Abstract
The standard procedure to increase microfluidic chip performance is to grow the number of parallel test systems on the chip. This process is accompanied by miniaturizing biochemical workflows and micromechanical elements, which is often a major challenge for both engineering fields. In this work, we show that it is possible to substantially increase the runtime performance of a microfluidic affinity assay for protein interactions by simultaneously engineering fluid logics and assay chemistry. For this, synergistic effects between the micro- and chemical architecture of the chip are exploited. The presented strategy of reducing the runtime rather than size and volume of the mechanical elements and biological reagent compartments, will, in general, be of importance for future analytical test systems on microfluidic chips to overcome performance barriers.
Keywords: Microfluidics, Chip Performance, Chemical and Micro-Architecture, Protein-Protein Interaction, High-Throughput Technology
Introduction
Microfluidic chip technologies have found a broad usage in various scientific fields.1,2 There is a steady demand for chips with higher performances and sensitivity in order to meet ever-more ambitious scientific goals. This is especially observed for microfluidics designed for systems biology approaches, where each chip is expected to provide a number of analytical test systems at genome- or proteome-wide scales. The demand for higher microfluidic chip performance is shared with their electrical analogs. Over the years, the performance of transistor logic has increased exponentially by increasing the number of transistors per area.3 There is a similar trend with microfluidic chips, but the design constraints are subtler. For example, the number of membrane valves per area on polydimethylsiloxane (PDMS) chips is increasing, but one cannot make the dimensions of the functional elements of the chip smaller than the object of study—whether a biological cell or the minimum volume required for a biological assay. To overcome this technical drawback, conceptually different control mechanisms have been developed to integrate biochemical assays on chips. In respect to parallelizing simple assays, emulsion4,5 and other oil-separation based microfluidic techniques6,7 have proved useful. The disadvantages of those platforms include their lower integration levels and complicated workflows.
A new strategy in the semiconductor industry to obtain higher chip performance is to redesign the chip micro-architecture.8 A prominent example for this is the multi-core design of microprocessors, where a given number of transistors per area are rewired with the goal of reducing the runtime of applications processed by the chip. This strategy can increase the performance of a chip without further size reduction of its structural elements. The process of redesigning the micro-architecture is always accompanied by adaption of the running application in order to fully exploit the capabilities of the new chip generation. Within a microfluidic approach, this would translate to redesign of the analytical assay, which we name hereafter the chemical architecture of a microfluidic chip. In fact, the redesign process of the chemical architecture offers additional potential to increase chip performance. Many microfluidic-implemented assays still resemble their bench top equivalent and performance aspects are not considered during the miniaturization process. In here, we tripled the performance of a highly integrated valve-based PDMS microfluidic chip designed for screening binary protein interactions by reducing the runtime of the chip application. The performance increase was achieved by solely reengineering the chemical and micro-architecture of the chip.
Materials and Methods
Chip Production
The layout of the two-layer PDMS chip was designed with AutoCAD (Autodesk) and printed onto emulsion photomasks (FineLine Imaging, Colorado). The mold for the flow circuit was manufactured in a two-layer process. AZ50-XT (DuPont, USA) was used to construct fluidic channels with a round channel profile and a height of 25 µm. Interlayer connectors were manufactured on top of the first AZ50-XT layer with the same photoresist. For this, first, a thin layer of AZ50-XT was spun for 1 min at 3500 rpm on the wafer to coat the wafer. We then again spun AZ40-nXT for 15 s at 500 rpm followed immediately by 30 s at 1000 rpm with a Laurell WS-400B-6NPP/Lite/10K. The mold was soft-baked for 3 min at 65 °C and 6 min at 115 °C. Rehydration occurred over the time interval of 1 d at a humidity level of ≥60%. After exposure, the AZ resist was developed and no post-bake was performed. Poles or differently named interlayer connectors with a total height of 70 µm were achieved with this procedure. Microchannels on the mold for the control circuit were fabricated with SU-8 (Microchem) and had a rectangular shape with a height of 25 µm. Molds were treated with chlorotrimethylsilane (Sigma Aldrich) prior to use. Sylgard-184 (Down Corning, USA) was used to replicate chips from the silicon molds within a process described before.
Construction of the linear cDNA templates with antibody tags
Gateway vectors (pDONR223) containing the complementary DNA (cDNA) encoding for S100A1 (OHS1770-932180), and S100B (OHS1770-9380380) (Open Biosystems) from Homo sapiens were used as entry templates for PCR amplification. Linear expression templates compatible with in vitro T7 E. coli expression systems (Promega) were constructed by a two-step extension PCR as described before.9 Antibody tags were included during the PCR amplification at the 5′ end. Primer sequences are given in the SI.
