Abstract
In multiple sclerosis (MS) pathogenic B cells likely act on both sides of the blood-brain barrier (BBB). However, it is unclear whether antigen-experienced B cells are shared between the CNS and the peripheral blood (PB) compartments. We applied deep repertoire sequencing of IgG heavy chain variable region genes (IgG-VH) in paired cerebrospinal fluid and PB samples from patients with MS and other neurological diseases to identify related B cells that are common to both compartments. For the first time to our knowledge, we found that a restricted pool of clonally related B cells participated in robust bidirectional exchange across the BBB. Some clusters of related IgG-VH appeared to have undergone active diversification primarily in the CNS, while others have undergone active diversification in the periphery or in both compartments in parallel. B cells are strong candidates for autoimmune effector cells in MS, and these findings suggest that CNS-directed autoimmunity may be triggered and supported on both sides of the BBB. These data also provide a powerful approach to identify and monitor B cells in the PB that correspond to clonally amplified populations in the CNS in MS and other inflammatory states.
Introduction
B cells are a heterogeneous population of lymphocytes with multifaceted roles in autoimmune pathology. Antigen-experienced (memory) B cells differentiate into antibody-secreting plasma cells and/or plasmablasts, serve as highly selective antigen-presenting cells (APCs) (1, 2), and regulate numerous effector functions of T and B cells (3). In the demyelinating disorder multiple sclerosis (MS), autoimmunity may be triggered by B cells that undergo antigen-dependent maturation within specialized lymphoid follicle-like structures in the meningeal and Virchow-Robin spaces of the CNS (4). In MS, B cells are also inherently polarized toward a proinflammatory response to activating stimuli, perhaps promoting antigen-nonspecific “bystander” activation of nearby immune cells (5). The dramatic clinical benefits observed by B cell depletion using therapeutic anti-CD20 monoclonal antibodies provide additional evidence that B cells are centrally involved in the MS disease process (6, 7). Clinical and radiographic effects are rapid in onset and precede changes in antibody production by CNS plasma cells, indicating that B cell depletion, and not reduction in pathogenic autoantibodies, is responsible (8, 9).
Cerebrospinal fluid (CSF) plasma cells have been shown to be the producers of soluble clonal IgG (10, 11), constituting the diagnostically important oligoclonal bands present in more than 95% of patients with MS (12). Although the antigenic targets of oligoclonal bands appear to be diverse and include autoantigens and viral antigens that could be plausibly considered as possibly pathogenic in MS, to what extent these clonal CSF signatures reflect the underlying disease process remains to be established (13–17).
In addition to identifying relevant target antigens of B cell and antibody responses in MS, understanding the repertoire of pathogenic B cells and answering questions of how they differentiate and become activated as well as where they are located in the CNS and peripheral immune system have become central issues in MS pathogenesis. It is known that CNS and CSF B cells are clonally expanded (13–15) and that their expressed immunoglobulin genes are somatically hypermutated (15–17) and IgG class switched (18, 19), indicating that antigen-driven responses can be generated and supported within the CNS. Remarkably, comparison of B cell receptor sequences in MS brain parenchyma, meningeal lymphoid follicles, and CSF indicates that the B cell repertoire overlaps across these compartments (20, 21).
To date, nothing is known about the dynamics of B cell migration between the CNS and peripheral compartments and in particular whether B cells that have undergone somatic hypermutation (SHM) within the CNS environment are also present in peripheral blood (PB). B cell repertoires, with a theoretical diversity that likely exceeds 1 × 1010, have only recently become accessible to interrogation by next-generation sequencing (22–25). To better understand the relationship between CSF and PB B cells and to assess the extent of affinity maturation within the CNS, we performed high-throughput sequencing of IgG heavy chain variable region (IgG-VH) repertoires following immunoglobulin heavy variable gene–unbiased (IGHV-unbiased) PCR amplification in patients with MS and other neurological diseases (ONDs). Using molecularly precise methods, we show for the first time to our knowledge an exchange of IgG-expressing B cells between the CNS and periphery. In addition, using extensive lineage analyses designed to visualize bicompartmental (i.e., CSF and PB) immunoglobulin lineage trees, we found evidence for robust affinity maturation of CNS-associated B cell clones on both sides of the blood-brain barrier (BBB).
Results
Sequencing data.
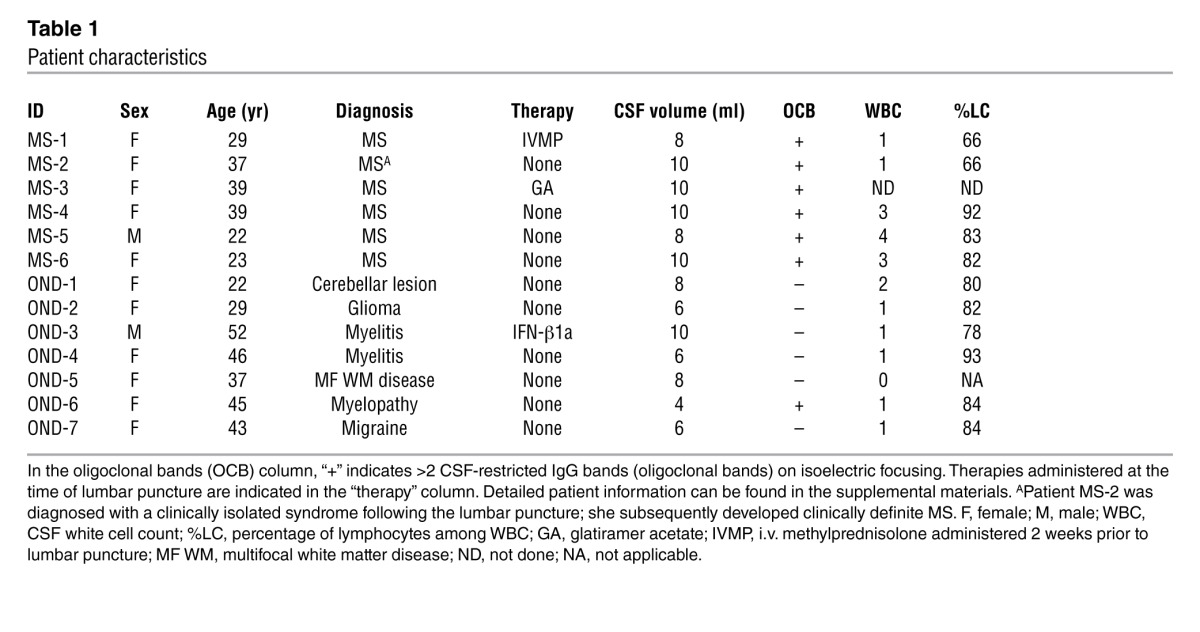
CSF and PB mononuclear cells were isolated from 6 patients with MS and 7 patients with ONDs. Presence of viable mRNA was confirmed by positive amplification of GAPDH mRNA from all CSF samples. CSF IgG-VH transcripts could be amplified by 5′-RACE from all patients with MS (n = 6) and from 4 out of 7 OND samples (Table 1 and see Supplemental Methods for detailed clinical descriptions; supplemental material available online with this article; doi: 10.1172/JCI63842DS1). Accordingly, all 6 patients with MS expressed IgG in their CSF, while 3 patients with ONDs did not. Mean CSF reads totaled 22,395 (SD ± 10,118) IgG-VH sequences from 10 patients (Supplemental Table 1). We obtained on average 399,966 (SD ± 113,192) IgG H chain sequences from the PB of 13 patients (Supplemental Table 1).
