Abstract
UDP-glucuronosyltransferase A1 (UGT2A1) is expressed in the lung and exhibits activity against polycyclic aromatic hydrocarbons (PAHs), suggesting UGT2A1 involvement in the local metabolism of PAH tobacco carcinogens. The goal of the present study was to investigate the importance of two additional UGT2A enzymes, UGT2A2 and UGT2A3, in tobacco carcinogen metabolism. Real-time polymerase chain reaction suggested that wild-type UGT2A2 had the highest expression in the breast, followed by trachea > larynx > kidney. A novel splice variant of UGT2A2 lacking exon 3 (termed UGT2A2Δexon3) was investigated, with UGT2A2Δexon3 expression determined to be 25–50% that of wild-type UGT2A2 in all tissues examined. UGT2A3 was determined to be well expressed in the liver and colon, followed by pancreas > kidney > lung > tonsil > trachea > larynx. Cell homogenates prepared from human embryonic kidney (HEK)293 cells overexpressing wild-type UGT2A2 (termed UGT2A2_i1) exhibited glucuronidation activity, as observed by reverse-phase ultra-pressure liquid chromatography, against 1-hydroxy-(OH)-pyrene, 1-naphthol, and hydroxylated benzo(a)pyrene metabolites, whereas homogenates prepared from HEK293 cells overexpressing UGT2A3 only showed activity against simple PAHs like 1-OH-pyrene and 1-naphthol. Activity assays showed the UGT2A2Δexon3 protein (termed UGT2A2_i2) exhibited no detectable glucuronidation activity against all substrates examined; however, coexpression studies suggested that UGT2A2_i2 negatively modulates UGT2A2_i1 activity. Both UGT2A2 and UGT2A3 exhibited no detectable activity against complex PAH proximate carcinogens, tobacco-specific nitrosamines, or heterocyclic amines. These data suggest that, although UGT2A1 is the only UGT2A enzyme active against PAH proximate carcinogens (including PAH diols), both UGTs 2A1 and 2A2 play an important role in the local detoxification of procarcinogenic monohydroxylated PAH metabolites.
Introduction
The UDP-glucuronosyltransferase (UGT) family of enzymes catalyzes the glucuronidation of a variety of endogenous compounds such as steroid hormones and bilirubin, as well as exogenous xenobiotics such as drugs and environmental carcinogens (Tephly and Burchell, 1990; Gueraud and Paris, 1998; Ren et al., 2000). The three members of the UGT2A family (UGTs 2A1, 2A2, and 2A3), similar to the UGT2B family members, are located on chromosome 4 and contain six exons. Similar to members of the UGT1A family, UGTs 2A1 and 2A2 are encoded by unique first exons joined to common exons 2–6 (Mackenzie et al., 2005; Sneitz et al., 2009). In contrast to the gene organization of UGT2A1 and UGT2A2, UGT2A3 has six unique exons and does not share common exons with any other UGT isoform (Mackenzie et al., 2005; Court et al., 2008).
Although the human UGT1A and UGT2B subfamilies have been characterized extensively, only a limited number of studies have analyzed UGT2A tissue expression and activity. UGT2A1 was initially cloned from olfactory RNA and was shown to exhibit glucuronidation activity against phenolic odorants, suggesting UGT2A1 involvement in the initiation and termination of olfactory stimuli (Jedlitschky et al., 1999). More recent studies have demonstrated UGT2A1 expression in a variety of aerodigestive, digestive, and respiratory tract tissues, including lung, trachea, tonsil, larynx, and colon tissues (Bushey et al., 2011). Using homogenates from UGT2A1-overexpressed human embryonic kidney (HEK)293 cells, UGT2A1 exhibited glucuronidation activity against a panel of polycyclic aromatic hydrocarbon (PAH) metabolites, including the proximate carcinogens 5-methylchrysense-1,2-diol, benzo(a)pyrene [B(a)P]-7,8-diol, and dibenzo(a,l)pyrene-11,12-diol (Bushey et al., 2011). These data suggest that UGT2A1 potentially plays an important role in the local metabolism of PAHs in target tissues for tobacco carcinogenesis. In addition, a well expressed UGT2A1 exon 3 deletion splice variant was recently identified; this novel splice variant was determined to have no detectable glucuronidation activity but negatively regulated wild-type UGT2A1 activity, likely through a protein-protein interaction (Bushey and Lazarus, 2012).
In a previous study, UGT2A2 mRNA expression was reported in fetal and adult nasal mucosa tissues, whereas no UGT2A2 expression was observed in the liver, fetal liver, lung, or fetal lung tissues (Sneitz et al., 2009). In the same study, UGT2A2 was histidine-tagged and overexpressed in a baculovirus system to investigate enzyme activity, with UGT2A2 exhibiting glucuronidation activity against a range of phenolic and estradiol substrates (Sneitz et al., 2009). In an additional study, UGT2A3 expression was shown to be highest in the small intestine, followed by the liver, colon, adipose, and pancreas (Court et al., 2008). UGT2A3, overexpressed in a baculovirus system, was shown to metabolize bile acids, including hyodeoxycholic acid, deoxycholic acid, ursodeoxycholic acid, and chenodeoxycholic acid (Court et al., 2008).
Functional protein domains have been elucidated for the UGTs, with the uridine 5′-diphosphoglucuronic acid (UDPGA) binding site conserved between UGT isoforms and found at the C terminus of the protein, whereas the substrate-binding domain is more variable and found at the N terminus of the protein (Burchell and Coughtrie, 1989; Meech and Mackenzie, 1998; Nagar and Remmel, 2006). With unique first exons, and differing amino acid sequences in the substrate recognition site of the enzyme, UGT2A1 and UGT2A2 would be hypothesized to exhibit divergent substrate specificities. However, UGT2A1 and UGT2A2 were reported to have overlapping substrate specificities, as both enzymes exhibited detectable O-glucuronidation activity against similar estradiol and phenolic substrates (Sneitz et al., 2009). By use of an alignment algorithm (Pearson and Lipman, 1988), the exon 1 nucleotide sequences of UGT2A1 and UGT2A2 exhibited 66% similarity, with an 84% similarity between the corresponding amino acids. Although no exon sharing occurs between UGT2A3 and other UGT2A enzymes, UGTs 2A1 and 2A3 also exhibit a high degree of sequence similarity in their first exons, exhibiting 56 and 70% similarity in nucleotide and amino acid sequences, respectively. The high degree of sequence similarity between UGT2A enzymes in the substrate recognition region of the enzymes supports the possibility of an overlap in substrate specificity.
On the basis of the observed UGT2A1 glucuronidation activity against PAHs and the sequence similarities between UGT2A enzymes, it was hypothesized that UGTs 2A2 and 2A3 exhibit glucuronidation activity against PAHs. In the present study, the expression of UGTs 2A2 and 2A3 was examined in a variety of human aerodigestive digestive and respiratory tract tissues, and their potential role in the metabolism of a variety of tobacco carcinogens and their metabolites was characterized. The role of a newly identified UGT2A2 splice variant isoform in UGT2A2 glucuronidation activity was also examined.
Materials and Methods
Chemicals and Materials.
Pfu High Fidelity DNA Polymerase was purchased from Agilent Technologies (Santa Clara, CA). All oligonucleotide polymerase chain reaction (PCR) primers were purchased from (Integrated Device Technology; Coralville, IA). Probes for real-time PCR experiments and real-time PCR gene expression assays were acquired from Applied Biosystems Inc. (ABI), Life Technologies (Carlsbad, CA). The pcDNA3.1/V5-His-TOPO mammalian expression vector kit, Superscript II RT kit, Lipofectamine 2000, Dulbecco’s phosphate-buffered saline, fetal bovine serum, penicillin-streptomycin, Geneticin (G418), and ampicillin were acquired from Invitrogen (Carlsbad, CA). The BCA protein assay kit was purchased from Pierce (Rockford, IL). The RNeasy kit, QIAquick gel extraction kit, Plasmid Mini kit, and Plasmid Maxi kit were all obtained from Qiagen (Valencia, CA). UDPGA, uridine 5′-diphospho (UDP)-glucose, UDP-galactose, UDP-N-acetylglucosamine, alamethicin, β-glucuronidase, dimethyl sulfoxide, 4-methylumbelliferone, 1-OH-pyrene, and 1-naphthol were all acquired from Sigma-Aldrich (St. Louis, MO). 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanol, N-nitrosonornicotine, N-nitrosoanabasine, N-nitrosoanatabine, nicotine, and 2-amino-1-methyl-6-phenylimidazo [4,5-b]-pyridine (PhIP) were purchased from Toronto Research Chemicals (Toronto, ON, Canada). 1-OH-B(a)P, 3-OH-B(a)P, 7-OH-B(a)P, 8-OH-B(a)P, 5-methylchrysene-1,2-diol, dibenzo(a,l)pyrene-11,12-diol, B(a)P-7,8-diol, and N-OH-PhIP were synthesized in the Organic Synthesis Core Facility at the Penn State College of Medicine (Hershey, PA). High-pressure liquid chromatography–grade ammonium acetate, acetonitrile, and agarose were purchased from Thermo Fisher Scientific (Pittsburgh, PA).
