Abstract
RNA is an important therapeutic target but information about RNA-ligand interactions is limited. Here we report a screening method that probes over 3,000,000 combinations of RNA motif-small molecule interactions to identify the privileged RNA structures and chemical spaces that interact. Specifically, a small molecule library biased for binding RNA was probed for binding to over 70,000 unique RNA motifs in a high throughput solution-based screen. The RNA motifs that specifically bind each small molecule were identified by microarray-based selection. In this library-versus-library or multidimensional combinatorial screening approach, hairpin loops (amongst a variety of RNA motifs) were the preferred RNA motif space that binds small molecules. Furthermore, it was shown that indole, 2-phenyl indole, 2-phenyl benzimidazole, and pyridinium chemotypes allow for specific recognition of RNA motifs. Since targeting RNA with small molecules is an extremely challenging area, these studies provide new information on RNA-ligand interactions that has many potential uses.
INTRODUCTION
RNA has diverse functions in cellular biology including encoding and translating protein, regulating the amount of protein expressed under different cellular conditions, and many others 1–4. In addition, RNA has been used as an artificial molecular switch to control cellular events such as RNA splicing and gene expression 5. Because of this, RNA is an attractive target for small molecules that serve as chemical genetics probes or therapeutics 6,7, as effectors of artificial gene circuits, orasanalytical tools 5,8.
Various studies have identified small molecules that bind RNA 6,9,10, however, the available information is sparse compared to the structural diversity of RNA in the transcriptome. One method that has been used to identify RNA structures that bind small molecules is Systematic Evolution of Ligands by Exponential Enrichment, or SELEX. In a SELEX experiment, aptamers (derived from an RNA library with a randomized region typically of >20 nucleotides)is identified that binds a small molecule with high affinity and specificity 11,12. Since the selected RNA is rather large, it is difficult to find it in genomic RNAs. However, there have been some excellent and notable cases in which the output of SELEX has been found in a biologically relevant RNA 13,14. A more common use of aptamer-small molecule interactions has been in the development of engineered cellular switches 5.
Another approach used to identify RNA-ligand interactions is high throughput screening (HTS) 6,7. In this approach, a single validated RNA probe or drug target is screened for binding to libraries of small molecules. Screening can be accomplished by using various techniques including Structure-Activity Relationships (SAR) by NMR spectroscopy 15, SAR by mass spectrometry 16,17, amongst others 18,19. Screening endeavors to find compounds that bind RNA, however, have much lower hit rates when compared to identifying small molecules that bind protein. Often, the hits identified are not specific for the RNA probed 6.
In an effort to develop a bottom-up rather than the traditional top-down approach to target RNA, we previously reported a method that merges the advantages of SELEX and of high throughput small molecule screening 20,21. This method probes chemical and RNA motif spaces simultaneously to identify selective interactions that can be used to target RNA. Termed 2-Dimensional Combinatorial Screening (2DCS), a library of small molecules is probed for binding to libraries of small RNA motifs that are likely to be present in a biologically important RNA. By using selection to identify the RNA motifs that bind each small molecule, the optimal RNA motif-small molecule partners are identified. These interactions are mined against RNA secondary structures in the transcriptome to design small molecules against a functionally important or toxic RNA. This approach has led to the development of small molecules that potently target several RNAs that contribute to disease, such as the myotonic dystrophies and Huntington’s disease 22–25.
In this report, we describe the development of an approach that allows for the facile identification of RNA motif-ligand interactions by merging solution-based HTS with microarray-based selection of the RNA motifs that bind a small molecule. This approach is high throughput and high content in that it probes millions of potential RNA motif-small molecule partners. Using this method, it was determined that members of a small molecule library have a significant bias for binding to RNA hairpin loops over thousands of other structures including internal loops, bulges, and base pairs. Analysis of the chemical space of the active small molecules reveals chemotypes that bias small molecules for recognition of RNA. This approach may have implications for the design of small molecules that modulate RNA function, which is an important yet an extremely challenging area.
RESULTS
High Throughput Screening and Microarray-Based Selections
We previously described a multidimensional combinatorial screening (MDCS) platform (also termed library-versus-library screening) that was developed to identify the optimal RNA motifs from a library of discrete secondary structures that bind small molecules (Figures 1b & 2). Termed 2-Dimensional Combinatorial Screening (2DCS), a microarray of small molecules is hybridized with a library of RNA motifs, such as internal loops, under conditions of high stringency 20,21. The randomized region of the RNA libraries is intentionally small (Figure 2) such that the structures have a high probability of occurring in biological RNAs 20,21. The bound motifs are harvested from the microarray via excision, amplified by RT-PCR, and identified by cloning and sequencing.
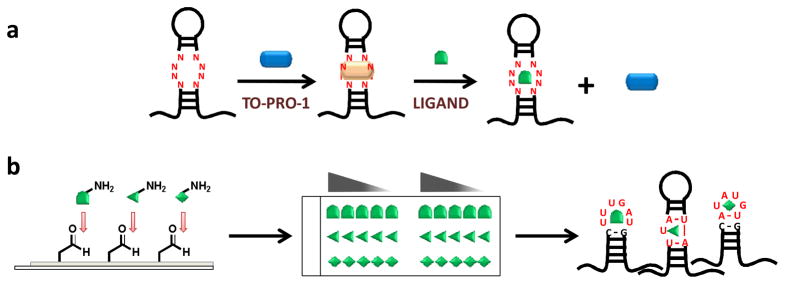
Figure 1.
The general procedure used to merge solution-based small molecule high throughput screening with microarray-based selection of RNAs that bind ligands. a, ligands are identified that bind to RNA motif libraries by using a TO-PRO-1 dye displacement assay. b, the RNA motifs that bind to each arrayed ligand are identified by microarray-based selection 20,21. Briefly, ligands identified that bind RNA from the dye displacement assay are conjugated to microarray surfaces and hybridized with RNA motif libraries under highly stringent conditions. Bound RNAs are harvested from the array by manual excision and sequenced.
