Abstract
Iron demand in bone marrow increases when erythropoiesis is stimulated by hypoxia via increased erythropoietin (EPO) synthesis in kidney and liver. Hepcidin, a small polypeptide produced by hepatocytes, plays a central role in regulating iron uptake by promoting internalization and degradation of ferroportin, the only known cellular iron exporter. Hypoxia suppresses hepcidin, thereby enhancing intestinal iron uptake and release from internal stores. While HIF, a central mediator of cellular adaptation to hypoxia, directly regulates renal and hepatic EPO synthesis under hypoxia, the molecular basis of hypoxia/HIF-mediated hepcidin suppression in the liver remains unclear. Here, we used a genetic approach to disengage HIF activation from EPO synthesis and found that HIF-mediated suppression of the hepcidin gene (Hamp1) required EPO induction. EPO induction was associated with increased erythropoietic activity and elevated serum levels of growth differentiation factor 15. When erythropoiesis was inhibited pharmacologically, Hamp1 was no longer suppressed despite profound elevations in serum EPO, indicating that EPO by itself is not directly involved in Hamp1 regulation. Taken together, we provide in vivo evidence that Hamp1 suppression by the HIF pathway occurs indirectly through stimulation of EPO-induced erythropoiesis.
Introduction
The increased production of rbc and thus increased O2-carrying capacity of blood represents a major adaptation to systemic hypoxia. This important physiologic response consists of cell type–specific changes that include increased erythropoietin (EPO) production in kidney and liver and enhanced iron uptake and utilization as well as adjustments in the BM microenvironment that facilitate erythroid progenitor maturation and proliferation (1). Hepcidin, encoded by the HAMP gene, is a hypoxia-regulated, small polypeptide produced in hepatocytes, which in its processed form consists of 25 amino acids and plays a central role in the maintenance of systemic iron homeostasis. It suppresses intestinal iron uptake and release from internal stores by facilitating the degradation and internalization of the only known iron exporter, ferroportin, which is expressed on the surface of enterocytes, hepatocytes, and macrophages. Chronic elevation of serum hepcidin, which often associates with inflammatory states, reduces ferroportin surface expression and produces hypoferremia. In contrast, constitutively low hepcidin production in the liver, e.g., due to genetic defects in intracellular signaling pathways that control hepcidin transcription, results in persistent hyperferremia and the development of hemochromatosis (2).
Central mediators of hypoxia-induced erythropoiesis are the O2-regulated basic helix-loop-helix transcription factors HIF-1 and HIF-2. They consist of an O2-sensitive α subunit (HIF-1α and HIF-2α, which is also known as endothelial PAS domain protein 1 [EPAS1]) and a constitutively expressed β subunit, HIF-β, which is also known as the aryl-hydrocarbon receptor nuclear translocator (ARNT). In vivo studies have identified HIF-2 as the main regulator of EPO (1), the glycoprotein that prevents apoptosis of erythroid progenitor cells and is essential for the maintenance of normal erythropoiesis and the increase in rbc production under hypoxia (3). The activity of HIF is controlled by O2-, iron-, and ascorbate-dependent dioxygenases, also known as prolyl-hydroxylase domain–containing proteins 1–3 (PHD1–3), which use 2-oxoglutarate as substrate for the hydroxylation of specific proline residues in HIF-α. Hydroxylation of HIF-α permits binding to the von Hippel–Lindau–E3 (VHL-E3) ubiquitin ligase complex, which results in proteasomal degradation of HIF-α (4).
Experimental studies in cell culture and in animals, as well as clinical data from patients with Chuvash polycythemia, who are homozygous for the VHL R200W mutation, support the notion that hepcidin synthesis involves the VHL/HIF/PHD axis (5–8). However, the molecular basis of its O2 dependence, in particular, the role of HIF in its regulation, is unclear. Genetic studies with iron-deficient mice in conjunction with transcriptional assays have suggested that Hif-1 activation in hepatocytes suppresses hepcidin (Hamp1) directly via hypoxia response element–dependent (HRE-dependent) mechanisms (8). However, this model is debated, and more recent in vitro experiments suggest that HIF does not function as a direct transcriptional repressor of HAMP (9, 10). A second model of hypoxia-induced hepcidin suppression involves iron-dependent signaling pathways that control HAMP transcription. Signaling through either HFE, which is mutated in patients with hereditary hemochromatosis, transferrin receptor 1 (TRFC) and transferrin receptor 2 (TFR2) (2, 11, 12), or hemojuvelin (HJV), which acts as a coreceptor for bone morphogenetic protein 6 (BMP6), increases hepcidin expression in a mothers against decapentaplegic homolog–dependent (SMAD-dependent) fashion (13–15). In vitro studies have shown that HIF induces furin, a proprotein convertase that cleaves HJV and generates soluble HJV, which in turn suppresses HAMP by competing for BMP6, thereby antagonizing signaling through membrane-bound HJV (16, 17). Similarly, transmembrane protease serine 6 (TMPRSS6), also known as matriptase-2, has been identified as HIF regulated and is predicted to blunt BMP6/HJV-mediated signals under hypoxia (18–20). A direct effect of EPO on HAMP transcription has also been postulated. Studies with primary hepatocytes and HepG2 cells have shown that EPO, in a dose-dependent fashion, is capable of regulating HAMP transcription via EPO receptor (EPOR) and CCAAT/enhancer-binding protein (C/EBP) α activation (21). A fourth model proposes that stimulation of erythropoiesis generates a BM-derived signal that suppresses Hamp1 in the liver (22). Growth differentiation factor 15 (GDF15), an iron- and O2-regulated (HIF-independent) member of the TGF-β superfamily, is secreted from maturing erythroblasts and suppresses HAMP transcription in primary human hepatocytes and hepatoma cells (23, 24). Since very high levels of serum GDF15 were found in patients with α- and β-thalassemia, it was proposed that GDF15 is the BM-derived factor that suppresses hepcidin under conditions of stimulated erythropoiesis (24). While high serum GDF15 levels were found in patients with syndromes of ineffective erythropoiesis (24–27), the association between serum GDF15 and serum hepcidin levels in other forms of anemia was less evident. This raised the possibility that GDF15 may be a marker of ineffective or apoptotic erythropoiesis. Nevertheless, its role in hepcidin regulation under physiologic or other pathologic conditions remains to be elucidated (for a recent review on this topic, see ref. 28).