Qualitative Determination of UGT2A2 and UGT2A3 Tissue Expression.
Reverse-transcription (RT)-PCR was completed to determine tissue-specific UGT2A2 and UGT2A3 expression. Pooled RNAs from the lung, larynx, trachea, breast, whole brain, prostate, cerebral cortex, kidney, and pancreas were obtained from either Clontech (Mountain View, CA) or Agilent Technologies. Adjacent normal liver, colon, floor of the mouth, tonsil, and esophagus tissues were obtained from the Penn State College of Medicine Tissue Bank (Hershey, PA), with RNA extracted from each tissue using an RNeasy kit. All qualitative RT-PCR experiments were performed using pooled RNA from at least three individuals for each tissue examined. RT reactions were completed using a Superscript II RT kit (Invitrogen, Carlsbad, CA), with cDNA corresponding to 100 ng of RNA used for each subsequent PCR reaction.
Full-length UGT2A2 was PCR amplified using Pfu polymerase and sense (5′-CCATAAGGGATTTTACCATGCCTAAG-3′; denoted as UGT2A2_S1) and antisense (5′-TTCTCTTTTTTTCTTCTTTCCTATCTTACC-3′) primers corresponding to nucleotides −17 to +9 and nucleotides +1584 to +1555, respectively, relative to the UGT2A2 translation start site. The nucleotide locations of all UGT2A2 primer sequences in this study are designated assuming that the UGT2A2 protein is eight amino acids shorter at the N terminus than the sequence currently described in GenBank (NM_001105677.2). This discrepancy was described previously by Sneitz et al. (2009) and likely has no consequence on the mature UGT2A2 protein sequence, due to the post-translational cleavage of the signal sequence at the N terminus. PCR parameters used to amplify UGT2A2 were as follows: an initial denaturing temperature of 94°C for 2 minutes, then 40 cycles of 94°C for 30 seconds, 58°C for 40 seconds, and 72°C for 2 minutes, followed by a final step of 72°C for 10 minutes. All PCR reactions were completed in a Bio-Rad Mycycler (Hercules, CA). RNAs from HEK293 cell lines overexpressing wild-type UGT2A2 and UGT2A2 lacking exon 3 (UGT2A2Δexon3) (see the Generation of HEK293 Cell Lines Overexpressing UGT2A Isoforms section described in the Materials and Methods below) were used as positive controls, whereas water was used as a negative control.
Tissues were screened for UGT2A3 expression using Pfu polymerase and exon 1-specific sense (5′-CATGAGGTCTGACAAGTCAGCTT-3′) and antisense (5′-CCTGTAGCTTCTTCATAAGCGTCTG-3′) primers corresponding to nucleotides −1 to +22 and nucleotides +418 to +394, respectively, relative to the UGT2A3 translation start site. PCR parameters used to amplify UGT2A3 were as performed for UGT2A2 using an annealing temperature of 58°C. RNA from a HEK293 cell line overexpressing UGT2A3 (see the Generation of HEK293 Cell Lines Overexpressing UGT2A Isoforms section described in the Materials and Methods below) was used as a positive control, whereas water was used as a negative control. UGT2A2 and UGT2A3 PCR products were gel purified using a QIAquick gel extraction kit and sequenced by dideoxy sequencing at the Penn State University Nucleic Acid Facility (State College, PA), with sequences compared with that described for UGT2A2 (NM_001105677.2) and UGT2A3 (NM_024743.3) in GenBank. To verify UGT2A2 or UGT2A3 expression in tissues following RT-PCR, reactions were run multiple times with positive and negative controls.
Quantitative Determination of UGT2A2 and UGT2A3 Tissue Expression.
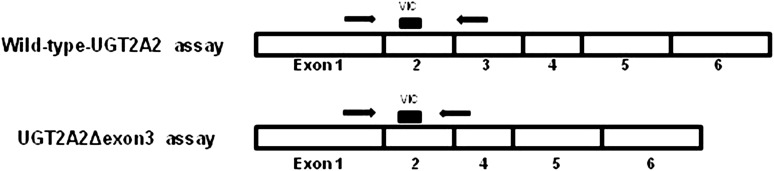
Real-time PCR experiments were completed to quantitatively determine relative UGT2A2 and UGT2A3 mRNA expression levels in tissues that were determined to express UGT2A2 or UGT2A3 by RT-PCR. Separate real-time PCR assays were designed to specifically and quantitatively detect wild-type UGT2A2 or UGT2A2Δexon3 transcripts (Fig. 1). A sense primer specific to UGT2A2 exon 1 (5′-GGAGAATGGAATTCATACTATAGCAAA-3′) and a 5′-labeled VIC probe (ABI) specific to UGT2A2 exon 2 (5′-TCCGAACATATTGGGATT-3′), corresponding to nucleotides +685 to +711 and +770 to +787, respectively, relative to the UGT2A2 translation start site, were used to detect both wild-type UGT2A2 and UGT2A2Δexon3 transcript expression. An antisense primer specific to UGT2A2 exon 3 (5′-TTACCTGAGCTCTGGATAAATTCTTC-3′), corresponding to nucleotides +899 to +874 relative to the UGT2A2 translation start site, was used to specifically detect wild-type UGT2A2. An antisense primer specific to the UGT2A2Δexon3 exons 2 and 4 junction (5′-TTTCCTTTGTATCTCCATAAAACCTTAG-3′), corresponding to nucleotides +890 to +863 relative to the UGT2A2Δexon3 start site, was used to specifically detect UGT2AΔexon3. cDNA corresponding to 20 ng of pooled RNA was used for each reaction. Assay specificity for wild-type UGT2A2 or UGT2A2Δexon3 was confirmed through agarose gel electrophoresis and dideoxy sequencing of real-time PCR products following amplification. The efficiency of each UGT2A2 real-time PCR assay was determined, and relative levels of wild-type UGT2A2 or UGT2A2Δexon3 were corrected for differences in assay efficiency, as described previously (Jones et al., 2012). Real-time PCR experiments to detect UGT2A3 tissue expression were completed using a UGT2A3-specific TaqMan gene expression assay (Hs00226904_m1; ABI, Carlsbad, CA) with cDNA corresponding to 20 ng of pooled RNA used for each UGT2A3 real-time PCR reaction.
Fig. 1.
Schematic of real-time PCR assays used to specifically detect wild-type UGT2A2 or UGT2A2Δexon3 transcripts. An identical forward primer and VIC-labeled probe (ABI) were used to detect both transcripts, and different antisense primers were designed to specifically amplify each UGT2A2 transcript. The antisense primer used to detect wild-type UGT2A2 was specific to exon 3, whereas the antisense primer used to detect UGT2A2Δexon3 was specific to the exon 2–4 junction. Assay specificity was validated by running the real-time PCR products on a 1% agarose gel and dideoxy sequencing.