Table 1.
Patient characteristics
Total numbers of IgG-VH sequencing reads were not different between MS and OND groups in PB or in CSF for the 10 individuals in whom CSF IgG-VH mRNA could be amplified (Supplemental Figure 1). IGHV and IGHJ germline segments and immunoglobulin heavy chain complementarity determining region 3 (H-CDR3) sequences could be called in 313,266 (SD ± 70,878) PBMC sequences and in 14,327 (SD ± 10,426) CSF sequences (Supplemental Table 1). IGHV usage was determined after removal of redundant IgG-VH (see Methods); resulting nonredundant sequence data sets contained on average 10,217 (SD ± 3,975) PBMC sequences and 273 (SD ± 200) CSF sequences. In CSF, MS samples contained more clonal IgG H chain sequences compared with OND samples (P = 0.0095; 2-tailed unpaired t test) (Supplemental Figure 1), suggestive of considerable B cell activation in MS CSF.
Related B cells are present on both sides of the BBB.
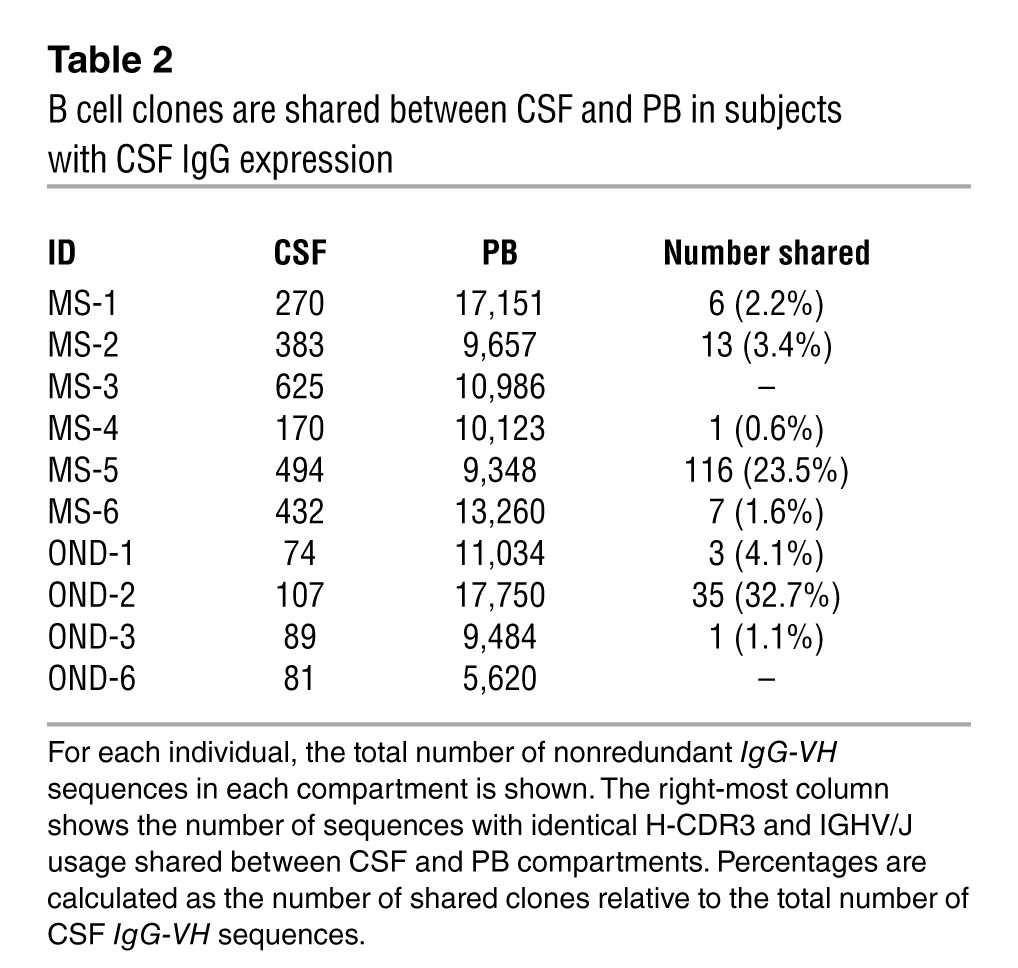
Among patients who express IgG in CSF, we identified IgG-VH sequences with identical H-CDR3 amino acid sequence and IGHV/IGH joining gene (IGHV/IGHJ) usage in paired CSF and PBMC IgG-VH data sets of 5 out of 6 patients with MS (MS-1, MS-2, MS-4, MS-5, and MS-6) and 3 out of 4 patients with ONDs (OND-1, OND-2, and OND-3) (Table 2); on average, 8.7% (range, 0.6% to 32.7%) of clonal CSF IgG-VH sequences could also be found in the PB in these patients (Table 2). No identical or related IgG-VH shared between CSF and PB were identified in 2 individuals, MS-3 and OND-6 (Table 2).
Table 2.
B cell clones are shared between CSF and PB in subjects with CSF IgG expression
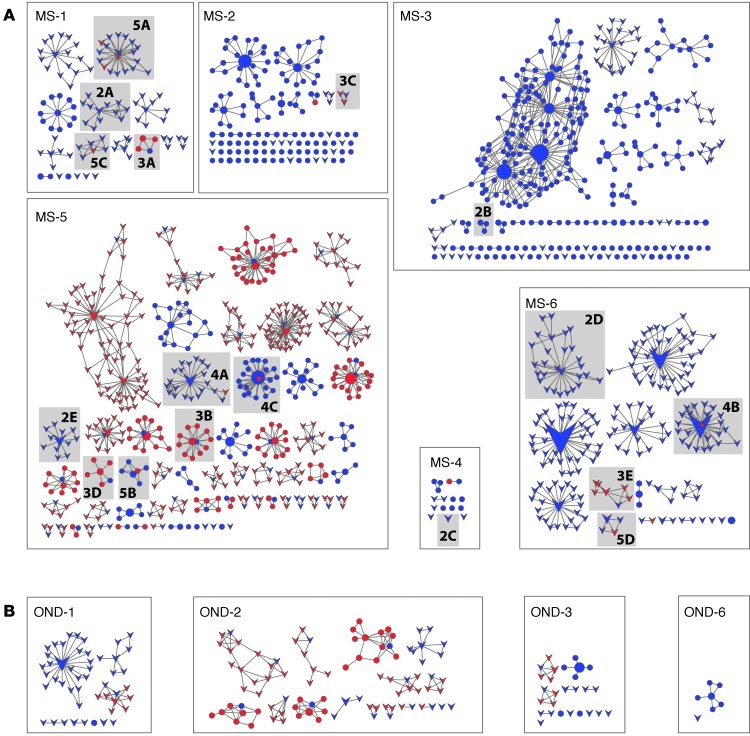
To expand our analyses beyond sequences with identical H-CDR3, IGHV, and IGHJ, we generated clusters of related CSF and PB IgG-VH using a distance metric approach. On average, there were more clusters of related IgG-VH sequences in patients with MS compared with those in patients with ONDs (Table 3). Resulting clusters of related IgG-VH present in CSF, or jointly present in CSF and PB, are visually represented as networks in Figure 1 (representative alignments of IgG-VH clusters are shown in Supplemental Figure 2; general information for clusters is shown in Supplemental Table 2). While these network representations are not suitable to depict clonal evolution, they provide a general visual impression of the complexity of IgG-VH clusters. Unconnected network nodes represent sequences that express identical H-CDR3 amino acid sequences but may still harbor mutational patterns in the IGHV-derived sequence portion.