To specifically dissect the role of the VHL/HIF/PHD axis and EPO in the hypoxic suppression of hepcidin in vivo, we have used a genetic approach to disengage Hif activation from Epo synthesis in mice. We utilized tamoxifen-inducible Cre/loxP-mediated recombination to activate Hif-1 and Hif-2 via ablation of Vhl while simultaneously inactivating Epo. We found that hypoxia/Hif-mediated suppression of Hamp1 required Epo inducibility and was associated with elevated serum Gdf15 levels. Including additional genetic models, we furthermore demonstrate that increased erythropoietic drive is required for Hamp1 suppression under conditions of hepatic Hif activation irrespective of serum Epo levels. Our genetic data establish that Hif activation in hepatocytes suppresses Hamp1 indirectly through Epo-mediated stimulation of erythropoiesis.
Results
Conditional inactivation of Vhl results in Hif-dependent hepcidin suppression.
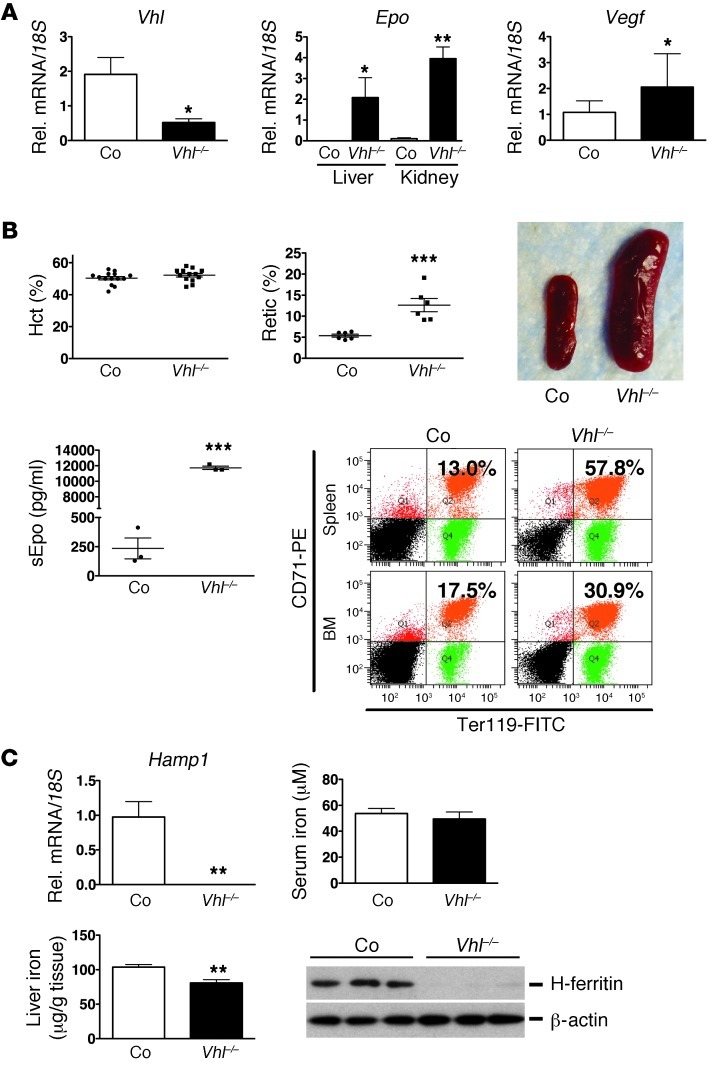
For the genetic dissection of hepcidin regulation by the HIF oxygen-sensing pathway, we established a model of acute Vhl inactivation using a globally expressed tamoxifen-inducible Cre-recombinase under the control of the ubiquitin c promoter, Ubc-cre/ERT2 (29). Although hepatocyte-specific Vhl inactivation via Cre-recombinase driven by the albumin promoter suppresses hepatic Hamp1 (8, 30), constitutive Hif activation in the liver has profound effects on glucose and fatty acid metabolism and results in sick animals that die from liver failure between the ages of 6 and 12 weeks (31), which makes the interpretation of hematologic data difficult and limits experimental options. To achieve efficient recombination in Ubc-cre/ERT2 mice that were homozygous for the Vhl floxed allele, 4 doses of tamoxifen were administered over a period of 7 days, followed by phenotypic analysis on day 8 (experimental time line is shown in Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI63924DS1). Tamoxifen administration resulted in an approximately 75% reduction in hepatic Vhl mRNA levels, stabilization of both Hif-1α and Hif-2α homologs, and increased expression of Hif target genes, such as Epo, Vegf, and divalent metal transporter 1 (Dmt1) (Figure 1A and Supplemental Figure 1). Recombination analysis by genomic PCR indicated efficient recombination in the kidney and in other organs (Supplemental Figure 1 and data not shown). Since hepatic and renal Epo synthesis were stimulated in Vhl–/– mutant mice, serum Epo levels were elevated to 11733 ± 217.0 pg/ml compared with Cre– control mice (235.3 ± 89.8 pg/ml). This resulted in increased formation of CD71hiTer119hi-positive erythroblasts in spleen and BM (in spleen 59.33% ± 2.93% for mutants vs. 18.2% ± 3.54% for control mice, and 32.2% ± 0.723% vs. 17.33% ± 3.48% in BM; n = 3 each), splenomegaly, and reticulocytosis (12.64% ± 1.57% for mutants vs. 5.37% ± 0.35% for controls, n = 6 each), all of which are consistent with increased erythropoietic activity (Figure 1B and Supplemental Figure 1). Hematocrit (Hct), hemoglobin (Hb), and rbc values in Vhl–/– mutants were not different from controls at day 8 after the first tamoxifen injection and were found to be increased at day 16; Hb values increased from 14.13 ± 0.186 g/dl in controls to 16.07 ± 0.73 g/dl in mutants on day 16, n = 3 each (Supplemental Figure 1 and data not shown). Hamp1 mRNA levels were undetectable in Vhl–/– livers by real-time PCR analysis (Figure 1C). While serum iron levels did not differ between mutants and Cre– controls, iron was decreased in Vhl–/– livers, which was associated with reduced ferritin heavy chain-1 (H-ferritin) levels (Figure 1C). This is expected, as the ferritin 5′ UTR contains an iron response element (IRE) that mediates translational inhibition in the presence of low intracellular iron. Taken together, these results demonstrate that acute global inactivation of Vhl results in increased erythropoietic activity and associates with decreased Hamp1 expression in the liver.