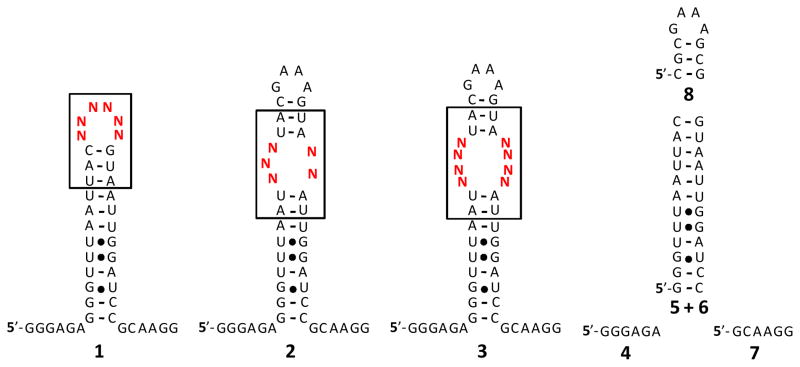
Figure 2.
The structures of the RNA motif libraries and the competitor oligonucleotides used in this study. N represents an equimolar mixture of A, C, G, and U. RNA motif libraries 1 – 3 contain 4,096, 1,024, and 65,536 members, respectively. Oligonucleotides 4 – 8 are used to compete off interactions that are common to all library members during selection experiments.
The direct microarray approach, however, is not amenable to more traditional small molecule screens. For example, high throughput screens that are completed under the Molecular Libraries Program at the National Institutes of Health (NIH) use solution-based screening to identify hit compounds that bind a target. Thus, in order to gain access to such resources and to increase the number of chemotypes and RNA motifs that are known to interact, we developed an approach that merges more standard high throughput, solution-based screens with microarray-based selection strategies (Figure 1). We then used this method to determine the features in RNA motifs (hairpins, internal loops, and bulges) and the features in small molecules that impart high affinity, selective binding. Specifically, the approach employs a solution-based dye displacement screening assay to identify small molecules that bind RNA motif libraries (Figure 1). The small molecules identified from this screen are then subjected to the previously described microarray-based MDCS selection.
Selection of a Dye for Solution-Based HTS
In order to complete a high throughput screen of RNA-ligand interactions, a read-out of binding is required. Previous studies have shown that dyes with emission properties that significantly change upon binding RNA can be used as probes in fluorescent indicator displacement (FID) assays to identify small molecules that bind nucleic acids 26–30. Examples include: a 2,7-disubstituted 9H-xanthen-9-one, or X2S, which has been used to study the binding of a library of pharmaceutically active compounds (LOPAC) to HIV Rev responsive element (RRE) RNA 30; TO-PRO-1 which has been used to study the binding of small molecule ligands to a variety of RNAs 28; and, Thiazole Orange (TO), which has been used to study the binding of threading intercalators to RNA 29.
In order to identify a dye that is optimal for our high throughput screening approach, we investigated the fluorescence properties of four unrelated dyes (TO-PRO-1, X2S, ethidium bromide, and SYBR green II) in the presence and absence of RNA library 1, which displays hairpin loops (Figure 2). The fluorescence intensity of TO-PRO-1, ethidium bromide, and SYBR green II increases in the presence of 1 while the fluorescence intensity of X2S decreases. Table 1 summarizes the EC50 for 1, or the concentration of 1 at which the half maximal change in fluorescence is observed, and the sensitivity, defined as the percentage change in fluorescence intensity at the EC50, for all four dyes. In summary, TO-PRO-1 binds 1 with the lowest EC50 (550 nM) and with excellent sensitivity (~450%). The EC50’s for ethidium bromide and X2S are slightly higher (1400 and 850 nM, respectively), however, their sensitivities are significantly decreased (~9-fold). Although the sensitivity of SYBR green II is the highest of the four dyes, no saturable binding to 1 was observed. (Please also see Supplementary Figure S1.)
Table 1.
EC50 and sensitivities of dyes for RNA library 1. The EC50 is the concentration of 1 required to observe half maximal change in fluorescence intensity. Sensitivity is the percentage change in fluorescence intensity at the EC50.
| Dye | EC50 (nM) | Sensitivity |
|---|---|---|
| TO-PRO-1 | 550 | ~450% |
| SYBR green II | >50,000 | >4,000% |
| Ethidium bromide | 1400 | ~60% |
| X2S | 850 | ~50% |
The fluorescence properties of dyes can also be affected by the presence of small molecules. We therefore completed two sets of experiments: dye was incubated individually with members of a small molecule library; and, dye was incubated individually with members of a small molecule library and 1. The results of these experiments are shown in Supplementary Figures S2–S5. Even though X2S and ethidium bromide bind 1 with relatively low EC50’s, their emission properties are significantly affected by the presence of small molecules, decreasing the signal to noise ratio. SYBR green II is less affected by the presence of small molecules globally; however, this is not the case for a subset of the compounds screened. In contrast, there is very little variability in the fluorescence intensity of TO-PRO-1 in the presence of small molecules. Thus, TO-PRO-1 was used to identify the features in RNA motifs and small molecules that impart binding affinity and selectivity.
Solution-Based High Throughput Screening
The general strategy of the HTS approach is illustrated in Figure 1. Three RNA motif libraries were used that include a 4,096-member 6-nucleotide hairpin library (1), a 1,024-member 3 × 2-nucleotide asymmetric internal loop library (2), and a 65,536-member 4 × 4-nucleotide symmetric internal loop library (3)(Figure 2). Each of these RNA motif libraries was chosen because they display small RNA motifs that are highly abundant in cellular RNAs. Thus, the specific small molecule-RNA motif partners identified from this approach could be utilized as lead ligands to modulate RNA function, provided a biologically important RNA that contains a targetable motif can be identified. Advances in RNA structure prediction and annotation from sequence provide relatively simple approaches to identify targetable motifs that are present in RNAs 31–33.
In our initial studies, the three RNA motif libraries (1 – 3, Figure 2) were screened for binding to small molecules that are biased for binding to RNA by using a TO-PRO-1 displacement assay in 384-well plate (Figure 1a). Previously, it was shown that chemical similarity-based virtual screening can be used to define a population of small molecules that are biased for binding to RNA 34,35. Various studies have identified privileged scaffolds that bind RNA including benzimidazoles 22,24,34,36–39, pentamidine 35,40–43, and 4′,6-diamidino-2-phenylindole (DAPI) 23,34,41, some of which are bioactive. Thus, a library of small molecules based on these scaffolds and therefore biased for binding RNA was constructed (43 compounds; Supplementary Table S1) 22,23. The compounds were generally restricted to contain an amino or imino group such that they could be anchored site-specifically onto aldehyde-functionalized agarose microarrays for selection of RNA motifs 21,44,45.