All real-time PCR experiments were completed in triplicate at the Penn State College of Medicine Functional Genomics Core Facility (Hershey, PA) using an ABI 7900HT Thermal Cycler, and real-time PCR data were analyzed using ABI Sequence Detection System 2.2 software. For all real-time PCR experiments, standard thermal cycling parameters were followed, and human large ribosomal protein (RPLPO) was used as a housekeeping gene (Hs99999902_m1). The RPLPO gene has been shown to have relatively low interindividual expression variability in lung and aerodigestive tract tissues (N. R. Jones and P. Lazarus, unpublished data), and multiple other studies analyzing mRNA expression in human tissues have also used RPLPO as an endogenous control (Maass et al., 2001; Bieche et al., 2007). Relative wild-type UGT2A2 or UGT2A3 expression levels were calculated using the ΔΔCt method, relative to the tissue that had the highest expression of wild-type UGT2A2 (breast) or UGT2A3 (liver) as described previously (Bushey et al., 2011). The relative expression of UGT2A2Δexon3 transcript was calculated using the same method relative to the amount of wild-type UGT2A2 transcript in each tissue, as wild-type UGT2A2 expression was found to be greater than UGT2A2Δexon3 expression in all of the tissues analyzed. The relative expression levels of wild-type UGT2A2 and UGT2A3 in the larynx, trachea, and kidney were also compared using the ΔΔCt method relative to wild-type UGT2A2 expression, following a correction made to account for differences in assay efficiency as described previously (Jones et al., 2012).
Generation of HEK293 Cell Lines Overexpressing UGT2A Isoforms.
Stable HEK293 cell lines overexpressing wild-type UGT2A2, UGT2A2Δexon3, and UGT2A3 were created similar to that described previously (Bushey et al., 2011). In brief, cell lines overexpressing wild-type UGT2A2 and UGT2A2Δexon3 were generated by RT-PCR using pooled trachea RNA. Following RT, the entire UGT2A2-coding region was amplified using the UGT2A2_S1 primer (described previously) and an anti-sense (5′-TGACAGGAAGAGGGTATAGTCAGC-3′) primer corresponding to nucleotides +1837 to +1814 relative to the UGT2A2 translation start site. Both wild-type UGT2A2 and UGT2A2Δexon3 PCR products were gel purified from the same PCR. Full-length UGT2A3 was amplified following RT-PCR from pooled pancreas RNA. Following RT, full-length UGT2A3 was amplified using sense (5′-TTGCAGATCAGTGTGTGAGGGAACTG-3′) and antisense (5′-CCCCATCAGGTCTTTCTTG-AATTTGG-3′) primers corresponding to nucleotides −31 to −6 and +1616 to +1591, respectively, relative to the UGT2A3 translation start site.
Wild-type UGT2A2, UGT2A2Δexon3, and UGT2A3 cDNA sequences were verified by direct dideoxy sequencing and cloned into a pcDNA3.1/V5-His-TOPO vector using standard protocols. After transformation using OneShot TOP10 competent Escherichia coli and a large-scale plasmid preparation, standard Lipofectamine protocols were used to generate HEK293 cell lines overexpressing the enzyme corresponding to wild-type UGT2A2 (termed UGT2A2_i1), UGT2A2Δexon3 (termed UGT2A2_i2), or UGT2A3. Cells were grown to 75% confluence in Dulbecco’s modified Eagle’s medium and supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and G418 (400 μg/ml). Cell homogenates were prepared as previously described in 1× Tris-buffered saline (25 mM Tris base, 138 mM NaCl, and 2.7 mM KCl, pH 7.4) (Dellinger et al., 2006; Sun et al., 2007; Chen et al., 2008b). Total RNA was extracted from each cell line using an RNeasy Mini kit following the manufacturer’s protocols. Total protein concentrations from cell line homogenates were determined using a BCA protein assay.
Determination of UGT2A2 or UGT2A3 Overexpression in Stable Cell Lines.
Currently, no readily available antibody exists for quantification of UGT2A2 or UGT2A3 in HEK293 overexpressing cell lines. A custom antibody specific for a UGT2A1 peptide sequence at the N terminus (Bushey et al., 2011) was tested for cross-reactivity against UGT2A2 and UGT2A3 protein. Up to 150 μg of UGT2A2_i1 and UGT2A3 protein lysates were used in a Western blot with the anti-UGT2A1 antibody (Open Biosystems, Huntsville, AL) at a 1:500 dilution. Fifty micrograms of UGT2A1 protein was loaded as a positive control, and 150 μg of HEK293 protein lysate was loaded as a negative control.
To examine relative wild-type UGT2A2, UGT2A2Δexon3, and UGT2A3 mRNA expression levels in HEK293 overexpressing cell lines, real-time PCR was completed using ABI TaqMan gene expression assays (Hs04195512_s1 for determination of UGT2A2 or UGT2A2Δexon3 expression, and Hs00226904_m1 for UGT2A3 expression). The ABI UGT2A2 gene expression assay is specific for UGT2A2 exon 1, enabling the assay to detect both wild-type UGT2A2 and UGT2A2Δexon3 transcripts. Glyceraldehyde 3-phosphate dehydrogenase (Hs99999905_m1) was used as a housekeeping gene, and all assays were completed as described previously using standard thermal cycling parameters. Relative levels of UGT2A overexpression in each stable cell line were calculated using the ΔΔCt method, and corrections were made to account for differences in the amplification efficiency of each assay (Jones et al., 2012).
Glucuronidation Activities of UGT2A2_i1, UGT2A2_i2, and UGT2A3.
Glucuronidation assays were performed using homogenates from HEK293 cell lines overexpressing UGT2A2_i1, UGT2A2_i2, and UGT2A3 as previously described (Fang et al., 2002; Dellinger et al., 2006; Sun et al., 2007; Chen et al., 2008b; Bushey et al., 2011). Briefly, after an initial incubation of 200 μg of total cell homogenate protein with 10 μg of alamethicin for 15 minutes on ice, glucuronidation reactions were performed in a final reaction volume of 25 μl at 37°C with 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 4 mM UDPGA, and between 6 μM and 1 mM substrate. Reactions were also completed using 4 mM UDP-glucose, 4 mM UDP-galactose, or 4 mM UDP-N-acetylglucosamine in place of UDPGA to determine the sugar specificities of UGT2A2 and UGT2A3. Reactions were stopped by the addition of 25 μl of acetonitrile and reaction mixtures were centrifuged at 16,100g for 10 minutes prior to the collection of supernatant.
Glucuronides were detected using ultra-pressure liquid chromatography (UPLC) on a Waters Acquity UPLC System (Milford, MA) as described previously (Sun et al., 2007; Balliet et al., 2009; Olson et al., 2009). The initial solvent gradients and UV absorbance wavelengths used to detect glucuronidation activity were described previously (Bushey et al., 2011). Quantification of glucuronide formation was completed as described previously (Dellinger et al., 2006; Dellinger et al., 2007; Sun et al., 2007; Chen et al., 2008a; Balliet et al., 2009; Bushey et al., 2011). The glucuronidation rate for each substrate was determined using at least eight substrate concentrations that encompassed the experimentally determined KM. Either β-glucuronidase treatment or mass spectrometry analysis was used to confirm all glucuronides. Various negative controls were used, including reactions with empty HEK293 cells and reactions with no substrate added to the reaction mixture. Substrates of UGT2A2 and UGT2A3 that had glucuronide formation below the limit of detection were incubated for 18 hours with 750 μg of total protein homogenate to confirm the lack of glucuronidation activity. For glucuronidation rate determinations, cell homogenate protein levels and incubation times for each substrate were determined experimentally to ensure that substrate utilization was less than 10% and to maximize levels of detection while in a linear range of glucuronide formation. The lower limit of glucuronide detection was determined experimentally using a 1-naphthol glucuronide standard.
Effect of UGT2A2_i2 Coexpression on UGT2A2_i1 Activity.
The pcDNA3.1/V5-His-TOPO wild-type UGT2A2 and pcDNA3.1/V5-His-TOPO UGT2A2Δexon3 vectors described previously were coexpressed in HEK293 cells to determine the functional effects of UGT2A2_i2 expression on UGT2A2_i1 activity. UGT2A2Δexon3 was transiently transfected in HEK293 cells stably overexpressing wild-type UGT2A2 in triplicate, with each transfection performed for 36 hours using 8 µg of the pcDNA3.1/V5-His-TOPO UGT2A2Δexon3 vector. The transfection efficiency was >95% for all transfection experiments as determined by visual inspection, confirming the expected >90% efficiency according to the manufacturer (Invitrogen). Following transfection, RNA was extracted and protein homogenate was prepared as described previously. In addition, RNA was extracted and protein homogenate was made from nontransfected cells stably overexpressing wild-type UGT2A2 alone.