Construction of the p-SNAP fusion templates
SNAP fusion proteins were obtained through molecular cloning. For this, the DNA sequences of S100A1, S100B, or eGFP (CloneTech) were amplified from the entry vectors by PCR. During the PCR procedure, a SbfI restriction site or BamHI together with a 6×His tag and serine linker were added to the 5′ and 3′ end, respectively. Primer sequences are given in the SI. One microgram of p-SNAP-tag (T7) plasmid vector (NEB) and linear PCR products of S100A1, S100B, or eGFP were subjected to restriction using SbfI and BamHI (Open Biosystems). After a 1-h incubation at 37 °C, the sample was subjected to QIAGEN MinElute Reaction Cleanup Kit. The followed 45 min ligation step was performed in a 3:1 insert to vector ratio (0.15:0.05 µg) with Quick Ligase (Open Biosystems). The ligation product was transformed with One Shot Top 10 Competent Cells (Invitrogen). Cells were plated and grown overnight on Luria Bertani (LB) agar plates with ampicillin. Single colonies were selected and then grown in LB broth spiked with 100 µg/µl ampicillin for 8 h and subjected to QIAGEN QIAprep Spin Miniprep Kit. cDNA in Miniprep was confirmed by Sanger sequencing (MCLAB).
Microarrays
Selected DNA expression templates with a concentration between 0.1 and 0.2 µg/µl in a 1.0% bovine serum albumin (BSA), 10 mM phosphate buffer (pH 7.2) solution were spotted onto epoxy coated glass slides (25 mm × 75 mm) (CEL Associates Online Storeroom). OmniGrid Micro microarray printer with a custom-made print-head holding 2 × 5 silicon pins (Parallel Synthesis Technologies) were used for contact printing of the cDNAs microarrays on epoxy glass slides. The humidity during the print was kept constant at 70%. BSA was utilized for visualization during the alignment process and to prevent covalent linkage of the target DNA to the epoxy functional groups. Microarrays were then aligned and heat bounded to the PDMS chip for 5 hours at 65 °C. Bound chips were used within one week and not stored for longer.
Synthesis of bovine serum albumin (BSA) conjugated to benzyl-guanin (BG)
High-purity BSA (Pierce, Rockford) was dissolved in deionized water to a concentration of 1.0 mg/ml. To obtain a 1:50 BSA to BG ratio 0.2 mg of BG-GLA-NHS (New England Biolabs, Ipswich) was dissolved in 10 µL of water free of N,N-dimethylformamid. Drops of the 10-µL BG-GLA-NHS solution were then slowly added under stirring to 1 ml of 1.0 mg/ml BSA solution and incubated overnight at room temperature. Aliquots were stored at −80 °C.
Chip operation and data evaluation
The microfluidic chip was controlled by a custom-made pressure system (for instructions see Stanford Microfluidic Foundry webpage). A programmed Matlab (Mathworks) interface was used for automation of the protein interaction runs. Single flush steps and conditions for both chemical architectures described in here are given in table format (SI-Table 1 and 2). Resulting protein interaction arrays still bound to the PDMS chip were scanned with an LS Reloaded laser microarray scanner (TCAN). Images were analyzed using GenePix v.6.0 software and downstream analysis was performed using Matlab 7.0.
Results
First-generation platform
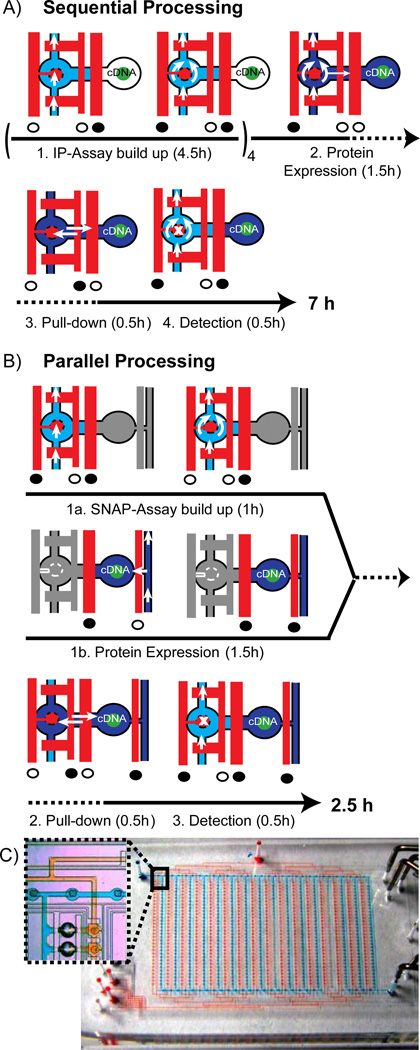
We started with a previously developed chip platform for testing binary biomolecular interactions. The generic chip design has been previously exploited to measure DNA-protein, RNA-protein, small molecule-protein, and protein-protein interactions (PPI).9 The chip integrates in hundreds of separated compartments four biochemical processes, i.e., (1) build-up of a pull-down assay on a glass surface, (2) in vitro expression of protein pairs from cDNA templates, (3) immuno-precipitation (IP) of the expressed proteins, and (4) detection of the protein interaction with a fluorescently labeled antibody (Ab). The workflow of the binary interaction screening technology is described in short. For testing hundreds of binary protein interactions on the chip, standard microarray spot technology is used first to create a cDNA microarray on an epoxy glass slide. A single microarray spot contains cDNAs of two proteins, which are named hereafter “bait” and “prey.” The bait refers to the protein, which is pulled down to the glass surface during the protein interaction assay and the prey refers to the protein, which is tested to co-precipitate. The multilayer PDMS chip is aligned and bonded onto the cDNA microarray. Figure 1A depicts the biochemical steps performed in one out of 640 parallel working unit cells on the chip. Each unit cell is divided into two sections, where the first section (left-handed on the pictograms) is assigned to the pull-down assay and the second section (right-handed on the pictograms) for the in vitro expression of bait and prey from the spotted cDNA templates. Further, Figure 1A shows the actuation cycles of the three different valves within the unit cells, which control the fluid flow during the four process steps of the protein interaction assay. An important design element within the chip platform is a round microfluidic valve named the button valve. The valve is located over the pull-down assay section within the