Figure 1. Inactivation of Vhl suppresses Hamp1.
(A) Shown are results from real-time PCR analysis of Vhl and Vegf mRNA levels in Vhl–/– livers (n = 3) and Epo mRNA levels in Vhl–/– kidneys and livers (n = 3); analysis was performed on day 8 after the first tamoxifen injection. Relative mRNA expression levels were normalized to 18S ribosomal RNA. (B) Global inactivation of Vhl induces erythropoiesis. Shown are individual Hct values (n = 14 and 13, respectively), reticulocyte counts (%) (n = 6 each), and serum Epo concentrations (sEpo) (n = 3 each) from control and mutant mice and a representative picture of a control and a Vhl–/– spleen. Lower right panel shows a representative FACS plot of CD71/Ter119 double-stained BM and spleen cells from an individual control mouse and Vhl mutant. Percentages of CD71hi/Ter119hi-positive cells (right upper quadrant) are indicated. (C) Shown are relative expression levels of Hamp1 mRNA in control and Vhl–/– livers (n = 5 and 3, respectively) and serum iron (n = 3 each) and liver iron concentrations (n = 7 and 4, respectively). H-ferritin protein levels in control and Vhl–/– livers were determined by immunoblot in 3 mice, β-actin served as loading control. Asterisks indicate a statistically significant difference when comparisons were made to the control group: *P < 0.05; **P< 0.01; ***P< 0.001. Shown are arithmetic mean values ± SEM. Co, Cre-negative littermate control; retic, reticulocytes.
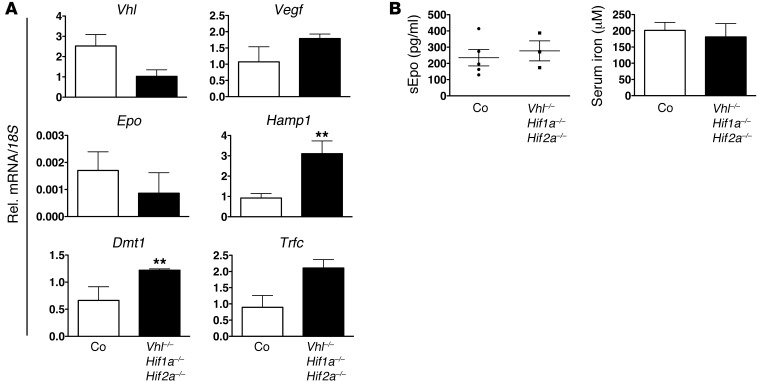
To investigate the role of Hif in VHL-associated hepcidin regulation, we generated mice that permitted global inactivation of both Hif-1α and Hif-2α in a Vhl-deficient background. Epo and Vegf were not significantly induced or decreased in Vhl/Hif-1/Hif-2–/– livers compared with Cre– littermate control mice (Figure 2A), which suggests (a) that their increased expression in Vhl–/– livers is Hif dependent and (b) that Hif-1 and Hif-2 do not participate in their transcriptional regulation under baseline, i.e., normoxic conditions. This is consistent with our previous studies in hepatocyte-specific Vhl-knockout mice, where we identified Hif-2 as the main regulator of hepatic Epo synthesis. With these studies, we demonstrated that inactivation of both Hif-1α and Hif-2α in a Vhl–/– background completely abrogated Epo induction (32). Consequently, serum Epo concentrations in Vhl/Hif-1/Hif-2–/– mice did not significantly differ from those in Cre– control mice (277.5 ± 61.66 pg/ml in mutants vs. 235.1 ± 50.69 pg/ml in controls; n = 3 and n = 5, respectively) (Figure 2B). The abrogation of Epo induction in triple mutants was associated with a statistically significant increase in Hamp1 mRNA levels in Vhl/Hif-1/Hif-2–/– livers compared with Cre– controls, P = 0.0035 for n = 3. In summary, our data establish that the suppression of Hamp1 in Vhl-defective livers requires Hif stabilization.
Figure 2. Hamp1 suppression in Vhl–/– livers is Hif dependent.
(A) Real-time PCR analysis of Vhl, Vegf, Epo, Hamp1, Dmt1, and Trfc expression in Vhl/Hif1a/Hif2a–/– livers. Relative mRNA expression levels were normalized to 18S ribosomal RNA. Bars represent arithmetic mean values ± SEM (n = 3). (B) Serum Epo concentrations and serum iron levels in Vhl/Hif1a/Hif2a–/– mice (n = 3). Circles and squares represent data points for individual mice. Error bars represent SEM. *P < 0.05; **P< 0.01, for comparisons with controls.
Hif-1 does not suppress hepcidin in Phd2–/– livers.
