Abstract
Most human tumors have abnormal numbers of chromosomes, a condition known as aneuploidy. The mitotic checkpoint is an important mechanism that prevents aneuploidy by restraining the activity of the anaphase-promoting complex (APC). The deubiquitinase USP44 was identified as a key regulator of APC activation; however, the physiological importance of USP44 and its impact on cancer biology are unknown. To clarify the role of USP44 in mitosis, we engineered a mouse lacking Usp44. We found that USP44 regulated the mitotic checkpoint and prevented chromosome lagging. Mice lacking Usp44 were prone to the development of spontaneous tumors, particularly in the lungs. Additionally, USP44 was frequently downregulated in human lung cancer, and low expression correlated with a poor prognosis. USP44 inhibited chromosome segregation errors independent of its role in the mitotic checkpoint by regulating centrosome separation, positioning, and mitotic spindle geometry. These functions required direct binding to the centriole protein centrin. Our data reveal a new role for the ubiquitin system in mitotic spindle regulation and underscore the importance of USP44 in the pathogenesis of human cancer.
Introduction
While aneuploidy has been observed in human tumors for more than a century, its role in the genesis of cancer continues to be a point of debate. Recent mouse data suggest that aneuploidy itself is capable of driving the development of tumors (1–3); however, only some mouse models that are prone to aneuploidy have increased tumor susceptibility (4). This suggests that there may be a select group of chromosome instability (CIN) genes that play a disproportionally important tumor-suppressive role that is not necessarily related to the magnitude of aneuploidy produced in mutant animals. To date, only a handful of such genes have been identified, many with limited evidence for direct importance for human tumors. To prevent errors in mitosis, a complex machinery known as the spindle assembly checkpoint, or mitotic checkpoint, acts to delay the onset of anaphase until all sister chromatids are properly attached to spindle microtubules (5, 6). Unoccupied kinetochores lead to the formation of an inhibitory complex, comprising MAD2, BUBR1, and BUB3, known as the mitotic checkpoint complex (MCC) (7). This complex inhibits the activity of the large, multi-subunit ubiquitin E3 ligase known as the anaphase-promoting complex (APC) through binding to the co-activating subunit CDC20 (APCCDC20) (8, 9). When all kinetochores are properly attached to spindle microtubules, the checkpoint becomes silenced and the MCC dissociates from APCCDC20, leading to UBCH10 (UBE2C)–dependent polyubiquitination and proteasomal degradation of cyclin B1 (CCNB1) and securin (PTTG1) — two proteins that inhibit the activity of the enzyme separase (ESPL1) (10, 11). Once liberated from this inhibition, separase cleaves the ring-like cohesin structures that connect replicated sister chromatids, leading to the onset of anaphase (12).
Two recent functional genetic screens identified the deubiquitinase USP44 as an important regulator of the mitotic checkpoint (13, 14). Depletion of USP44 leads to unscheduled silencing of the mitotic checkpoint, increasing the risk of entry into anaphase prior to the complete attachment of all chromosomes to the mitotic spindle. A model was proposed in which the checkpoint was silenced through a mechanism involving UbcH10-dependent polyubiquitination of CDC20 that leads to the dissociation of the MCC (14, 15). USP44 is thought to counteract this process by deubiquitinating CDC20, thereby maintaining the complex between the MCC and CDC20 (16).
To address the physiological relevance of USP44 and to clarify its role in mitosis, we engineered a mouse with a deletion of the Usp44 gene. In contrast to most other mitotic regulators, USP44 is not essential for cell viability or mouse embryogenesis, as Usp44-homozygous-null mice were born at Mendelian frequencies and were overtly healthy. Cells from Usp44-null animals have cellular and biochemical features consistent with a role for USP44 in regulating the mitotic checkpoint. Surprisingly, however, correction of the checkpoint defect through pharmacologic inhibition of APC/C failed to correct chromosome mis-segregation. We describe a novel interaction that is required in order to prevent chromosome lagging between USP44 and the centriole protein centrin. We observe that a pool of USP44 localizes to the centrosome and that cells lacking USP44 have abnormal spindle geometry due to abnormal centrosome separation and positioning. At the physiological level, mice lacking USP44 are highly prone to spontaneous tumors, particularly in the lungs. We also found significant reductions in USP44 expression in human bronchial adenocarcinomas, and strikingly, these reductions strongly predicted a poor prognosis. These data establish a novel role for USP44 in centrosome functioning and mitotic spindle formation and identify USP44 as a key tumor suppressor.
Results
Usp44 loss causes chromosome mis-segregation and aneuploidy.
To study the role of USP44 in mice, we inserted LoxP sites flanking exon 1 such that, in the presence of Cre recombinase, it would be excised in its entirety (Figure 1A). This exon encodes approximately 75% of the residues of USP44 (aa 1–475 based on NM_001206851.1), including the N-terminal zinc finger domain and the catalytic cysteine. To generate the null allele (Usp44–), we crossed Usp44+/h mice with the protamine-Cre (Prm-Cre) strain to excise exon 1 in the male germline (17). To generate homozygous-null animals, we intercrossed Usp44+/– mice. Surprisingly, Usp44–/– animals were viable, were obtained at near-Mendelian frequencies, and had no substantial changes in growth patterns compared with Usp44+/– or wild-type mice (Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI63084DS1). Usp44-null animals were able to interbreed and produce normal litter sizes. Despite extensive efforts (see Methods), we were unable to identify an antibody that can detect endogenous USP44 by immunoblotting. The deletion of exon 1 was therefore verified by Southern blotting in ES cells, as well as RT-PCR and direct sequencing in MEFs (Supplemental Figure 1, C–F). On this basis, we conclude that USP44 is not essential for mouse development.
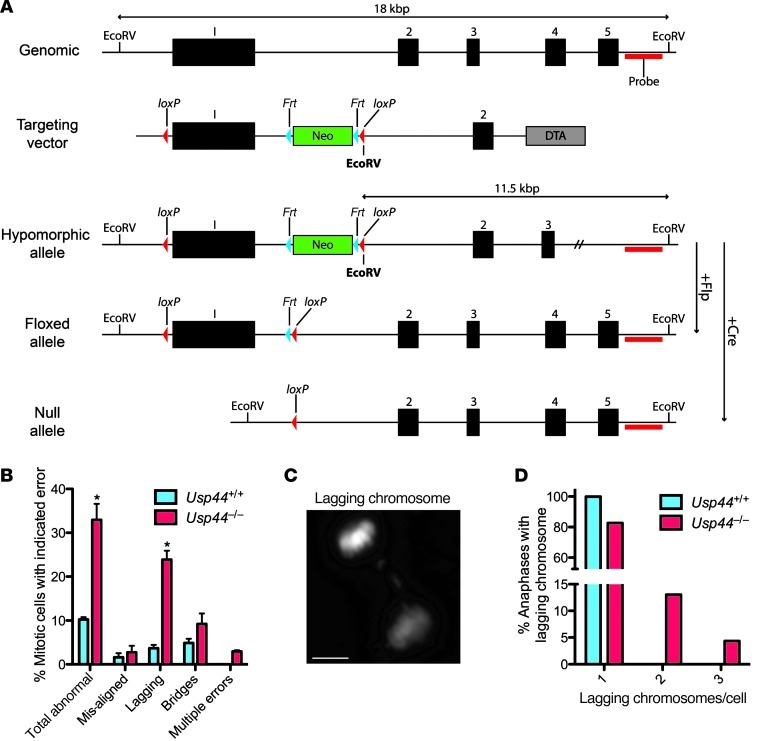
Figure 1. USP44 loss leads to chromosome mis-segregation.
(A) Strategy for targeting the Usp44 gene in mice. The Usp44 gene with indicated EcoRV sites, location of the Southern probe, the targeting vector with NeoR, Frt and loxP sites, and the resulting hypomorphic floxed and null alleles. (B) MEFs of the indicated genotypes were transduced with H2B-YFP to visualize chromosomes and followed through mitosis by live-cell microscopy. Values represent the mean ± SEM of 3 independent lines per genotype (n = 137 Usp44+/+ and n = 91 Usp44–/–). (C) Still image of lagging chromosome captured from live-cell time-lapse microscopy from Usp44–/– MEFs transduced with H2B-YFP. Scale bar: 5 μm. (D) Cells encountering a lagging chromosome were scored for the number of chromosomes involved in the error. Graph represents the percent incidence of each error in the overall population of cells observed. *P < 0.05, 2-tailed unpaired t test.