Table 3.
Clusters of related IgG-VH sequences indicate robust B cell activation in MS
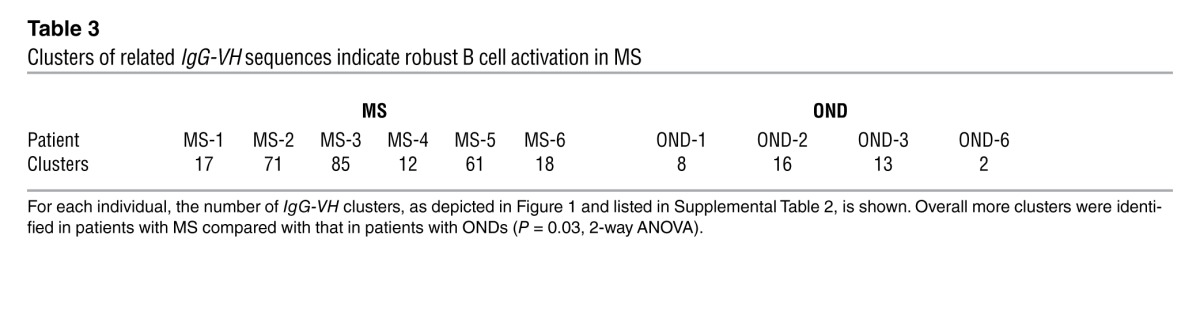
Figure 1. Clusters of B cells expressing identical and/or related IgG-VH are shared between CSF and BP.
Closely related IgG-VH sequences from (A) patients with MS and (B) patients with ONDs were clustered and colored according to their compartment (blue circles or arrowheads indicate CSF; red circles or arrowheads indicate PB) and depicted as networks. Only clusters containing at least one CSF node (i.e., IgG-VH clone) are shown. Directly connected nodes differ in their H-CDR3 by 1 amino acid; node sizes are proportional to the total number of identical H-CDR3 sequences identified. Clusters shaded in gray are presented as lineage trees in Figures 2–5, as indicated by labels with figure number and panel designation. Overall, IGHV4 germline segments are preferentially used in MS CSF; round nodes (circles) indicate clusters of related IgG-VH using IGHV4; v-shaped nodes (arrowheads) indicate clusters using all other IGHV subfamilies. The smallest nodes represent 2 IgG-VH sequences; the largest node (patient MS-6) represents 4,278 IgG-VH sequences.
Most individuals expressing closely related or clonal IgG-VH in their CSF and PB displayed a predominance of CSF clones in these clusters (MS-1, MS-2, MS-4, MS-6, OND-1, OND-2, and OND-3), suggesting their origin in the CNS. However, 2 patients, 1 with active MS (MS-5) and another with a low-grade spinal cord glioma (OND-2), displayed predominant PB IgG-VH representation in bicompartmental clusters (Figure 1), suggesting that affinity-matured B cells derived in the periphery had secondarily “seeded” the CNS. Another patient with stable MS (MS-3) displayed signs of highly active but entirely CNS-restricted B cell activity.
Clonal evolution of B cells on both sides and across the BBB.
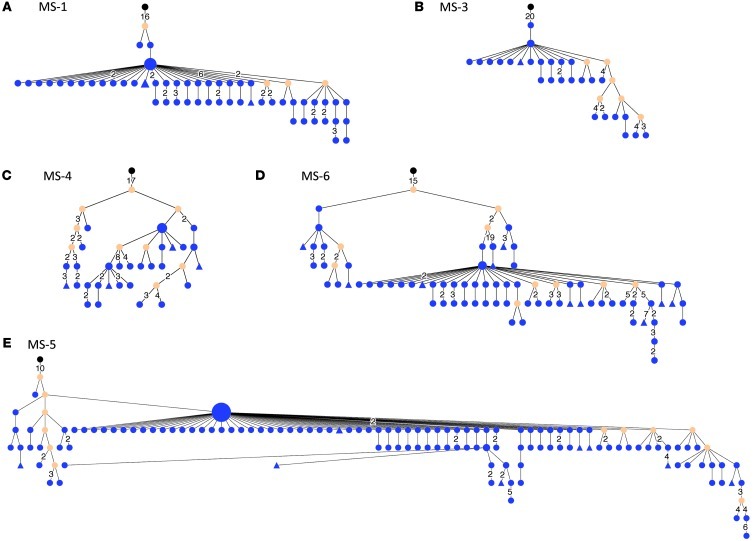
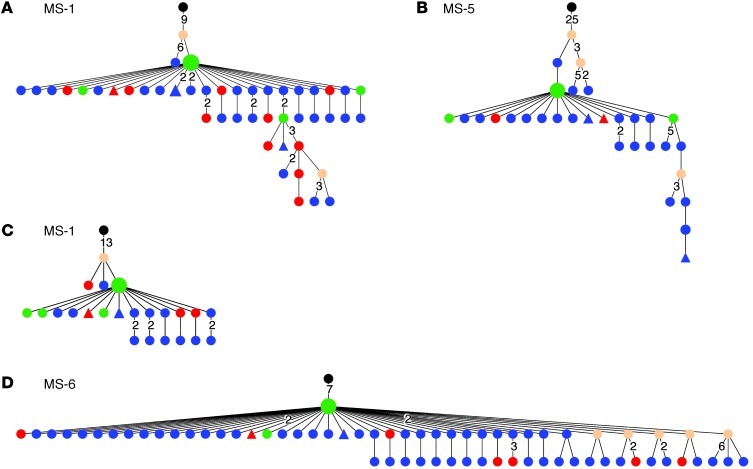
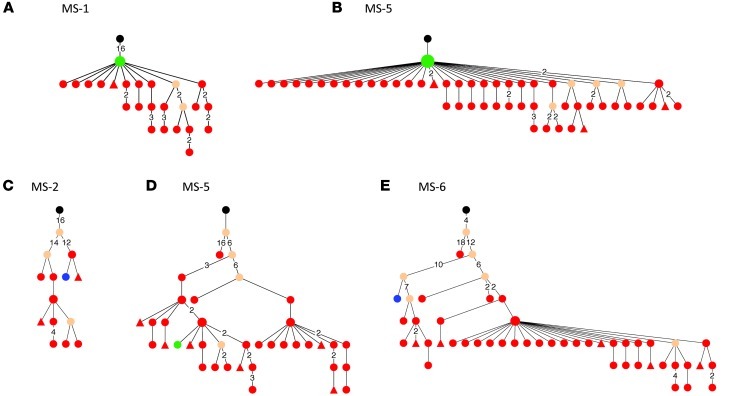
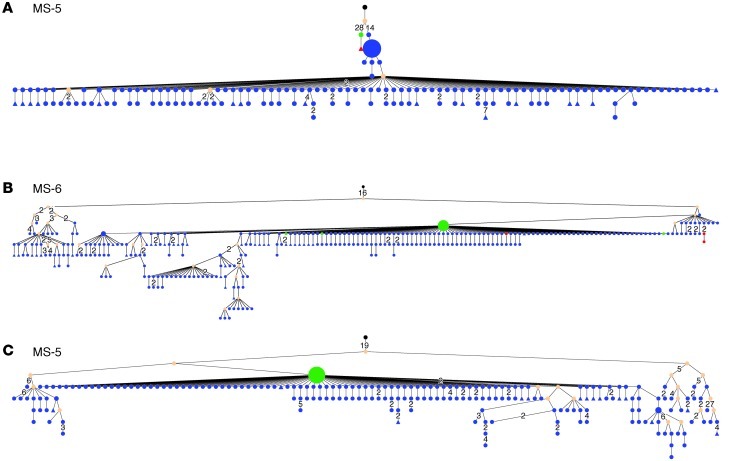
To better understand lineage development of B cells that participate in affinity maturation inside the CNS and in exchange across the BBB, we performed extensive analyses using ClustalW (26) for multiple alignments and IgTree software (27) to generate lineage trees from clusters of related IgG-VH. Only IgG-VH sequences ranging at least from the 5′ end of the H-CDR1 to the 3′ end of the H-CDR3 were included in the lineage tree pipeline, thus taking into account the IgG-VH sequence portions that in combination contain the highest level of diversity. A total of 303 lineage trees were generated, i.e., 1 tree per cluster as shown in Figure 1 (representative trees are shown in Figures 2–5). As expected, complex clusters seen in Figure 1 also yielded complex lineage trees, and most single-node clusters resulted in lineage trees with 2 or more mutational steps distance from the putative germline sequence, as represented in Figure 1A (MS-4, cluster 2C) and Figure 2C. We identified numerous B cell lineages containing identical IgG-VH in both compartments (Figures 3–5, green nodes), suggesting direct clonal exchange across the BBB. Focusing on MS samples, we identified 4 scenarios of clonal relationships among the lineage trees. (a) We found robust affinity maturation of B cells inside the CNS, with some lineages containing IgG-VH of numerous developmental stages and large nodes of identical sequences, suggesting intrathecal clonal expansion (Figure 2); this is the most commonly encountered scenario found in both patients with MS and patients with ONDs, but it was found overall more frequently in patients with MS. (b) We also found extensive trees representing histories of clonal B cell diversification in the periphery with