The level of UGT2A2Δexon3 expression following transfection was first assessed by RT-PCR using Pfu polymerase and a sense (5′-GGGCTTTCTACAAAGAACTAGG-3′) primer specific for UGT2A2 exon 1 and an antisense (5′-CCATAGGGACTCCGTGTTAAAT-3′) primer specific to exon 4, with the sense and antisense primers corresponding to nucleotides +317 to +338 and nucleotides +1168 to +1147, respectively, relative to the UGT2A2 translation start site. RT was completed using 2 µg of RNA and oligo(dT), with cDNA corresponding to 100 ng of RNA used in subsequent PCR reactions. PCR was completed using an initial denaturing temperate of 94°C for 2 minutes, 40 cycles of 94°C for 30 seconds, 57°C for 45 seconds, and 72°C for 2 minutes, followed by a final cycle of 10 minutes at 72°C. Following RT-PCR, the UGT2A2 real-time PCR assay described previously (see Fig. 1) was used to quantitatively determine relative wild-type UGT2A2 and UGT2A2Δexon3 expression following transfection. cDNAs corresponding to 20 ng of RNA from 1) cells overexpressing wild-type UGT2A2 and 2) cells overexpressing wild-type UGT2A2 that were transiently transfected with UGT2A2Δexon3 were used for real-time PCR reactions. Relative expression levels were corrected for differences in assay efficiency, as described previously (Jones et al., 2012). Levels of wild-type UGT2A2 and UGT2A2Δexon3 expression were calculated relative to wild-type UGT2A2 expression in the nontransfected cells (set to 1.0 as a reference). Real-time PCR reactions were completed as described previously, with RPLPO used as the control gene.
Glucuronidation assays were completed to determine the functional effects of UGT2A2_i2 coexpression on UGT2A2_i1 activity. UGT2A2_i1-mediated glucuronidation activity was determined against the PAHs 1-OH-pyrene and 8-OH-B(a)P using a substrate concentration at the previously determined KM for UGT2A2_i1 [100 µM for 1-OH-pyrene; 300 µM for 8-OH-B(a)P]. Glucuronidation assays were completed as described previously, with 200 µg of protein homogenate used for each activity assay. For glucuronidation rate determinations, cell homogenate protein level and incubation times for each substrate were chosen so that substrate utilization was less than 10%.
Data Analysis and Statistics.
Three independent experiments were completed for UGT2A2 and UGT2A3 kinetic analyses in this study. Kinetic constants were calculated using GraphPad Prism 5 software (GraphPad Software, Inc., San Diego, CA). Vmax and KM values for each substrate examined were calculated by graphing the glucuronide product formed versus the substrate concentration and then using the Michaelis-Menten equation. All kinetics data were transformed into a linear Eadie-Hofstee plot to visually confirm that a simple Michaelis-Menten mechanism was followed. The Student’s t test was used to compare kinetic values of UGT2A isoforms after correcting for relative UGT2A overexpression in each stable cell line, and to compare activity following UGT2A2_i2 coexpression with UGT2A2_i1 following normalization for relative wild-type UGT2A2 expression.
Results
Expression of UGTs 2A2 and 2A3 in Human Tissues.
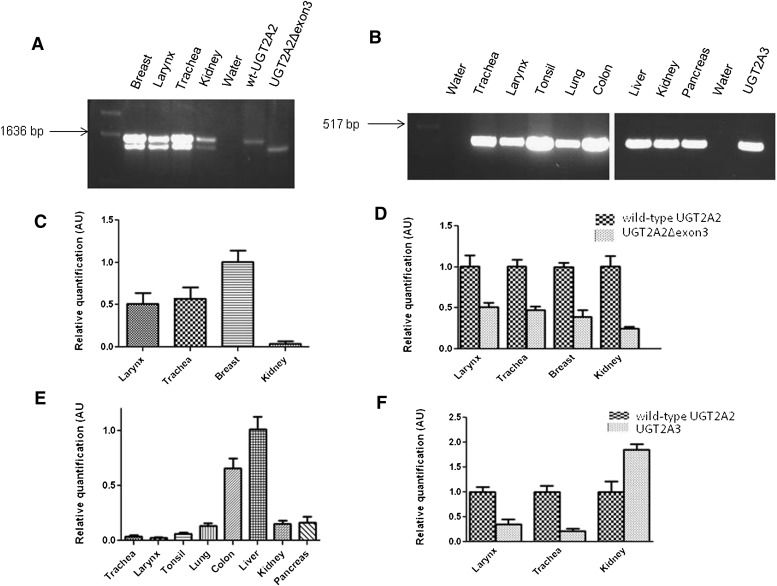
UGT2A2 was previously reported to be expressed in fetal and adult nasal mucosa tissues (Sneitz et al., 2009), whereas UGT2A3 was reported in several tissues including the small intestine, liver, colon, pancreas, kidney, and stomach (Court et al., 2008; Sneitz et al., 2009). In the current study, a more comprehensive analysis of UGT2A2 and UGT2A3 expression was performed, focusing on UGT2A enzyme expression in aerodigestive and respiratory tract tissues. Initially, pooled RNA samples from multiple tissues were analyzed qualitatively by RT-PCR for UGT2A2 expression. As shown in Fig. 2A, UGT2A2 was well expressed in the breast, larynx, and trachea, with some expression also observed in the kidney. Two distinct UGT2A2 products were amplified in these tissues using PCR primers specific for the amplification of full-length UGT2A2. Following gel extraction and sequencing, it was determined that, in addition to wild-type UGT2A2 (NM_001105677.2), a novel UGT2A2 transcript lacking exon 3 (UGT2A2Δexon3) was also PCR-amplified. The deletion of this exon creates a transcript that is 132 nucleotides shorter than wild-type UGT2A2, with the open reading frame of the gene remaining intact. No UGT2A2 expression was observed following multiple RT-PCR attempts in any of the other tissues examined, including the whole brain, lung, tonsil, colon, pancreas, prostate, cerebral cortex, floor of the mouth, esophagus, and liver (data not shown).
Fig. 2.
Qualitative and quantitative determination of UGT2A2 and UGT2A3 tissue expression. (A) initial characterization of UGT2A2 expression in multiple human tissues using RT- PCR and primers specific for amplification of the full-length UGT2A2 transcript. RNAs from HEK293 cell lines overexpressing wild-type (WT) UGT2A2 and UGT2A2Δexon3 were used as positive controls, with water used as a negative control. Two distinct transcripts were observed, and following gel extraction and sequencing of the PCR products, it was determined that both wild-type UGT2A2 and a novel UGT2A2 exon 3 deletion splice variant were amplified. (B) initial screening of UGT2A3 expression in multiple human tissues using RT-PCR. Primers specific for exon 1 of UGT2A3 were used to determine UGT2A3 tissue expression. RNA from a HEK293 cell line overexpressing UGT2A3 was used as a positive control, and water was used in place of cDNA as a negative control. For the determination of both UGT2A2 and UGT2A3 expression, tissue RNAs pooled from at least three individuals were used and the cDNA equivalent of 100 ng RNA was used for each reaction. (C) RNAs from tissues exhibiting UGT2A2 expression in A were used with a wild-type UGT2A2–specific real-time PCR assay (see Fig. 1) to quantitatively determine relative wild-type UGT2A2 expression. Wild-type UGT2A2 expression was calculated using the ΔΔCt method, relative to wild-type UGT2A2 expression in the breast. (D) relative UGT2A2Δexon3 mRNA expression in each tissue was determined using both assays described in Fig. 1. Expression data were corrected to account for differences in the amplification efficiency of each assay (Jones et al., 2012). Relative UGT2A2Δexon3 expression in each tissue was determined by comparing levels of UGT2A2Δexon3 to levels of wild-type UGT2A2 expression (set to 1.0 as the reference). (E) RNAs from tissues exhibiting UGT2A3 expression in B were used with a UGT2A3-specific real-time PCR assay to quantitatively determine UGT2A3 tissue expression. UGT2A3 mRNA expression was calculated using the ΔΔCt method relative to UGT2A3 expression in the liver. (F) relative expression levels of wild-type UGT2A2 and UGT2A3 were compared in the three tissues that were determined to express both UGT2A transcripts. Expression levels were normalized to wild-type UGT2A2 expression (set to 1.0 as a reference) in each tissue following correction for the amplification efficiency of each assay. For all real-time PCR data, results are expressed as the mean ± S.D. of triplicates, and results were normalized to RPLPO mRNA expression in each tissue. cDNA corresponding to 20-ng RNA was used for each real-time PCR reaction. AU, arbitrary unit.