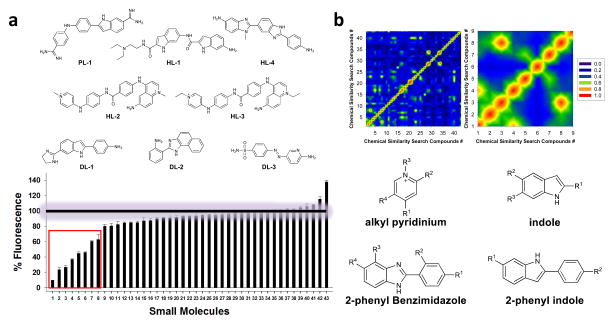
The RNA libraries were incubated with TO-PRO-1, and then the ligand of interest was added at 100 μM. If the ligand binds the RNA target, then a decrease in fluorescence intensity is observed due to dye displacement. Using this method, eight compounds that bind 1 – 3 were identified, affording a hit rate of 19% (Figure 3a). One compound is similar to pentamidine (pentamidine-like, PL; PL-1), four compounds are Hoechst-like (HL; HL-1, HL-2, HL-3 and HL-4), and three compounds are DAPI-like (DL; DL-1, DL-2 and DL-3) (Figure 3a). In order to ensure that ligands were not aggregating (forming self-structure) and then displacing TO-PRO-1, the screen was repeated in the presence of Igepal detergent. No significant change in TO-PRO-1 displacement was observed for all compounds except for two, indicating that ligand aggregation is generally not occurring (Supplementary Figure S6). (Neither of the two compounds was one of the four ligands for which the preferred RNA motif space was determined.)
Figure 3.
Solution-based screen and chemoinformatic analysis of the hit compounds. a, solution-based screen identifies small molecules that bind 1 – 3. Top, the structures of the top eight ligands identified. Bottom, data for screening the entire library for binding 1. A “% Fluorescence” of 100% indicates that the compound does not displace TO-PRO-1 while “% Fluorescence” below 100% indicates that the small molecule binds the RNA and displaces TO-PRO-1. b, Chemoinformatic analysis of the hit compounds. Top left, two dimensional plot of the Tanimoto scores for each small molecule as compared to every library member. Top, right, a close-up view of the plot to the left for the top eight hit compounds. Bottom, results of common scaffold analysis of the top eight hit ligands reveals features in chemical space that facilitate RNA binding.
In order to assess the quality of the hit rate of our RNA-focused library relative to a library that is unbiased for binding RNA and to demonstrate the scalability of the screening method, we applied the TO-PRO-1 displacement assay to the 1280-member LOPAC (Supplementary Figures S7 – S9). The LOPAC library provided only 13 hits, or a hit rate of 1.0%. Thus, by using a library enriched in chemotypes for binding RNA, a 19-fold increase in the hit rate is obtained.
Chemoinformatic Analysis Identifies Privileged Chemotypes
The chemotypes in the entire library of compounds, including the eight hit compounds, were then analyzed for structural similarity (Figure 3b). The analysis was completed by comparing the shape-based similarity (Tanimoto) scores 46 of every compound to each other. Tanimoto scores range from 0 – 1.0 and quantitatively assign the shape similarity between two compounds (where a score of 1.0 indicates complete shape similarity).
When considering the entire library, some compounds are highly similar to each other; however, most have only modest or moderate similarity. For the hit compounds, there is an increase in their similarity to each other. The average Tanimoto similarity score of each compound in the library to all the others is 0.29 ± 0.12, while the average Tanimoto similarity score between the eight hit compounds is 0.43 ± 0.15. Thus, there is a slight enrichment in the features that are similar within the hit compounds; that is, the hit compounds are more similar to each other than the entire library is similar to itself.
In addition, the structures of the eight hit compounds (Figure 3a) were analyzed for common scaffolds according to the method of Clark and Labute 47. The common scaffolds include indole, 2-phenyl indole, 2-phenyl benzimidazole, and pyridinium groups (Figure 3b). Information on the chemotypes that are biased for selective recognition of RNA is important for the development of RNA-focused small molecule libraries. Thus, small molecule collections that contain chemotypes that allow for selective RNA recognition should have a higher hit rate when screened for binding to RNA targets compared to non-RNA focused libraries, such as LOPAC.
Microarray-based Selection of RNA-Ligand Interactions
The solution-based screen identifies lead small molecules that bind RNA. However, hit compounds can be selected that bind to the randomized region or to the constant regions in the RNA motif libraries (Figure 2). In order to identify the small molecules that bind to the randomized region in the RNA, a secondary screening assay is completed. In this secondary assay, the eight hit compounds from the TO-PRO-1 displacement assay were conjugated onto a microarray surface and probed for binding to RNA motifs under highly stringent conditions.
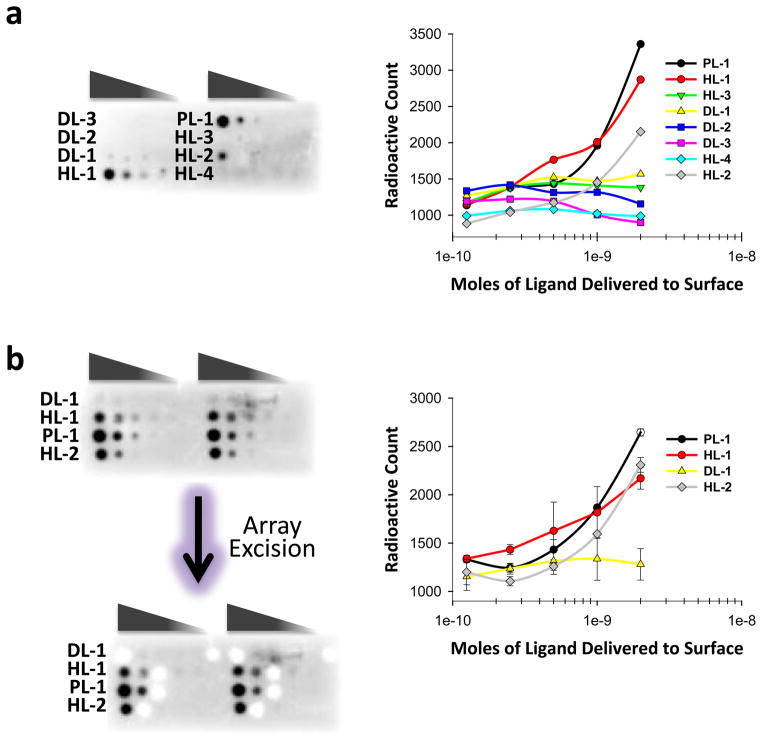
Specifically, arrays were incubated simultaneously with 32P-labeled RNA motif libraries 1–3 in the presence of a large excess (>1000-fold) of unlabeled competitor oligonucleotides 4 – 8 and d(AT)11 and d(GC)11 (Figure 2). Thus, the RNA motifs that bind to each small molecule are selected, and small molecules that bind the constant regions are eliminated. Good signal is observed for PL-1, HL-1, HL-2, and DL-1, indicating that they bind specifically to the randomized regions in 1 – 3 even under highly stringent conditions (Figure 4a). In contrast, HL-3, DL-2, DL-3 and HL-4 failed to give signal over background (Figure 4a). These compounds are unable to bind the randomized regions under highly stringent conditions and likely bind to the constant regions.
