Abstract
Organic anion transporter-1 (OAT1) mediates the body’s disposition of a diverse array of environmental toxins and clinically important drugs. Therefore, understanding the regulation of this transporter has profound clinical significance. We had previously established that OAT1 undergoes constitutive internalization from and recycling back to the cell surface and that acute activation of protein kinase C (PKC) inhibits OAT1 activity by reducing OAT1 cell-surface expression through accelerating its internalization from cell surface to intracellular compartments. However, the underlying mechanisms are poorly understood. In the current study, we provide novel evidence that acute activation of PKC significantly enhances OAT1 ubiquitination both in vitro and ex vivo. We further show that ubiquitination of cell-surface OAT1 increases in cells transfected with dominant negative mutant of dynamin-2, a maneuver blocking OAT1 internalization, which suggests that OAT1 ubiquitination proceeds before OAT1 internalization. Mass spectroscopy has revealed that ubiquitination of OAT1 consists of polyubiquitin chains, primarily through lysine 48 linkage. Transfection of cells with the dominant negative mutant of ubiquitin Ub-K48R, which prevents the formation of Lys48-linked polyubiquitin chains, abolishes PKC-stimulated OAT1 ubiquitination and internalization. Together, our findings demonstrate for the first time that Lys48-linked polyubiquitination is essential for PKC-regulated OAT1 trafficking.
Introduction
The organic anion transporter (OAT) family mediates the body’s disposition of a diverse array of environmental toxins and clinically important drugs, including anti-HIV therapeutics, antitumor drugs, antibiotics, antihypertensives, and anti-inflammatories (You, 2002; Dantzler and Wright, 2003; Srimaroeng et al., 2008; Ahn and Nigam, 2009; VanWert et al., 2010). Therefore, understanding the regulation of these transporters has profound clinical significance.
Ten OATs (OAT1–10) have been cloned, and their expressions have been identified in distinct tissues and cell membranes. In the kidney, OAT1 and OAT3 use a tertiary transport mechanism to move organic anions across the basolateral membrane into the proximal tubule cells for subsequent exit across the apical membrane into the urine for elimination. Through this tertiary transport mechanism, Na+/K+-ATPase maintains an inwardly directed (blood-to-cell) Na+ gradient. The Na+ gradient then drives a sodium dicarboxylate cotransporter, sustaining an outwardly directed dicarboxylate gradient that is used by a dicarboxylate/organic anion exchanger—namely, OAT—to move the organic anion substrate into the cell. This cascade of events indirectly links organic anion transport to metabolic energy and the Na+ gradient, allowing the entry of a negatively charged substrate against both its chemical concentration gradient and the electrical potential of the cell (You, 2002; Dantzler and Wright, 2003; Srimaroeng et al., 2008; Ahn and Nigam, 2009; VanWert et al., 2010).
All of the cloned OATs share several common structural features, including 12 transmembrane domains flanked by intracellular amino and carboxyl termini, multiple glycosylation sites localized in the first extracellular loop, and multiple potential phosphorylation sites. Investigations by our laboratory on the structure–function relationship of OATs have revealed that glycosylation is necessary for the targeting of these transporters to the plasma membrane (Tanaka et al., 2004).
The amount of OATs at the cell surface is critical for their drug transport activity. We previously showed that members of OAT family constitutively internalize from and recycle back to the cell surface, and that inhibition of OAT activity by acute activation of protein kinase C (PKC) results from an accelerated internalization of these transporters from the cell surface to intracellular compartments without affecting the total expression of the transporters (Zhang et al., 2008). However, the effect of the mechanisms of PKC on OAT internalization and function is largely unknown. PKC-induced direct phosphorylation has been reported for other membrane proteins, yet our results showed that a range of PKC activators failed to elevate the phosphorylation level of OATs under various experimental conditions (You et al., 2000). This suggests that direct phosphorylation of OATs is unlikely to be the cause for PKC-induced inhibition of OAT activity.
Recently, modification of receptors and channels by ubiquitin conjugation has emerged as the major regulatory mechanism of internalization, intracellular sorting, and turnover of these membrane proteins (Miranda et al., 2005; Kumar et al., 2007; Zhou et al., 2007; Varghese et al., 2008; Bomberger et al., 2009). Ubiquitin moiety can be recognized by the components of plasma membrane internalization and endosomal sorting machinery.
Ubiquitin is a highly conserved 76-amino-acid protein that forms an isopeptide bond between its C-terminal glycine and a lysine residue on the target protein. Each ubiquitin moiety harbors seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63), allowing for the formation of ubiquitin chains linked through its internal lysine residues. Therefore, a substrate can be modified by different types of ubiquitin conjugation: monoubiquitination (conjugation of one single ubiquitin to one single lysine on the substrate), multiubiquitination (conjugation of several monoubiquitin molecules to multiple lysine residues on the substrate), and polyubiquitination (extended polyubiquitin chain). In addition, a polyubiquitin chain can bear different linkages such as Lys48 linkage (Lys48 in the ubiquitin serves as a base for chain elongation) and Lys63 linkage (Lys63 in the ubiquitin serves as a base for chain elongation). It has been shown that different types and linkages of ubiquitination have different physiologic outcomes for the ubiquitinated substrate (Kolodziejski et al., 2002; Miranda et al., 2005; Kumar et al., 2007; Zhou et al., 2007; Bomberger et al., 2009). Our study investigated the role of ubiquitination in PKC-regulated OAT1 trafficking.