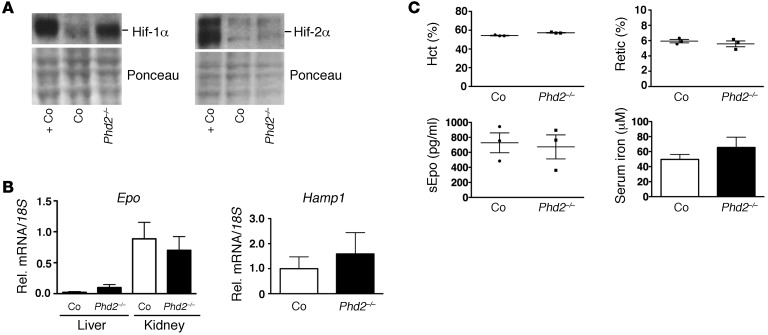
Since hepatic Hif-1 has been shown to participate in the regulation of Hamp1 in iron-deficiency anemia (8), we made use of a mouse model that specifically activates hepatic Hif-1 in a genetic background that is WT for Vhl. We generated mice with hepatocyte-specific inactivation of Phd2. Phd2, also known as EglN1, is the major Hif prolyl-4-hydroxylase that targets Hif-α subunits for hydroxylation and subsequent proteasomal degradation under normoxia. Phd2 inactivation in hepatocytes (EglN12lox/2lox;Albumin-cre) resulted in stabilization of Hif-1α, but not of Hif-2α (Figure 3A), which is consistent with previous reports (33). Surprisingly, Hamp1 expression levels in Phd2-deficient livers did not change compared with controls (Figure 3B). Also, hepatocyte-specific Phd2 inactivation did not increase Epo mRNA levels, nor did it stimulate erythropoietic BM activity, as an increase in blood reticulocytes and Hct was not observed (reticulocyte count: 5.93% ± 0.22% vs. 5.58% ± 0.381%; Hct: 54.33% ± 0.33% and 57.33% ± 0.33%, respectively, n = 3 each) (Figure 3, B and C). This is consistent with our previous observation that hepatic Epo synthesis is predominantly Hif-2 regulated (32). Our findings indicate that Hif-1α stabilization alone is not sufficient to suppress Hamp1 in hepatocytes. Furthermore, analysis of Phd2-knockout mice suggests that Hif-associated Hamp1 suppression is linked to Hif-2–dependent stimulation of erythropoiesis.
Figure 3. Hepatocyte-specific inactivation of Phd2 does not suppress Hamp1.
(A) Hif-1α and Hif-2α protein levels in Phd2–/– livers. Ponceau staining is used to assess for equal protein loading. +Co, positive control sample obtained from Vhl–/– livers. (B) Hepatocyte-specific inactivation of Phd2 does not increase Epo mRNA and does not suppress Hamp1 mRNA levels in Phd2–/– livers. Shown are relative mRNA expression levels normalized to 18S ribosomal RNA in mutant and control livers. Corresponding renal Epo mRNA levels are shown for comparison (n = 3). (C) Hct, reticulocyte counts, and serum Epo and serum iron levels in control and Phd2 mutant mice (n = 3 each). Shown are mean values ± SEM. For statistical analysis, mutants were compared with controls.
Hif-mediated hepcidin suppression requires Epo inducibility.
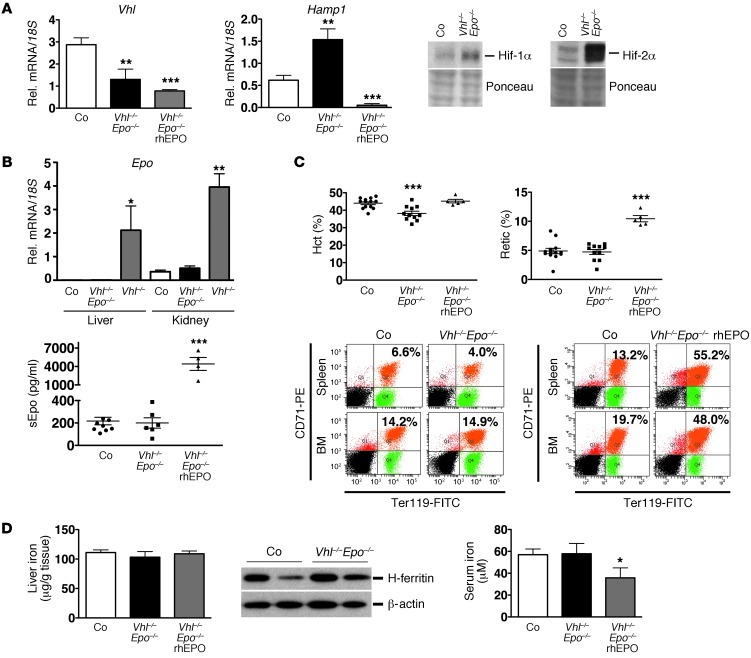
Our findings in liver-specific Phd2-knockout mice suggested that hepatic Hif-2 activation and/or Hif-2–stimulated erythropoiesis led to the suppression of Hamp1 in Vhl-deficient livers. To determine whether Hif-induced Hamp1 suppression in Vhl–/– mice is dependent on the ability to synthesize Epo, we generated a genetic mouse model in which Hif activation can be dissociated from Epo synthesis and Epo-induced erythropoietic activity. In this model, both Hif-1 and Hif-2 are activated in hepatocytes and other cell types without any concomitant increase in renal and hepatic Epo production. For this purpose, we bred the Epo-2lox allele into the Vhl-2lox background and generated mice that permitted global, tamoxifen-inducible, and concurrent Vhl and Epo gene inactivation (Vhl2lox/2lox;Epo2lox/2lox;Ubc-cre/ERT2). While Vhl ablation in Vhl/Epo–/– mice resulted in stabilization of hepatic and renal Hif-1α and Hif-2α (Figure 4A) as well as increased Vegf and Dmt-1 mRNA levels (Supplemental Figure 2C), Epo mRNA was not induced in the liver and kidney (Figure 4B, hepatic and renal Epo mRNA levels in Vhl–/– mice are shown for comparative purposes). Serum Epo levels in Vhl/Epo–/– mice were slightly decreased (200.1 ± 46.6 pg/ml vs. 217.3 ± 33.4 pg/ml in control animals, n = 6 and 10, respectively), but not significantly different from control mice, while rbc numbers and Hct and Hb values were decreased compared with controls (Figure 4C and Supplemental Figure 2A). The rbc numbers and Hct and Hb values decreased further over time; at day 16 after the first tamoxifen injection, mean Hct in Vhl/Epo–/– mutants was 24.03% ± 1.937%, mean Hb was 6.5 ± 0.59 g/dl, and mean rbc count was 5.01 ± 0.41 M/μl (n = 3), which is consistent with hypoproliferative anemia that develops in mice with global Epo inactivation (34). Despite Hif-1α and Hif-2α stabilization in the liver, Hamp1 was no longer suppressed and increased significantly, by approximately 3-fold, P = 0.0032, n = 6 (Figure 4A). These findings indicate that (a) Hamp1 is not directly regulated by either Hif-1 or Hif-2 and (b) that its suppression is dependent on the induction of Epo synthesis, while acute Epo deficiency increases its transcription.