only single-clone connections to the CSF (Figure 3). Among this pattern, there are lineages with the shared clone being represented directly after the putative germline sequence; this may indicate that a certain clone left the CNS to be further modified in the periphery (Figure 3, A and B). Conversely, we also identified IgG-VH clusters in which the shared or CSF clone appears further down the lineage, which may support seeding of B cells clones into the CNS (Figure 3, C–E). (c) We also observed lineages with predominantly CSF-derived and few PB-derived IgG-VH; these may represent clonally related B cells that have emigrated into the CNS for further affinity maturation with only traces remaining in the periphery (Figure 4). (d) A small number of trees were found with more balanced distribution of CSF and PB IgG-VH clones (Figure 5); these lineages may suggest ongoing exchange of B cells across the BBB or parallel affinity maturation in both compartments.
Figure 2. In MS, B cell receptors are subject to extensive intrathecal SHM.
Nucleotide sequences, represented by clusters shown in Figure 1, were selected from the IgG-VH sequence database and used to generate lineage trees using IgTree software (see Methods). (A–E) Representative trees for CSF-restricted clusters for patients (A) MS-1, (B) MS-3, (C) MS-4, (D) MS-6, and (E) MS-5. The corresponding IGHV, IGHJ, and most common H-CDR3 AA sequences are listed in Supplemental Table 2. In the lineage trees, each round node represents at least one unique IgG-VH sequence ranging from at least the 5′ end of H-CDR1 to the 3′ end of H-CDR3; larger nodes represent up to hundreds of identical sequences. Putative germline sequences were determined using SoDA ( https://dulci.org/soda/; ref. 36) and are labeled as black, and hypothetical intermediates calculated by IgTree are labeled as beige. The numbers represent mutational steps between nodes; only mutational steps >1 are indicated; thus, unlabeled branches represent a single mutation. Triangular nodes contain 2 or more singleton sequences in leaves.
Figure 5. Lineage trees of MS IgG-VH suggest ongoing B cell exchange across the BBB.
IgG-VH lineages suggestive of ongoing B cell exchange across the BBB for patient (A and C) MS-1, (B) MS-5, and (D) MS-6. These lineages could also reflect affinity maturation occurring in both compartments in parallel. Blue nodes represent CSF-derived IgG-VH sequences, red nodes represent PB-derived IgG-VH sequences, and green nodes represent identical sequences found in both compartments. For additional information, see the legend of Figure 2.
Figure 3. Select B cells undergoing SHM in the periphery of patients with MS are also present in the CSF.
Shown are IgG-VH lineages with predominantly PB-derived IgG-VH, (A and B) suggestive of B cell migration from the CNS to the PB or (C–E) seeding from the PB into the CNS. Blue nodes represent CSF-derived IgG-VH sequences, red nodes represent PB-derived IgG-VH sequences, and green nodes represent identical sequences found in both compartments. For additional information, see the legend of Figure 2.
Figure 4. B cells undergoing intrathecal SHM in patients with MS can also be found in the periphery.
IgG-VH lineage trees with predominantly CSF-derived IgG-VH sequences for patient (A and C) MS-5 and (B) MS-6. These lineages are suggestive of B cell migration from the PB into the CNS with traces of the clusters remaining in the PB and with extensive intrathecal B cell SHM. Blue nodes represent CSF-derived IgG-VH sequences, red nodes represent PB-derived IgG-VH sequences, and green nodes represent identical sequences found in both compartments. For additional information, see the legend of Figure 2.
IGHV usage in CSF and PB differs in MS compared with that in ONDs.