Materials and Methods
Membrane impermeable biotinylation reagent NHS-SS-biotin and streptavidin agarose beads were purchased from Pierce (Rockford, IL). COS-7 cells and Dynamin (Dyn)-2/K44A mutant were purchased from the American Type Culture Collection (Manassas, VA). cDNA for hemagglutinin (HA)-tagged mutant ubiquitin HA-UbΔG was generously provided by Dr. N. Tony Eissa of the Department of Medicine, Baylor College of Medicine, Houston, TX. cDNAs for HA-tagged wild-type ubiquitin and HA-tagged ubiquitin mutants HA-K48R and HA-K63R were generously provided by Dr. Cam Patterson of the Carolina Cardiovascular Biology Center, University of North Carolina (Chapel Hill, NC). Mouse anti-myc antibody (9E10) and mouse anti-HA antibody 12CA5 were purchased from Roche Diagnostics Corporation (Indianapolis, IN). Mouse anti-ubiquitin antibody P4D1 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-ubiquitin antibody FK2 was purchased from Enzo Life Sciences (Farmingdale, NY). PKC activator phorbol 12-myristate 13-acetate (PMA) and all other reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Cultures.
COS-7 cells stably expressing human OAT1 (tagged with epitope myc at its carboxyl terminus for immunodetection) (Zhang et al., 2008) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 0.2 mg/ml G418, 10% fetal bovine serum, penicillin/streptomycin (100 U/ml), and glucose (100 mg/ml).
Ubiquitination Assay.
Cells expressing OAT1-myc were treated with dimethyl sulfoxide (DMSO) or 1 μM PMA at 37°C for indicated time periods. Treated cells were lysed with lysis buffer I (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 5 mM EDTA, and 1 mM NaF) freshly added with 1% of proteinase inhibitor cocktail and 20 mM N-ethylmaleimide (a deubiquitination inhibitor). OAT1 was then immunoprecipitated with anti-myc antibody, followed by immunoblotting with anti-ubiquitin antibody P4D1 or anti-myc antibody 9E10.
Preparation of Rat Kidney Slices.
Male Sprague-Dawley rats were euthanized by CO2 inhalation, and the kidneys were immediately placed into freshly oxygenated ice-cold saline. Tissue slices (<0.5 mm; 5–10 mg wet weight) were cut with a Stadie-Riggs microtome and maintained in ice-cold modified Cross and Taggart saline (95 mM NaCl, 80 mM mannitol, 5 mM KCl, 0.74 mM CaCl2, and 9.5 mM Na2PO4, pH 7.4) for ubiquitination studies.
Cell Surface Biotinylation.
The cell-surface expression level of OAT1 was examined using the membrane-impermeable biotinylation reagent NHS-SS-biotin. Cells were plated in six-well plates. Each well of cells was incubated with 1 ml of NHS-SS-biotin (0.5 mg/ml in phosphate-buffered saline [PBS]) in two successive 20-minute incubations on ice with very gentle shaking. The reagent was freshly prepared each time. After biotinylation, each well was briefly rinsed with 3 ml of PBS/CM containing 100 mM glycine then incubated with the same solution for 30 minutes on ice to ensure complete quenching of the unreacted NHS-SS-biotin. The cells were then lysed on ice for 1 hour in 400 μl of lysis buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100 with 1/100 protease inhibitor cocktail). The cell lysates were cleared by centrifugation at 16,000g at 4°C. We then added 50 μl of streptavidin-agarose beads to the supernatant to isolate cell membrane proteins. OAT1 was detected in the pool of surface proteins by immunoblotting using an anti-myc antibody 9E10.
To determine ubiquitinated OAT1 in the biotinylated protein fraction, cell-surface proteins were biotinylated. OAT1 was immunoprecipitated with anti-myc antibody. Immunoprecipitated proteins were eluted from the beads with 1% SDS (37°C, 15 minutes). Next, SDS was diluted to 0.1%, and the eluted OAT1 proteins were incubated with streptavidin-agarose beads, followed by immunoblotting using anti-ubiquitin antibody P4D1 or anti-myc antibody.
Internalization Assay.
For the internalization assay, we followed the procedure previously established in our laboratory (Zhang et al., 2008). OAT1-expressing cells underwent biotinylation with 0.5 mg/ml sulfo-NHS-SS-biotin at 4°C. Following biotinylation, OAT1 internalization was initiated by incubating the cells (37°C) in PBS containing either 1 μM PMA or vehicle for designated periods of time. Residual cell-surface biotin was stripped by incubating cells 3 times for 20 minutes with freshly prepared 50 mM MesNa in NT buffer (150 mM NaCl, 1 mM EDTA, 0.2% bovine serum albumin, 20 mM Tris, pH 8.6). Cells were lysed in lysis buffer with protease inhibitor cocktail. Biotinylated (internalized) proteins were separated from nonbiotinylated proteins by streptavidin pull-down from equivalent amounts of cellular proteins, followed by immunoblotting with anti-myc antibody.
Purification of Ubiquitinated OAT1 by Sequential Immunoprecipitation.
Six 10-cm-diameter dishes of COS-7 cells expressing OAT1 were treated with 1 μM PMA for 30 minutes at 37°C. Treated cells were solubilized with lysis buffer I with 3% proteinase inhibitor cocktail and 40 mM N-ethylmaleimide. The total cell lysate was precleared with protein G-agarose for 2 hours at 4°C, followed by immunoprecipitation with anti-myc antibody that was cross-linked to protein G beads with dimethyl pimelimidate (Pierce). After an extensive wash with lysis buffer I containing 500 mM NaCl, the immunoprecipitated proteins were eluted with 1% SDS and immunoprecipitated again with anti-ubiquitin antibody FK2, which was cross-linked to protein G beads with dimethyl pimelimidate. After the second immunoprecipitation, 10% of precipitated proteins were analyzed by immunoblotting with anti-ubiquitin antibody P4D1 or anti-myc antibody, and 90% by Coomassie Blue staining.