Figure 4. Hif-mediated Hamp1 suppression is Epo dependent.
(A). Hepatic Vhl and Hamp1 mRNA levels in control, Vhl/Epo–/–, and Vhl/Epo–/– mice treated with rhEPO (n = 6, 6, and 5, respectively). Right panel shows Hif-1α and Hif-2α protein levels in Vhl/Epo–/– livers. Ponceau staining is used to assess for equal protein loading. (B) Epo levels in control, Vhl/Epo–/–, and Vhl–/– livers and kidneys (n = 6, 6, and 3, respectively). Bottom panel shows serum Epo concentrations in control, Vhl/Epo–/–, and Vhl/Epo–/– mice treated with rhEPO (n = 10, 6, and 4, respectively). (C) Hct and reticulocyte counts in control, Vhl/Epo–/–, and rhEPO-treated Vhl/Epo–/– mice and representative FACS analysis plot of CD71/Ter119-stained BM and spleen cells from 1 control and 1 mutant mouse. Percentages of CD71hiTer119hi-positive cells are indicated in the right upper quadrant. (D) Liver (n = 8, 4, and 5, respectively) and serum iron concentrations (n = 10, 6, and 4, respectively) in control, Vhl/Epo–/–, and Vhl/Epo–/– mice treated with rhEPO, and H-ferritin protein levels in control and Vhl/Epo–/– livers. β-actin served as loading control. Shown are mean values ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, for comparisons of mutants with controls. Vhl/Epo–/– (rhEPO), Vhl/Epo double-mutant mice treated with rhEPO.
Since we established Epo dependence of VHL-associated Hamp1 regulation, we asked whether administration of recombinant EPO was able to overcome the genetic Epo deficiency and could restore Hamp1 suppression in Vhl/Epo–/– livers. For this, we administered 200 IU of recombinant human EPO (rhEPO) to Vhl/Epo–/– mice i.p. every day for 3 days prior to mouse phenotyping. While a rise in rbc numbers was not seen 4 days after the initiation of rhEPO treatment (Figure 4C), reticulocyte counts increased from 4.91% ± 0.61% to 10.44% ± 0.53% (n = 10 and 5, respectively), and CD71hiTer119hi-positive erythroblasts increased from 16.42% ± 5.28% to 45% ± 3.35% in spleen and from 16.5% ± 1.25% to 43.36% ± 2.78% in BM (n = 10 and 5, respectively) (Figure 4C and Supplemental Figure 2). Restoration of erythropoietic activity suppressed Hamp1 in Vhl/Epo–/– livers (Figure 4A). Taken together, our data provide genetic evidence that Hamp1 suppression in Vhl–/– livers requires intact Epo synthesis and is not directly dependent on Hif-1 and/or Hif-2 activation in hepatocytes.
Hif-mediated hepcidin suppression requires erythropoietic activity and associates with increased serum Gdf15 levels.
Although our genetic data established a clear role for Epo in the regulation of Hamp1, it was unclear whether Epo effects on Vhl–/– hepatocytes were direct, e.g., via EpoR activation, or whether Hif-mediated Hamp1 suppression was dependent on Epo-induced erythropoietic activity (22). To investigate the role of erythropoiesis in the regulation of Hamp1 in this model, Vhl2lox/2lox;Ubc-cre/ERT2 animals were pretreated with carboplatin (Cp) to achieve efficient BM suppression. Cp-treated and vehicle-treated mice were analyzed 1 day after the final tamoxifen injection (day 8). We found comparable increases in hepatic and renal Epo mRNA levels in both Cp-treated and vehicle-treated Vhl–/– mice, which suggested similar degrees of recombination in both groups (Figure 5A). Despite the presence of very high serum Epo levels (6911 ± 276.9 pg/ml), reticulocyte counts were severely reduced in Cp-treated Vhl–/– mice (0.66% ± 0.08% in Cp-treated Vhl–/– mice vs. 5.64% ± 0.65% in vehicle-treated controls, n = 4 and 3, respectively), which is consistent with robust inhibition of erythropoietic activity by Cp (Figure 5A). Most strikingly, the strong induction of renal and hepatic Epo synthesis in Cp-treated Vhl–/– animals was not associated with Hamp1 suppression, but with significantly increased Hamp1 mRNA levels, P = 0.0044 for n = 3 and 6 for control (Figure 5A). Taken together, these findings indicate that Epo-dependent induction of erythropoiesis is required for the suppression of Hamp1 in Vhl–/– mice.
Figure 5. Hif-associated Hamp1 suppression requires erythropoietic activity.
(A) Renal and hepatic Epo (n = 3, 5, and 3, respectively) in control mice and Vhl–/– mutants with or without Cp treatment and liver Hamp1 RNA levels (n = 6, 4, and 3, respectively) in nontreated control, Cp-treated control, and Cp-treated Vhl–/– mutants. Lower panels show Hct, reticulocyte counts, (n = 3 and 4, respectively), serum Epo (n = 3 and 5, respectively), and spleen to body weight ratios in nontreated control and Cp-treated Vhl–/– mice (n = 3 each). (B) Hamp1 mRNA levels in control and Hif2a/Pax3-cre (P3) mutants exposed to chronic hypoxia (10% O2 for 10 days) (n = 3 each) and in thalassemic mice (th3/th3) and control littermates (+/+) (n = 4 each). Shown are mean values ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, for comparisons with control group or comparison with normoxia. Cp, mice pretreated with Cp; Hx, treatment with 10% O2 for 10 days.