unit cells. Actuation of the button valve is able to protect underlying reactive glass or chemical surfaces from fluids. The button valves can thus be exploited within process 1 to sequentially build up the pull-down chemistry on the area under the button. The pull-down chemistry is explained in detail below. Introducing an S30 E. coli extract to the unit cells, which is guided to the chamber aligned over the cDNA spot, started process 2. Bait and prey proteins are in vitro expressed when the extract fills the chamber by pressing out air through the PDMS. For process 3, the control logics are set to allow diffusion of both proteins to the pulled-down section in each separated unit cell. At the end of process 3, the button valve is actuated and is thus used to mechanically trap proteins in the pull-down area. Finally, in process 4, all non-bound material is flushed out from the unit cells and protein interactions are detected with a fluorescently labeled antibody. For this, the button valve is shortly lifted to allow diffusion of the antibody over the pull-down area. In the first chip generation, the four integrated processes were performed sequentially on chip. The corresponding runtime for the processes one to four were 4.5, 1.5, 0.5, and 0.5 hours, respectively (see Figure 1A).
Figure 1.
Process sequence of the protein interaction assay and corresponding flow logics within one unit cell on the sequential (A) and parallel operating (B) microfluidic chip. Blue and red denote the flow layer and control layer of the multilayer PDMS chip. Full and open circles below the control channels indicate the actuated and released state of the valves, respectively. Time lines are given below the pictograms of the corresponding micro-architecture. A) Only one flow circuit is implemented in the first-generation chip. Chemicals to build up the IP assay (light blue), to express bait and prey proteins from spotted cDNA’s (dark blue) and a fluorescently labeled Ab (light blue again) to detect PPI’s are flushed sequentially. The button valve is located on the left-hand side of the unit cell. Process 1 has to be circled four times in order to specifically deposit an Ab in the pull-down area (marked with an “x” in process 4). White arrows indicate the flow direction of the fluids, whereas the double arrows in process 3 indicate passive diffusion of proteins from the place of synthesis to the pull-down area. B) An additional flow circuit was implemented on the next-generation chip to process the first two steps of the PPI assay in parallel (grayed out sections in the state pictograms denote the parallel process). C) Overview image of the next-generation chip with 640 unit cells shown in B. The two independent flow circuits are visualized with red and blue food colors. Inlet shows a zoom version of the inter-layer connector structure for bridging the flow circuits together with two unit cells.
Reengineering of the micro-architecture
In first phase of the reengineering phase, we concentrated on the micro-architecture of the chip, whereas in the second phase, we concentrated on the chemical architecture. To decrease the overall runtime of the protein interaction assay, we took the engineering approach to process step 1 and 2 in parallel on the chip. This is possible since both processes are not dependent on each other. In order to achieve the parallel workflow, the unit cells of the microfluidic chip were rewired, as shown in Figure 1B. Distinct from the previous design, a second entry point to the unit cells was created. The second entry point allowed the address of each unit cell from two independent flow circuits. Both flow circuits are shown in an overall chip view in Figure 1C. The first flow circuit is used to sequentially deliver fluids required to spatially build up the pull-down assay on the glass surface. The second flow circuit is used to fill the chamber within the unit cell containing the cDNA with S30 E. coli extract. Thus, process 2 can be started in parallel to process 1.
The two independent flow circuits were achieved by using “via” elements. Vias are interlayer connectors, bridging the flow streams at fluid cross-junction and cause minimal increase of the overall chip area.10 On the PDMS chip, vias are holes in the thin PDMS membrane between the two layers of the flow and control circuit. A detailed photolithographic protocol for production of the molds and PDMS chips is given in the materials and methods section. To obtain reliable prototyping results for vias the corresponding microstructures on the molds were constructed with positive rather than negative photoresist.10 This had the advantage of obtaining microstructures with a hydrophilic surface property and avoiding a closing of the vias with thin layers of PDMS during the prototyping process.
The functionality of the chip with parallel processing micro-architecture is demonstrated by in vitro expression and pull-down of GFP. For this, cDNA of GFP tagged with 6×His at the C-terminus was spotted on an epoxy glass slide and subjected to standard chip run as described above. The scan image of the pull-down array after the chip run is shown in Figure SI-1.