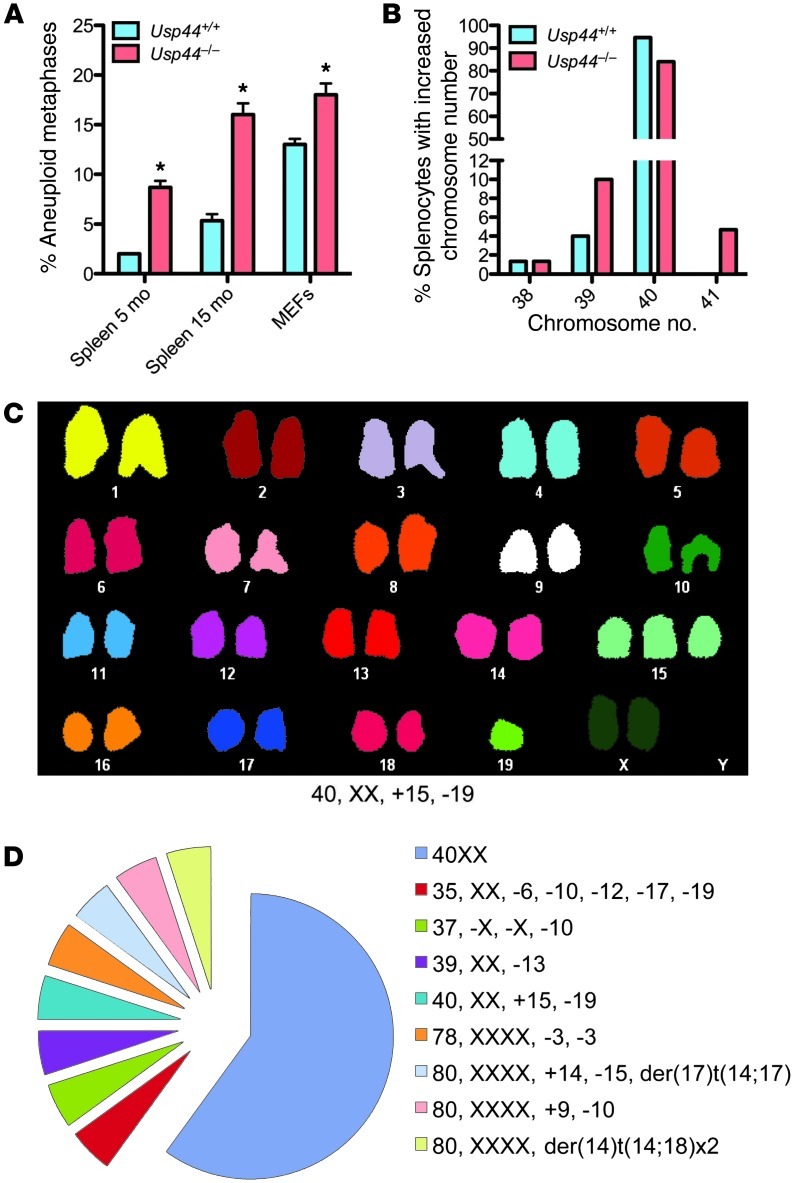
To understand the role of USP44 in mitotic progression, we followed Usp44-null and wild-type MEFs through unperturbed mitoses by live-cell fluorescence microscopy. Chromosomes were visualized through the expression of a fusion between histone H2B and yellow fluorescent protein (H2B-YFP) (18). We found a significant increase in chromosome segregation errors in the absence of USP44 compared with controls, with the vast majority involving chromosome lagging (Figure 1, B and C). The majority of cells with errors had 1–3 lagging chromosomes (Figure 1D). We next examined whether this chromosome mis-segregation resulted in aneuploidy by performing metaphase chromosome counts on Giemsa-stained metaphases following short-term culture in colcemid. We found a significant increase in aneuploidy in passage 5 Usp44–/– MEFs compared with controls (Figure 2A). We observed no chromosome breaks or gaps in the Giemsa-stained chromosomes, nor by spectral karyotyping (SKY), indicating that the defect involves the loss or gain of whole chromosomes (Figure 2, C and D). Aneuploidy was also seen in splenocytes in vivo, with rates that increased with age (Figure 2, A and B). To investigate whether these mitotic defects might result from a dominant negative function of residual truncated USP44 protein resulting from the use of an alternative start site, we cloned a cDNA corresponding to a potential ORF using the next in-frame methionine in exon 2 as the new start site (Supplemental Figure 2A). This construct (USP44C25HA) encodes an approximately 25-kDa protein representing the C terminus of USP44. To examine its potential impact on mitosis, we cloned it into the lentiviral vector TSiN and transduced wild-type MEFs. In repeated experiments, we noted that the fragment was not efficiently expressed compared with full-length USP44 (Supplemental Figure 2B), indicating a potential inherent instability of this fragment of USP44. Additionally, cells expressing USP44C25HA exhibited no increase in mitotic errors compared with control MEFs (Supplemental Figure 2C). In contrast, we previously observed that overexpression of full-length USP44 results in an increase in mitotic errors in MEFs (19). These data strongly suggest that any residual USP44 fragment is likely present in small amounts and has no dominant negative effect on mitosis. Taken together, these data suggest that USP44 is required in order to prevent chromosome mis-segregation and that the loss of this function results in aneuploidy.
Figure 2. USP44 loss leads to whole-chromosome aneuploidy in vitro and in vivo.
(A) Chromosome counts were performed on MEFs and splenocytes of the indicated genotype at the indicated ages. Values represent the mean ± SEM of 3 independent lines or animals. n = 150 for each genotype. (B) Incidence of splenocytes from A with the indicated numbers of chromosomes at 15 months. The graph represents the percent incidence of cells with each chromosome count out of the entire cohort of cells observed. (C) Representative spectral karyotype (SKY) from Usp44–/– MEFs showing pseudo-diploidy. Note that despite the normal chromosome number (i.e., 40), there is trisomy 15 and monosomy 19. (D) Complete SKY data from C. n = 20. *P < 0.05, 2-tailed unpaired t test.
USP44 has a checkpoint-independent role in regulating chromosome segregation.
To examine the impact of USP44 loss on checkpoint functioning, we studied the ability of Usp44-null MEFs to initiate and maintain checkpoint-dependent mitotic arrest. After the addition of nocodazole, H2B-YFP–expressing MEFs were marked in early prophase and followed by use of time-lapse microscopy. We observed a significant decrease in the duration of mitotic arrest in Usp44-null MEFs compared with controls, consistent with a reinforcing role in the mitotic checkpoint (Figure 3A). In the normal progression through mitosis, cells rarely would need to maintain a checkpoint arrest lasting hours. Certain defects in the mitotic checkpoint (e.g., Bub1b [BUBR1] or Mad2l1 [MAD2] loss), however, have a direct effect on the timing of mitosis through the premature activation of APC (20). We followed H2B-YFP–expressing MEFs through unchallenged mitoses and measured the time required to progress from prophase to anaphase. We observed a significant mitotic acceleration in MEFs lacking USP44 compared with wild-type MEFs (Figure 3B). This prompted us to examine the amount of MAD2 bound to CDC20 in mitotic MEFs. Consistent with prior reports, we found an average reduction of 57% in the recovery of MAD2 from CDC20 immunoprecipitates in Usp44-null cells compared with controls (Supplemental Figure 3, A and B). There were no changes in the overall levels of the APC substrates cyclin B1 or securin or of the APC components CDC20, CDC27, or BUBR1 in immunoblots (Supplemental Figure 4, A and B). We did observe an increase in the levels of MAD2, and we hypothesized that this may have been a cellular response in an attempt to compensate for USP44 loss. Examination of Usp44–/–Mad2l1+/– MEFs suggested that this was not the case, however, as USP44 loss did not exacerbate the chromosome instability phenotype resulting from reduced MAD2 (Supplemental Figure 4, C–E). We further found no evidence of cyclin B1 destabilization in fixed or living cells by microscopy (Supplemental Figure 5, A and B). This may reflect the modest (~50%) reduction in MAD2 association with CDC20, as degradation of cyclin B is not overtly accelerated in MAD2L1+/– human or mouse cells despite clear mitotic defects (21).