In order to gain additional insight into the role of Hif in the suppression of Hamp1 under hypoxic conditions and its relation to erythropoietic activity, we compared mice with hypoproliferative anemia (Hif2a/Pax3-cre [P3] mutants) that were exposed to chronic hypoxia with mice with hyperproliferative anemia (th3/th3 thalassemia mutants). Anemia in th3/th3 mutants is due to the elimination of both β-Hb chains. Th3/th3 mice stabilize Hif-α in liver and kidney and are characterized by high serum Epo levels, high erythropoietic activity, iron overload, and substantial suppression of liver hepcidin (35). In contrast to th3/th3 mice, P3 mutants lack the ability to induce renal Epo in response to acute and chronic hypoxic stimuli. P3 mutant mice develop severe hypoproliferative anemia at baseline, are characterized by Hif-α stabilization in kidney and liver, and display a blunted erythropoietic response when exposed to chronic hypoxia (10% O2) for 10 days (30). We found that Hamp1 was not suppressed in P3 mutant livers compared with Cre– control littermates under conditions of chronic hypoxia (Figure 5B), which is in contrast with th3/th3 mutants examined under baseline conditions. These findings in mice with 2 different forms of severe chronic anemia are in support of the notion that erythropoietic activity regulates Hamp1 suppression under conditions of chronic hypoxia and/or Hif activation.
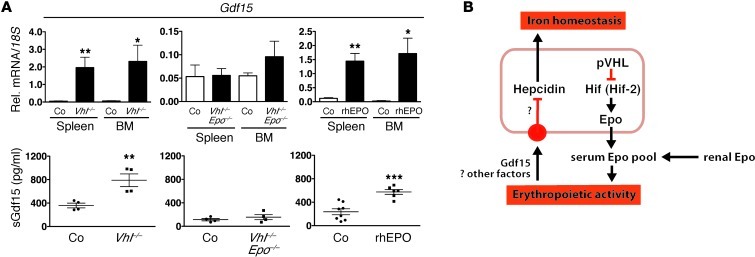
Since GDF15 has been proposed to be involved in hepcidin suppression, at least under conditions of ineffective erythropoiesis, such as in patients with thalassemia syndromes (28), we examined serum Gdf15 levels in Vhl–/– and Vhl/Epo–/– mice and in WT mice injected with rhEPO. We found that serum Gdf15 levels were increased in Vhl–/– mice (789 ± 108.5 pg/ml vs. 359.1 ± 40.16 pg/ml) but not in Vhl/Epo double mutants compared with controls. A similar degree of Gdf15 increase was found when WT mice were treated with 3 daily injections of human recombinant EPO at a dose of 200 IU each (Figure 6). EPO treatment was associated with hepcidin suppression (data not shown). Elevated serum Gdf15 levels correlated with increased Gdf15 mRNA expression in total cell isolates from spleen and BM and in Ter119-positive cells purified by immunomagnetic separation (Figure 6 and Supplemental Figure 3A). In contrast with Gdf15, mRNA levels of twisted gastrulation homolog 1 (Twsg1), an erythrokine that has been shown to regulate hepcidin in vitro (36), did not change in Vhl mutants compared with littermate controls (Supplemental Figure 3B). Taken together, our data indicate that Gdf15 may participate in the suppression of Hamp1 in Vhl–/– mice.
Figure 6. Elevation of serum Gdf15 in Vhl–/– mice is Epo dependent.
Gdf15 mRNA levels in total spleen and BM cell isolates and corresponding serum Gdf15 levels in pg/ml. (A) Left panels, Vhl–/– mutants and Cre– littermate controls (n = 3 and 4, respectively for mRNA analysis; for serum analysis, n = 4 each); middle panels, Vhl/Epo–/– mice and Cre– littermate controls (n = 4 each); right panels, WT mice treated with rhEPO or with vehicle (for mRNA analysis, n = 3 and 4, respectively; for serum analysis, n = 6 and 8, respectively). Shown are mean values ± SEM: *P < 0.05; **P< 0.01; ***P < 0.001, for comparisons of mutants with controls. (B) Schematic depiction of Hif’s role in the regulation of hepcidin transcription in hepatocytes. C–, Cre-negative littermate control mice or vehicle-treated WT mice.
Discussion
To dissect the role of Hif in the regulation of hepcidin in vivo, we have generated a mouse model that permits dissociation of Epo synthesis from Hif activation. From using this model, we provide genetic evidence that Hamp1 suppression requires Epo-induced erythropoiesis and is not directly regulated by either Hif-1 or Hif-2. We furthermore show that the ability of the BM to respond to elevated serum Epo levels with increased rbc production determines whether Hamp1 is suppressed under conditions of Hif activation in the liver.
The importance of pO2 in the regulation of hepcidin has been well established in cell-culture models, in animal experiments, and in humans who were exposed to hypobaric hypoxia (7, 37). Its hypoxic regulation involves the VHL/HIF/PHD oxygen-sensing pathway, as shown in mouse models (8) and in patients with Chuvash polycythemia, a form of familial secondary erythrocytosis that associates with low serum hepcidin levels (5, 6). Chuvash patients are homozygous for specific non–tumor-causing germ line mutations in the VHL tumor suppressor. These mutations impair the ability to efficiently degrade Hif-α under normoxia (5, 38). In line with laboratory findings in Chuvash patients is the Hif-dependent decrease of Hamp1 in Vhl–/– livers. Although Hif acts as an O2-sensitive transcription factor, a direct transcriptional role for Hif was not evident from our studies, which is consistent with recently reported findings in hepatoma cell lines (9). While Hif-1 binding to the Hamp1 promoter has been reported (8), stabilization of Hif-1α alone in Phd2–/– hepatocytes did not result in a transcriptional repression of Hamp1. In this model, Hif-1 activation occurs without the induction of Epo synthesis, which is seen when Hif-2α is stabilized (32). Furthermore, the notion that Hamp1 is not directly regulated by Hif-1 is consistent with genome-wide ChIP analysis in breast cancer cells, which indicates that HIF transcription factors are very unlikely to act as direct transcriptional repressors (39).