Protein Identification by Liquid Chromatography–Tandem Mass Spectrometry.
The gel bands of interest were reduced, carboxymethylated with iodoacetamide, digested with trypsin. Peptides were extracted, solubilized in 0.1% trifluoroacetic acid, and analyzed by nano-liquid chromatography–tandem mass spectrometry (LC-MS/MS) using a rapid separation liquid chromatography system (Dionex, Sunnyvale, CA) interfaced with a Velos-LTQ-Orbitrap (Thermo Fisher Scientific, San Jose, CA). Samples were loaded onto a self-packed 100 μm × 2 cm trap packed with Magic C18AQ, 5-μm 200 A (Michrom Bioresources Inc., Auburn, CA) and washed with buffer A (0.2% formic acid) for 5 minutes with flow rate of 10 μl/min. The trap was brought in line with the homemade analytical column (Magic C18AQ, 3 μm 200 A, 75 μm × 50 cm), and peptides were fractionated at 300 nl/min with a 90-minute linear gradient of 2 to 45% buffer B (0.2% formic acid, acetonitrile). Mass spectrometry data were acquired using a data-dependent acquisition procedure, with a cyclic series of a full scan acquired in Orbitrap with resolution of 60,000 followed by MS/MS scans (acquired in linear ion trap) of 20 most intense ions with a repeat count of two and the dynamic exclusion duration of 60 seconds.
The LC-MS/MS data were searched against the ENSEMBL human database using a local version of the Global Proteome Machine (GPM USB; Beavis Informatics Ltd, Winnipeg, MB, Canada) with carbamidoethyl on cysteine as the fixed modification and ubiquitination on lysine (+114 Da) as well as oxidation of methionine and tryptophan as the variable modifications using a 10-ppm precursor ion tolerance and a 0.4-Da fragment ion tolerance. Polyubiquitin sites were identified on ubiquitin lysine residues.
Electrophoresis and Immunoblotting.
Protein samples were resolved on 7.5% SDS-PAGE minigels and electroblotted on to polyvinylidene difluoride membranes. The blots were blocked for 1 hour with 5% nonfat dry milk in PBS–0.05% Tween-20, washed, and incubated for 1 hour at room temperature with appropriate primary antibodies followed by horseradish peroxidase–conjugated secondary antibodies. The signals were detected by the SuperSignal West Dura Extended Duration Substrate kit (Pierce). Nonsaturating, immunoreactive protein bands were quantified by scanning densitometry with the FluorChem 8000 imaging system (Alpha Innotech Corporation, San Leandro, CA).
Data Analysis.
Each experiment was repeated a minimum of three times. The statistical analysis was from multiple experiments. Statistical analysis was performed using Student’s paired t tests. P < 0.05 was considered statistically significant.
Results
PKC-Induced Ubiquitination of OAT1 by Endogenous Ubiquitin.
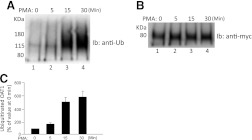
To examine whether OAT1 undergoes ubiquitination, COS-7 cells stably expressing OAT1 were treated with or without PMA, a PKC activator, for different periods of time. OAT1 was then immunoprecipitated by anti-myc antibody (myc was tagged to OAT1), followed by immunoblotting with anti-ubiquitin antibody. As shown in Fig. 1A, OAT1 ubiquitination was barely detectable under the basal condition. Interestingly, PMA treatment induced a time-dependent ubiquitination of the transporter with maximum ubiquitination around 15 to 30 minutes. Moreover, when the immunoblot was reprobed with anti-myc antibody, it was shown that these short-term treatments did not produce any major change in the amount of OAT1 (Fig. 1B). The PMA-induced OAT1 ubiquitination was blocked by staurosporine, a general PKC inhibitor (Fig. 2A), without affecting the total amount of OAT1 (Fig. 2B), demonstrating the specific involvement of PKC in OAT1 ubiquitination. The ubiquitin-immunoreactive signal displayed a smeary band centered ∼140 kDa, ∼60 kDa larger than the size of OAT1 (∼80 kDa). Given that each ubiquitin molecule is ∼8 kDa, OAT1 most likely is either multiubiquitinated or polyubiquitinated.
Fig. 1.
Time dependence of PMA-induced OAT1 ubiquitination in COS-7 cells. (A) OAT1-expressing COS-7 cells were treated with PKC-activator PMA (1 μM) for the time periods as indicated. Treated cells were then lysed, and OAT1 was immunoprecipitated with anti-myc antibody, followed by immunoblotting (Ib) with anti-ubiquitin antibody. (B) the same immunoblot as seen in A was reprobed by anti-myc antibody. (C) densitometry plot of results from (A) as well as from other experiments. The values are mean ± S.E. (n = 3).
Fig. 2.
PKC specificity of OAT1 ubiquitination in COS-7 cells. (A) OAT1-expressing COS-7 cells were treated with PKC-activator PMA (1 μM) in the presence or absence of the PKC-inhibitor staurosporine (St, 2 μM) for 30 minutes. The treated cells were then lysed, and OAT1 was immunoprecipitated with anti-myc antibody, followed by immunoblotting (Ib) with anti-ubiquitin antibody. (B) the same immunoblot as seen in (A) was reprobed by anti-myc antibody. (C) densitometry plot of results from (A) as well as from other experiments. The values are mean ± S.E. (n = 3).