Reengineering of the chemical architecture
The runtime of a protein interaction assay on a chip with parallel processing micro-architecture compared to a run on a chip with sequentially processing micro-architecture under the same conditions would only be reduced by 1.5 h, i.e., the runtime of process 2. It is obvious that the rate-limiting step in the overall runtime of the protein interaction assay is process 1. In the following, we developed a new chemical architecture of the protein interaction assay to fully exploit the parallel-processing micro-architecture.
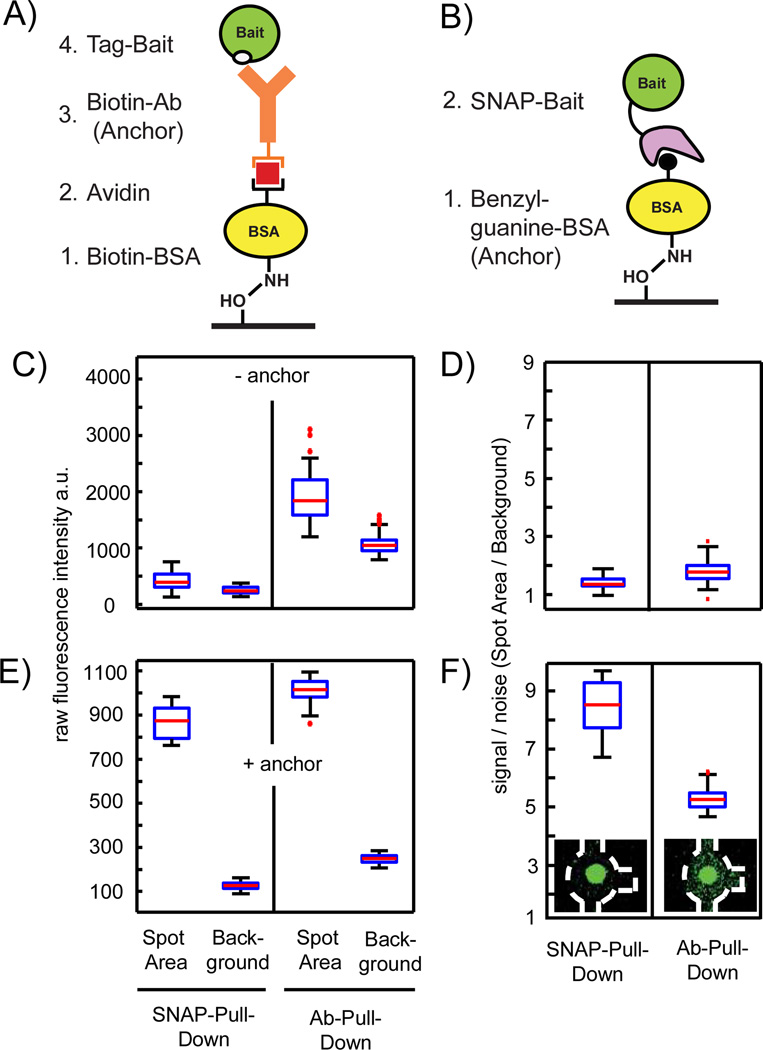
The chemical architecture of the first chip generation comprising the antibody-based pull-down assay is shown in Figure 2A. For anchoring the antibody on the epoxy glass surface, biotinylated BSA (b-BSA), and avidin are used. To achieve a spatial depositing of the antibody only on the glass area under the button valve, four flush cycles including buffer exchanges in between each cycle are required. Table SI-1 shows in detail the valve actuation state of the button valve for the build-up steps of the pull-down. Direct deposition of the pull-down antibody on an epoxy or other reactive glass surfaces would decrease the runtime of process 1. This approach, however, led to unspecific protein adsorption to the glass surfaces and thus to inferior results for the IP assay.
Figure 2.
Redesign process of the chemical architecture of the PPI assay on the chip. A–B) Antibody and SNAP tag pull-down strategies for the bait protein onto an epoxy glass surface, respectively. C) Control measurement to determine background signals from the chemical architecture of the pull-down assay by omitting the anchor molecules. Raw fluorescence intensities are measured within one chip for the fluorescence probe S-GFP-H (1 mg/ml) after flushing through the channels containing either unit cells with BSA-biotin/Avidin/BSA-biotin or BSA on the pull-down area under the button. D) Corresponding signal-to-noise levels (spot area/background) to the data in C. E) Repeat of the control experiment with anchor molecules. Raw fluorescence intensities are measured within one chip for the fluorescence probe S-GFP-H (1 mg/ml) after flushing over the fully assembled IP and SNAP pull-down arrays. F) Corresponding signal-to-noise levels to the data in E. Insets showing exemplarily fluorescent images of the corresponding unit cell on the chip (outlined with the dashed white line) after the S-GFP-H was pulled down. A total of 160 individual pull-down areas per condition were evaluated for all boxplots.