Although in cell lines, HIF has been reported to induce matriptase-2 (TMPRSS6) and furin, 2 proteases that modulate HAMP expression by blunting BMP6/HJV signaling (16, 19), hepatic Hif activation without the concomitant increase in Epo transcription does not suppress Hamp1, which would argue against a regulatory role of furin and matriptase 2 in our model. This notion is furthermore supported by a lack of increase in Tmprss6 and furin mRNA levels in Vhl–/– mice (Supplemental Figure 3D). The ability to synthesize Epo was an absolute requirement for Hamp1 suppression despite constitutive Hif-1 and Hif-2 activation in the liver. Our data also argue against a direct role for Epo in the regulation of Hamp1 and suggest that Hamp1 suppression in Vhl–/– livers is independent of hepatic EpoR activation. In our model of global Vhl deficiency, Epo synthesis is strongly enhanced in liver and kidney, and paracrine or autocrine activation of hepatocyte EpoR is unlikely to be involved in Hamp1 regulation in vivo. During preparation of this manuscript, Mastrogiannaki and colleagues reported that treatment with anti-Epo blocking serum raised Hamp1 mRNA levels in hepatocyte-specific Vhl/Hif1a-knockout mice (Albumin-cre model) to levels similar to those found in vehicle-injected control mice (40). This observation together with our findings is in contrast to in vitro data from human hepatoma cells and primary hepatocytes, where EPO has been shown to regulate HAMP in a dose-dependent manner through activation of its receptor (21). While this discrepancy could be a reflection of differences between experimental approaches, i.e., cell-culture studies versus whole animal models, our data are consistent with findings in severely anemic mice (anemia was induced by phlebotomy), which are characterized by low Hamp1 expression (22). In their report, Pak and colleagues investigated whether anemia itself, elevated serum Epo, or erythropoietic activity was required for Hamp1 suppression. Treatment of anemic mice with Cp, doxorubicin, or a noncytotoxic Epo-blocking Ab inhibited erythropoiesis and raised Hamp1 levels above normemic control levels (22), suggesting that Hamp1 is not directly regulated by either Epo or tissue hypoxia under anemic conditions, but rather by a signal that is associated with increased erythropoietic activity. In keeping with the findings by Pak and colleagues in Cp-, doxorubicin- and anti-Epo Ab–treated mice, Hamp1 levels were also markedly increased in Vhl/Hif-1/Hif-2–/–, Vhl/Epo–/–, and Cp-treated control and Vhl–/– mice, which are characterized by diminished or inhibited erythropoietic activity.
Serum iron has been shown to regulate hepcidin synthesis. Acute depletion of iron results in Hamp1 suppression involving matriptase-2 (41), whereas iron loading increases Hamp1 via TFR2-, HJV-, BMP6-, and HFE-mediated signals (42). In the context of iron-deficiency anemia Hif-1α and Hif-2α are stabilized in Epo-producing tissues, primarily in kidney, but also in the liver, depending on the severity of anemic hypoxia. This results in an increase of serum Epo levels and the suppression of hepcidin (1). Hif-2 is the main regulator of both renal and hepatic Epo synthesis under hypoxic conditions (30, 32) and does not appear to be directly involved in the regulation of Hamp1 in hepatocytes (40). While we did not observe changes in serum iron levels in mice with global Vhl deficiency at the time point our analyses were performed, we found that hepatic H-ferritin levels were reduced, which is suggestive of a decrease in intracellular free iron. The ferritin 5′ UTR contains an IRE that mediates translational inhibition in the presence of low intracellular iron. It is of interest to point out that H-ferritin reduction was dependent on the ability to synthesize Epo, but not on Vhl status nor the presence of stabilized Hif-α (H-ferritin was not reduced in Vhl/Epo–/– livers). However, in certain VHL-deficient renal cancer cell lines, H-ferritin and the labile iron pool were decreased (43). It is likely that the changes in H-ferritin levels seen in Vhl–/– livers are a consequence of enhanced erythropoietic activity and iron utilization. Nevertheless, we cannot exclude that altered intracellular iron levels have contributed to the regulation of Hamp1 in our model. While liver tissue has not been examined, serum iron and ferritin levels are decreased in Chuvash patients and in individuals sojourning at high altitude for 10–12 days (5400 m) (6, 37). However, time-course analysis showed that the decrease of serum hepcidin in subjects ascending to high altitude was rapid and preceded changes in serum ferritin and transferrin saturation. This observation suggests that iron-independent systemic signals must play a major role in the physiologic regulation of hepcidin under hypoxic conditions (44). This notion is supported by clinical observations in patients with β-thalassemia, who have low serum hepcidin levels in the presence of iron overload (45).