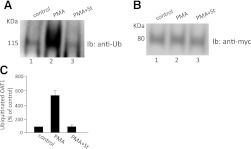
A similar experiment was then performed with rat kidney slices (Fig. 3). Upon PMA treatment, dramatic increase of OAT1 ubiquitination was detected. Again, such ubiquitination was blocked by PKC inhibitor staurosporine (Fig. 3A). These results suggest that OAT1, regardless of species origin, undergoes PKC-regulated ubiquitination both in cultured cells and in rat kidney tissue.
Fig. 3.
PKC specificity of OAT1 ubiquitination in the rat kidney. (A) rat kidney slices were treated with PKC-activator PMA (1 μM) in the presence or absence of the PKC-inhibitor staurosporine (St, 2 μM) for 30 minutes. The treated slices were then lysed, and OAT1 was immunoprecipitated with anti-OAT1 antibody, followed by immunoblotting (Ib) with anti-ubiquitin antibody P4D1. (B) the same immunoblot as seen in (A) was reprobed by anti-OAT1 antibody. (C) densitometry plot of results from (A) as well as from other experiments. Values are mean ± S.E. (n = 3).
PKC-Induced Ubiquitination of OAT1 by Ectopically Expressed Ubiquitin.
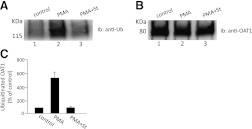
OAT1-expressing cells were transiently transfected with HA-tagged ubiquitin. Transfected cells were treated with PMA in the presence and absence of PKC inhibitor staurosporine. OAT1 was then immunoprecipitated, followed by immunoblotting with anti-HA antibody (to detect HA-tagged ubiquitin). The HA immunoreactivity was dramatically increased in the samples treated with PMA, and was significantly decreased in the presence of staurosporine (Fig. 4A), indicating that the conjugation of HA-ubiquitin to OAT1 is PKC-dependent.
Fig. 4.
PMA-induced OAT1 ubiquitination by ectopically expressed ubiquitin. (A) OAT1-expressing COS-7 cells were transiently transfected with HA-tagged ubiquitin. The transfected cells were treated with PMA (1 μM) in the presence or absence of the PKC-inhibitor staurosporine (St, 2 μM) for 30 minutes. OAT1 was then immunoprecipitated by anti-myc antibody, followed by immunoblotting (Ib) with anti-HA antibody. (B) the same immunoblot as seen in A was reprobed by anti-myc antibody. (C) densitometry plot of results from (A) as well as from other experiments. The values are mean ± S.E. (n = 3).
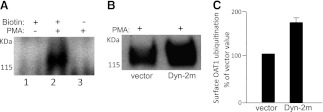
OAT1 Ubiquitination at the Cell Surface.
To determine whether OAT1 ubiquitination occurs at the cell surface, we developed a two-step procedure to ensure that only cell-surface OAT1 ubiquitination was detected (Fig. 5A). Briefly, cell surface proteins were labeled with or without the membrane-impermeable reagent NHS-SS-biotin. Labeled cells were lysed. OAT1 was then immunoprecipitated, with subsequent streptavidin pull-down to isolate cell surface OAT1. Immunoblotting of this streptavidin pull-down with anti-ubiquitin antibody showed no signal for ubiquitin in cells not labeled with biotin. However, in cells labeled with biotin, the ubiquitin signal dramatically increased in the presence of PMA. This result demonstrated that the two-step procedure detected only cell surface OAT1 ubiquitination.
Fig. 5.
Cell-surface OAT1 ubiquitination. (A) cell-surface proteins of OAT1-expressing cells were labeled with or without the membrane-impermeable reagent biotin. The cells were treated with or without PMA for 30 minutes. The labeled cells were lysed. OAT1 was then immunoprecipitated with subsequent streptavidin pull-down to isolate cell-surface OAT1, followed by immunoblotting with anti-ubiquitin antibody. (B) OAT1-expressing cells were transfected with the cDNA encoding dominant negative mutant of dynamin-2 (Dyn-2m, empty vector as control). The transfected cells were treated with PMA (1 μM) for 30 minutes. Cell-surface OAT1 was isolated by cell-surface biotinylation, as described in (A), followed by immunoblotting with anti-ubiquitin antibody. (C) densitometry plot of results from (B) as well as from other experiments. Surface OAT1 in PMA-treated cells was expressed as the percentage of surface OAT1 in control cells. The values are mean ± S.E. (n = 3).
Using this procedure, we next examined the effect of a dominant negative mutant of dynamin-2 (Dyn-2m) on OAT1 ubiquitination. We previously had demonstrated that OAT1 internalization occurred partly through a Dyn- and clathrin-dependent pathway and that such internalization was blocked in cells transfected with a Dyn-2m. As a result, OAT1 accumulated at the cell surface (Zhang et al., 2008). In the current experiment, we transfected OAT1-expressing cells with Dyn-2m (or empty vector as control). Transfected cells were treated with PMA for 30 minutes, and cell surface proteins were biotinylated, followed by the same procedure shown in Fig. 5A for the detection of cell-surface OAT1 ubiquitination. Our results showed that ubiquitinated OAT1 was significantly elevated in cells transfected with Dyn-2m (Fig. 5, B and C), a maneuver that blocked OAT1 internalization. These data indicate that OAT1 is preferentially ubiquitinated when present at the cell surface and that ubiquitination of OAT1 precedes its internalization.
Identification of Ubiquitination Linkage of OAT1.