A different chemical architecture to decrease the runtime of process 1 is shown in Figure 2B. Here, the pull-down assay is comprised of a SNAP tag, which is a small protein named O6-alkylguanine-DNA transferase fused to a bait protein11 rather than an antibody tag sequence. The SNAP protein binds covalently to benzyl-guanosine (BG), which can be deposited as an anchor group onto the surface. SNAP tags have two advantages over antibodies: they lower cross-contamination risks among the microfluidic unit cells owing to covalent attachment and they prevent nonspecific pull-downs by antibodies. To shorten the runtime of the build-up process of the pull-down assay, the SNAP substrate BG was coupled to BSA (BG-BSA). This is analogous to the first step in the IP chemical architecture for deposition of an antibody on epoxy glass surfaces (see Figure 2A). To optimize the anchor concentration for the pull-down assay, different BG to BSA ratios between 1:10 and 1:500 were synthesized. To determine the pull-down efficiency of the different BSA-BG constructs, the anchors were deposited on the epoxy glass slide under the button valve area in different sections on one chip. Local deposition of BSA-BG was achieved by passivating the epoxy glass surface with BSA alone. The epoxy groups on the area under the button-valve area were protected by actuation of the valve. The different BSA-BG anchor constructs were sequentially flushed to different sections on the chip and coupled to the protected epoxy groups by release of the button valve. The pull-down efficiency of the anchor was tested with a fluorescence probe protein, i.e., GFP containing a SNAP and 6×His tag (S-GFP-H) at the N- and C-terminus, respectively. The strongest fluorescence signal after flushing a 1 mg/ml S-GFP-H solution over the pull-down array with different BSA to BG anchor molecules was observed for a BSA:BG ratio of 1:50. Lower BSA:BG ratios have shown clearly lower GFP fluorescence signals, were BSA:BG ratios over 1:100 had the same fluorescence as observed for the BSA-BG anchor 1:50 tended. Solutions with higher BSA:BG ratios tend to precipitate in the channels.
To compare background signals in the SNAP to antibody pull-down chemistry, first, a control experiment was performed, in which the anchor for both chemical architectures were omitted. For this, one chip was used and divided into two halves, wherein each half, the SNAP and antibody chemical architecture was built up. S-GFP-H was flushed through the chip under the same conditions used for a protein interaction run, including button actuation in the presence of the GFP probe. In general, to evaluate microarray images of the microfluidic chip, the median fluorescence intensities of the pull-down areas on the glass slide, IO and a local background value, IB were extracted from the images. The pull-down areas corresponded to the button-valve areas (diameter of 25 µm) and could clearly be visualized in the images (see insets in Figure 2F), were the local background value was defined as a cycle area surrounding the pull-down spot with twice the area of the spot. Figure 2C shows the raw fluorescence intensity values measured for S-GFP-H within the button and background area without antibody and BG anchor. The corresponding signal-to-noise ratios, which are the fluorescence intensity of the area under the button divided by the local background, are given in Figure 2D. Raw fluorescence intensities and signal–to-noise ratios on the detection area below the button valve are more increased on the no-antibody rather than for the no-BG control. A somewhat higher S-GFP-H fluorescence on the detection area in comparison to the background area is expected owing to actuation of the button valve during the presence of the S-GFP-H within the channels, which led to a small non-specific adsorption event. One reason for the higher background noise on the IP chemical architecture is the multilayer design of the pull-down assay, where each layer can incorporate and adsorb nonspecifically probe molecules.
Next, the experiment was repeated under the same conditions including the pull-down anchors for the IP and SNAP chemical architecture. Figure 2E and F shows the raw fluorescence signal obtained from the button-valve and background areas for S-GFP-H after pull-down to the surface. The absolute fluorescence values for the S-GFP-H protein on the pull-down areas are comparable, indicating similar pull-down efficiency for S-GFP-H with BSA:BG (1:50) and antibody (anti 6×His). In difference, the background fluorescence intensities obtained for S-GFP-H on SNAP pull-down areas was lower than for S-GFP-H on antibody pull-down areas. This result is in agreement with the no-anchor controls. Thus, the fluorescence signal–to-noise ratio for S-GFP-H on pull-down areas with the SNAP chemical architecture is increased compared signal-to-noise ratios for S-GFP-H on pull-downs areas with IP chemical architecture.