RT-PCR using primers specific for the amplification of UGT2A3 exon 1 (Fig. 2B) showed UGT2A3 expression in the trachea, larynx, tonsil, lung, colon, liver, kidney, and pancreas. The full-length UGT2A3 cDNA (NM_024743.3) was also amplified in these tissues, but in contrast to UGT2A2, no UGT2A3 splice variants were observed (data not shown). No UGT2A3 expression was observed following multiple RT-PCR attempts in the whole brain, cerebral cortex, floor of the mouth, breast, esophagus, and prostate (data not shown).
A custom UGT2A2 real-time PCR assay (see Fig. 1) was developed to specifically and quantitatively detect wild-type UGT2A2 and UGT2A2Δexon3 expression in tissues that were shown to express UGT2A2 by RT-PCR. Wild-type UGT2A2 expression was highest in the breast (1.0 ± 0.09, set as the reference), followed by the trachea (0.57 ± 0.10) > larynx (0.51 ± 0.11) > kidney (0.05 ± 0.02; Fig. 2C). The UGT2A2 real-time assay was also used to determine the relative expression of UGT2A2Δexon3 in comparison with wild-type UGT2A2 expression in each tissue. In each tissue analyzed, wild-type UGT2A2 expression (set at 1.0 as the reference) was greater than UGT2A2Δexon3 expression, with the UGT2A2Δexon3:wild-type UGT2A2 expression ratio similar in the larynx (0.51 ± 0.06), trachea (0.47 ± 0.05), and breast (0.39 ± 0.08), and slightly lower in the kidney (0.25 ± 0.02; Fig. 2D).
Real-time PCR was also used to quantify relative UGT2A3 expression in tissues where UGT2A3 expression was detected by RT-PCR. UGT2A3 expression was found to be highest in the liver (set as the reference at 1.0 ± 0.12) followed by the colon (0.65 ± 0.09) > pancreas (0.17 ± 0.05) ≥ kidney (0.15 ± 0.03) ≥ lung (0.14 ± 0.02) > tonsil (0.06 ± 0.01) > trachea (0.04 ± 0.01) > larynx (0.02 ± 0.01; Fig. 2E). UGT2A3 expression was lower than wild-type UGT2A2 expression in the larynx (0.36 ± 0.09) and trachea (0.22 ± 0.05), but was higher than wild-type UGT2A2 expression in the kidney (1.85 ± 0.12; Fig. 2F).
Enzyme Activities of UGTs 2A2 and 2A3 Against Tobacco Carcinogens.
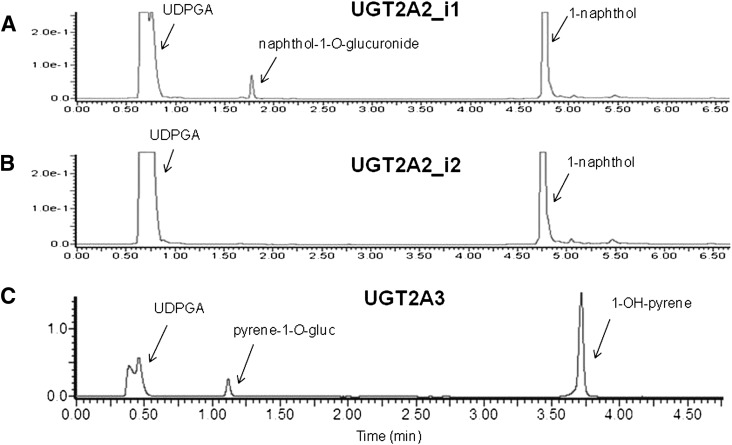
With mRNA expression of both UGT2A2 and UGT2A3 in tobacco carcinogen target tissues, and UGT2A1 previously shown to exhibit activity against PAH metabolites (Bushey et al., 2011), experiments were performed to examine the glucuronidation activities of UGTs 2A2 and 2A3 against various tobacco carcinogens, including PAHs, tobacco-specific nitrosamines (TSNAs), and heterocyclic amines (HCAs). Using homogenates of HEK293 cells overexpressing UGT2A enzymes, wild-type UGT2A2 (UGT2A2_i1) was shown to exhibit activity against 1-naphthol, with a 1-naphthol-O-glucuronide peak observed at a retention time of 1.8 minutes and a 1-naphthol substrate peak observed at 4.8 minutes on UPLC (Fig. 3A). In addition to 1-naphthol, UGT2A2_i1 exhibited glucuronidation activity against a panel of PAHs, including 1-OH-pyrene, 1-OH-B(a)P, 3-OH-B(a)P, 7-OH-B(a)P, and 8-OH-B(a)P. No UGT2A2_i1 activity was detected against more complex PAH proximate carcinogens including 5-methylchrysene-1,2-diol, B(a)P-7,8-diol, or dibenzo(a,l)pyrene11,12-diol; using 1-naphthol-glucuronide as the test substrate, the lower limit of glucuronide detection for the Waters Acquity UPLC System used in the present analysis was 20 pmol. In similar glucuronidation assays, UGT2A2 splice variant protein (UGT2A2_i2) exhibited no detectable glucuronidation activity against 1-naphthol (Fig. 3B); only a 1-naphthol substrate peak was observed (retention time of 4.8 minutes). No detectable glucuronidation activity was exhibited for UGT2A2_i2 against all PAHs tested, including the six PAHs metabolized by UGT2A2_i1 (data not shown). UGT2A2_i2 also exhibited no detectable activity against the common UGT substrate 4-methylumbelliferone (Uchaipichat et al., 2004) (data not shown).
Fig. 3.
Representative UPLC chromatograms of UGT2A2 and UGT2A3 glucuronidation activities against PAH substrates. (A) UPLC trace of 1-naphthol-O-glucuronide formation following 1-naphthol incubation with UGT2A2_i1 overexpressing cell homogenate. A naphthol-1-O-glucuronide peak was observed at 1.8 minutes, and a 1-naphthol substrate peak was observed at 4.8 minutes. (B) no detectable 1-naphthol-O-glucuronide was observed following incubation of UGT2A2_i2 overexpressing cell homogenate with 1-napthhol, as shown by a representative UPLC chromatogram with only a 1-napthol substrate peak at 4.8 minutes. (C) UPLC trace of pyrene-1-O-glucuronide formation after 1-OH-pyrene incubation with UGT2A3 overexpressing cell homogenate. A glucuronide peak was observed at 1.1 minutes and a 1-OH-pyrene peak was observed at 3.7 minutes.
UGT2A3 exhibited detectable glucuronidation activity against the simple PAH 1-OH-pyrene, with a pyrene-1-O-glucuronide peak observed at 1.1 minutes and a 1-OH-pyrene substrate peak observed at 3.7 minutes on UPLC (Fig. 3C). UGT2A3 also exhibited glucuronidation activity against 1-naphthol; however, no UGT2A3 activity was detected against any of the other PAHs analyzed, including 1-OH-B(a)P, 3-OH-B(a)P, 7-OH-B(a)P, 8-OH-B(a)P, 5-methylchrysene-1,2-diol, B(a)P-7,8-diol, and dibenzo(a,l)pryrene-11,12-diol (data not shown).
UGT2A2_i1, UGT2A2_i2, and UGT2A3 all exhibited no detectable glucuronidation activity against a panel of TSNAs and HCAs including 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol, N-nitrosoanabasine, N-nitrosoanatabine, N-nitrosonornicotine, PhIP, and N-OH-PhIP, and exhibited no activity against nicotine (data not shown). In addition, no glucuronidation activity was detected for any of the three UGT isoforms against any of the tested substrates using alternate sugars including UDP-glucose, UDP-galactose, and UDP-N-acetylglucosamine in place of UDPGA (data not shown).