As mentioned previously, different types of ubiquitination (mono- versus polyubiquitination via different types of linkages) have different physiologic outcomes for the ubiquitinated substrate. To identify the ubiquitination type of OAT1, we employed a sequential immunoprecipitation procedure to purify ubiquitinated OAT1. Briefly, we treated OAT1-expressing cells with the PKC activator PMA for 30 minutes. Treated cells were then lysed. OAT1 was immunoprecipitated, followed by a second immunoprecipitation using ubiquitin-specific antibody to purify ubiquitinated OAT1 (normal IgG as negative control). The ubiquitinated OAT1 was then separated on SDS-PAGE, followed by Coomassie Blue-staining. A Coomassie Blue-stained broad band centered ∼140 kDa was dissected for mass spectrometric analysis, and a sufficient amount of information regarding the peptides that originated from ubiquitin sequences was obtained. Among various peptides identified from ubiquitin, which cover all its seven lysine residues, only Lys48-linked ubiquitin was detected (Table 1). These results indicate that OAT1 is ubiquitinated mainly through Lys48-linked polyubiquitin chains.
TABLE 1.
Ubiquitinated peptides of ubiquitin
Modified amino acid is shown in boldface. Periods represent the beginning and end of the tryptic peptide.
| Identification | Peptidesa | Charge |
|---|---|---|
| Ubiquitin | R.LIFAGK48*QLEDGR.T | +2 |
| R.LIFAGK48*QLEDGR.T | +3 |
An asterisk denotes Gly-Gly modification of the amino group of the lysine side chain.
The Role of Lys48-Linked Polyubiquitin Chains in PKC-Regulated OAT1 Ubiquitination.
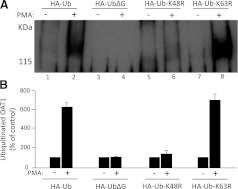
To explore the role of Lys48-linked polyubiquitin chains in PKC-regulated OAT1 ubiquitination, we employed four HA-tagged-ubiquitin constructs: wild-type ubiquitin Ub; mutant ubiquitin UbΔG (Kolodziejski et al., 2002), which has the C-terminal Gly residue deleted and therefore prevents the conjugation of ubiquitin to OAT1; mutant ubiquitin Ub-K48R (Li et al., 2007), which has the invariant Lys in position 48 mutated to Arg and prevents the formation of Lys48-linked polyubiquitin chains; and mutant ubiquitin Ub-K63R (Li et al., 2007), which has the invariant Lys in position 63 mutated to Arg and prevents the formation of Lys63-linked polyubiquitin chains. The cDNA encoding these ubiquitin constructs were transfected into OAT1-expressing cells. The transfected cells were treated with or without PMA. OAT1 was then immunoprecipitated, followed by immunoblotting with anti-HA antibody.
Our results (Fig. 6) showed that PMA treatment resulted in a strong HA-immunoreactive signal in both Ub- and Ub-K63R-transfected cells, suggesting that a significant amount of Ub and Ub-K63R was incorporated into OAT1. In contrast, the same treatment resulted in almost no detection of HA-immunoreactive signal in both UbΔG- and Ub-K48R-transfected cells. There was no major change in the total amount of OAT1 immunoprecipitated under these conditions (not shown). These results strongly support our mass spectrometric data that majority of OAT1 ubiquitination consists of Lys48-linked polyubiquitin chains.
Fig. 6.
Effects of wild-type ubiquitin and ubiquitin mutants on OAT1 ubiquitination. (A) cDNA for HA-tagged wild-type ubiquitin (Ub), ubiquitin mutants UbΔG, Ub-K48R, and Ub-K63R were transfected into COS-7 cells, respectively, followed by treatment with or without PMA for 30 minutes. The treated cells were lysed. OAT1 was immunoprecipitated by anti-myc antibody, followed by immunoblotting with anti-HA antibody. (B) densitometry plot of results from (A) as well as from other experiments. Ubiquitinated OAT1 in PMA-treated cells was expressed as the percentage of that found in control cells. The values are mean ± S.E. (n = 3).
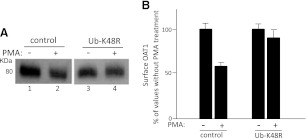
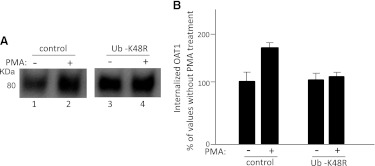
The Role of Lys48-Linked Polyubiquitin Chains in PKC-Regulated OAT1 Surface Expression and Internalization.
We previously had demonstrated that acute activation of PKC inhibits OAT1 activity by reducing OAT1 surface expression through accelerating internalization of this transporter from the cell surface to intracellular compartments (Zhang et al., 2008). To explore the role of Lys48-linked polyubiquitin chains of OAT1 in this process, we examined PKC-regulated OAT1 surface expression and internalization in cells transfected with Ub-K48R. As shown in Fig. 7, PMA treatment resulted in a significant decrease in OAT1 surface expression in control cells, whereas this PMA effect was substantially reversed in Ub-K48R-transfected cells. Similarly, PMA treatment resulted in a significant increase in OAT1 internalization in control cells, whereas this PMA effect was substantially reversed in Ub-K48R-transfected cells (Fig. 8). These data indicate that Lys48-linked polyubiquitin chains of OAT1 are essential for PKC-regulated OAT1 surface expression and internalization.
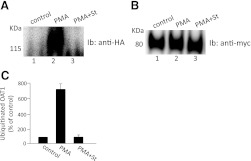
Fig. 7.