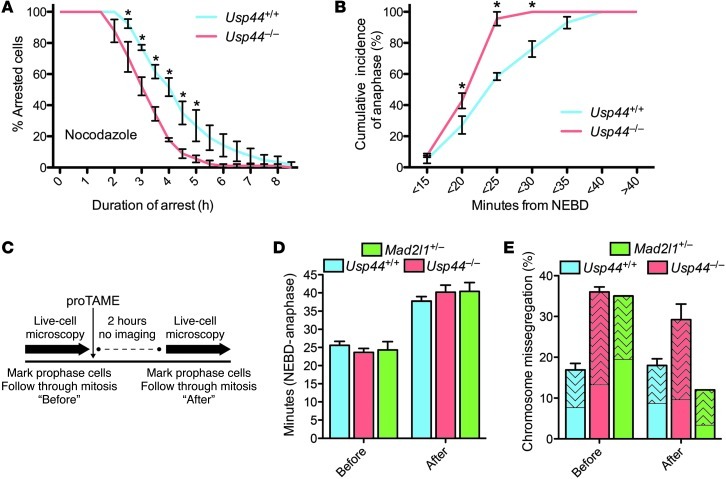
Figure 3. Correction of the checkpoint defect in Usp44-null cells does not prevent mitotic errors.
(A) MEFs of the indicated genotype were transduced with H2B-YFP, followed by live-cell microscopy. Mitotic arrest duration was determined by calculating the time from nuclear envelope breakdown (NEBD) to chromosome decondensation. Graph represents the mean percentage ± SEM of cells remaining in mitosis at each time point (3 lines per genotype). Total n = 50–90 cells for each genotype per condition. (B) The interval between NEBD and anaphase was determined in the absence of nocodazole for MEFs of the indicated genotypes. The graph represents the cumulative incidence of anaphase at each time point (mean ± SEM of 3 lines). Total n = 50–60 cells for each genotype. (C) Experimental scheme for D and E. MEFs expressing H2B-YFP were marked in prophase and followed by time-lapse microscopy to determine mitotic duration and chromosome mis-segregation. Two hours after the addition of proTAME, new prophase cells were selected and similarly examined. (D) Duration of mitosis for Usp44+/+, Usp44–/–, and Mad2l1+/– MEFs imaged before and after proTAME addition. The graph represents the mean ± SEM for 3 lines per genotype. Total n = 25–35 cells per time period. (E) The effect of proTAME on chromosome mis-segregation in Usp44+/+ (n = 67 before, 83 after), Usp44–/– (n = 65 cells per time period from 3 MEF lines), and Mad2l1+/– MEFs (n = 20–25 cells per time period, 1 MEF line). The patterned area indicates the proportion of cells encountering at least one lagging chromosome. The graph represents the mean ± SEM. *P < 0.05, 2-way ANOVA.
We next sought to establish whether the mitotic defects would be corrected by providing Usp44-null MEFs with additional time to achieve proper microtubule-kinetochore attachments. To accomplish this, we used a recently described small molecule inhibitor of APC (proTAME, ref. 22). At high doses, this inhibitor leads to complete metaphase arrest, consistent with inhibition of APC. We incubated cells with a low dose (6 μM) of proTAME, such that anaphase was delayed by 12–18 minutes without inducing mitotic errors (Figure 3, C and D). Consistent with a prior report, this strategy was able to correct the mitotic defects seen in Mad2l1+/– MEFs down to levels seen in wild-type MEFs (compare with Figure 1B), validating the approach (Figure 3E; ref. 22). Surprisingly, despite a delay in anaphase onset, APC inhibition was unable to prevent chromosome mis-segregation in Usp44-null cells. We conclude that while there is a clear yet modest defect in mitotic checkpoint activity in the absence of USP44, this does not seem to be the primary mechanism behind the chromosome instability seen in cells or tissues lacking this enzyme.
A novel USP44-centrin complex is required in order to prevent chromosome mis-segregation.
Lagging chromosomes result from aberrant attachments of spindle microtubules to kinetochores, most commonly those resulting from a single kinetochore attached to both spindle poles — an error known as a merotelic attachment. Increased levels of lagging chromosomes may therefore result from either defects in the correction of these errors, which depends on the Aurora B “error correction pathway,” or an increased rate of formation of erroneous attachments (23–25). We found no defects in the level, localization, or activity of Aurora B (AURKB) toward the substrate CENP-A in Usp44-null MEFs (Supplemental Figure 6, A and B). We therefore suspected that there was an increased rate of formation of merotelic attachments. Recent work revealed a connection between spindle defects and merotelic attachments (26, 27), leading us to examine the integrity of the centrosome and mitotic spindle. We found a significant number of Usp44–/– MEFs with abnormal spindle geometry, with the spindle poles not lying perpendicular to the metaphase plate (Figure 4, A and B). This corresponded to abnormal placement of centrosomes away from the perpendicular axis in these cells. The abnormal spindles were characterized as having an acute angle between the spindle and the metaphase plate, with nearly 30% of Usp44-null cells having an angle of less than 82° compared with 6% in wild-type MEFs (Supplemental Figure 7, A and B). The cells with abnormal spindle geometry also had a significant reduction in the inter-pole distance (Figure 4C).
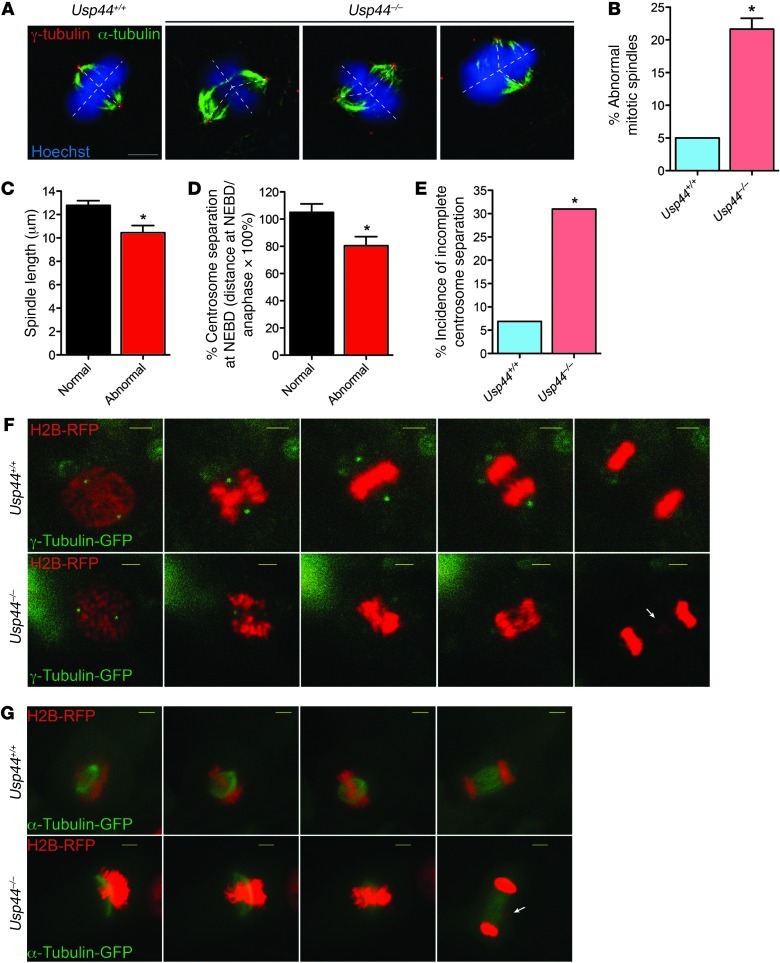
Figure 4. USP44 is required for proper centrosome separation and spindle geometry.
(A) Confocal immunofluorescence microscopy was performed on MEFs of the indicated genotypes using antibodies against the indicated proteins. Scale bar:5 μm. (B) Incidence of spindle abnormalities in MEFs of the indicated genotypes. Graph represents the mean ± SEM for 3 lines per genotype, with at least n = 20 cells per line. (C) Comparison of the inter-pole distance, as defined as the distance between γ-tubulin–positive centrosomes, in metaphases with normal versus abnormal spindle geometry. Images collected by confocal immunofluorescence microscopy. Graph represents the mean ± SEM for n = 12–13 metaphases per group regardless of genotype. (D) MEFs were transduced with γ-tubulin–CFP and H2B-RFP and followed through unperturbed mitoses with live-cell microscopy. The inter-pole distance was calculated at nuclear envelope breakdown (NEBD) and the onset of anaphase in Usp44+/+ (n = 31) and Usp44–/– cells (n = 26), and the data are expressed as the ratio between these values. Graph represents the mean ± SEM for cells with normal (n = 45) versus abnormal (n = 14) chromosome segregation, regardless of genotype. (E) The incidence of incomplete centrosome separation (inter-pole distance at NEBD <75% that at anaphase onset) was determined in MEFs of the indicated genotype from B. The graph represents the percent incidence of cells with incomplete centrosome separation in the entire cohort of cells observed. (F and G) Examples of MEFs with normal or abnormal centrosome separation (F) or spindle geometry (G) as seen by live-cell microscopy. Scale bars: 5 μm. *P < 0.05, 2-tailed unpaired t test.