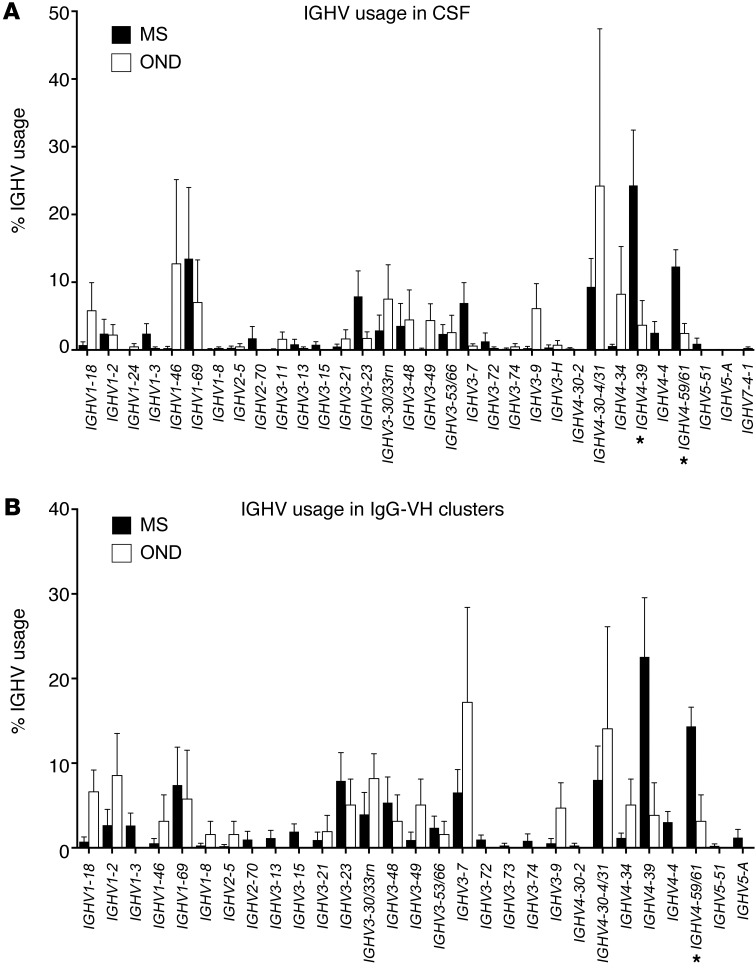
Analyses of IGHV usage in nonredundant CSF IgG-VH data sets identified IGHV4-39 (P = 0.04, resampling-based permutation test) and IGHV4-59 or IGHV4-61 (P = 0.01) to be significantly overrepresented among MS CSF IgG-VH (Figure 6 and Supplemental Table 3). Comparisons of IGHV usage in clusters of MS and OND B cells contributing to the CSF revealed IGHV4-59 or IGHV4-61 to be significantly overrepresented in MS (P = 0.02) (Figure 6 and Supplemental Table 4); IGHV4-39 and IGHV4-4 are marginally overrepresented in MS B cell clusters (P = 0.06 and P = 0.07, respectively). In contrast, in PB IGHV3-23 and IGHV3-74 were overrepresented (P = 0.04 and P = 0.02, respectively) (Supplemental Figure 3 and Supplemental Table 5) in patients with MS compared with patients with ONDs. Overall, we confirm biased usage of IGHV4 germline segments in the CSF of patients with MS as shown by others before (13, 28).
Figure 6. Preferential usage of IGHV4 germline segments by IgG-expressing CSF B cells in MS.
(A) Averages of IGHV germline segment usage in (A) MS CSF compared with that in OND CSF and (B) B cell clusters in MS CSF compared with MS PBMCs. (A) IgG-VH using IGHV4-39 (P = 0.04, resampling-based permutation test) and IGHV4-59 or IGHV4-61 (P = 0.01) are significantly overrepresented in MS CSF (also see Supplemental Table 3) compared with OND CSF. (B) Comparisons of IGHV usage in clusters of MS and OND B cells contributing to the CSF reveal overrepresentation of IGHV4-59 or IGHV4-61 in MS (P = 0.02)(also see Supplemental Table 4). IGHV4-39 and IGHV4-4 are marginally overrepresented (P = 0.06 and P = 0.07, respectively). IGHV3-53/66, IGHV3-53 or IGHV3-66; IGHV3-30/33rn, IGHV3-30 or IGHV3-33; IGHV4-59/61, IGHV4-59 or IGHV4-61; IGHV4-30-4/31, IGHV4-30-4 or IGHV4-31. Values shown in the graph are mean ± SEM. *P < 0.05.
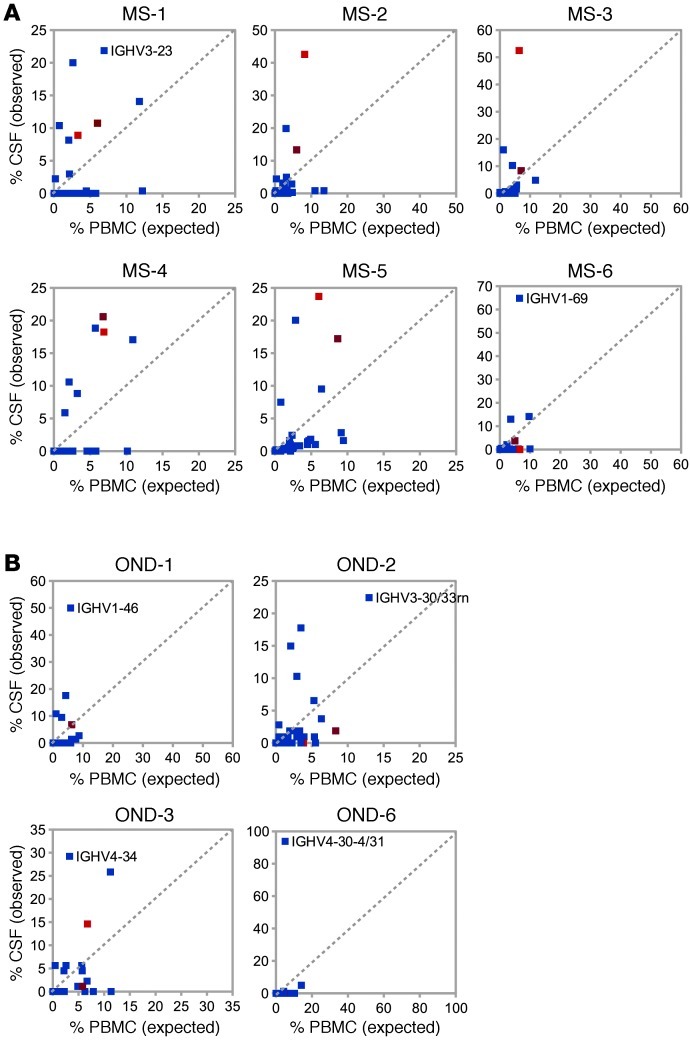
If the B cell repertoire was fully equilibrated between the periphery and the CNS, an identical distribution of IGHV usage would be expected and a plot of IGHV usage in PBMCs (x axis) versus that in CSF (y axis) would place all IGHV sequences along a diagonal. However, this is not the case, because IGHV4-39 and IGHV4-59 or IGHV4-61 sequences stand out as overrepresented in most MS CSF samples studied here (Figure 7A). Only 1 individual in the MS group (MS-6) did not show overutilization of IGHV4-39 or IGHV4-59 or IGHV4-61 in CSF; instead, this patient showed a dominant usage of IGHV1-69 (Figure 7A). In contrast, IGHV4-39 is present and appears overutilized in the CSF of only 1 patient with OND (OND-3; Figure 7B). Taken together, these data suggest that B cells expressing certain IGHV4 segments predominate within the CNS compartment of patients with MS.
Figure 7. Direct comparisons of IGHV germline segment usage in CSF and PB visually confirm overrepresentation of IGHV4 germline segments in MS CSF.
IGHV segment usage in PB (expected) plotted against usage in CSF (observed) in (A) MS and (B) OND. Proportions (%) of IGHV segments in PB were plotted against relative proportions of CSF IGHV. Each graph represents an individual patient. IGHV4-39 frequencies are in light red and IGHV4-59 or IGHV4-61 frequencies are in purple. IGHV most frequently observed in the CSF are labeled if different from IGHV4-39 and IGHV4-59 or IGHV4-61.