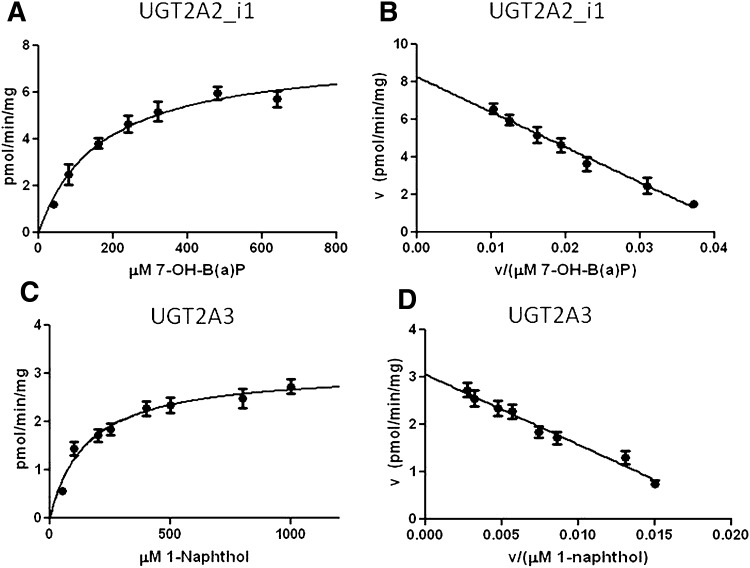
Representative Michaelis-Menten and Eadie-Hofstee kinetics curves are shown for UGT2A2_i1 activity against 7-OH-B(a)P (Fig. 4, A and B, respectively) and UGT2A3 activity against 1-naphthol (Fig. 4, C and D, respectively). The Eadie-Hofstee transformation established linearity for all reactions and ensured that they followed simple Michaelis-Menten kinetics. Similar properties were observed for all UGT2A2/UGT2A3–glucuronidated substrates examined (data not shown). UGT2A2_i1 exhibited approximately 10-fold higher levels of activity as determined by Vmax/KM against the simple PAH substrates, 1-OH-pyrene and 1-naphthol, as compared with hydroxylated B(a)P metabolites (Table 1). No readily available antibody exists to detect UGT2A2 or UGT2A3 protein, and a custom-designed UGT2A1 antibody was shown to exhibit no cross reactivity toward UGT2A2 or UGT2A3 (data not shown). Using real-time PCR to normalize for mRNA expression differences in the UGT2A cell lines, a comparison between UGT2A2_i1 and UGT2A3 glucuronidation activities suggests that UGT2A2_i1 has significantly higher activity (Vmax/KM) against 1-OH-pyrene (P < 0.005) and 1-naphthol (P < 0.01), with UGT2A2_i1 exhibiting an approximately 40-fold higher Vmax/KM against 1-OH-pyrene and a 10-fold higher Vmax/KM against 1-naphthol (Table 1).
Fig. 4.
Representative enzyme kinetics curves for UGT2A2_i1 and UGT2A3 activity against PAH substrates. (A) representative Michaelis-Menten curve for UGT2A2_i1 activity against 7-OH-B(a)P. (B) Eadie-Hofstee transformation of UGT2A2_i1 activity against 7-OH-B(a)P. (C) representative Michaelis-Menten curve for UGT2A3 activity against 1-naphthol. (D) Eadie-Hofstee transformation of UGT2A3 activity against 1-naphthol.
TABLE 1.
Enzyme kinetics summary of UGT2A2_i1 and UGT2A3 activities against PAHs. Vmax, KM, and Vmax/KM represent the mean of three independent experiments.
| Substrate | UGT2A2_i1a |
UGT2A3b |
||||
|---|---|---|---|---|---|---|
| KM | Vmax | Vmax/KM | KM | Vmax | Vmax/KM | |
| μM | pmol/min/mg | μl/min/mg | μM | pmol/min/mg | μl/min/mg | |
| 1-OH-pyrene | 100 ± 15 | 38 ± 2.1 | 0.38 ± 0.03 | 363 ± 38 | 5.5 ± 0.2 | 0.01 ± 0.001* |
| 1-Naphthol | 106 ± 15 | 26 ± 1.5 | 0.25 ± 0.04 | 148 ± 18 | 3.1 ± 0.2 | 0.02 ± 0.003* |
| 1-OH-B(a)P | 279 ± 40 | 8.2 ± 0.8 | 0.03 ± 0.004 | BLD | ||
| 3-OH-B(a)P | 332 ± 54 | 7.4 ± 0.5 | 0.02 ± 0.002 | BLD | ||
| 7-OH-B(a)P | 240 ± 29 | 8.5 ± 0.4 | 0.04 ± 0.008 | BLD | ||
| 8-OH-B(a)P | 287 ± 37 | 9.3 ± 0.6 | 0.03 ± 0.002 | BLD | ||
| 5-Methylchrysene-1,2-diol | BLD | BLD | ||||
| B(a)P-7,8-diol | BLD | BLD | ||||
| Dibenzo(a,l)pyrene-11,12-diol | BLD | BLD | ||||
BLD, below limit of detection.
Data expressed as milligrams of total protein homogenate, corrected for relative wild-type UGT2A2 expression as determined by real-time PCR.
Data expressed as milligrams of total protein homogenate, corrected for relative UGT2A3 expression as determined by real-time PCR.
P < 0.01 versus UGT2A2_i1 activity.
Effect of UGT2A2_i2 Expression on UGT2A2_i1 Activity.
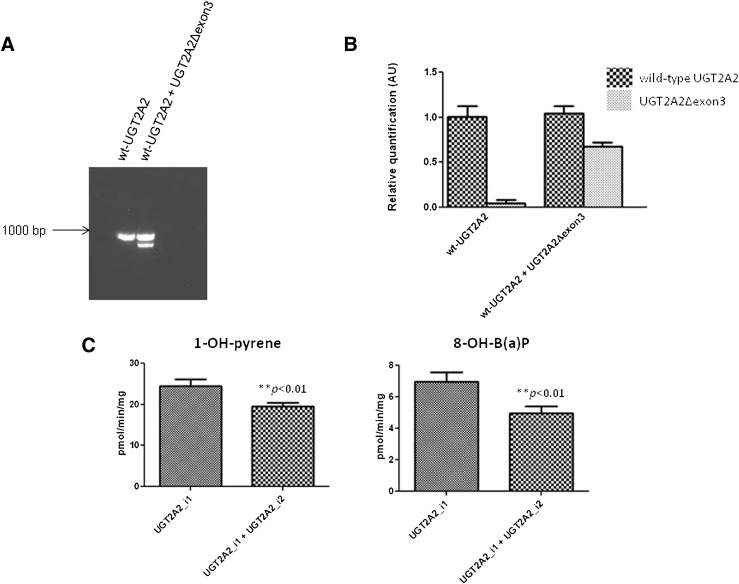
To examine the effect of UGT2A2_i2 coexpression on UGT2A2_i1 glucuronidation activity, a vector containing UGT2A2Δexon3 was transfected into HEK293 cells stably overexpressing wild-type UGT2A2. Using primers specific for exons 1 and 4 of UGT2A2, two distinct amplicons (850 and 700 bp) were observed after RT-PCR of the total RNA of cells following transfection with UGT2A2Δexon3; only a single amplicon (850 bp) was observed using the total RNA from nontransfected control cells (Fig. 5A). Using real-time PCR to quantify UGT2A2 expression, the relative expression ratio of UGT2A2Δexon3:wild-type UGT2A2 following transfection was determined to be 0.61 ± 0.08 (Fig. 5B). Using homogenates from UGT2A2Δexon3-transfected versus untransfected UGT2A2_i1–overexpressing cells, a significant ∼25% decrease in UGT2A2_i1–mediated glucuronidation of 1-OH-pyrene and 8-OH-B(a)P was observed following UGT2A2_i2 coexpression (P < 0.01 for both substrates; Fig. 5C).
Fig. 5.
Effect of UGT2A2_i2 coexpression on UGT2A2_i1 activity against PAH substrates. (A) a representative RT-PCR gel showing UGT2A2Δexon3 coexpression with wild-type (WT) UGT2A2 following transient transfection into HEK293 cells stably overexpressing UGT2A2_i1. No UGT2A2Δexon3 transcript was detected using RNA from the untransfected cells. The cDNA equivalent of 100-ng RNA was used for each RT-PCR reaction. (B) real-time PCR was completed using the assay described in Fig. 1 to determine relative wild-type UGT2A2 and UGT2A2Δexon3 levels following transient transfection. cDNA corresponding to 20 ng of RNA was used in each reaction, with levels of wild-type UGT2A2 and UGT2A2Δexon3 expression determined relative to wild-type UGT2A2 expression in the untransfected cells. As expected, only trace amounts of UGT2A2Δexon3 were detected in the untransfected UGT2A2_i1 overexpressing cells. (C) effect of UGT2A2_i2 coexpression on UGT2A2_i1 activity against 1-OH-pyrene and 8-OH-B(a)P using a substrate concentration at the previously determined KM for UGT2A2. Glucuronidation rates were corrected to account for differences in the relative level of UGT2A2_i1 expression in the untransfected cells and cells transiently transfected with UGT2A2_i2. AU, arbitrary unit.