Recently, lagging chromosomes were found to occur at an increased frequency in cells experiencing delayed centrosome separation (28). Therefore, to further investigate the relationship between the spindle anomalies and chromosome mis-segregation, we performed live-cell microscopy on MEFs expressing fluorescent γ-tubulin (TUBG1), in addition to fluorescent histone H2B, to allow monitoring of centrosome and DNA movements. In cells that successfully completed mitosis, centrosomes largely separated prior to nuclear envelope breakdown (NEBD), achieving an inter-centrosome distance of nearly 100% of the distance seen at anaphase onset (Figure 4D). In contrast, cells that experience a defect in anaphase chromosome movements (mostly lagging chromosomes) had a significant reduction in the degree of prior centrosome separation, achieving on average only 80% of the inter-pole distance determined at anaphase onset (Figure 4D). To help explain the increased chromosome mis-segregation rate seen in the absence of USP44, we next evaluated the incidence of incomplete centrosome separation (defined as <75% separation at NEBD) between wild-type and Usp44-null MEFs. There was a substantial and significant increase in the rate of incomplete centrosome separation in Usp44–/– compared with wild-type MEFs (Figure 4, E and F). To examine the connection between abnormal spindle morphology and mitotic errors, we monitored cells expressing α-tubulin–RFP (TUBA1A) and H2B-GFP. We observed that an average of 26% of Usp44-null cells developed an abnormal spindle, compared with 15% in wild-type controls. However, most cells that developed abnormal spindles in the early phases of mitosis eventually achieved a metaphase plate that appeared largely normal — with the spindle-poles perpendicular to the DNA (Figure 4G). This suggests that the abnormal spindle geometry seen in experiments using fixed cells represent intermediate forms that are resolved prior to anaphase. Compiling the data from 118 cells from 6 cell lines of either genotype, there was a very strong association between abnormal spindle morphology and eventual anaphase chromosome movement errors, with 88% of those with abnormal spindles going on to experience an error (Figure 4G and Supplemental Figure 7C). Conversely, 87% of those cells that experienced an error had an abnormal spindle shape prior to the error. Taken together, these data demonstrate that USP44 is required for proper centrosome separation and spindle geometry. Defects in these processes that result from USP44 loss highly predispose cells to mitotic chromosome mis-segregation and cause mitotic errors and aneuploidy.
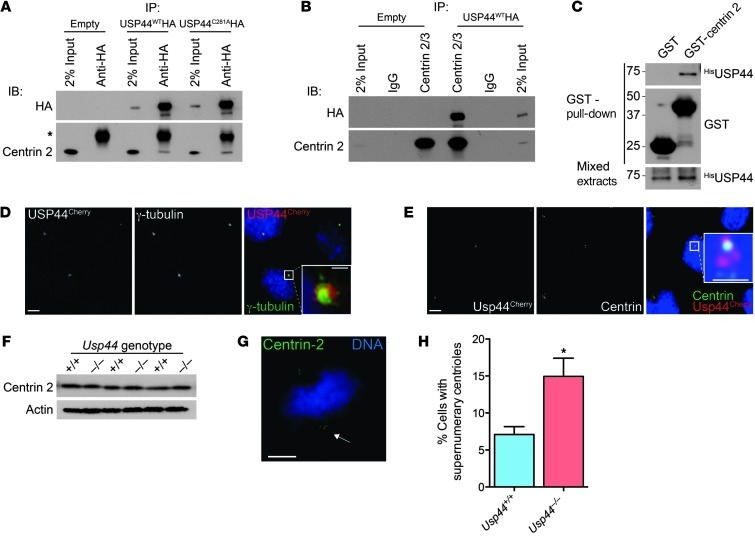
Recently, a large deubiquitinating enzyme (DUB) proteomics database was generated ( http://falcon.hms.harvard.edu/ipmsmsdbs/comppass.html) that lists the centriole protein centrin 2 (CETN2) as a candidate interacting protein of human USP44. We therefore tested the ability of endogenous centrin to interact with USP44 by co-immunoprecipitation. We observed that centrin 2 is specifically recovered with either wild-type USP44 (USP44WTHA) or catalytic mutant USP44 (USP44C281AHA; Figure 5A). These results were verified by the finding that USP44WTHA was recovered specifically in centrin 2/3 immunoprecipitates (Figure 5B). The binding of USP44 with centrin was also confirmed by GST pull-down from bacterially expressed 6xHIS-USP44 and GST–centrin 2, demonstrating that the interaction is direct (Figure 5C). We next examined whether USP44 colocalizes with centrosomes in cells expressing a fusion between USP44 and the fluorescent protein mCherry (USP44Cherry). In methanol-fixed MEFs, there was obvious colocalization of USP44Cherry with the centrosome marker γ-tubulin and centrin (Figure 5E and Supplemental Figure 7D). The colocalization was most prominent in interphase cells, with little USP44 seen at the centrosome in mitotic cells (data not shown), consistent with our prior observation that levels of exogenous USP44 are very low in mitosis (19). Although we found no change in the level of centrin 2 in immunoblots (Figure 5F), Usp44-null MEFs had a significant increase in cells with 3 centrioles at one pole (Figure 5, G and H). We also noted some highly disordered centriole configurations in cells overexpressing USP44Cherry, perhaps contributing to the chromosome mis-segregation seen in these cells (Supplemental Figure 7D and ref. 19). We conclude that USP44 forms a complex with centrin 2/3 and localizes to centrosomes.
Figure 5. USP44 is required for timely centrosome separation.
(A and B) MEFs transduced with the indicated USP44 constructs were subjected to immunoprecipitation using the indicated antibodies. Precipitates were immunoblotted using antibodies against the indicated proteins. Asterisk indicates IgG light chain. (C) N-terminally 6xHis-tagged USP44, GST, or GST–centrin 2 was expressed in bacteria, and resulting extracts were mixed, followed by affinity purification with glutathione agarose. Precipitates were immunublotted using the indicated antibodies. (D and E) Confocal immunofluorescence microscopy was performed on methanol-fixed MEFs transduced with USP44Cherry using antibodies against the indicated proteins. Scale bar: 5 μm; insets, 1 μm. (F) The level of centrin 2/3 was determined in asynchronous MEFs of the indicated genotypes. Extracts were immunoblotted using antibodies for the indicated proteins. (G) Example of an Usp44-null cell with a supernumerary centrin signal at one spindle pole. Scale bar: 5 μm. (H) Incidence of one or more extra centrin signals in at least one spindle pole in MEFs of the indicated genotypes. Graph represents the mean ± SEM from 3 lines per genotype, 20 cells per line. *P < 0.05, unpaired t test.
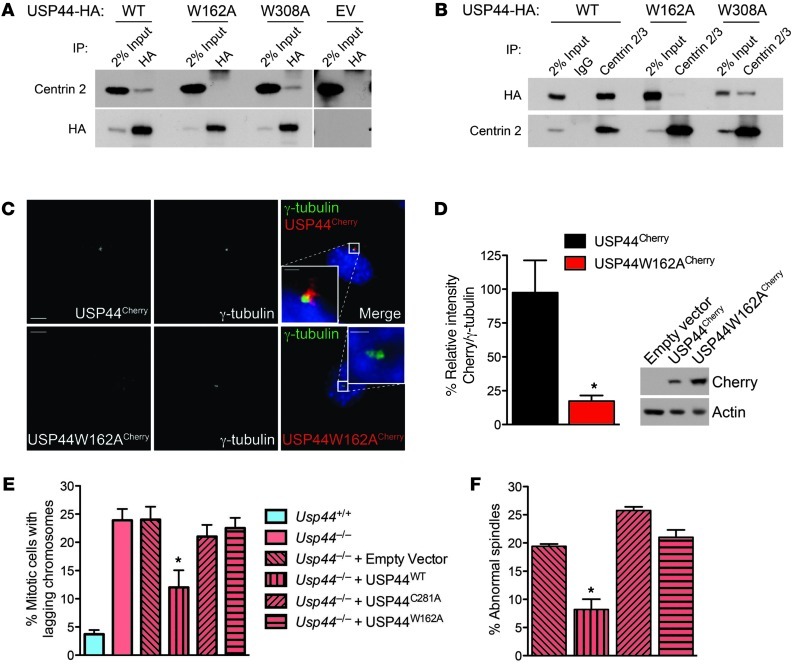
A conserved centrin-binding domain (CBD) has been defined in the centrin partners XPC and SFI1 (Supplemental Figure 8, A and B, and refs. 29–31). We scanned the sequence of USP44 and found two putative CBDs that are highly conserved in mammals (Supplemental Figure 8A). To confirm the centrin-binding capacity of these motifs, we prepared USP44 mutants (USP44W162AHA and USP44W308AHA) and examined their ability to co-precipitate with centrin. We found a profound reduction in centrin binding to USP44W162AHA, whereas there was only a slight reduction in binding to USP44W308AHA (Figure 6, A and B). We also observed a dramatic reduction in the localization of USP44W162ACherry to the centrosome, consistent with an important role for centrin binding in this localization (Figure 6, C and D). We next tested the ability of these mutant constructs to correct lagging chromosomes seen in Usp44-null MEFs. In contrast to the rescue observed with USP44WT, transduction with empty virus, USP44C281A, or USP44W162A was unable to affect the rate of lagging chromosomes despite equal levels of expression (Figure 6E and Supplemental Figure 2B). Similarly, complementation of Usp44-null MEFs with USP44WTHA restored spindle geometry, whereas USP44C281AHA or USP44W162AHA could not (Figure 6F). USP44W162A retained catalytic activity toward di-Ub chains in vitro, making catalytic inactivity an unlikely cause of its failure to rescue chromosome lagging (Supplemental Figure 9, A and B). Taken together, these data demonstrate that USP44 forms a complex with centrin through a highly conserved motif and that both DUB activity and the ability to form a complex with centrin are required in order to prevent chromosome lagging.