Effect of ubiquitin mutant Ub-K48R on PMA-regulated OAT1 surface expression. (A) OAT1-expressing cells were transfected with Ub-K48R (or vector as control), followed by treatment with or without PMA (1 μM) for 30 minutes. The treated cells underwent cell-surface biotinylation. Biotinylated proteins were isolated with streptavidin beads and analyzed by immunoblotting with an anti-myc antibody. (B) densitometry plot of results from (A) as well as from other experiments. Surface OAT1 in PMA-treated cells was expressed as the percentage of surface OAT1 in control cells. The values are mean ± S.E. (n = 3).
Fig. 8.
Effect of ubiquitin mutant Ub-K48R on PMA-regulated OAT1 internalization. (A) COS-7 cells were transfected with OAT1 (1 μg) and Ub-K48R (4 μg) (or vector as control). OAT1 internalization (15 minutes) was then determined as described under Materials and Methods, followed by immunoblotting using anti-myc antibody. (B) densitometry plot of results from (A) as well as from other experiments. The internalized surface OAT1 in PMA-treated cells was expressed as the percentage of internalized surface OAT1 in control cells. The values are mean ± S.E. (n = 3).
Discussion
The OAT family mediates the body disposition of a diverse array of environmental toxins and clinically important drugs. Therefore, understanding the regulation of these transporters has profound clinical significance. Short-term (acute) regulation of OAT activity is particularly important when the body must deal with rapidly changing amounts of substances as a consequence of variable intake of drugs, fluids, and meals as well as metabolic activity.
The amount of OATs at the cell surface is critical for their drug transport activity. We previously had established that OAT1 undergoes constitutive internalization from and recycling back to the cell surface and that acute activation of PKC inhibits OAT1 activity by reducing OAT1 cell-surface expression through accelerating its internalization from the cell surface to intracellular compartments without affecting the recycling and the total expression of the transporter (Zhang et al., 2008). Owing to this characteristic, PKC inhibition of OAT1 activity is usually moderate (30–50%) because there is always a certain amount of OAT1 at the cell surface resulting from the recycling, even with the maximum stimulation of PKC. In vivo, regulation at such a moderate scale may play an important role in providing quick and efficient fine-tuning in the body’s response to environmental changes. However, the mechanisms underlying PKC-regulated OAT1 trafficking are largely unknown. In our current study, we provide the first demonstration that a topology-specific polyubiquitination of OAT1 plays an essential role in this process.
We first examined OAT1 ubiquitination in COS-7 cells stably expressing this transporter. Although native systems that endogenously express transporters are great assets to identify the endogenous stimuli controlling the transporter function, heterologous expression systems are useful for asking mechanistic questions with regard to understanding the relationship between transporter trafficking and regulation. COS-7 cells offer several useful advantages for study of the cloned OAT. 1) These cells were directly derived from the kidney and have been very useful in understanding other renal transport processes and cellular functions, including organic cation transport (Nagai et al., 2006; Zhang et al., 2002). 2) This cell line does not express endogenous OATs. Therefore, expression of OAT1 in COS-7 cells allows us to dissect the transport characteristics of OAT1 in a relevant mammalian system without the possibly confounding effects of other OATs. 3) They possess endogenous PKC and PKA signaling pathways and provide a good experimental model system for studying the regulatory mechanisms underlying many transport processes (Cobb et al., 2002; Kazanietz et al., 2001). 4) The transport characteristics of OAT1 in COS-7 cells are in good agreement with those observed in other systems (Miller, 1998; Shuprisha et al., 2000; Zhang et al., 2008).
Using these cells, we obtained new data showing that under basal condition, OAT1 was not significantly ubiquitinated. However, acute activation of PKC by PMA induced a time-dependent increase in OAT1 ubiquitination (Fig. 1), and PMA-induced ubiquitination was blocked by PKC inhibitor staurosporine (Fig. 2), confirming that this PMA effect is indeed PKC-dependent. The result was further confirmed with naturally occurring OAT1 from rat kidney (Fig. 3), providing the physiologic relevance for our study.
We previously had demonstrated that OAT1 internalizes partly through a Dyn- and clathrin-dependent pathway and that such internalization was blocked in cells transfected with a dominant negative mutant of Dyn-2. As a result, OAT1 accumulated at the cell surface (You et al., 2000). In our current study, we show (Fig. 5) that blocking OAT1 internalization with dominant negative mutant of Dyn-2 correlates with the accumulation of ubiquitinated OAT1 in the plasma membrane, suggesting that ubiquitination is an early event that occurs at the cell surface before OAT1 internalization.
It has been shown that different types of ubiquitination (mono- versus polyubiquitination via different types of linkages) have different physiologic outcomes for the ubiquitinated substrate. For example, while monoubiquitination is required for down-regulation of the epidermal growth factor (EGF) receptor (Haglund et al., 2003a, 2003b), Lys63-linked ubiquitin chains have been shown to contribute to endocytosis of the TrkA nerve growth factor receptor (Geetha et al., 2005). Although IFNAR1 internalization requires both Lys48 and Lys63 linkages, PRLr internalization relies mainly on Lys63-conjugated chains (Kumar et al., 2007; Varghese et al., 2008). To identify the specific types of OAT1 ubiquitination, we performed mass spectrometric analysis, one of the most powerful approaches to identify post-translational modifications of numerous proteins. For this purpose, we developed sequential immunoprecipitation procedures to ensure that a high purity of ubiquitinated OAT1 was used. The mass spectrometric results indeed confirmed that ubiquitin and OAT1 were the two predominant proteins detected in our sample, providing evidence that ubiquitin was conjugated to OAT1 rather than to an OAT1-associated protein. Furthermore, our mass spectrometric analysis identified several peptide fragments from ubiquitin protein, which covered all its seven lysine residues. Among these fragments, only Lys48-linked ubiquitin was detected (Table 1). These results indicate that OAT1 is ubiquitinated mainly through Lys48-linked polyubiquitin chains.
