248:158–165 [DOI] [PubMed] [Google Scholar]
- Lin HL, Myshkin E, Waskell L, Hollenberg PF. (2007) Peroxynitrite inactivation of human cytochrome P450s 2B6 and 2E1: heme modification and site-specific nitrotyrosine formation. Chem Res Toxicol 20:1612–1622 [DOI] [PubMed] [Google Scholar]
- Miwa GT, West SB, Huang MT, Lu AY. (1979) Studies on the association of cytochrome P-450 and NADPH-cytochrome c reductase during catalysis in a reconstituted hydroxylating system. J Biol Chem 254:5695–5700 [PubMed] [Google Scholar]
- Nash T. (1953) The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem J 55:416–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oezguen N, Kumar S, Hindupur A, Braun W, Muralidhara BK, Halpert JR. (2008) Identification and analysis of conserved sequence motifs in cytochrome P450 family 2. Functional and structural role of a motif 187RFDYKD192 in CYP2B enzymes. J Biol Chem 283:21808–21816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem 239:2379–2385 [PubMed] [Google Scholar]
- Oneta CM, Lieber CS, Li J, Rüttimann S, Schmid B, Lattmann J, Rosman AS, Seitz HK. (2002) Dynamics of cytochrome P4502E1 activity in man: induction by ethanol and disappearance during withdrawal phase. J Hepatol 36:47–52 [DOI] [PubMed] [Google Scholar]
- Patten CJ, Thomas PE, Guy RL, Lee M, Gonzalez FJ, Guengerich FP, Yang CS. (1993) Cytochrome P450 enzymes involved in acetaminophen activation by rat and human liver microsomes and their kinetics. Chem Res Toxicol 6:511–518 [DOI] [PubMed] [Google Scholar]
- Peterson JA, Ebel RE, O’Keeffe DH, Matsubara T, Estabrook RW. (1976) Temperature dependence of cytochrome P-450 reduction. A model for NADPH-cytochrome P-450 reductase:cytochrome P-450 interaction. J Biol Chem 251:4010–4016 [PubMed] [Google Scholar]
- Pratt-Hyatt M, Lin HL, Hollenberg PF. (2010) Mechanism-based inactivation of human CYP2E1 by diethyldithocarbamate. Drug Metab Dispos 38:2286–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Backes WL. (2012) Formation of P450 · P450 complexes and their effect on P450 function. Pharmacol Ther 133:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Eyer M, Backes WL. (2010) Functional interactions between cytochromes P450 1A2 and 2B4 require both enzymes to reside in the same phospholipid vesicle: evidence for physical complex formation. J Biol Chem 285:8942–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami-Hodjegan A, Tucker GT. (2007) Simulation and prediction of in vivo drug metabolism in human populations from in vitro data. Nat Rev Drug Discov 6:140–148 [DOI] [PubMed] [Google Scholar]
- Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. (2003) An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci USA 100:13196–13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EE, Spatzenegger M, Halpert JR. (2001) A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys 395:57–68 [DOI] [PubMed] [Google Scholar]
- Subramanian M, Low M, Locuson CW, Tracy TS. (2009) CYP2D6-CYP2C9 protein-protein interactions and isoform-selective effects on substrate binding and catalysis. Drug Metab Dispos 37:1682–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Tam H, Zheng H, Tracy TS. (2010) CYP2C9-CYP3A4 protein-protein interactions: role of the hydrophobic N terminus. Drug Metab Dispos 38:1003–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermilion JL, Coon MJ. (1978) Purified liver microsomal NADPH-cytochrome P-450 reductase. Spectral characterization of oxidation-reduction states. J Biol Chem 253:2694–2704 [PubMed] [Google Scholar]
- Walsky RL, Astuccio AV, Obach RS. (2006) Evaluation of 227 drugs for in vitro inhibition of cytochrome P450 2B6. J Clin Pharmacol 46:1426–1438 [DOI] [PubMed] [Google Scholar]
- Wang H, Tompkins LM. (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9:598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Hamdane D, Shen AL, Choi V, Kasper CB, Pearl NM, Zhang H, Im SC, Waskell L, Kim JJ. (2011) Conformational changes of NADPH-cytochrome P450 oxidoreductase are essential for catalysis and cofactor binding. J Biol Chem 286:16246–16260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Im SC, Waskell L. (2007) Cytochrome b5 increases the rate of product formation by cytochrome P450 2B4 and competes with cytochrome P450 reductase for a binding site on cytochrome P450 2B4. J Biol Chem 282:29766–29776 [DOI] [PubMed] [Google Scholar]
- Zhang H, Kenaan C, Hamdane D, Hoa GH, Hollenberg PF. (2009) Effect of conformational dynamics on substrate recognition and specificity as probed by the introduction of a de novo disulfide bond into cytochrome P450 2B1. J Biol Chem 284:25678–25686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sridar C, Kenaan C, Amunugama H, Ballou DP, Hollenberg PF. (2011) Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: a charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome p450-reductase complex. J Pharmacol Exp Ther 338:803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]