Figure 4.
Multidimensional combinatorial screening (MDCS) selection of RNA motifs that bind to small molecules. a, image of a microarray displaying the eight hit compounds after hybridization with radioactively labeled 1 – 3 and unlabeled 4 – 8 (Figure 2) 20,21,48 and a plot of the corresponding data. Only PL-1, HL-1, HL-2, and DL-1 bind to 1 – 3 under these highly stringent conditions and were subjected to microarray-based selections. b, image of a microarray displaying PL-1, HL-1, HL-2, and DL-1 after hybridization with radioactively labeled 1–3 and unlabeled competitors 4 – 8 and a plot of the corresponding data. The error bars are the standard deviations.
To ensure the interactions of PL-1, HL-1, HL-2, and DL-1 are specific to the RNA libraries, arrays displaying PL-1, HL-1, HL-2, and DL-1 were hybridized with 1 in the presence of a large excess of tRNA (500 times the number of moles of compound delivered to the surface and 8,000 times the concentration of 1; Supplementary Figure S10). Excellent signal was still observed for all compounds. Subsequently, compounds PL-1, HL-1, HL-2, and DL-1 were subjected to microarray-based selection using the oligonucleotides competitors described above (Figure 2) 20,21,48. Three rounds of selection were completed prior to sequencing the RNA motifs that bind each ligand. As shown in Figure 4b, RNAs that bind to each compound were cleanly excised from the microarray surface. RT-PCR amplification, cloning, and sequencing identified the bound RNAs.
Identification of Privileged RNA Space
An initial statistical analysis was completed to determine if the small molecules are biased for binding one particular RNA motif library. Due to the differences in the number of library members in each library and biases that may be introduced during RT-PCR amplification or cloning, the starting population of RNAs applied to the microarray surface was subjected to RT-PCR amplification, cloning, and sequencing. The representation of each library in the sequencing data was used as baseline in the statistical analysis of the results from our selection. Our statistical analysis is completed using a population comparison. For example, the proportion of hairpin loops in the sequencing data of the starting library is compared to the proportion of hairpin loops in the selected RNAs. This difference is then used to calculate statistical significance, reported as a Z-score. The results of this analysis are present in Table 2. All four small molecules have a strong bias for binding to RNA hairpins that are derived from 1 (positive Zobs) and a bias against members in libraries 2 and 3 (negative Zobs). These general trends were also observed when the individual selections were analyzed.
Table 2.
Statistical analysis of selected RNAs for binding PL-1, HL-1, HL-2 and DL-1.a
| RNA | All | PL-1 | HL-1 | HL-2 | DL-1 |
|---|---|---|---|---|---|
| Zobs | Zobs | Zobs | Zobs | Zobs | |
| 1 | 6.63 | 5.52 | 6.35 | 4.76 | 3.90 |
| 2 | −3.80 | −2.34 | −4.31 | −3.03 | −1.65 |
| 3 | −5.12 | −3.34 | −3.27 | −2.44 | −3.27 |
| p-value | p value | p-value | p-value | p-value | |
| 1 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 |
| 2 | 0.0001 | 0.0193 | 0.0001 | 0.0024 | 0.0989 |
| 3 | 0.0001 | 0.0008 | 0.0011 | 0.0147 | 0.0011 |
A negative Zobs represents selection against a given interaction (for 2 and 3) whereas a positive Zobs represents selection for a given interaction (for 1). The p-values were computed from Zobs and describe the confidence in the selection for or against an RNA library.
In order to precisely define the RNA motif space preference for each ligand, the selected RNA sequences were analyzed by using a statistical approach 49,50. In this approach, the populations of RNA motifs that comprise the starting library are compiled, and the occurrence rate of each feature in the library is compared to the occurrence rate of that feature in the selected RNA motifs. By comparing these two populations, the relative enrichment for a specific feature in RNA motif space for binding to a ligand can be computed. This enrichment is assigned a statistical significance, or a Z-score and the corresponding two-tailed p-value.
Statistical analysis was completed using a method that we describe as Structure-Activity Relationships Through Sequencing, or StARTS 49,51. Briefly, the RNA-Privileged Space Predictor program (RNA-PSP, v. 2.0) 50 extracts the nucleotides that are derived from the randomized positions in selected RNAs. The sequences and the features within them are analyzed to determine statistical biases relative to the entire library. The individual features are assigned a Z-score. Each RNA can have multiple privileged features for binding a ligand. Thus, the Z-scores for all features that occur with ≥95% confidence are summed to afford the Sum Z-score 49–51. Previous studies have shown that the RNA motifs with the highest Sum Z-scores bind to a ligand with the highest affinity while ones with lower Sum Z-scores bind more weakly 49,51.
A Venn diagram was derived based on the statistically significant features that have a confidence level of ≥99% in RNA hairpin loops selected to bind PL-1, HL-1, HL-2, and DL-1 (when four bases are defined and two are N’s; Supplementary Figure S11). These features most commonly have AU, UA, or AG steps. Compounds PL-1, HL-1, and DL-1 have overlapping RNA hairpin loop space that includes many different orientations of AU or UA steps. RNA motif space that is unique for HL-1, HL-2, and DL-1 generally have G’s in hairpin loops while unique space for PL-1 typically has UU or AA steps.