Other diversifying elements in the CNS compartment.
There was no significant difference in overall distance from germline IGHV or H-CDR3 length between the MS and OND samples (Supplemental Figure 4).
Discussion
The sequencing depth achieved in this study permitted identification of related or identical B cells on both sides of the BBB for the first time to our knowledge. Clustering of related IgG-VH sequences followed by generation of lineage trees permitted visualization of clonal diversification in relation to their compartmental origin. Closely related B cells, existing on both sides of the BBB, were not exclusively found in patients with MS but were also detected in some patients with ONDs, including inflammatory conditions and other etiologies that are not primarily inflammatory in origin. As expected, we did not find IgG-VH transcripts in the CSF of all patients studied. It is likely that in the healthy state B cell migration across the BBB must be limited or nonexistent but is initiated or magnified during the course of diverse CNS disorders. In MS, this connection may be distinctive in 2 important ways: CSF B cell repertoires appear to be biased in their usage of certain IGHV germline segments, and there is strong evidence for robust B cell activation. These results lend support to the concept that highly active immune responses involving a select pool of B cells are centrally involved in the immunopathology of MS.
In 5 patients with MS (MS-1, MS-2, MS-4, MS-5, and MS-6), we identified common IgG-VH sequences existing both in the CSF and PB. However, our data also suggest that only a select few B cells migrate between both compartments and are retained inside the CNS compartment to undergo clonal expansion, clonal diversification, or both. In most of the patients with MS studied, there was evidence for significant CSF-restricted B cell activation, reinforcing earlier work indicating that B cells can undergo extensive affinity maturation within the CNS compartment (4, 13–19). One patient (MS-3), clinically stable and on disease-modifying therapy, was found to have highly active intrathecal B cell diversification without sharing of B cell clones with the periphery. Conversely, in another patient (MS-5), an extensive and diverse CSF/PB connection was observed. This individual had an acute inflammatory white matter lesion, noted on MRI 1 week prior to the CSF sampling, that might have facilitated de novo migration of B cell clones between the periphery and the nervous system.
Our results show an intimate connection between B cell repertoires located on both sides of the BBB. In MS, this connection appears to be biased toward clones utilizing IGHV4-39 and IGHV4-59 or IGHV4-61 germline segments, confirming and extending earlier, less comprehensive studies, reporting overrepresentation of some IGHV4 segments in MS lesions (13) and CSF (28). Although the cause of the IGHV4 bias in MS is not known, it is possible that these B cell receptors provide a particularly suitable scaffold for relevant target antigens in MS.
Importantly, extensive lineage analyses of IgG-VH clusters revealed direct clonal exchange across the BBB and histories of clonal diversification, suggesting that in MS driving forces for activation of disease-associated B cells operate both in the periphery and in the CNS and possibly in the two compartments in parallel. Our data indicate that B cell exchange across the BBB can be bidirectional and raise the possibility that in MS B cell–mediated autoimmunity can be triggered and supported on both sides of the BBB.
An important challenge for future work will be to determine whether PB B cells that share target specificity with their related CSF B cells are similar or different in terms of their potential contributions to the disease process. For example, IgG-expressing B cells in the periphery may function as APCs (1, 2), while their clonally related B cells in the CNS may further differentiate into antibody-secreting plasma cells or plasmablasts (10, 11, 21, 29, 30). Recently, IL-10–producing CD27+ memory B cells were described in humans (3), and studies in the MS disease model EAE assign critical regulatory functions to this B cell population (31). These findings raise the possibility that regulatory B cells existing on either side of the BBB may also play a role in MS immunopathology, in this case by inhibiting rather than promoting deleterious proinflammatory responses.
The use of molecular IGHV profiles as diagnostic and/or prognostic biomarkers, as probes to identify MS autoantigens, or as targets for novel therapies has considerable potential. With respect to the first possibility, clusters of related CSF B cells could represent diagnostic markers for underlying chronic humoral immune processes in the CNS, similar to oligoclonal banding of CSF immunoglobulins but with the advantage of molecular precision amenable to longitudinal tracking. Larger cohort studies using the technology described in this study are warranted to determine whether an identifiable IGHV usage bias also exists in PB that could be diagnostically useful. Identification of antigenic targets of B cells and antibodies using certain IGHV segments would represent a major step forward toward elucidating the underlying autoimmune etiology of MS. Last, IGHV-based targeting of B cells may be envisioned as a more precise and possibly individualized application to MS immune therapy. In this approach, clones that have undergone extensive somatic diversification within the CNS environment could be identified and specifically targeted, and, in theory, highly selective disruption of the inflammatory process in MS might be achieved even if the underlying autoantigen is not identified.
Methods
Sample generation.
PB and CSF were obtained from 6 patients with MS and 7 patients with ONDs. General information on each individual is summarized in Table 1, and more detailed descriptions can be found in the supplemental materials. Except for patient MS-3, who participated for research only, all patients underwent lumbar punctures for diagnostic workup for suspected demyelinating inflammatory disease. All 6 patients with MS met the commonly applied diagnostic criteria (latest revision) for MS (32), whereas none of the patients with ONDs did.
PBMCs were isolated using Ficoll paque density centrifugation and cryopreserved using standard protocols. CSF cells were spun down and frozen at –80°C. Total RNA from 10 × 106 PBMCs was isolated using the AllPrep Kit (Qiagen) and reverse transcribed using the SMARTer RACE cDNA Amplification Kit (Clontech). cDNA was amplified using the SMARTer RACE Universal 5′ Primer Mix (Clontech) (long, 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′; short, 5′-CTAATACGACTCACTATAGGGC-3′) and a validated IgG isotype-specific 3′ primer (5′-GGGAAGACSGATGGGCCCTTGGTGG-3′). To enable 454 sequencing, the SMARTer RACE Universal short primer and the IgG isotype-specific primer also contained the GS FLX Titanium Primer B or A sequence, respectively, for the Lib-L chemistry (454 Sequencing, Roche). In addition, 6 base multiplex identifier sequences were added to the IgG isotype-specific PCR primer to enable unique assignment of sequencing reads. PCR amplification was performed at 95°C for 5 minutes; 95°C for 30 seconds, 65°C for 30 seconds, 72°C for 1 minute for 33 cycles; and 72°C for 7 minutes. Products were purified using AMPure XP beads (Beckman Coulter Genomics) and quantified using a Quant-iT PicoGreen dsDNA Kit (Invitrogen). This 5′-RACE approach allowed for unbiased amplification of IgG-VH regions. All CSF samples were confirmed to contain viable mRNA by amplification of GAPDH cDNA.
Sequencing.
Purified PCR products were diluted to 1 × 109 molecules per μl and uniquely barcoded samples were pooled. The final sample pools were subjected to emulsion PCR and unidirectional sequencing (from the IgG isotype-specific primer end) on a 2-region sequencing run using the GS FLX Titanium Lib-L chemistry (454 Sequencing, Roche). The relationship between sampling depth and repertoire resolution was determined previously by simulated subsampling of reference V-gene distributions: a target depth of 10,000 usable reads was determined for V-gene analysis and a target depth of 100,000 usable reads was determined for V(D)J analysis (33).
Sequence analysis.