Figure 6. The USP44-centrin complex is required in order to prevent chromosome mis-segregation.
(A and B) MEFs transduced with HA-tagged wild-type USP44 (WT), USP44W162A (W162A), USP44W308A (W308A), or empty vector (EV) were subjected to immunoprecipitation using the indicated antibodies. Precipitates were immunoblotted using antibodies against the indicated proteins. (C) Confocal immunofluorescence microscopy was performed on methanol-fixed MEFs transduced with USP44WTCherry or USP44W162ACherry using antibodies against the indicated proteins. Scale bar: 5 μm; insets, 1 μm. (D) Quantitation of USP44Cherry and USP44W162ACherry at centrosomes. Images were captured with identical settings, and the cherry signal was normalized to that of γ-tubulin. n = 11 cells from each genotype. Graph represents the mean ± SEM. (E) MEFs of the indicated genotype were transduced with empty vector or the indicated USP44 constructs followed by live-cell microscopy. Chromosomes were visualized through the expression of H2B-YFP. The graph represents the mean ± SEM of 3 lines per genotype. n = 71–137 total cells per condition. (F) Cells treated as in E were fixed and stained with α- and γ-tubulin, followed by confocal microscopy. Spindles with poles off axis were scored as abnormal. The graph represents the mean ± SEM of 3 lines per genotype. *P < 0.05, 2-tailed unpaired t test.
USP44 suppresses tumorigenesis.
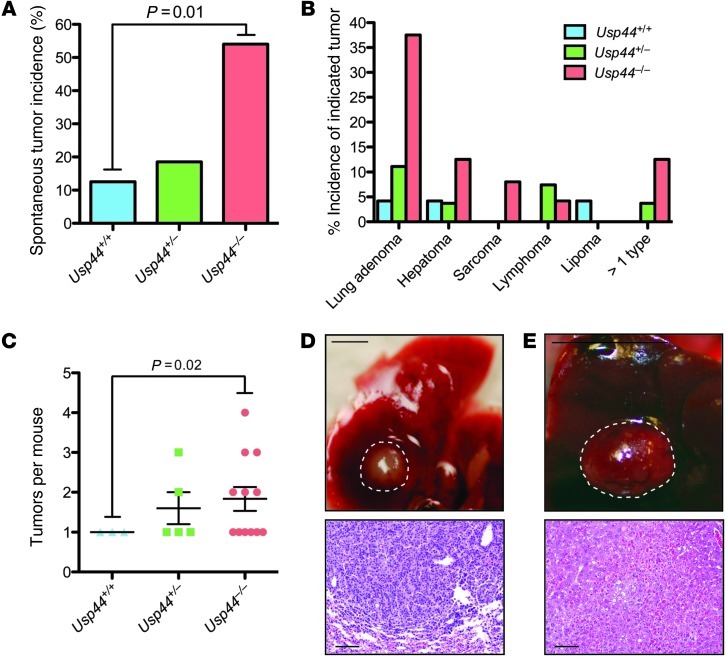
We generated cohorts of Usp44+/+, Usp44+/–, and Usp44–/– mice and aged them for 15 months to determine the incidence of spontaneous tumors. There was a dramatic increase in tumorigenesis in the Usp44-null group, with 12 of 24 (50%) having at least one tumor type (Figure 7A), and a significant increase in tumor burden in Usp44-null animals, with some having 2–4 tumors (Figure 7, B and C). The tumor spectrum observed was diverse, with 4 distinct histologic types observed (Figure 7, B, D, and E). Strikingly, there was a 9-fold increase in the number of lung tumors in Usp44-null animals, with histology consistent with bronchiolar adenomas. While aneuploidy was not observed in the normal tissues, 5 of the 10 lung tumors from Usp44-null mice had substantial losses or gains at either chromosome 4 or 7 by fluorescence in situ hybridization on paraffin-embedded sections (Supplemental Figure 10A). We conclude that Usp44 is a tumor suppressor gene that, when deleted, leads to tumorigenesis in the context of increased aneuploidy.
Figure 7. USP44 is a tumor suppressor.
A. Mice of the indicated genotypes (n = 27, 27, and 25 for Usp44+/+, Usp44+/–, and Usp44–/–, respectively), were sacrificed at 15 months of age and were dissected and scored for visible tumors. Graph represents the percent incidence of spontaneous tumors in the overall cohorts. The P value was calculated using Fisher’s exact test. (B) Tumor spectrum seen in mice from A. All tumor classifications were confirmed by histopathology. The graph represents the percent incidence of each tumor type in the overall cohorts. (C) Graph represents the total number of tumors per mouse, regardless of histology (mean ± SEM) including all that had at least 1 tumor. The P value was calculated using a 2-tailed 1-sample t test. (D and E) Gross (scale bars: 1 mm) and microscopic (scale bars: 50 μm) appearance of representative lung adenoma (D) and liver hepatoma (E) from Usp44-null mice.
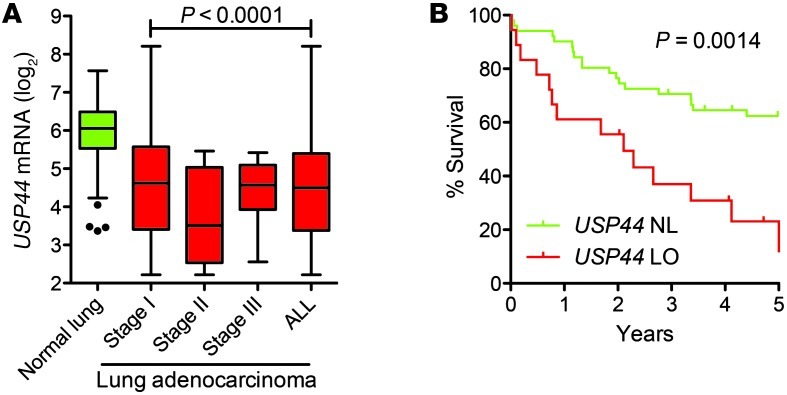
Given the striking incidence of lung adenomas in Usp44-null animals, we next asked whether there was an association between USP44 downregulation and human adenocarcinoma. To accomplish this, we queried a microarray expression dataset from patient specimens included in the Mayo Clinic Lung Specimen Registry. Expression data were obtained from 194 fresh-frozen resected lung specimens consisting of tumor and adjacent normal lung tissues derived from 155 patients with stages I–III lung adenocarcinoma. There was a statistically significant reduction in the expression of USP44 in all stages of disease compared with normal lung as assessed by analyzing 100 normal and 94 tumor tissues of mixed stage and outcome (Figure 8A; P < 0.0001). To assess the potential clinical significance of USP44 in these tumors, we analyzed survival in this patient cohort. As there is a significant association of stage and outcome, we limited the analysis to the group of patients with stage I disease (n = 69). When dividing patients into USP44-low (<27th percentile) versus normal (28–100th percentile), we found that low levels of USP44 had a highly significant negative impact on overall survival (P = 0.0014; Figure 8B). To test the frequency of aneuploidy in lung adenocarcinoma, we performed FISH for chromosomes 7 and 8 on archived paraffin sections in a selection of cases with normal or low levels of USP44 mRNA using FISH. Compared with the results from adjacent normal lung tissues, nearly all tumor samples had substantial aneuploidy, with the majority exhibiting chromosome gains with up to 10 copies of the analyzed chromosomes (Supplemental Figure 10B). These results demonstrate that aneuploidy is very common in human adenocarcinoma of the lung, but suggest that reduced USP44 expression may be only one of the ways through which this genetic instability is achieved. Taken together, our results indicate that reduced expression of USP44 is common in patients with lung adenocarcinoma and that its reduction is associated with a more aggressive tumor biology and poor clinical outcome.