The Affinities of RNA-Ligand Interactions
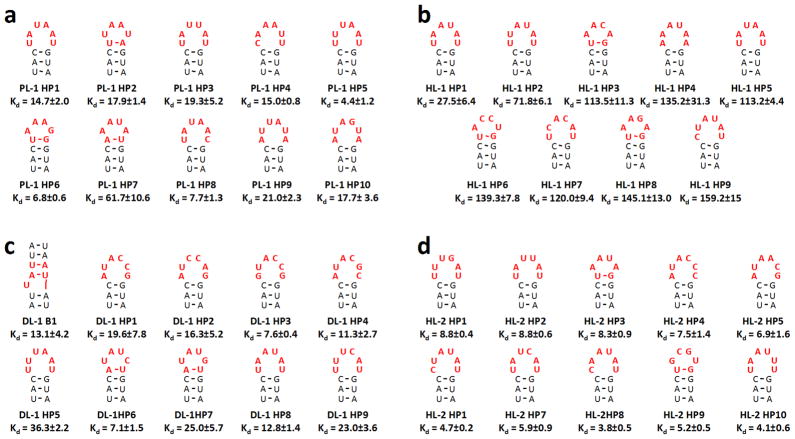
The RNA motifs with the highest Sum Z-scores for binding each ligand are shown in Figure 5 and were studied for binding to the corresponding small molecule ligand. (The secondary structures shown in Figure 5 were predicted by free energy minimization using the RNA structure program 31.) Since each small molecule used in this particular MDCS study is fluorescent, binding constants were determined in solution by measuring the change in fluorescence intensity of the small molecule as a function of RNA concentration. In addition, the affinities of the ligands for the starting libraries were also determined to measure the enrichment in affinity that the selection strategy provides.
Figure 5.
Secondary structures and affinities of the selected RNA motif-ligand interactions. The secondary structures shown were predicted by free energy minimization using the RNA structure program 31. The RNA motif-ligand partners that were subjected to binding assays were predicted to have the highest affinities based on analysis by Structure-Activity Relationships Through Sequencing (StARTS) 49–51. The red letters in the secondary structures indicate nucleotides that are derived from the randomized region of the libraries; the secondary structure shown is from the boxed nucleotides in Figure 2. All dissociation constants (Kd’s) are reported in micromolar. PL-1, HL-1, HL-2, and DL-1 do not bind oligonucleotide 8 (Figure 2; GAAA tetraloop) or the starting libraries 1 – 3 (Kd >200 μM). a, the RNA motifs selected to bind PL-1. b, the RNA motifs selected to bind HL-1. c, the RNA motifs selected to bind DL-1. d, the RNA motifs selected to bind HL-2. In the RNA identifiers, “B” indicates a bulge while “HP” indicates a hairpin. Each experiment was completed in triplicate, and the error bars are the standard deviations.
Saturable binding was observed for each of the selected RNA motifs. The range of affinities observed for each small molecule is different. For example, PL-1, DL-1 and HL-2 have dissociation constants in the low micromolar range while HL-1 binds slightly weaker to its selected RNAs with affinities ranging from 30 to 160 μM (Figure 5). In contrast, no saturable binding was observed for any of the ligands with the starting RNA motif libraries 1–3 (Figure 2), indicating Kd’s ≫ 200 μM. Oligonucleotide 8, which mimics the GAAA hairpin in RNA libraries 2 and 3 (Figure 2 and Supplementary Figures S22 – S25), has a Kd ≫ 200 μM while tRNA has a Kd ≫ 2400 μM(Supplementary Figure S27).
The affinities of the selected RNA motifs for the corresponding small molecules in these studies are weaker than those previously observed for aminoglycoside derivatives used in 2DCS selections 20,21. However, they are similar to those observed for a series of benzimidazoles 51, including a rigid benzimidzaole derivative that binds to and modulates the function of the Hepatitis C Virus (HCV) Internal Ribosomal Entry Site (IRES) 36. This compound binds to an asymmetric internal loop in IRES with mid-micromolar affinity and inhibits propagation 36.
Since our initial screen was completed in solution, it is possible that binding affinity is affected by conjugation to the slide surface. That is, immobilization of the compound through the amino or imino group could affect molecular recognition. Therefore, we studied the binding of a DL-1 derivative that mimics the structure of the compound when displayed on an array surface to four RNAs selected to bind DL-1 (DL-1 B1, DL-1 HP1, DL-1 HP2, and DL1-HP3; Figure 5 and Supplementary Figure S28). The free amine in DL-1 was reacted with benzyaldehyde via reductive amination to afford DL-1 Benzyl. For DL-HP1 and DL-HP2, the affinities for DL-1 and DL-1 Benzyl are the same or within error. The change in the affinities of DL-1 B1 and DL-1 HP2 for the two compounds is modest (13±4 and 37±7 μM for DL-1 B1; 8±1 and 34±6 μM for DL-1 HP3). Similar results were previously observed for the RNA motifs that prefer to bind 6′-N-5-hexynoate kanamycin A. The affinities of 6′-N-5-hexynoate kanamycin A and kanamycin A were comparable.20
Interestingly, no signal for HL-3 was observed on the microarray despite the fact that it is very similar to HL-2. In order to determine if this observation was an artifact of the microarray selection, the affinity of HL-3 for the RNAs selected for HL-2 were determined as described above. No binding of HL-3 was observed to any of the RNAs, in good agreement with the observations from the microarray experiment. Furthermore, these studies indicate that subtle differences in chemical structure can significantly affect RNA binding affinity.
DISCUSSION
Recent studies have established that RNA plays critical and varied roles in disease. Small molecules that bind these RNAs and modulate function/toxicity could serve as chemical genetics probes or therapeutics. Lead compounds could be developed based on the preferences of small molecules for RNA secondary structural motifs. At present, however, such information is sparse and has hampered the development of compounds targeting RNA.
Herein, we have utilized a modified version of two-dimensional combinatorial screening to quickly identify chemotypes in small molecules that bind RNA and the RNA motif preferences for binding small molecules. There are immediate uses for this information. First, transcriptomes can be mined to identify RNAs that have the targetable motifs identified from our studies. These ligands could then be tested for modulating the function of the corresponding RNAs. Second, chemically diverse small molecule libraries that are biased for binding RNA could be constructed using the privileged chemotypes defined herein. Libraries that are currently screened for binding RNA are generally biased for modulating protein function, thus yielding much lower hit rates for RNA targets. We will use this approach to screen larger, more chemically diverse small molecule libraries, which will undoubtedly define additional chemotypes that impart affinity for RNA and additional RNA motifs that are preferred by small molecules.