For all reads, IGHV and IGHJ segment usage was determined by probabilistic classification among NCBI Blastn solutions to an “ImMunoGeneTics” (IMGT) reference database (24). CDR boundaries and amino acid composition were determined by aligning all translated reading frames to IMGT-VDJ segment–trained, Kabat column-annotated hidden Markov models (HMM) with HMMER (Version 3) (24); the HMMER functions used were hmmalign and hmmscore. Reads not passing all criteria (uniquely classified IGHV, IGHJ, and H-CDR3) were filtered from subsequent analysis. For the purposes of generating nonredundant IGHV usage data sets from each CSF and PBMC sample, H-CDR3s were extracted from translated HMM-aligned multiple sequence alignments that spanned FR3-CDR3-FR4 (24). V-segment profiles were constructed from normalized counts of segment usage in unique clonal rearrangement events. Clonal nonredundancy was established by considering only one representative from a set of reads bearing the same V-segment, J-segment, H-CDR3 length, and H-CDR3 amino acid composition that differed by fewer than 2 residues from any other H-CDR3 in the data set (33). Thus, IgG-VH sequences were identified as being related based on the fact that IGHV and IGHJ usage are stable features within clonally related B cells and that H-CDR3 is highly specific to clonally related B cells and its length is virtually invariable within groups of clonally related B cells. IgG-VH expressed by B cells in both CSF and PBMCs was identified by direct identification of IgG-VH with identical H-CDR3 amino acid sequence and usage of the same IGHV and IGHJ segments in CSF and PBMC sequence data sets.
Clustering of IgG H chain V regions.
The availability of large numbers of IgG-VH sequences prompted us to further evaluate the complexity within clusters of clonally related sequences present in CSF only or shared between PB and CSF compartments. IgG-VH sequence clusters were generated using a Levenshtein distance of 0 or 1 between their H-CDR3 amino acid sequences (34). Focusing on the H-CDR3 takes advantage of the “fingerprint-like” quality of the V-D-J junction containing sequence elements of IGHV, IGHD, and IGHJ germline segments and a defined length that remains nearly invariable during affinity maturation. By selecting sequences based on their translated H-CDR3 sequence, sequences with silent mutations in the H-CDR3 were considered as identical for clustering purposes. Clustering sequences based on a Levenshtein distance of 0 or 1 ensured clustering of related sequences with identical H-CDR3 (distance = 0) as well as those with some additional diversity in the H-CDR3 (distance = 1) (Supplemental Figure 5). Features of resulting clusters including the most frequently observed H-CDR3 amino acid sequences are shown in Supplemental Table 2. Our clustering approach identified sequences that belong to clonally related lineages, as shown by multiple alignments of amino acid sequences contained within clusters (for examples, see Supplemental Figure 2) and subsequently by more extensive lineage analyses. Resulting H-CDR3–based clusters were represented in Cytoscape (version 2.8.3, ref. 35) as nodes colored by their compartment (CSF = blue, PBMC = red), and connections were represented as linear edges between them to provide a visual overview of some of the diversity contained within IgG-VH clusters.
Lineage trees of IgG-VH clusters.
As shown in Figure 1, a large number of clusters that were composed of clonally related IgG-VH sequences were identified, permitting an analysis of clonal B cell evolution in clusters limited to the CSF and also in those that connect CSF with PB. We used IgTree (27) (provided by Ramit Mehr, Bar-Ilan University, Ramat-Gan, Israel) to display B cell lineages contained within each cluster. Importantly, the IgTree software takes into account the possibility that not all intermediate sequences of B cell lineages may have been identified.
Groups of IgG-VH sequences representative of each cluster and each H-CDR3 represented per cluster were selected from the data set. To generate biologically relevant lineage trees of productive IgG-VH rearrangements, only IgG-VH sequences spanning at least from the 5′ end of H-CDR1 to the 3′ end of H-CDR3 with a contiguous reading frame were considered; thus, sequences containing frame shift–inducing indels were automatically removed. For each cluster, the putative germline sequence, including the V-D-J junction, was determined using SoDA ( https://dulci.org/soda/; ref. 36) and used as tree root for lineage tree calculations. Multiple alignments of cluster-derived IgG-VH sequences and the corresponding germline sequence were performed using ClustalW 2.1 (26). To be able to include compartment information for each tree node, we visualized lineage trees as circles using a hierarchical layout in Cytoscape (version 2.8.3; ref. 35). IgG-VH exclusively found in CSF are labeled as blue, those in PB are labeled as red, and those present in both compartments are labeled as green in the figures. Putative germline sequences are in black in the figures and are located at the root of the tree. Unknown intermediates calculated by IgTree are shown in beige in the figures. The size of tree nodes are proportional to the number of identical sequences contained within each node. Given that we used an unbiased approach to amplify IgG-VH, large nodes may represent sequences derived from either clonally expanded B cells or plasma cells. We frequently encountered large tree nodes containing many branches or leafs; to minimize misinterpreting sequencing errors for SHM, we assembled singleton leaf sequences as triangular nodes, and only display leaves as round if they contain 2 or more identical sequences.
Statistics.
In order to test for overrepresentation of IGHV segments among CSF, PB, or B cell clusters, we implemented a simulation-based evaluation of the significance of the differences in IGHV segment usage between patients with MS and ONDs. Under the null hypothesis that a certain IGHV segment was used at the same proportion in MS and ONDs, we exhaustively enumerated the data set configurations [exhaustive C(10,6) combination tests, also referred to as resampling-based permutation test]; resampling P values were generated and are provided where necessary, and P values of less than 0.05 were considered significant. Given the limited sample size we have limited our investigation to the analysis of segment usage using relative proportions. Comparisons of IGHV usage in CSF and B cell clusters considered only those patients expressing IgG in their CSF; comparisons of IGHV usage were made between all patients with MS and ONDs.
Study approval.
The studies involving humans presented here were reviewed and approved by the UCSF Committee on Human Research.
Supplemental materials.
Patient descriptions and supplemental MRI images (see Supplemental Methods) as well as additional figures (Supplemental Figures 1–5) and tables (Supplemental Tables 1–5) can be found in the supplemental materials.
Supplementary Material
Acknowledgments
The authors express their gratitude to the individuals who agreed to serve as subjects for this study. We thank Ramit Mehr, Bar-Ilan University, Ramat-Gan, Israel, for providing IgTree and Mark Segal, UCSF, for expert advice on statistics. These studies were supported by grants from Pfizer Inc., The Nancy Davis Foundation, and the NIH (R01NS026799; R01NS049477; K02NS072288). H.C. von Büdingen was also supported by an endowment from the Rachleff Family Foundation. C.J. van Belle was a FCMSC Research Scholar and was supported by a PACCTR Short-Term Fellowship from UCSF.
Footnotes
Conflict of interest: Tracy C. Kuo, Marina Sirota, Jacob Glanville, Gabriella Huerta, Dilduz Telman, Purnima D. Sundar, and David R. Cox are employees of Pfizer Inc.
Citation for this article: J Clin Invest. 2012;122(12):4533–4543. doi:10.1172/JCI63842.