Discussion
The role of the UGT2A subfamily in overall metabolism has been relatively understudied when compared with members of the UGT1A and UGT2B families. It was previously reported that UGT2A1 is expressed in the lung, larynx, trachea, and colon and is active against PAH proximate carcinogens found in tobacco smoke, suggesting that UGT2A1 could play a role in local PAH carcinogen metabolism (Bushey et al., 2011). The current study expands the focus to an assessment of the potential role of two additional UGT2A family members, UGT2A2 and UGT2A3, in tobacco carcinogen metabolism.
This is the first study to examine UGT2A2 and UGT2A3 expression in aerodigestive and respiratory tract tissues, with UGT2A2 determined to be expressed in the larynx and trachea and UGT2A3 found to be expressed in the lung, trachea, larynx, colon, and tonsil. A previous study investigating UGT2A2 expression in a limited number of tissues reported UGT2A2 expression only in fetal and adult nasal mucosa tissues (Sneitz et al., 2009). Adult lung and liver tissues were previously screened for UGT2A2 expression, and in agreement with our results, no UGT2A2 expression was observed in these tissues (Sneitz et al., 2009). High UGT2A3 expression was observed in the liver, colon, pancreas, and kidney in the present study, which is consistent with results reported in the single previous study analyzing UGT2A3 expression in human tissues (Court et al., 2008). Our results suggest modest UGT2A3 expression in the lung, using RNA pooled from five Caucasian individuals; this is in contrast to results from two previous studies suggesting no detectable UGT2A3 mRNA expression in individual lung specimens (Court et al., 2008; Sneitz et al., 2009). These conflicting results could be due to interindividual variability in UGT2A3 expression, mRNA quality issues, or a lack of homogeneity between different lung specimens. Additional experiments directly comparing UGT2A1, UGT2A2, and UGT2A3 mRNA levels would help further clarify the tissue-specific expression level of each UGT2A enzyme.
Previous studies have demonstrated UGT2A2 glucuronidation activity against 1-OH-pyrene and 1-naphthol (Sneitz et al., 2009). Whereas UGT2A2 was found to exhibit glucuronidation activity against hydroxylated B(a)P metabolites in the present study, UGT2A3 activity was limited to simple PAHs like 1-OH-pyrene and 1-naphthol. No detectable glucuronidation activity was observed against proximate PAH carcinogens, including 5-methylchrysene-1,2-diol, B(a)P-7,8-diol, and dibenzo(a,l)pyrene-11,12-diol, for either enzyme. This is in contrast with results described previously for UGT2A1, which was active against all PAHs tested (Bushey et al., 2011). Similar to that observed previously for UGT2A1 (Bushey et al., 2011), UGTs 2A2 and 2A3 exhibited no detectable glucuronidation activity against TSNAs, HCAs, or nicotine, suggesting that all UGT2A enzymes lack the capacity for N-glucuronidation.
UDPGA is the primary sugar donor for UGT1A- and UGT2B-mediated glucuronidation (Meech and Mackenzie, 2010); however, the sugar specificity of the UGT2A family had never been investigated. Multiple UGTs have been shown to use alternate sugars other than UDPGA as cosubstrates in metabolic reactions. For example, UGT3A1 has been reported to use UDP-N-acetylglucosamine as its preferential sugar donor (Mackenzie et al., 2008), and UGT3A2 has been reported to use both UDP-glucose and UDP-xylose in glycosidation reactions (MacKenzie et al., 2011). UGT2B7 has also been reported to use UDP-glucose and UDP-xylose in the metabolism of hyodeoxycholic acid (Mackenzie et al., 2003). In the current study, no detectable enzyme activity was observed for either UGT2A2 or UGT2A3 when alternate sugars, including UDP-N-acetylglucosamine, UDP-glucose, and UDP-galactose, were used in glucuronidation assays. Recently completed studies have also shown that UGT2A1 is unable to metabolize substrates using these alternate sugars (R. T. Bushey and P. Lazarus, unpublished data), suggesting that the entire UGT2A family requires UDPGA as the cosubstrate for glucuronidation activity.
UGT2A1 and UGT2A2 are encoded by common exons 2–6 and share a high degree of sequence similarity in exon 1. The KM values of UGT2A1 and UGT2A2 are similar for 1-OH-pyrene [91 μM for UGT2A1 (Bushey et al., 2011) versus 100 μM for UGT2A2] and hydroxylated B(a)P metabolites [247–308 μM for UGT2A1 (Bushey et al., 2011) versus 240–332 μM for UGT2A2]. Somewhat higher KM values (P < 0.001) were observed for UGT2A3 activity against 1-OH-pyrene (363 μM) and 1-naphthol (148 μM) as compared with that observed for UGT2A1 [91 μM for 1-OH-pyrene and 30 μM for 1-naphthol (Bushey et al., 2011)]. These data suggest that, although UGT2A1 is the only UGT2A enzyme active against PAH proximate carcinogens, including PAH-diols, both UGTs 2A1 and 2A2 play an important role in the local detoxification of monohydroxylated PAH metabolites within aerodigestive and respiratory tissues. Monohydroxylated PAHs are known to be procarcinogenic (Hecht, 1999), suggesting that UGTs 2A1 and 2A2 may play a role in preventing PAH-mediated carcinogenesis in target tissues.
Prevalent single-nucleotide polymorphisms (SNPs) have been identified for a variety of UGTs, with many of these polymorphisms having a significant functional effect on glucuronidation activity. A prevalent (>1%) nonsynonymous UGT2A3 SNP was previously characterized; the UGT2A3 was determined to cause no significant change in UGT2A3 activity against bile acids (Court et al., 2008). Previous studies of UGT2A1 identified two prevalent non-synonymous polymorphisms that cause significant changes in enzyme activity. A conservative UGT2A1Lys75Arg polymorphism in the substrate recognition region of the enzyme was determined to cause a statistically significant 25% decrease in activity, whereas a nonconservative UGT2A1Gly308Arg polymorphism in a conserved region necessary for UDPGA binding within UGT2A1 was found to completely ablate enzyme activity (Bushey et al., 2011). With UGT2A1 and UGT2A2 sharing common exons 2–6, the UGT2A1Gly308Arg polymorphism is also found in UGT2A2 and would also be hypothesized to significantly alter UGT2A2 activity. Through a screening of HapMap, additional nonsynonymous UGT2A2-coding SNPs with a minor allele frequency greater than 1.0% were determined to include UGT2A2Ala58Val and UGT2A2Val392Ile variants (Gibbs, 2003). Functional studies analyzing the effects of these SNPs on UGT2A2 glucuronidation activity must be performed to better assess their potential role in altering UGT2A2 activity against PAH metabolites.
The present study is also the first to identify and investigate a novel UGT2A2 exon-deleted splice variant, UGT2A2_i2, which was determined by real-time PCR to be expressed at levels ranging from ∼25% to 50% of wild-type UGT2A2 in multiple tissues. Activity assays suggested that UGT2A2_i2 exhibited no detectable glucuronidation activity in any substrate examined. Coexpression studies suggested that UGT2A2_i2 negatively regulates UGT2A2_i1 activity. These studies were performed using a cell-line model encompassing a transient cotransfection of UGT2A2Δexon3 into a cell line stably overexpressing wild-type UGT2A2, with the ratio of UGT2A2Δexon3:wild-type UGT2A2 in this system (0.61) closely approximating the ratio observed in multiple human tissues. These results are similar to the effect of the exon 3 splice variant recently discovered for UGT2A1, with the UGT2A1_i2 isoform both lacking enzyme activity and acting as a negative regulator of UGT2A1 glucuronidation activity, likely via protein-protein interactions (Bushey and Lazarus, 2012). Future experiments must be performed to determine whether a protein-protein interaction occurs between UGT2A2_i1 and UGT2A2_i2, as well as whether UGT2A1_i2 and UGT2A2_i2 can hetero-oligomerize with other UGT2A isoforms. These studies will provide important information on the potential regulation of UGT2A-mediated glucuronidation activity against PAHs by UGT2A1_i2 and UGT2A2_i2, as expression of these variants could have implications on UGT2A detoxification mechanisms in vivo.