METHODS
High Throughput Screening of RNA-Ligand Interactions
Prior to screening, the EC50’s of TO-PRO-1 to the RNA motif libraries were determined. Briefly, the RNA library of interest (1 – 3, Figure 2)was folded in in 1X Screening Buffer 1 (SB1; 8 mM Na2HPO4, pH7.0, 185 mM NaCl, 0.1 mM EDTA) at 60 °C for 5 min followed by slowly cooling to room temperature on the bench top. BSA was then added to a final concentration of 40 μg/mL to afford 1X Screening Buffer 2 (SB2). The RNA was titrated into 100 nM TO-PRO1 prepared in 1X SB2, and the fluorescence intensity was measured after each addition using an excitation wavelength of 485 nm and an emission wavelength of 528 nm. The resulting curves were fit to a one-site binding model. The EC50 are: 1, 544 ± 67 nM; 2,239 ± 38 nM; and 3,558 ± 54 nM.)
The concentration of RNA library that corresponded to the EC50’s for binding TO-PRO-1 was used in the TO-PRO-1 displacement assay. The RNA libraries (1, 2 or 3) were folded as described above. TO-PRO-1 was then added to a final concentration of 100 nM. The RNA/TO-PRO-1 mixture was incubated at room temperature for 15 min. Then, 10 μL of this solution was dispensed into each well of a black 384-well plate (Greiner Low-Volume 784076) using an Aurora Discovery FRD-1B liquid dispenser. A 100 nL aliquot of a 10 mM stock of each small molecule was pinned into each well using Biomek NXP Laboratory Automation Workstation that was equipped with a 384-pin head. The solution was incubated at room temperature for 15 min. Fluorescence intensity was measured on an Envision 2104 Multilabel Plate Reader (Perkin Elmer) with an excitation wavelength of 485/14 nm, an emission wavelength of 528/25, and a 505 nm cut-off mirror. The change in fluorescence was normalized to a percentage response (%Res) according to equation 1 (eq. 1):
| (eq.1) |
where I represents the fluorescence intensity of each sample, Ĩ−ve represents the median of the fluorescence intensity of the negative control raw data, and Ĩ+ve represents the median of the fluorescence intensity of the positive control raw data. Results are summarized in Supplementary Figures S2 – S9.
Chemoinformatic Analysis
The chemotype similarity of every compound compared to each other was determined using shape Tanimoto scores. Shape Tanimoto scores were calculated by Instant JChem (JChem 5.8.0, 2012, ChemAxon, http://www.chemaxon.com). Chemical substructures of the top eight small molecules (Figure 3b) were generated by NCGC Automatic R-group analysis program(Tripod Development; http://tripod.nih.gov/?p=46) 47.
Construction of Small Molecule Microarrays
Microarrays were constructed as previously described 20,21,48–56. Compounds that were identified as hits from the dye-displacement assay were immobilized onto aldehyde-functionalized microarray via a reductive amination reaction. Serial dilutions of compounds were prepared in 75% DMSO in NANOpure water. A 400 nL aliquot of each serial dilution was then spotted onto the surface (five 1:5 dilutions beginning with 5 mM compound). A negative control for non-specific binding of RNA to the slide surface was generated by delivering 400 nL of 75% DMSO in NANOpure water to the slide surface. The spotted microarray was placed in a humidity chamber for 3 h. The resulting imine was reduced with a solution of 4:1 1X phosphate-buffered saline (PBS): ethanol containing 32 mM NaCNBH3 for 3 min at room temperature. Slides were then washed with 0.1% sodium dodecyl sulfate (SDS; 3 × 5 min) and water (5 × 5 min)and allowed to dry to a thin film at room temperature.
RNA Selection Procedures
The RNA libraries (1, 2, 3) were radioactively labeled by runoff transcription using an RNAMaxx transcription kit (Stratagene). Half the concentration of cold ATP per the manufacturer’s protocol and 10 μL of [α-32P]ATP (3000 Ci/mol; PerkinElmer) were used.
Small molecule microarrays were pre-equilibrated with 1X SB2 for 5 min at room temperature. Radioactively labeled internal loop library (1, 2 and 3; 50 pmol each) and competitor oligonucleotides (4–8; 50 nmol each; Figure 2) were annealed separately in 1X SB1 at 60 °C for 5 min and allowed to slowly cool on the bench top. Folded RNAs were mixed together in a total volume of 400 μL, and BSA was added to a final concentration of 40 μg/mL. The mixture was pipetted onto the slide and evenly distributed across the slide surface with a custom-cut sheet of Parafilm. Slides were hybridized at room temperature for 30 min. After the 30 min hybridization period, the slides were washed by submersion in 30 mL of 1X SB2 for 30 min with gentle agitation. This step was repeated three times. Excess buffer was removed from the slide surface, and the slides were left to dry on the bench top.
The arrays were exposed to a phosphorimager screen and imaged using a Typhoon 9410 variable mode imager. The image was used as a template to identify spots that bound RNA and to mechanically remove them from the surface. A 400 nL aliquot of NANOpure water was added to the spot to be excised. After 30 s, excess water was pipetted from the surface (most is absorbed), and the gel at that position was excised.
RT-PCR Amplification
The agarose containing bound RNAs was placed into a thin-walled PCR tube with 16 μL of NANOpure water, 2 μL of 10X RQ DNase I Buffer, and 2 units of RQ DNase I (Promega). The tube was vortexed, centrifuged for 4 min at 8000 × g, and then incubated at 37 °C for 2 h. The reaction was quenched by addition of 2 μL of 10X DNase Stop Solution (Promega), and the sample was incubated at 65 °C for 10 min to inactivate the DNase. This solution was used for reverse transcription-polymerase chain reaction (RT-PCR) amplification, which was completed as previously described 57. Aliquots of the RT-PCR reactions were checked every five cycles starting at cycle 25 on a denaturing 15% polyacrylamide gel stained with ethidium bromide or SYBR Gold (Invitrogen).
Multiple Rounds of Selection
The selection procedure was repeated three times with all eight lead small molecules (PL-1, HL-1, HL-2, HL-3, HL-4, DL-1, DL-2, and DL-3) in one spot to enrich the RNA pool. The final round of selection was completed by spotting the small molecules individually as serial dilutions (Figure 4).