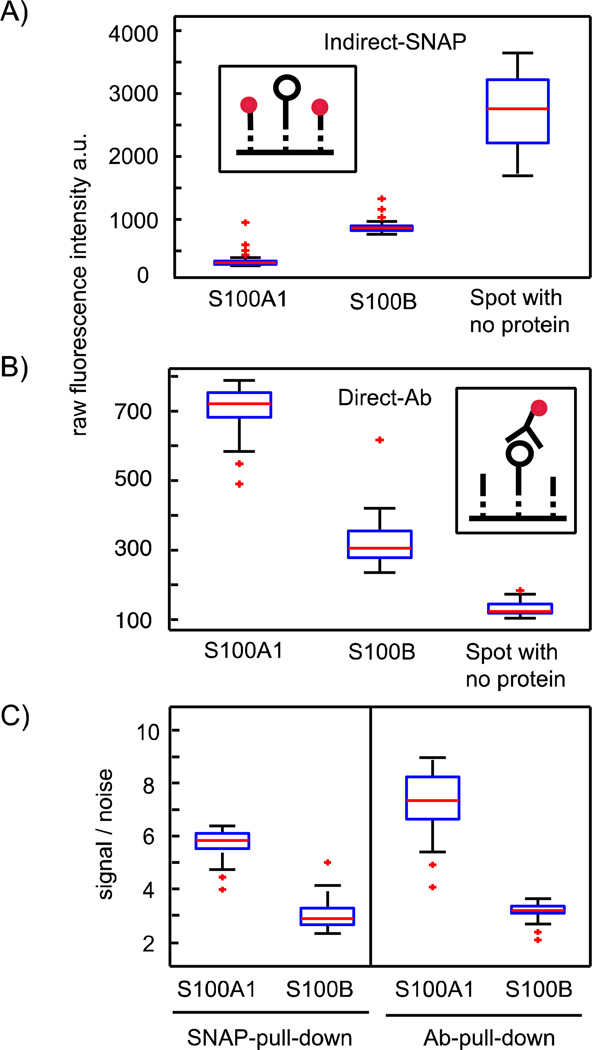
The covalent attachment of SNAP-tagged proteins offers an additional advance within a protein interaction assay. Protein interactions are determined with a fluorescently labeled antibody specific for the tag encoded in the prey sequence. The detection signal has to be normalized to the total amount of bait molecules on the glass surface under the button area. Thus, the normalization of fluorescence signals for protein interactions measured on the chip resembles the standard two-color image normalization method of DNA microarrays.12 The concentration of the bait within the IP chemical architecture was previously determined with the help of an additional fluorescently labeled antibody. In turn, the bait proteins must contain two antibody tags, i.e., one for the pull-down and one for the concentration determination on the glass surface in the area of the button valve. Thus, in total, three antibodies are needed in order to obtain the protein interaction data, when including the detection antibody. The intense use of antibodies is expensive and can lead to cross-contaminations and steric hindrance on the binding surfaces of interacting proteins. With the SNAP chemical architecture, however, it is possible to indirectly determine the bait concentration by measuring the proportion of unoccupied to occupied BG-anchors after pull-down of the bait. This is only possible with the covalent SNAP strategy, since competition between a fluorescence-probe molecule and the bait protein for the BG anchor places will not occur, as it is the case for antibodies. To test and compare the indirect and direct normalization method for the bait concentration, we expressed the human calcium binding proteins S100A1 and S100B on the chip with SNAP chemical architecture. In addition to the N-terminal SNAP tag, both test proteins carried a 6×His tag at the C-terminus. Protein expression levels for the pulled-down proteins were determined either with a fluorescently labeled antibody on one-half of the chip or with S-GFP-H on the other half of the chip. Figure 3A and B shows the fluorescence signals of both probe molecules on areas with pulled-down S100A1, S100B, or no protein. Bait concentrations on the pulled-down areas after in vitro expression on the chip are then determined by referencing the fluorescence signals of the areas with pulled-down proteins to the controls, which are areas in unit cells without expressed and pulled-down proteins. Bait surface concentrations for S100A1 and S100B from direct and indirect measuring methods are shown Figure 3C as signal-to-noise values. For direct comparison, values obtained from the indirect method are inverted. Clearly, Figure 3C demonstrates that both referencing methods lead to similar results for bait surface concentration of S100A1 and S100B under the same conditions. Thus, the indirect normalization method for baits on the SNAP chemical architecture can be applied in protein interaction assays on the chip and can replace the intense use of antibodies on the IP chemical architecture.
Figure 3.
Comparison between the indirect and direct detection method of bait proteins on the SNAP pull-down area after in vitro expression. A) The inset illustrates the indirect methods to determine bait concentrations on the pull-down areas. The black open circle denotes the bait, dotted lines denote the BG anchor places, and red dots denote the fluorescence probe. The probe is able to bind the remaining free BG anchor places after the bait is pulled down. The boxplot shows the raw fluorescence data (λem = 509 nm) of the fluorescence probe, S-GFP-H (1 mg/ml), on the pull-down areas in unit cells with either in vitro expressed S100A1, or S100B, or no protein. B) The inset illustrates the direct methods to determine bait concentrations on the pull-down areas. The black open circle denotes the bait, dotted lines denote the BG anchor places, and red dots denote a fluorescently labeled antibody specific for a bait tag. The boxplot shows the raw fluorescence data (λem = 647 nm) for the fluorescently labeled antibody specific for the bait-tag sequence in detection areas in unit cells with either in vitro expressed S100A1, or S100B, or no protein (Ab-detection: anti-6×His). C) Signal-to-noise ratio obtained from the two reference methods in A and B, where, for the indirect method, the value was inverted. For all boxplots, 48 single measurements on the chip were used and all raw fluorescence data points were correct by subtraction of the local background.