The mass spectrometric data that OAT1 was ubiquitinated mainly through Lys48-linked polyubiquitin chains was independently confirmed through the use of a series of ubiquitin mutants (Fig. 6). The mutant Ub-K63R, which is defective in forming Lys63-linked polyubiquitin chains, incorporated into OAT1 as efficiently as that of wild-type ubiquitin, whereas mutant Ub-K48R, which is defective in forming Lys48-linked polyubiquitin chains, had the minimum incorporation into OAT1.
The strong evidence of the importance of Lys48-linked polyubiquitination for OAT1 trafficking came from our studies using dominant negative ubiquitin mutant Ub-K48R. Interfering with Lys48-linked polyubiquitination impeded the PKC-induced down-regulation of OAT1 surface expression (Fig. 7) and the PKC-accelerated OAT1 internalization (Fig. 8).
It is important to note that our entire study investigated the role of ubiquitination in short-term PKC regulation (PMA treatment of 30 minutes) on OAT1 trafficking and function. We had previously demonstrated (Zhang et al., 2008) that short-term treatment of OAT1-expressing cells with PMA (30 minutes) inhibits OAT1-mediated transport by reducing OAT1 cell-surface expression through accelerating its internalization from the cell surface to intracellular compartments without affecting the total expression of the transporter, suggesting that degradation of OAT1 (through either proteasome or lysosome) does not happen during the 30 minutes of PMA treatment. This is similar to several other ubiquitination-regulated proteins for which ubiquitination regulates endocytosis without affecting their degradation (Mukhopadhyay and Riezman, 2007). It would be really interesting to explore, in the future, whether long-term treatment with PMA (2 to 8 hours) would affect OAT1 degradation.
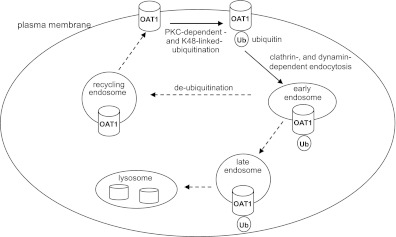
In conclusion, ubiquitination has been shown to regulate internalization, postinternalization sorting, and degradation of other membrane proteins. Our current investigation focuses on the first step: internalization. The major findings from our studies are (Fig. 9) that 1) activation of PKC promotes OAT1 ubiquitination both in vitro and ex vivo, 2) OAT1 ubiquitination mainly occurs at the cell surface through Lys48-linked polyubiquitin chains, and 3) Lys48-linked polyubiquitination plays an essential role in PKC-regulated OAT1 trafficking.
Fig. 9.
The role of ubiquitination in OAT1 internalization. Dashed arrows indicate the steps that were not explored in our current study.
To the best of our knowledge, this report is the first mechanistic study to demonstrate a topology-specific role of ubiquitination in the regulation of any drug transporters. What would be the physiologic implication of our studies? Abnormal OAT1 trafficking may contribute to the impaired drug elimination in bilateral ureteral obstruction (BUO). BUO is a serious and common clinical condition, and an important cause of acute renal failure (Seldin and Giebisch, 2000; Villar et al., 2005). It has been shown (Villar et al., 2005) that in rats with BUO the elimination of drugs is impaired, partly due to a redistribution of OAT1 from the cell surface to the intracellular compartment. In BUO, angiotensin II has an elevated level of expression (Klahr, 1998; Klahr and Morrissey, 2002; Seldin and Giebisch, 2000). We previously reported that angiotensin II inhibits OAT1 activity through activation of PKC in cultured cells (Li et al., 2009). Therefore, the high level of angiotensin II in rats with BUO may inhibit OAT1 activity through PKC-regulated OAT1 ubiquitination and trafficking. Our current studies may provide important insight into the molecular, cellular, and clinical bases underlying BUO.
Finally, while the limited coverage of OAT1-derived peptides did not allow us to identify the ubiquitin-accepting lysine residues on OAT1 using mass spectrometry, while the current manuscript was being reviewed we resorted to a different approach—site-directed mutagenesis—and we succeeded in identifying three lysine residues on OAT1 that serve as ubiquitin acceptors (a separate manuscript has been submitted). Our work paves the way for the investigation of the ubiquitin regulation of other OAT isoforms and related drug transporters.
Abbreviations
- BUO
bilateral ureteral obstruction
- Dyn
dynamin
- HA
hemagglutinin
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- OAT
organic anion transporter
- PBS
phosphate-buffered saline
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- Ub
ubiquitin
Authorship Contributions
Participated in research design: Zhang, You.
Contributed reagents: Patterson.
Conducted experiments: Zhang, Li.
Performed data analysis: Zhang, You.
Wrote or contributed to the writing of the manuscript: Zhang, You.
Footnotes
This work was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01-DK60034] (to G.Y.), and National Institutes of Health National Institute of General Medical Sciences [R01-GM079123 and R01-GM097000] (to G.Y.).