Cloning and Sequencing
The RT-PCR products were cloned into the corresponding site of the pGEM®-T vector. Sequencing was completed by Functional Biosciences, Inc..
Statistical Analysis of Selected RNAs
Analysis of the selected RNA libraries after multiple rounds of selection was completed by calculating Zobs and p-values. In this analysis, the selected RNAs are compared to the RNAs in the entire starting library. The statistical significance parameter Zobs was then calculated according to eq. 2 and eq. 3 49–51:
| (eq.2) |
| (eq.3) |
where n1 is the size of Population 1 (the selected RNAs); n2 is the size of Population 2 (the starting library); p1 is the observed proportion of Population 1 (the selected RNAs)that displays the feature of interest; and p2 is the observed proportion for Population 2 (the starting library) that displays the feature of interest. Zobs is manually converted to the corresponding two tailed p-value, which represents the confidence level that a feature in the selected RNA sequences is preferred by the ligand and did not occur randomly. The number of RNAs in the starting library and the number of selected RNA sequences are summarized in Supplementary Table S2.
Binding Affinity Measurements
Dissociation constants were determined using an in-solution, fluorescence-based assay 20,21,48–51,54,55,58. A selected RNA or RNA mixture was folded as described above. Then, PL-1, HL-1, HL-2 or DL-1 was added to a final concentration of 100 nM, 1000 nM, 1000 nM or 1000 nM, respectively. Serial dilutions (1:2) were then completed in 1X SB2 containing the corresponding concentration of small molecule. The solutions were incubated for 15 min at room temperature and then transferred to a well of a black 96-well plate. Fluorescence intensity was measured using a Bio-Tek FLx800 plate reader. The change in fluorescence intensity as a function of RNA concentration was fit to the following equation (eq. 4)59:
| (eq.4) |
where I is the observed fluorescence intensity;I0 is the fluorescence intensity in the absence of RNA; Δε is the difference between the fluorescence intensity in the absence of RNA and in the presence of infinite RNA concentration and is in units of M−1; [FL]0 is the concentration of compound;[RNA]0 is the concentration of the selected RNA; and Kt is the dissociation constant. Representative binding curves are shown in Supplementary Figures S22 – S28.
Supplementary Material
Acknowledgments
We thank Raman Parkesh for chemical similarity searches. This work was funded by the National Institutes of Health grant R01GM079235. MDD is a Camille and Henry Dreyfus Teacher-Scholar and a Research Corporation Cottrell Scholar.
Footnotes
ADDITIONAL INFORMATION
Supplementary Information accompanies this paper at http://www.nature.com/naturecommunications.
Contributions
M. D. and T. T. conceived and designed the experiments, analysed the data, and co-wrote the paper; and T. T. performed the experiments.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 3.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 4.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat Rev Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 5.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 6.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 7.Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 8.Seetharaman S, Zivarts M, Sudarsan N, Breaker RR. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat Biotechnol. 2001;19:336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- 9.Chow CS, Bogdan FM. A Structural Basis for RNAminus signLigand Interactions. Chem Rev. 1997;97:1489–1514. doi: 10.1021/cr960415w. [DOI] [PubMed] [Google Scholar]
- 10.Gallego J, Varani G. Targeting RNA with small-molecule drugs: therapeutic promise and chemical challenges. Acc Chem Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 11.Joyce GF. In vitro evolution of nucleic acids. Curr Opin Struct Biol. 1994;4:331–336. doi: 10.1016/s0959-440x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 12.Klug SJ, Famulok M. All you wanted to know about SELEX. Mol Biol Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 13.Carlson CB, Vuyisich M, Gooch BD, Beal PA. Preferred RNA binding sites for a threading intercalator revealed by in vitro evolution. Chem Biol. 2003;10:663–672. doi: 10.1016/s1074-5521(03)00147-9. [DOI] [PubMed] [Google Scholar]
- 14.Lato SM, Ellington AD. Screening chemical libraries for nucleic-acid-binding drugs by in vitro selection: a test case with lividomycin. Mol Divers. 1996;2:103–110. doi: 10.1007/BF01718707. [DOI] [PubMed] [Google Scholar]
- 15.Yu L, et al. Discovery of aminoglycoside mimetics by NMR-based screening of Escherichia coli A-site RNA. J Am Chem Soc. 2003;125:4444–4450. doi: 10.1021/ja021354o. [DOI] [PubMed] [Google Scholar]
- 16.Griffey RH, Hofstadler SA, Sannes-Lowery KA, Ecker DJ, Crooke ST. Determinants of aminoglycoside-binding specificity for rRNA by using mass spectrometry. Proc Natl Acad Sci U S A. 1999;96:10129–10133. doi: 10.1073/pnas.96.18.10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swayze EE, et al. SAR by MS: a ligand based technique for drug lead discovery against structured RNA targets. J Med Chem. 2002;45:3816–3819. doi: 10.1021/jm0255466. [DOI] [PubMed] [Google Scholar]
- 18.Mei HY, et al. Inhibition of self-splicing group I intron RNA: high-throughput screening assays. Nucleic Acids Res. 1996;24:5051–5053. doi: 10.1093/nar/24.24.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mei HY, et al. Discovery of selective, small-molecule inhibitors of RNA complexes--I. The Tat protein/TAR RNA complexes required for HIV-1 transcription. Bioorg Med Chem. 1997;5:1173–1184. doi: 10.1016/s0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 20.Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD. A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem Biol. 2007;2:745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- 21.Disney MD, et al. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 22.Parkesh R, et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database & chemical similarity searching. J Am Chem Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A, et al. Chemical correction of pre-mRNA splicing defects associated with sequestration of muscleblind-like 1 protein by expanded r(CAG)-containing transcripts. ACS Chem Biol. 2012;7:496–505. doi: 10.1021/cb200413a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pushechnikov A, et al. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MM, Pushechnikov A, Disney MD. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tse WC, Boger DL. A fluorescent intercalator displacement assay for establishing DNA binding selectivity and affinity. Acc Chem Res. 2004;37:61–69. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]