Next-generation chip with reengineered chemical and micro-architecture
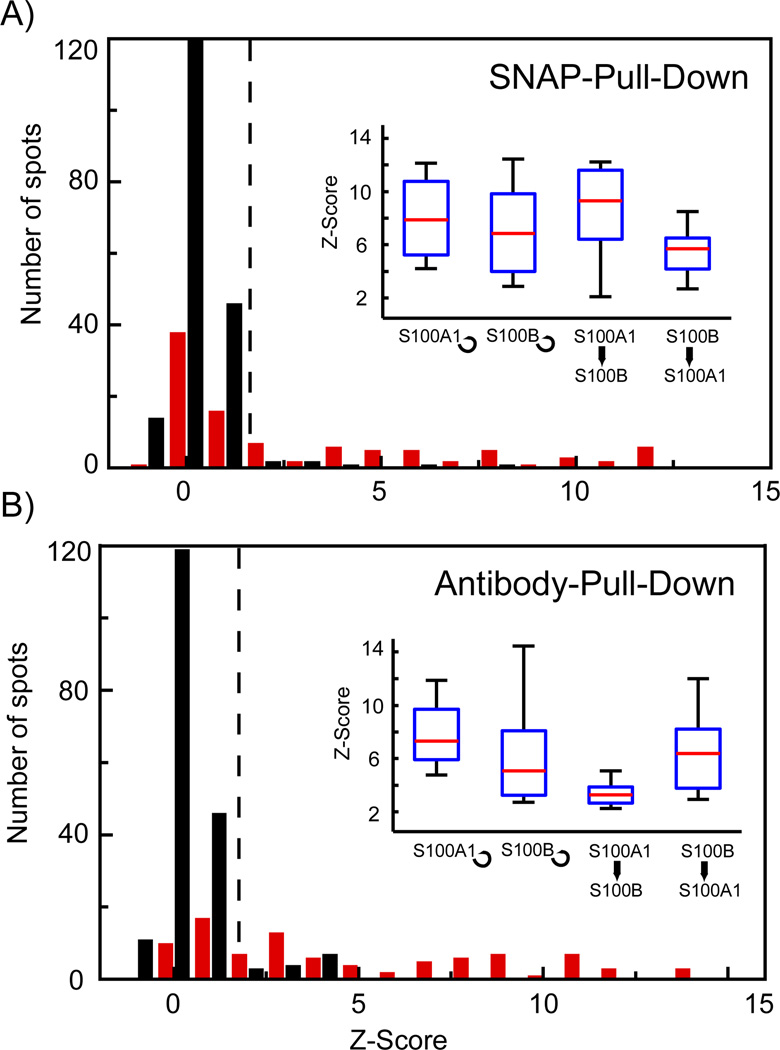
After functional establishment of a new chemical architecture of the pull-down assay for the protein interaction screening on the chip, we compared in the last step a protein interaction assay performed on a chip with parallel-processing architecture and SNAP chemical architecture to an assay performed under the same conditions on the sequentially operating chip with IP chemical architecture. For this, we used four positive protein interactions comprised of the proteins S100A1 and S100B. Both proteins interact with each other13 and form homo-dimers (self-interactions) 14,15,15. The corresponding cDNAs for the bait with SNAP and the antibody tag (His tag) and prey with only the antibody tag (c-Myc) were synthesized and spotted on a microarray. Spots on the microarray contained either two cDNAs for the bait and prey, no DNA, or only one cDNA of the bait, or prey. The latter two-spot variances are used as the negative control set. The positive set with two spotted cDNAs comprised in total 96 samples (24 repeats for each interaction) and the negative control set comprised 192 samples (48 no-bait, 48 no-prey, 96 no-cDNA). Figure 4A and B shows the Z-Score histogram for the PPI assays from a parallel operating microfluidic chip with SNAP chemical architecture and from a sequentially operating microfluidic chip with IP chemical architecture, respectively. The Z-Score is calculated as follows, where x̄ is the mean normalized fluorescence intensity of the control experiments, σ is the standard deviation of the control set, and IN is the normalized fluorescence intensity. Fluorescence values are normalized by subtraction of the local background and division of the fluorescence signal indicating the bait concentration using the pull-down dependent normalization method described above. The Z-Score indicates by how many standard deviations the fluorescence signal of a positive PPI measurement is above the mean of the control experiments. About half of the Z-Scores measured for the positive interaction set (red bars) in both experiments were higher than the Z-Scores for the control set (black bars); however, Z-Scores for the other half were not. Low protein expression levels can explain this result. Co-expression of S100A1 and S100B in one unit cell on chip resulted in a lower and more variable expression level as it was observed for the single proteins in separate unit cells. This was consistently observed for both platforms. In order to compare the protein interaction results from the two chip generations, we used only the fraction of protein interaction with Z-Score higher than 2. This cut-off value (dotted line in Figure 4A and B) has been previously used in the analysis to identify positive biomolecular interactions on the first-generation chip.16 The insets of Figure 4 then show the median Z-Scores (red line in the boxes) for the four tested protein interactions in boxplot format. Interestingly, we find higher Z-Scores for all tested interactions on the chip platform with the SNAP than with the IP chemical architecture. The large error and deviation for the interaction Z-Scores on both platforms are highly dependent on the proteins examples and are expected to decrease for different protein samples. But more importantly, the interaction result shows that SNAP chemical architecture in conjugation with the parallel processing micro-architecture on the chip is equally functional.
Figure 4.