Figure 8. Reduced expression of USP44 is a marker of poor prognosis in human adenocarcinoma at the lung.
(A) Level of USP44 mRNA (log2) measured by microarray in lung adenocarcinomas of the indicated stages compared with adjacent normal tissue. n = 100 normal lung specimens and 69, 12, and 13 stage I, II, and III tumors respectively. Graph represents a Tukey plot, with the bar representing the median, the box representing the 25th and 75th percentiles, and the whiskers representing 1.5 times the interquartile distance. The P value was calculated using a 2-tailed unpaired t test. (B) Kaplan-Meier survival curve for stage I tumors (n = 69) with normal (NL; >28th percentile) or low (LO; <27th percentile) USP44 mRNA. The P value was calculated with the log-rank test.
Discussion
The deubiquitinase USP44 was previously identified in two functional genomic screens designed to uncover novel regulators of the mitotic checkpoint. Several key questions remained unanswered, however, including the physiologic relevance of USP44, whether its loss leads to in vivo chromosome instability, whether the enzyme has other functions in mitosis, and whether these functions provide antitumor protection in vivo. Using a mouse genetic approach, we here provide direct evidence that Usp44 is a tumor suppressor in mice and humans, with a particularly important role in lung cancer. We confirm the role of USP44 in modulating the mitotic checkpoint, though surprisingly, this function appears not to be its most essential function. Rather, we found an important role for USP44 deubiquitinase activity in regulating centrosome positioning and spindle geometry through a direct interaction with the centriole protein centrin. This is a previously undescribed function involving the ubiquitin-proteasome system that, when disrupted, leads to chromosome mis-segregation, aneuploidy, and tumorigenesis in vivo.
USP44 and the mitotic checkpoint.
We observe that USP44 is required for efficient mitotic checkpoint signaling, consistent with the reports of others using RNAi. The fact that cells maintain arrest for some time in the presence of nocodazole suggests that initiation of checkpoint activity is intact, but that this signal is prone to early decay. The effect is modest, however, suggesting that APC restraint is not entirely lost in the absence of USP44. An effect of similar magnitude was observed in cells overexpressing the E2 conjugating enzyme UBCH10, which is proposed to ubiquitinate CDC20 (32). These two results are consistent with an antagonistic role between the two enzymes as proposed by the Elledge group (14). Despite the clear evidence of accelerated anaphase in cells lacking USP44, we found no evidence of accelerated degradation of cyclin B1. This is not surprising in light of the modest (~50%) reduction in MAD2 association with CDC20, as degradation of cyclin B is not overtly accelerated in MAD2L1+/– human or mouse cells despite the presence of clear mitotic defects (21). Finally, there also may be redundancy in the system, with other DUBs taking over the essential checkpoint-related functions of USP44 upon its loss.
USP44 and the centrosome.
Here we describe a new function for USP44 in regulating centrosome separation and positioning. We identify and map a new interaction between USP44 and centrin that is required to prevent chromosome lagging. The direct interaction between USP44 and centrin likely serves to localize USP44 to the centrosome, as this localization is disturbed when the centrin-binding motif is mutated. USP44 does not appear to prevent proteasomal degradation of centrin, as we found no changes in the level of centrin in Usp44-null MEFs, though redundancy with other DUBs remains possible. The use of centrin as a centrosomal targeting mechanism may indicate that the substrate of USP44 is in close proximity to centrin, though further insights will require identification of the relevant substrate(s). The function of USP44 at the centrosome at least in part involves regulating the events surrounding centrosome separation, as its loss leads to an increase in cells with incomplete centrosome separation. Recent work from the Cimini laboratory demonstrates that incomplete centrosome separation strongly predisposes cells to develop merotelic attachments, as the mitotic spindle microtubules that emanate from abnormally positioned centrosomes cannot efficiently probe the kinetochore landscape (28). This leads us to conclude that the lagging chromosomes seen in cells lacking USP44 are the result of incomplete centrosome separation. In experiments using α-tubulin–GFP, we observed that that USP44 loss caused cells to have difficulties in spindle geometry early in mitosis, though most cells eventually formed a reasonable metaphase plate. However, the cells with disturbed spindle geometry early in mitosis rarely were successful in distributing their chromosomes properly, even though the spindle geometry appeared to have improved by anaphase. These data suggest that proper centrosome separation and early spindle assembly are essential for high-fidelity chromosome segregation.
The detailed events surrounding centrosome separation are not completely understood. In part, this process is triggered by the kinase NEK2A, which phosphorylates the proteins C-NAP1 (CEP250) and rootletin (CROCC), which are thought to constitute the inter-centrosomal linker (33, 34). The phosphorylation of these substrates leads to their eventual loss from the linker, allowing centrosome movement that is dependent on kinesin Eg5 (KIF11) (35). Several mitotic regulators, including UBCH10 (UBE2C) (32), UBE2S (36), APC (37), and cyclin B1 (38), localize to the centrosome prior to or during mitosis, though the exact role for this localization is not clear. Knockdown of UBCH10 and/or UBE2S leads to centrosome duplication errors in Drosophila S2 cells (36). Most interestingly, the depletion of these Ub E2 enzymes also led to spindle defects, with abnormal geometry particularly seen in cells with low levels of UBE2S, where spindle pole positioning abnormalities were found. It is possible that a cycle of ubiquitination and deubiquitination of critical centrosome substrates is also required in order to coordinate these processes. The spindle abnormalities seen in our study also have similarities to those seen in cells following depletion of proteins making up the Emi/NuMA/dynein/dynactin complex, also known as the END network (39, 40). Knockdown of any component of this network leads to similar centrosome positioning and spindle abnormalities. Interestingly, an important role of this network includes the “anchoring” of APC at the spindle pole, a function that is thought to limit the activity of APC in early mitosis (41). It is tempting to speculate based on our data that restraining APC activity may be also important for the normal positioning of the centrosome.
USP44 and cancer.
We have found that animals lacking USP44 are highly prone to spontaneous tumors, particularly of the lung. While other DUBs have been implicated as tumor suppressors based on mutations found in humans, USP44 is the first to our knowledge whose loss leads to spontaneous tumors in mice (42). While it is reasonable to conclude that the tumorigenesis in Usp44-null mice is the result of aneuploidy, our results do not exclude the possibility that there are other mechanisms by which USP44 suppresses tumors. It is noteworthy, however, that many other models carrying mutant CIN gene alleles also have increased rates of lung adenoma formation (1). There is a strong association between lung cancer and aneuploidy (43). Recently, empiric evidence supporting the long-held belief that aneuploidy increases rates of loss of heterozygocity (LOH) was provided from studies using mice with reduced levels of the checkpoint protein BUB1 crossed with animals heterozygous for TP53 or APCmin (44). Further studies are needed to explore the role of USP44 in this process and the timing of its loss in the development of lung tumors in vivo.
Methods
Generation of Usp44-null mice and other mouse strains.