- 27.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. A simple, high-resolution method for establishing DNA binding affinity and sequence selectivity. J Am Chem Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 28.Asare-Okai PN, Chow CS. A modified fluorescent intercalator displacement assay for RNA ligand discovery. Anal Biochem. 2011;408:269–276. doi: 10.1016/j.ab.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnamurthy M, Schirle NT, Beal PA. Screening helix-threading peptides for RNA binding using a thiazole orange displacement assay. Bioorg Med Chem. 16:8914–8921. doi: 10.1016/j.bmc.2008.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Umemoto S, Nakatani K. Fluorescent indicator displacement assay for ligand-RNA interactions. J Am Chem Soc. 2010;132:3660–3661. doi: 10.1021/ja100089u. [DOI] [PubMed] [Google Scholar]
- 31.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Disney MD, Childs JL, Turner DH. Hoechst 33258 selectively inhibits group I intron self-splicing by affecting RNA folding. Chembiochem. 2004;5:1647–1652. doi: 10.1002/cbic.200400159. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Li Z, Pilch DS, Leibowitz MJ. Pentamidine inhibits catalytic activity of group I intron Ca. LSU by altering RNA folding. Nucleic Acids Res. 2002;30:2961–2971. doi: 10.1093/nar/gkf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seth PP, et al. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J Med Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 37.He YYJ, Wu B, Robinson D, Sprankle K, Kung P-P, Lowery K, Mohan V, Hofstadler S, Swayze EE, Griffey R. Synthesis and evaluation of novel bacterial rRNA-binding benzimidazoles by mass spectrometry. Bioorg Med Chem Lett. 2004;14:695–699. doi: 10.1016/j.bmcl.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 38.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho J, Rando RR. Specific binding of Hoechst 33258 to site 1 thymidylate synthase mRNA. Nucleic Acids Res. 2000;28:2158–2163. doi: 10.1093/nar/28.10.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Tidwell RR, Leibowitz MJ. Inhibition of in vitro splicing of a group I intron of Pneumocystis carinii. J Eukaryot Microbiol. 1994;41:31–38. doi: 10.1111/j.1550-7408.1994.tb05931.x. [DOI] [PubMed] [Google Scholar]
- 41.Miletti KE, Leibowitz MJ. Pentamidine inhibition of group I intron splicing in Candida albicans correlates with growth inhibition. Antimicrob Agents Chemother. 2000;44:958–966. doi: 10.1128/aac.44.4.958-966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Bell A, Perlman PS, Leibowitz MJ. Pentamidine inhibits mitochondrial intron splicing and translation in Saccharomyces cerevisiae. RNA. 2000;6:937–951. doi: 10.1017/s1355838200991726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dufva M, Petronis S, Jensen LB, Krag C, Christensen CB. Characterization of an inexpensive, nontoxic, and highly sensitive microarray substrate. Biotechniques. 2004;37:286–292. 294–296. doi: 10.2144/04372MT02. [DOI] [PubMed] [Google Scholar]
- 45.Afanassiev V, Hanemann V, Wolfl S. Preparation of DNA and protein micro arrays on glass slides coated with an agarose film. Nucleic Acids Res. 2000;28:E66. doi: 10.1093/nar/28.12.e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haigh JA, Pickup BT, Grant JA, Nicholls A. Small molecule shape-fingerprints. J Chem Inf Model. 2005;45:673–684. doi: 10.1021/ci049651v. [DOI] [PubMed] [Google Scholar]
- 47.Clark AM, Labute P. Detection and assignment of common scaffolds in project databases of lead molecules. J Med Chem. 2009;52:469–483. doi: 10.1021/jm801098a. [DOI] [PubMed] [Google Scholar]
- 48.Tran T, Disney MD. Two-dimensional combinatorial screening of a bacterial rRNA A-site-like motif library: defining privileged asymmetric internal loops that bind aminoglycosides. Biochemistry. 2010;49:1833–1842. doi: 10.1021/bi901998m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velagapudi SP, Seedhouse SJ, Disney MD. Structure-activity relationships through sequencing (StARTS) defines optimal and suboptimal RNA motif targets for small molecules. Angew Chem Int Ed Engl. 2010;49:3816–3818. doi: 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paul DJ, Seedhouse SJ, Disney MD. Two-dimensional combinatorial screening and the RNA Privileged Space Predictor program efficiently identify aminoglycoside-RNA hairpin loop interactions. Nucleic Acids Res. 2009;37:5894–5907. doi: 10.1093/nar/gkp594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velagapudi SP, Seedhouse SJ, French J, Disney MD. Defining the RNA internal loops preferred by benzimidazole derivatives via 2D combinatorial screening and computational analysis. J Am Chem Soc. 2011;133:10111–10118. doi: 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seedhouse SJ, Labuda LP, Disney MD. The Privileged Chemical Space Predictor (pcsp): A computer program that identifies privileged chemical space from screens of modularly assembled chemical libraries. Bioorg Med Chem Lett. 2010;20:1338–1343. doi: 10.1016/j.bmcl.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Disney MD, Childs-Disney JL. Using selection to identify and chemical microarray to study the RNA internal loops recognized by 6′-N-acylated kanamycin A. Chembiochem. 2007;8:649–656. doi: 10.1002/cbic.200600569. [DOI] [PubMed] [Google Scholar]
- 54.Aminova O, Paul DJ, Childs-Disney JL, Disney MD. Two-dimensional combinatorial screening identifies specific 6′-acylated kanamycin A- and 6′-acylated neamine-RNA hairpin interactions. Biochemistry. 2008;47:12670–12679. doi: 10.1021/bi8012615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Childs-Disney JL, Disney MD. A simple ligation-based method to increase the information density in sequencing reactions used to deconvolute nucleic acid selections. RNA. 2008;14:390–394. doi: 10.1261/rna.576908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Labuda LP, Pushechnikov A, Disney MD. Small molecule microarrays of RNA-focused peptoids help identify inhibitors of a pathogenic group I intron. ACS Chem Biol. 2009;4:299–307. doi: 10.1021/cb800313m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bevilacqua JM, Bevilacqua PC. Thermodynamic Analysis of an RNA Combinatorial Library Contained in a Short Hairpin. Biochemistry. 1998;37:15877–15884. doi: 10.1021/bi981732v. [DOI] [PubMed] [Google Scholar]
- 58.Tran T, Disney MD. Molecular recognition of 6′-N-5-hexynoate kanamycin A and RNA 1x1 internal loops containing CA mismatches. Biochemistry. 2011;50:962–969. doi: 10.1021/bi101724h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Rando RR. Specific binding of aminoglycoside antibiotics to RNA. Chem Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.