Comparison between protein-protein interaction measurements performed on microfluidic chips with SNAP or IP chemical architecture. A and B) Histograms of the Z-Scores obtained for the self- and cross-interactions of S100A1 and S100B on chips with SNAP and IP chemical architecture, respectively. Red and black bars denote the positive protein interaction set and negative control set. The positive-interaction set comprised 24 repeats for each of the four test interactions and the control set comprised 192 spots including no bait, no prey, and no protein controls. Insets in A and B show boxplots for the separated tested protein interactions with a Z-Score higher than 2, which is used as threshold value for identification of positive protein-protein interactions (dotted line in the histograms).
Conclusion
Together, the redesign of the chemical and micro-architecture of the microfluidic chips for testing protein-protein interactions led to a significant reduction of its runtime. In detail, by processing the integrated biochemical steps of the pull-down assay and the in vitro expression of the bait and prey proteins in parallel, we could reduce the runtime of the application by 1.5 h. Further, the redesigned chemical architecture of the PPI platform led to a reduction of 2.5 h. The gain in runtime is due to the reduction of steps in the buildup phase of the protein pull-down assay. After the redesign, the processes 1 and 2 of the chemical architecture both had the same length of 1.5 h. Therefore, no lag time is created when processes 1 and 2 are started in parallel on the chip. The duration of the protein interaction detection step has not been changed. In summary, the total runtime for the PPI assay is 2.5 h on the next-generation microfluidic chip platform compared to 6.5 h on the first-generation chip. 9,17 The total number of unit cells (640) and its size (~0.22 mm2) was unchanged during the redesign process. Therefore, a performance increase by a factor of about 3 is calculated from the previous to the next chip generation. Attended with the redesign of the chemical architecture was a sensitivity gain of protein interaction assay by a factor of two. Reduction of the dependence of the antibodies within the assay led to the signal-to-noise increase.
Microfluidic chip performance increase is not always possible by minimizing the biochemical assays to smaller volumes. The challenges in engineering for small volumes include signal-to-noise problems that arise in standard detection technologies. For the binary protein interaction chip platform, a reduction of the reaction volume to improve performance would not be possible without major changes of the optical detection technology; however, we have shown here that dramatic performance improvement can be obtained by coupled redesign of architecture and chemistry. Future microfluidic chip designs will include a much stronger interplay between chemical and micro-architecture to overcome volume restrictions.
Supplementary Material
ACKNOWLEDGMENTS
Funding Sources
The National Institutes of Health and Howard Hughes Medical Institute funded this study.
We thank Lance Martin and Aaron Streets for experimental suggestions and comments on the manuscripts.
Abbreviations
- PDMS
polydimethylsiloxane
- PPI
protein-protein interaction
- IP
immuno-precipitation
- cDNA
complementary deoxyribonucleic acid
- GFP
green fluorescence protein
- NHS
N-hydroxysuccinimide
- BG
benzyl-guanosine
- BSA
bovine serum albumin
- His
histidine
Footnotes
Supporting Information. Material and methods, including chip production, chip operation, experimental details, and image analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 2.Melin J, Quake SR. Annu Rev Biophys Biomol Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 3.Moore EG. Electronics. 1965;38:114–118. [Google Scholar]
- 4.Thorsen T, Roberts RW, Arnold FH, Quake SR. Phys Rev Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 5.Song H, Ismagilov R. J Am Chem Soc. 2003;125:14613–14619. doi: 10.1021/ja0354566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heyries KA, Tropini C, VanInsberghe M, Doolin C, Petriv OI, Singhal A, Leung K, Hughesman CB, Hansen CL. Nat Meth. 2011;8:649–651. doi: 10.1038/nmeth.1640. [DOI] [PubMed] [Google Scholar]
- 7.Shen F, Du W, Kreutz JE, Fok A, Ismagilov RF. Lab Chip. 2010;10:2666. doi: 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borkar S. San Diego, California, USA: 2007. [Google Scholar]
- 9.Maerkl SJ, Quake SR. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 10.Kartalov EP, Walker C, Taylor CR, Anderson WF, Scherer A. Proc Natl Acad Sci USA. 2006;103:12280–12284. doi: 10.1073/pnas.0602890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Proc Natl Acad Sci USA. 2004;101:9955–9959. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalon D, Smith SJ, Brown PO. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, O'Hanlon D, Heizmann CW, Marks A. Exp. Cell Res. 1999;246:501–509. doi: 10.1006/excr.1998.4314. [DOI] [PubMed] [Google Scholar]
- 14.Rustandi RR, Baldisseri DM, Inman KG, Nizner P, Hamilton SM, Landar A, Landar A, Zimmer DB, Weber DJ. Biochemistry. 2002;41:788–796. doi: 10.1021/bi0118308. [DOI] [PubMed] [Google Scholar]
- 15.Smith SP, Shaw GS. Structure. 1998;6:211–222. doi: 10.1016/s0969-2126(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 16.Fordyce PM, Gerber D, Tran D, Zheng J, Li H, DeRisi JL, Quake SR. Nat Biotechnol. 2010;28:970–975. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber D, Maerkl SJ, Quake SR. Nat Meth. 2009;6:71–74. doi: 10.1038/nmeth.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.