Unlike other mammalian genomes, the mouse Usp44 gene is annotated to consist of two exons of nearly equal size that are predicted to encode a protein of 505 amino acids lacking an active site cysteine. Using the human gene as a template, however, the genomic sequence of mouse chromosome 10 is predicted to encode a second Usp44 ORF consisting of 5 exons translated to produce a protein with 709 amino acids, including a full catalytic triad. This predicted USP44 protein is 79% identical and 86% similar to human USP44. The expression of this “alternate,” more homologous transcript was verified by cloning the full-length mouse USP44 from cDNA generated from mouse kidney RNA, as well as by quantitative real-time PCR demonstration of the widespread presence of the “alternate” transcript in other mouse tissues. We therefore based our design for the hypomorphic and null alleles of Usp44 on this alternate transcript, which we believe to encode the bona fide Usp44 gene. The construction of the Usp44 targeting vector was based upon the multipurpose targeting strategy described previously (45) and was carried out entirely using recombinational cloning techniques as described in ref. 46. Briefly, the Usp44 gene region was cloned into the targeting vector pDTA.4b via gap repair from a bacterial artificial chromosome derived from AB2.2-ES cells (bMQ collection) obtained from Source Bioscience/Geneservice. The orphan loxP site was then introduced approximately 100 bp upstream of exon 1 by first inserting a loxP-Pgk-em7-Neo-loxP cassette from plasmid PL452, followed by Cre-mediated excision of the neo ORF, leaving only a single loxP site. We then inserted an Frt-Pgk-em7-Neo-Frt-loxP cassette from plasmid PL451 into the intronic region approximately 100 bp downstream of exon 1. The Neo ORF is in reverse orientation with respect to the Usp44 gene to allow the generation of a hypomorphic allele. The integrity of the resulting targeting construct was tested functionally by establishing the ability of the Cre recombinase to eliminate kanamycin resistance in bacteria (as successful excision removes the Pgk-em7-Neo construct in addition to exon 1), as well as through sequence confirmation of several regions of the construct. The construct was then linearized with ApaI and was introduced into murine ES cells by electroporation. G418-resistant colonies were screened for homologous recombination by Southern blotting using a probe amplified from a 750-bp region that lies 3′ to the engineered region on chromosome 10. Clones with correct recombination were karyotyped prior to blastocyst injection. Following germline transmission, the floxed allele was generated by mating Usp44+/h animals with those carrying the Flpe recombinase in all tissues (Jackson Laboratory, stock 007844). The null allele was generated through mating with mice expressing Cre in the male germline (Prm-Cre; Jackson Laboratory, stock 003328). Analysis of the Usp44 gene reveals a possible alternative start site in exon 2 that would theoretically produce a 190-residue protein lacking the conserved N-terminal zinc finger domain that is thought to be important for ubiquitin binding (47) and the cysteine that is essential for all non-JAMM/MPN class isopeptidases (48). Mad2l1+/– mice were a gift of Robert Benezra (Memorial Sloan-Kettering Cancer Center, New York, New York, USA) (49).
Evaluation of USP44 antibodies and confirmation of Usp44 deletion.
In an attempt to detect USP44 by immunoblotting or immunofluorescence, we screened all 7 published and commercially available USP44 antibodies, raised polyclonal antisera using 6 distinct immunogens (3 peptide sequences and 3 protein fragments of 50–100 residues) in a total of 10 rabbits, and raised 6 monoclonal antibodies against distinct peptide epitopes. Commercial USP44 antibodies tested included F-15, N-14, and S-14 (Santa Cruz Biotechnology Inc.), GTX87933 (GeneTex), SAB4503322 and HPA026543 (Sigma-Aldrich), and 15521-1-AP (Proteintech). USP44 polyclonal antisera were also provided by Kwang-Hyun Baek (CHA Stem Cell Institute, College of Medicine, CHA University, CHA General Hospital, Seoul, Republic of Korea) (50). All commercial and custom-produced antisera were tested in human and mouse cells and screened by the ability to (a) recognize a band at the expected size (75–80 kDa) and (b) detect a band of increased intensity of the appropriate size in cells overexpressing mouse or human USP44-HA and by (c) failure to detect a 75- to 80-kDa band in Usp44-null MEFs. No antibody satisfied these criteria. The deletion of exon 1 was therefore verified by Southern blotting, direct sequencing, and by RT-PCR on RNA from MEFs (Supplemental Figure 1, A–E).
Cell culture, reagents, and cloning.
Cells used in this study include MEFs that were generated from day 13.5 embryos (51). All experiments were performed on primary MEFs between passages 2 and 5, with the exception of CDC20 immunoprecipitation (Supplemental Figure 3), which was performed using mitotic MEFs immortalized with SV40 large T antigen (52). All MEFs were cultured as described previously (32). For all experiments, at least 3 independent MEF lines were used. Chromosome counts on MEFs were performed as previously described (44, 53). Spectral karyotypic analysis and FISH were performed by the Mayo Clinic Cytogenetics Core Facility, using the protocol, reagents, instrumentation, and software from Applied Spectral Imaging. Cell synchronization was performed as described previously (54). Generation, use, and quantitation of cyclin B1–venus were performed as described (55). The C-terminally tagged USP44WTHA and USP44Cherry constructs were previously described (19). The USP44 mutant constructs were generated using the GeneArt mutagenesis kit (Invitrogen). All USP44, α-tubulin, and γ-tubulin constructs were cloned into the lentiviral vector pTSiN, and virus production was performed as described previously (56). MEFs were transduced by culturing with a 1:1 mixture of fresh medium and virus supernatant with Polybrene (4 μg/mg final concentration) for 24–72 hours. All experiments were performed on polyclonal pools of transduced cells. For the experiments with proTAME, MEFs were imaged for 2 hours to determine the pretreatment (“before”) mitotic duration and chromosome mis-segregation rate. Following this period of observation, the APC inhibitor proTAME (a gift from Randy King, Harvard Medical School, Boston, Massachusetts, USA) was added (6 μM), followed by a 2-hour period to allow the drug to take effect. Cells were then monitored again for mitotic timing and chromosome mis-segregation (“after”).
SDS-PAGE, antibodies, immunoblotting, and immunoprecipitation.
Antibodies used in this study included anti-HA (clone 3F10; Roche Applied Science); centrin 2/3 (20H5; ref. 57); centrin 2 (Proteintech); γ-tubulin (Abcam); BubR1 (catalog 612503), Cdc27 (catalog 610454), and Mad2 (catalog 610678, BD Biosciences); BubR1 (51), Mad2, and securin (Abcam); phospho–histone H3(Ser10) (Millipore, 06-570); cyclin B1 (SC-245) and CDC20 (SC-8358; Santa Cruz Biotechnology Inc.); and human anti-centromere antibody (Antibodies Inc.). We screened all 7 published and commercially available USP44 antibodies and raised polyclonal antisera using 6 distinct immunogens (3 peptide sequences and 3 protein fragments of 50–100 residues) in a total of 10 rabbits. All resulting antisera were screened for the ability to detect a protein of the appropriate molecular weight (75–80 kDa) that increases in intensity and overlaps with HA-tagged USP44. No antisera satisfied these criteria. Commercial USP44 antibodies tested included F-15, N-14, and S-14 (Santa Cruz Biotechnology); GTX87933 (GeneTex); SAB4503322 and HPA026543 (Sigma-Aldrich); and 15521-1-AP (Proteintech). USP44 polyclonal antisera were also provided by Kwang-Hyun Baek (50). Immunoblotting was performed using standard procedures. Immunoprecipitation was performed as described previously (58). Codon-optimized murine USP44 was cloned into pET3a with an N-terminal 6xHis tag (HisUSP44) and was introduced into the bacterial strain BL21. GST-tagged human centrin 2 was previously described (59). Extracts of induced BL21 cultures were mixed and subjected to affinity purification with glutathione agarose (Pierce), followed by immunoblotting. The identity and migration of HisUSP44 was further verified by mass spectrometry.
USP44 activity assay.
USP44WTHA, USP44C281AHA, and USP44W162AHA were immunoprecipitated from MEFs in the absence of any protease inhibitors; washed thoroughly; and resuspended in buffer containing 50 mM HEPES, 150 mM NaCl, and 2 mM DTT. Equal amount of beads were transferred to separate tubes to which 500 ng di-Ub of the indicated linkage type was added. After a 30-minute incubation at room temperature, 2× Laemmli SDS sample buffer was added to stop reactions, and the mixtures were immediately boiled for 5 minutes. Samples were resolved on a 15% polyacrylamide gel and immunoblotted for ubiquitin and HA to verify equal enzyme loading.
Immunofluorescence and fluorescence live-cell microscopy.
Live-cell imaging experiments were performed as previously described in detail (18, 54). A lentiviral construct encoding YFP-tagged H2B (pTSIN-H2B-YFP) was used to visualize chromosomes by fluorescence microscopy. Indirect immunofluorescence was performed as described (58, 60). A laser scanning microscope (LSM 510 v3.2SP2; Zeiss) with Axiovert 100M (Zeiss) with a c-Apochromat ×100 oil immersion objective was used to analyze immunostained cells and capture representative images. For quantification of cyclin BCerulean and USP44Cherry levels at each mitotic stage, 10 or more cells were analyzed per each of 3 MEF lines. The mean fluorescence intensity was determined after background subtraction of images transformed to 8-bit grayscale using ImageJ software (NIH). Imaging of centrosomes was performed by transducing cells with γ-tubulin–CFP and H2B-RFP, followed by live-cell microscopy. Cells were marked in G2 (two centrosomes) and followed through mitosis. For the calculation of inter-pole distances, only those images in which the two centrosomes were visible in the same focal plane were used, to avoid the need for z-stack images, as the imaging frequency required for this analysis often led to mitotic abortion. Imaging of the mitotic spindle in living cells was performed by transducing MEFs with α-tubulin–GFP and H2B-RFP, followed by live-cell microscopy.