References
- Ahn SY, Nigam SK. (2009) Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol Pharmacol 76:481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomberger JM, Barnaby RL, Stanton BA. (2009) The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem 284:18778–18789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BR, Ruiz F, King CM, Fortenberry J, Greer H, Kovacs T, Sorscher EJ, Clancy JP. (2002) A(2) adenosine receptors regulate CFTR through PKA and PLA(2). Am J Physiol Lung Cell Mol Physiol 282:L12–L25 [DOI] [PubMed] [Google Scholar]
- Dantzler WH, Wright SH. (2003) The molecular and cellular physiology of basolateral organic anion transport in mammalian renal tubules. Biochim Biophys Acta 1618:185–193 [DOI] [PubMed] [Google Scholar]
- Geetha T, Jiang J, Wooten MW. (2005) Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol Cell 20:301–312 [DOI] [PubMed] [Google Scholar]
- Haglund K, Di Fiore PP, Dikic I. (2003a) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci 28:598–603 [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. (2003b) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5:461–466 [DOI] [PubMed] [Google Scholar]
- Kazanietz MG, Caloca MJ, Aizman O, Nowicki S. (2001) Phosphorylation of the catalytic subunit of rat renal Na+, K+-ATPase by classical PKC isoforms. Arch Biochem Biophys 388:74–80 [DOI] [PubMed] [Google Scholar]
- Klahr S. (1998) Obstructive nephropathy. Kidney Int 54:286–300 [PubMed] [Google Scholar]
- Klahr S, Morrissey J. (2002) Obstructive nephropathy and renal fibrosis. Am J Physiol Renal Physiol 283:F861–F875 [DOI] [PubMed] [Google Scholar]
- Kolodziejski PJ, Musial A, Koo JS, Eissa NT. (2002) Ubiquitination of inducible nitric oxide synthase is required for its degradation. Proc Natl Acad Sci USA 99:12315–12320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KG, Barriere H, Carbone CJ, Liu J, Swaminathan G, Xu P, Li Y, Baker DP, Peng J, Lukacs GL, et al. (2007) Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J Cell Biol 179:935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, Patterson C. (2007) Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest 117:3211–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Duan P, You G. (2009) Regulation of human organic anion transporter 1 by ANG II: involvement of protein kinase Calpha. Am J Physiol Endocrinol Metab 296:E378–E383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DS. (1998) Protein kinase C regulation of organic anion transport in renal proximal tubule. Am J Physiol 274:F156–F164 [DOI] [PubMed] [Google Scholar]
- Miranda M, Wu CC, Sorkina T, Korstjens DR, Sorkin A. (2005) Enhanced ubiquitylation and accelerated degradation of the dopamine transporter mediated by protein kinase C. J Biol Chem 280:35617–35624 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315:201–205 [DOI] [PubMed] [Google Scholar]
- Nagai K, Takikawa O, Kawakami N, Fukao M, Soma T, Oda A, Nishiya T, Hayashi M, Lu L, Nakano M, et al. (2006) Cloning and functional characterization of a novel up-regulator, cartregulin, of carnitine transporter, OCTN2. Arch Biochem Biophys 452:29–37 [DOI] [PubMed] [Google Scholar]
- Seldin DW, Giebisch G. (2000) The Kidney: Physiology and Pathophysiology, 3rd ed, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- Shuprisha A, Lynch RM, Wright SH, Dantzler WH. (2000) PKC regulation of organic anion secretion in perfused S2 segments of rabbit proximal tubules. Am J Physiol Renal Physiol 278:F104–F109 [DOI] [PubMed] [Google Scholar]
- Srimaroeng C, Perry JL, Pritchard JB. (2008) Physiology, structure, and regulation of the cloned organic anion transporters. Xenobiotica 38:889–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Xu W, Zhou F, You G. (2004) Role of glycosylation in the organic anion transporter OAT1. J Biol Chem 279:14961–14966 [DOI] [PubMed] [Google Scholar]
- VanWert AL, Gionfriddo MR, Sweet DH. (2010) Organic anion transporters: discovery, pharmacology, regulation and roles in pathophysiology. Biopharm Drug Dispos 31:1–71 [DOI] [PubMed] [Google Scholar]
- Varghese B, Barriere H, Carbone CJ, Banerjee A, Swaminathan G, Plotnikov A, Xu P, Peng J, Goffin V, Lukacs GL, et al. (2008) Polyubiquitination of prolactin receptor stimulates its internalization, postinternalization sorting, and degradation via the lysosomal pathway. Mol Cell Biol 28:5275–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar SR, Brandoni A, Anzai N, Endou H, Torres AM. (2005) Altered expression of rat renal cortical OAT1 and OAT3 in response to bilateral ureteral obstruction. Kidney Int 68:2704–2713 [DOI] [PubMed] [Google Scholar]
- You G. (2002) Structure, function, and regulation of renal organic anion transporters. Med Res Rev 22:602–616 [DOI] [PubMed] [Google Scholar]
- You G, Kuze K, Kohanski RA, Amsler K, Henderson S. (2000) Regulation of mOAT-mediated organic anion transport by okadaic acid and protein kinase C in LLC-PK(1) cells. J Biol Chem 275:10278–10284 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Hong M, Duan P, Pan Z, Ma J, You G. (2008) Organic anion transporter OAT1 undergoes constitutive and protein kinase C-regulated trafficking through a dynamin- and clathrin-dependent pathway. J Biol Chem 283:32570–32579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Evans KK, Wright SH. (2002) Molecular cloning of rabbit organic cation transporter rbOCT2 and functional comparisons with rbOCT1. Am J Physiol Renal Physiol 283:F124–F133 [DOI] [PubMed] [Google Scholar]
- Zhou R, Patel SV, Snyder PM. (2007) Nedd4-2 catalyzes ubiquitination and degradation of cell surface ENaC. J Biol Chem 282:20207–20212 [DOI] [PubMed] [Google Scholar]