Acknowledgments
The authors thank the Penn State Cancer Institute’s Organic Synthesis Core for supplying various PAH carcinogens used in this study. They also thank the Functional Genomics Core at the Penn State University College of Medicine for equipment used for real-time PCR and genotyping analysis. Finally, they thank the Nucleic Acid Facility at the University Park campus of Penn State University for DNA sequencing services.
Abbreviations
- HCA
heterocyclic amine
- HEK
human embryonic kidney
- PAH
polycyclic aromatic hydrocarbon
- PhIP
2-amino-1-methyl-6-phenylimidazo [4,5-b]-pyridine
- RT-PCR
reverse-transcription polymerase chain reaction
- SNP
single-nucleotide polymorphism
- TSNA
tobacco-specific nitrosamines
- UDPGA
uridine 5′-diphosphoglucuronic acid
- UGT
UDP-glucuronosyltransferase
- UPLC
ultra-pressure liquid chromatography
Authorship Contributions
Participated in research design: Bushey, Lazarus.
Conducted experiments: Bushey, Dluzen.
Contributed new reagents or analytic tools: Bushey, Dluzen, Lazarus.
Performed data analysis: Bushey, Lazarus.
Wrote or contributed to the writing of the manuscript: Bushey, Dluzen, Lazarus.
Footnotes
This work was supported by the National Institutes of Health National Institute of Dental and Craniofacial Research [Grant R01-DE13158]; the National Institutes of Health National Cancer Institute [Grant R01-CA164366]; and the Pennsylvania Department of Health’s Health Research Formula Funding Program [Grant 4100038714].
References
- Balliet RM, Chen G, Gallagher CJ, Dellinger RW, Sun D, Lazarus P. (2009) Characterization of UGTs active against SAHA and association between SAHA glucuronidation activity phenotype with UGT genotype. Cancer Res 69:2981–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bièche I, Narjoz C, Asselah T, Vacher S, Marcellin P, Lidereau R, Beaune P, de Waziers I. (2007) Reverse transcriptase-PCR quantification of mRNA levels from cytochrome (CYP)1, CYP2 and CYP3 families in 22 different human tissues. Pharmacogenet Genomics 17:731–742 [DOI] [PubMed] [Google Scholar]
- Burchell B, Coughtrie MW. (1989) UDP-glucuronosyltransferases. Pharmacol Ther 43:261–289 [DOI] [PubMed] [Google Scholar]
- Bushey RT, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. (2011) Characterization of UDP-glucuronosyltransferase 2A1 (UGT2A1) variants and their potential role in tobacco carcinogenesis. Pharmacogenet Genomics 21:55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey RT, Lazarus P. (2012) Identification and functional characterization of a novel UGT2A1 splice variant: Potential importance in tobacco-related cancer susceptibility. J Pharmacol Exp Ther 343:712–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Dellinger RW, Gallagher CJ, Sun D, Lazarus P. (2008a) Identification of a prevalent functional missense polymorphism in the UGT2B10 gene and its association with UGT2B10 inactivation against tobacco-specific nitrosamines. Pharmacogenet Genomics 18:181–191 [DOI] [PubMed] [Google Scholar]
- Chen G, Dellinger RW, Sun D, Spratt TE, Lazarus P. (2008b) Glucuronidation of tobacco-specific nitrosamines by UGT2B10. Drug Metab Dispos 36:824–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH, Hazarika S, Krishnaswamy S, Finel M, Williams JA. (2008) Novel polymorphic human UDP-glucuronosyltransferase 2A3: cloning, functional characterization of enzyme variants, comparative tissue expression, and gene induction. Mol Pharmacol 74:744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger RW, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. (2007) Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis 28:2412–2418 [DOI] [PubMed] [Google Scholar]
- Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. (2006) Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab Dispos 34:943–949 [DOI] [PubMed] [Google Scholar]
- Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, Marques MM, Lazarus P. (2002) Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res 62:1978–1986 [PubMed] [Google Scholar]
- Gibbs RA, International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796 [DOI] [PubMed] [Google Scholar]
- Guéraud F, Paris A. (1998) Glucuronidation: a dual control. Gen Pharmacol 31:683–688 [DOI] [PubMed] [Google Scholar]
- Hecht SS. (1999) Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91:1194–1210 [DOI] [PubMed] [Google Scholar]
- Jedlitschky G, Cassidy AJ, Sales M, Pratt N, Burchell B. (1999) Cloning and characterization of a novel human olfactory UDP-glucuronosyltransferase. Biochem J 340:837–843 [PMC free article] [PubMed] [Google Scholar]
- Jones NR, Sun D, Freeman WM, Lazarus P. (2012) Quantification of Hepatic UDP glucuronosyltransferase 1A splice variant expression and correlation of UDP glucuronosyltransferase 1A1 variant expression with glucuronidation activity. J Pharmacol Exp Ther 342:720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass N, Hojo T, Ueding M, Lüttges J, Klöppel G, Jonat W, Nagasaki K. (2001) Expression of the tumor suppressor gene Maspin in human pancreatic cancers. Clin Cancer Res 7:812–817 [PubMed] [Google Scholar]
- Mackenzie P, Little JM, Radominska-Pandya A. (2003) Glucosidation of hyodeoxycholic acid by UDP-glucuronosyltransferase 2B7. Biochem Pharmacol 65:417–421 [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. (2005) Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 15:677–685 [DOI] [PubMed] [Google Scholar]
- MacKenzie PI, Rogers A, Elliot DJ, Chau N, Hulin JA, Miners JO, Meech R. (2011) The novel UDP glycosyltransferase 3A2: cloning, catalytic properties, and tissue distribution. Mol Pharmacol 79:472–478 [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Rogers A, Treloar J, Jorgensen BR, Miners JO, Meech R. (2008) Identification of UDP glycosyltransferase 3A1 as a UDP N-acetylglucosaminyltransferase. J Biol Chem 283:36205–36210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meech R, Mackenzie PI. (1998) Determinants of UDP glucuronosyltransferase membrane association and residency in the endoplasmic reticulum. Arch Biochem Biophys 356:77–85 [DOI] [PubMed] [Google Scholar]
- Meech R, Mackenzie PI. (2010) UGT3A: novel UDP-glycosyltransferases of the UGT superfamily. Drug Metab Rev 42:45–54 [DOI] [PubMed] [Google Scholar]
- Nagar S, Remmel RP. (2006) Uridine diphosphoglucuronosyltransferase pharmacogenetics and cancer. Oncogene 25:1659–1672 [DOI] [PubMed] [Google Scholar]
- Olson KC, Dellinger RW, Zhong Q, Sun D, Amin S, Spratt TE, Lazarus P. (2009) Functional characterization of low-prevalence missense polymorphisms in the UDP-glucuronosyltransferase 1A9 gene. Drug Metab Dispos 37:1999–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Murphy SE, Zheng Z, Lazarus P. (2000) O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1- (3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos 28:1352–1360 [PubMed] [Google Scholar]
- Sneitz N, Court MH, Zhang X, Laajanen K, Yee KK, Dalton P, Ding X, Finel M. (2009) Human UDP-glucuronosyltransferase UGT2A2: cDNA construction, expression, and functional characterization in comparison with UGT2A1 and UGT2A3. Pharmacogenet Genomics 19:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Sharma AK, Dellinger RW, Blevins-Primeau AS, Balliet RM, Chen G, Boyiri T, Amin S, Lazarus P. (2007) Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab Dispos 35:2006–2014 [DOI] [PubMed] [Google Scholar]
- Tephly TR, Burchell B. (1990) UDP-glucuronosyltransferases: a family of detoxifying enzymes. Trends Pharmacol Sci 11:276–279 [DOI] [PubMed] [Google Scholar]
- Uchaipichat V, Mackenzie PI, Guo XH, Gardner-Stephen D, Galetin A, Houston JB, Miners JO. (2004) Human udp-glucuronosyltransferases: isoform selectivity and kinetics of 4-methylumbelliferone and 1-naphthol glucuronidation, effects of organic solvents, and inhibition by diclofenac and probenecid. Drug Metab Dispos 32:413–423 [DOI] [PubMed] [Google Scholar]