Gene expression profiling of lung tumors.
All patients with a likely lung tumor who were undergoing wedge resection were approached for enrollment in the Mayo Lung Specimen Registry. Samples were acquired following receipt of informed consent and were completely de-identified for subsequent analysis. Using the Mayo Clinic Lung Specimen Registry, lung specimens (n = 194) resected from non-neoplastic and tumor tissues from 155 patients with lung adenocarcinoma (AD) between 1997 and 2007 were selected. Formalin-fixed, paraffin-embedded (FFPE) H&E sections from the corresponding surgical specimens were reviewed, and the diagnoses were confirmed according to the 2004 WHO classification of tumors. Neoadjuvant therapy was not given to any patient included in this study. Lung tumor cells and non-neoplastic cells were collected by macrodissection to assure high tumor content (>50%). Total RNA from samples obtained by macrodissection was isolated using the RNeasy kit (QIAGEN). The quality and quantity of these RNA samples were controlled by the Agilent Bioanalyzer and a NanoDrop spectrophotometer. Total RNA (1.2 μg) was labeled according to the standard Affymetrix protocol, and labeled cRNA hybridized to the Affymetrix U133PLUS2 array. The GEO accession number for the microarray data in this article is GSE40791.
GC-RMA expression values (log2 transformed) were calculated by the open source software package R ( http://www.r-project.org/). Survival analysis was performed using the “survival” package, and the graph was prepared using GraphPad Prism. We postulated that tumors with higher expression levels of USP44 would have longer survival times. Setting a threshold for high and low USP44 at multiple points within the 20–35 percentile range produced significant association with survival time (P ≤ 0.05, log-rank test). We selected a threshold (27 percentile) that produced maximum separation in survival curves between the high- and low-USP44-expressing tumors.
Statistics.
All statistics were calculated using GraphPad Prism. The tests used include the 2-tailed, unpaired t-test; the 2-way ANOVA; Fisher’s exact test; the 2-tailed, 1-sample t-test; and the log-rank test where indicated in the figure legends. P values ≤ 0.05 were considered statistically significant. All P values are depicted in the figures or in the figure legends.
Study approval.
All experiments involving animals were approved by the IACUC of the Mayo Clinic (protocol A25510). All human samples were obtained following receipt of informed consent. The use of human disease samples was performed following the approval of the Mayo Clinic Institutional Review Board (IRB study ID 09-00476).
Supplementary Material
Acknowledgments
We would like to thank Darren Baker, Richard Bram, Sajjad Hussain, Liviu Malureanu, and Robin Ricke for critical reading of the manuscript. We would like to further thank Liviu Malureanu for assistance with microscopy and Darren Baker for assistance with mouse dissection and histology. We thank Randy King for the gift of proTAME. We would also like to thank the members of the van Deursen laboratory for thoughtful discussions. This work was supported by a Howard Hughes Medical Institute Physician Scientist Early Career Award (to P.J. Galardy) and by funds from the Mayo Foundation (to P.J. Galardy).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(12):4362–4374. doi:10.1172/JCI63084.
See the related Commentary beginning on page 4325.
References
- 1.Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24(9):457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10(2):102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11(1):25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Kops GJ. The kinetochore and spindle checkpoint in mammals. Front Biosci. 2008;13:3606–3620. doi: 10.2741/2953. [DOI] [PubMed] [Google Scholar]
- 6.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 7.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154(5):925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7(9):644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 9.Baker DJ, Dawlaty MM, Galardy P, van Deursen JM. Mitotic regulation of the anaphase-promoting complex. Cell Mol Life Sci. 2007;64(5):589–600. doi: 10.1007/s00018-007-6443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Townsley FM, Aristarkhov A, Beck S, Hershko A, Ruderman JV. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci U S A. 1997;94(6):2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6(4):455–466. doi: 10.1016/S0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 12.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293(5533):1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 13.Song EJ, et al. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24(13):1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stegmeier F, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446(7138):876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 15.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446(7138):921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10(12):1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A. 1997;94(26):14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179(2):255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, van Deursen J, Galardy PJ. Overexpression of ubiquitin specific protease 44 (USP44) induces chromosomal instability and is frequently observed in human T-cell leukemia. PLoS One. 2011;6(8):e23389. doi: 10.1371/journal.pone.0023389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7(1):45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Michel L, Diaz-Rodriguez E, Narayan G, Hernando E, Murty VV, Benezra R. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci U S A. 2004;101(13):4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng X, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18(4):382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ricke RM, van Deursen JM. Correction of microtubule-kinetochore attachment errors: mechanisms and role in tumor suppression. Semin Cell Dev Biol. 2011;22(6):559–565. doi: 10.1016/j.semcdb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16(17):1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Lampson MA, Cheeseman IM. Sensing centromere tension: aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21(3):133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4(8):e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23(3):401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JR, Ryan ZC, Salisbury JL, Kumar R. The structure of the human centrin 2-xeroderma pigmentosum group C protein complex. J Biol Chem. 2006;281(27):18746–18752. doi: 10.1074/jbc.M513667200. [DOI] [PubMed] [Google Scholar]
- 30.Kilmartin JV. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol. 2003;162(7):1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Sanz J, Yang A, Blouquit Y, Duchambon P, Assairi L, Craescu CT. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006;273(19):4504–4515. doi: 10.1111/j.1742-4658.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- 32.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188(1):83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Adamian M, Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol Biol Cell. 2006;17(2):1033–1040. doi: 10.1091/mbc.E05-10-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayor T, Hacker U, Stierhof YD, Nigg EA. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J Cell Sci. 2002;115(pt 16):3275–3284. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- 35.Mardin BR, et al. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol. 2010;12(12):1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106(43):18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim AH, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136(2):322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailly E, Pines J, Hunter T, Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistant compartment and with the centrosome. J Cell Sci. 1992;3(4):529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- 39.Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol. 2010;191(6):1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morales-Mulia S, Scholey JM. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol Biol Cell. 2005;16(7):3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ban KH, et al. The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev Cell. 2007;13(1):29–42. doi: 10.1016/j.devcel.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 42.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8(11):1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 43.Gawrychowski J, Lackowska B, Gabriel A. Prognosis of the surgical treatment of patients with non-small cell lung cancer (NSCLC)—relation to DNA ploidy. Eur J Cardiothorac Surg. 2003;23(6):870–877. doi: 10.1016/S1010-7940(03)00144-1. [DOI] [PubMed] [Google Scholar]
- 44.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16(6):475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawlaty MM, van Deursen JM. Gene targeting methods for studying nuclear transport factors in mice. Methods. 2006;39(4):370–378. doi: 10.1016/j.ymeth.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Malureanu LA. Targeting vector construction through recombineering. Methods Mol Biol. 2011;693:181–203. doi: 10.1007/978-1-60761-974-1_11. [DOI] [PubMed] [Google Scholar]
- 47.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124(6):1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 48.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101(6):635–645. doi: 10.1016/S0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 50.Suresh B, et al. K48- and K63-linked polyubiquitination of deubiquitinating enzyme USP44. Cell Biol Int. 2010;34(8):799–808. doi: 10.1042/CBI20090144. [DOI] [PubMed] [Google Scholar]
- 51.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36(7):744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 52.Todaro GJ, Green H. Cell growth and the initiation of transformation by SV40. Proc Natl Acad Sci U S A. 1966;55(2):302–308. doi: 10.1073/pnas.55.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160(3):341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeganathan KB, Malureanu L, van Deursen JM. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438(7070):1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 55.Malureanu L, et al. Cdc20 hypomorphic mice fail to counteract de novo synthesis of cyclin B1 in mitosis. J Cell Biol. 2010;191(2):313–329. doi: 10.1083/jcb.201003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miest T, Saenz D, Meehan A, Llano M, Poeschla EM. Intensive RNAi with lentiviral vectors in mammalian cells. Methods. 2009;47(4):298–303. doi: 10.1016/j.ymeth.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109(pt 13):3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- 58.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19(1):764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Craig TA, Benson LM, Bergen HR. Metal-binding properties of human centrin-2 determined by micro-electrospray ionization mass spectrometry and UV spectroscopy. J Am Soc Mass Spectrom. 2006;17(8):1158–1171. doi: 10.1016/j.jasms.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 60.Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci. 2001;114(pt 24):4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.