References
- 1.Bar-Or A, et al. Immunological memory: contribution of memory B cells expressing costimulatory molecules in the resting state. J Immunol. 2001;167(10):5669–5677. doi: 10.4049/jimmunol.167.10.5669. [DOI] [PubMed] [Google Scholar]
- 2.Harp CT, et al. Memory B cells from a subset of treatment-naive relapsing-remitting multiple sclerosis patients elicit CD4(+) T-cell proliferation and IFN-gamma production in response to myelin basic protein and myelin oligodendrocyte glycoprotein. Eur J Immunol. 2010;40(10):2942–2956. doi: 10.1002/eji.201040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwata Y, et al. Characterization of a rare IL-10-competent B-cell subset in humans that parallels mouse regulatory B10 cells. Blood. 2011;117(2):530–541. doi: 10.1182/blood-2010-07-294249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serafini B, Rosicarelli B, Magliozzi R, Stigliano E, Aloisi F. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 2004;14(2):164–174. doi: 10.1111/j.1750-3639.2004.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Or A, et al. Abnormal B-cell cytokine responses a trigger of T-cell-mediated disease in MS? Ann Neurol. 2010;67(4):452–461. doi: 10.1002/ana.21939. [DOI] [PubMed] [Google Scholar]
- 6.Hauser SL, et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358(7):676–688. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 7.Kappos L, et al. Ocrelizumab in relapsing-remitting multiple sclerosis: a phase 2, randomised, placebo-controlled, multicentre trial. Lancet. 2011;378(9805):1779–1787. doi: 10.1016/S0140-6736(11)61649-8. [DOI] [PubMed] [Google Scholar]
- 8. Cross AH, Stark JL, Lauber J, Ramsbottom MJ, Lyons JA. Rituximab reduces B cells and T cells in cerebrospinal fluid of multiple sclerosis patients. J Neuroimmunol. 2006. 180 (1-2): 63 70 10.1016/j.jneuroim.2006.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin Mdel P, et al. Depletion of B lymphocytes from cerebral perivascular spaces by rituximab. Arch Neurol. 2009;66(8):1016–1020. doi: 10.1001/archneurol.2009.157. [DOI] [PubMed] [Google Scholar]
- 10.Obermeier B, et al. Matching of oligoclonal immunoglobulin transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nat Med. 2008;14(6):688–693. doi: 10.1038/nm1714. [DOI] [PubMed] [Google Scholar]
- 11.von Büdingen HC, Gulati M, Kuenzle S, Fischer K, Rupprecht TA, Goebels N. Clonally expanded plasma cells in the cerebrospinal fluid of patients with central nervous system autoimmune demyelination produce “oligoclonal bands”. J Neuroimmunol. 2010;218(1–2):134–139. doi: 10.1016/j.jneuroim.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Walsh MJ, Tourtellotte WW, Roman J, Dreyer W. Immunoglobulin G, A, and M--clonal restriction in multiple sclerosis cerebrospinal fluid and serum--analysis by two-dimensional electrophoresis. Clin Immunol Immunopathol. 1985;35(3):313–327. doi: 10.1016/0090-1229(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 13.Baranzini SE, Jeong MC, Butunoi C, Murray RS, Bernard CC, Oksenberg JR. B cell repertoire diversity and clonal expansion in multiple sclerosis brain lesions. J Immunol. 1999;163(9):5133–5144. [PubMed] [Google Scholar]
- 14.Owens GP, Ritchie AM, Burgoon MP, Williamson RA, Corboy JR, Gilden DH. Single-cell repertoire analysis demonstrates that clonal expansion is a prominent feature of the B cell response in multiple sclerosis cerebrospinal fluid. J Immunol. 2003;171(5):2725–2733. doi: 10.4049/jimmunol.171.5.2725. [DOI] [PubMed] [Google Scholar]
- 15.Qin Y, Duquette P, Zhang Y, Talbot P, Poole R, Antel J. Clonal expansion and somatic hypermutation of V(H) genes of B cells from cerebrospinal fluid in multiple sclerosis. J Clin Invest. 1998;102(5):1045–1050. doi: 10.1172/JCI3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith-Jensen T, Burgoon MP, Anthony J, Kraus H, Gilden DH, Owens GP. Comparison of immunoglobulin G heavy-chain sequences in MS and SSPE brains reveals an antigen-driven response. Neurology. 2000;54(6):1227–1232. doi: 10.1212/WNL.54.6.1227. [DOI] [PubMed] [Google Scholar]
- 17.Colombo M, et al. Accumulation of clonally related B lymphocytes in the cerebrospinal fluid of multiple sclerosis patients. J Immunol. 2000;164(5):2782–2789. doi: 10.4049/jimmunol.164.5.2782. [DOI] [PubMed] [Google Scholar]
- 18.Sandberg-Wollheim M, Turesson I. Lymphocyte subpopulations in the cerebrospinal fluid and peripheral blood in patients with multiple sclerosis. Scand J Immunol. 1975;4(8):831–836. doi: 10.1111/j.1365-3083.1975.tb03724.x. [DOI] [PubMed] [Google Scholar]
- 19.Ritchie AM, et al. Comparative analysis of the CD19+ and CD138+ cell antibody repertoires in the cerebrospinal fluid of patients with multiple sclerosis. J Immunol. 2004;173(1):649–656. doi: 10.4049/jimmunol.173.1.649. [DOI] [PubMed] [Google Scholar]
- 20.Lovato L, et al. Related B cell clones populate the meninges and parenchyma of patients with multiple sclerosis. Brain. 2011;134(pt 2):534–541. doi: 10.1093/brain/awq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obermeier B, et al. Related B cell clones that populate the CSF and CNS of patients with multiple sclerosis produce CSF immunoglobulin. J Neuroimmunol. 2011;233(1–2):245–248. doi: 10.1016/j.jneuroim.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd SD, et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184(12):6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glanville J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106(48):20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein JA, Jiang N, White RA 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324(5928):807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Barak M, Zuckerman NS, Edelman H, Unger R, Mehr R. IgTree: creating Immunoglobulin variable region gene lineage trees. J Immunol Methods. 2008;338(1–2):67–74. doi: 10.1016/j.jim.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Owens GP, et al. VH4 gene segments dominate the intrathecal humoral immune response in multiple sclerosis. J Immunol. 2007;179(9):6343–6351. doi: 10.4049/jimmunol.179.9.6343. [DOI] [PubMed] [Google Scholar]
- 29.von Büdingen HC, Harrer MD, Kuenzle S, Meier M, Goebels N. Clonally expanded plasma cells in the cerebrospinal fluid of MS patients produce myelin-specific antibodies. Eur J Immunol. 2008;38(7):2014–2023. doi: 10.1002/eji.200737784. [DOI] [PubMed] [Google Scholar]
- 30.Owens GP, et al. Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid. Ann Neurol. 2009;65(6):639–649. doi: 10.1002/ana.21641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber MS, et al. B-cell activation influences T-cell polarization and outcome of anti-CD20 B-cell depletion in central nervous system autoimmunity. Ann Neurol. 2010;68(3):369–383. doi: 10.1002/ana.22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polman CH, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glanville J, et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci U S A. 2011;108(50):20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levenshtein VI. Binary codes capable of correcting deletions, insertions, and reversals. Soviet Physics Dokl. 1966;10(8):707–710. [Google Scholar]
- 35.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volpe JM, Cowell LG, Kepler TB. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics. 2006;22(4):438–444. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.