Recently, GDF-15 and TWSG1 have been proposed to be erythroblast-derived factors, although not erythroblast specific, that mediate hepcidin suppression under conditions of increased erythropoietic activity (24, 36). In particular, high levels of serum GDF15 associate with ineffective erythropoiesis and may reflect a certain type of BM stress or erythroblast apoptosis (28). The role of GDF15 in hepcidin regulation under physiologic conditions and in other pathologic settings, however, is unclear and has been debated. Whereas Twsg1 mRNA expression levels did not change in BM and spleen from Vhl–/– mice, Gdf15 mRNA levels were elevated and were associated with increased serum concentrations of Gdf15. Although Gdf15 serum levels in Vhl–/– mice were much lower (increased by approximately 2-fold over control) than those reported in β-thalassemia patients (mean of 66,000 pg/ml, ref. 24), our data from Hep3B cells exposed to smaller doses of recombinant Gdf15 support the hypothesis that Gdf15 may have contributed to Hamp1 suppression in Vhl–/– mice. We found that recombinant murine Gdf15 suppressed HAMP in Hep3B cells at a concentration of 750 pg/ml (Supplemental Figure 3C). This is in contrast to previous reports where higher doses of GDF15 were needed to achieve HAMP suppression in human HuH-7 hepatoma cells and in primary hepatocytes, while low-dose GDF15 treatment increased HAMP in these cells (24). The molecular basis of these differences in GDF15 dose responses is not clear and warrants further investigations. While we cannot completely exclude that Hif activation in hepatocytes modulates their response to Gdf15, elevation of serum Gdf15 in Vhl–/– mice is not likely to result from Hif activation, as a similar degree of increase was found when WT mice were treated with human recombinant EPO, which was also associated with Hamp1 suppression. Studies in humans have not yet demonstrated a significant association between suppression of hepcidin levels and serum GDF15 levels following EPO administration (46), which may relate to the EPO doses used, study size, complexity of regulation, and species-dependent differences in hepcidin regulation. In the context of iron-deficiency anemia, Tanno and colleagues reported that GDF15 serum levels were not elevated (47), while in a report by Lakhal and colleagues, patients with low serum iron had elevated GDF15 levels compared with iron-replete controls (mean of 1048 pg/ml vs. 542 pg/ml) (23). Similarly, increased serum GDF15 levels were found following DFO treatment, suggesting iron-dependent regulation (23). Temporary increases in serum GDF15 levels were also observed following ascent to high altitude, which associated with increases in serum EPO (44).
In summary, here we have used genetic means to dissect the role of HIF and EPO in the regulation of hepcidin and have shown that Hif-associated suppression of Hamp1 occurs indirectly through Epo-induced erythropoiesis and may involve Gdf15 (Figure 6B). Our data have implications for targeted therapies that aim at exploiting the VHL/HIF/PHD axis for the treatment of anemia and disorders of iron homeostasis.
Methods
Generation of mice and genotyping.
The generation and genotyping of Vhl, Epo, Hif1a, and Hif2a (Epas1) conditional alleles as well as Albumin-cre and Ubc-cre/ERT2 transgenes have been described elsewhere (29, 32, 34). Inducible Cre-mediated global inactivation of Vhl, Hif-1α, Hif-2α, and/or Epo was achieved by generating mice that were homozygous for the Vhl, Hif1a, Hif2a, and/or Epo conditional alleles and expressed a tamoxifen-inducible Cre-recombinase under control of the ubiquitin c promoter Ubc-cre/ERT2 (29). The following genotypes were generated: (a) Vhl2lox/2lox;Ubc-cre/ERT2, (b) Vhl2lox/2lox;Hif1a2lox/2lox;Hif2a2lox/2lox;Ubc-cre/ERT2, and (c) Vhl2lox/2lox;Epo2lox/2lox;Ubc-cre/ERT2, referred to as Vhl–/–, Vhl/Hif-1/Hif-2–/–, or Vhl/Epo–/– after completion of tamoxifen treatment. For the temporary activation of the Ubc-cre/ERT2 transgenic system, mice received 4 i.p. injections of tamoxifen (Sigma-Aldrich) administered every other day at a concentration of 10 mg/ml (∼1.5 mg/mouse). Tamoxifen was dissolved in a mixture of 10% (by volume) ethanol and 90% (by volume) sunflower oil. Mice were phenotyped on day 8 after the first tamoxifen injection (outline of experimental protocol is shown in Supplemental Figure 1). Hepatocyte-specific inactivation of Phd2 (EglN1) was achieved by generating mice that expressed the Albumin-cre transgene and that were homozygous for the Phd2 conditional allele (EglN12lox/2lox;Albumin-cre). Cre-negative (Cre–) littermates from the same breeding pair were used as controls in all experiments. The generation and characterization of Hif2a/Pax3-cre mutant mice, referred to as P3 mutants, and thalassemic mice (th3/th3 mutants) has been described elsewhere (30, 35). Stefano Rivella (Weill Medical College of Cornell University, New York, New York, USA) provided liver mRNAs from th3/th3 mutants and littermate controls.
Phenotypic analysis of mutant mice.
Hcts were determined by microcapillary tube centrifugation or with a Hemavet 950 analyzer (Drew Scientific). Serum Epo levels were determined by ELISA (R&D Systems); serum and liver iron concentrations were measured using the Iron Assay Kit from BioVision; serum Gdf15 concentrations were measured by ELISA (MyBioSource, LLC). Reticulocyte counts were determined by FACS analysis of whole blood stained with thiazole orange following the manufacturer’s instructions (Sigma-Aldrich). For FACS analysis of BM and spleen-derived erythroid precursor cells, 1 × 106 BM or spleen cells were incubated with PE-conjugated anti-transferrin receptor protein 1 (CD71) or FITC-conjugated anti-Ter119 monoclonal antibodies (BD Biosciences — Pharmingen) as previously described (48). For the analysis of Gdf15 mRNA levels, Ter119-positive cells were isolated from BM and spleen by immunomagnetic separation (Ter119-MicroBeads; Miltenyi Biotech). rhEPO (Amgen) was dissolved in 0.1 ml water and injected i.p. at a dose of 200 IU every day for 3 days prior to mouse phenotyping. For the pharmacologic inhibition of erythropoiesis, mice received a single i.p. injection of Cp (Sigma-Aldrich), 2.5 mg dissolved in 0.25 ml water 1 day before the first tamoxifen injection. Control animals received 0.25 ml of normal saline. For studies under conditions of chronic hypoxia, mice were exposed to 10% O2 for 10 days in an animal hypoxia chamber (Biospherix Ltd) and analyzed immediately after hypoxia treatment.
DNA, RNA, and protein analysis.
For genotyping, tail DNA was isolated according to Laird et al. (49). Other tissue DNA was isolated with DNeasy Blood and Tissue Kit according to the manufacturer’s instructions (QIAGEN). RNA was isolated using RNeasy Mini Kit according to the manufacturer’s protocol (QIAGEN). For real-time PCR analysis, 1 μl of cDNA was subjected to PCR amplification on an ABI 7300 platform using either SYBR Green PCR Master Mix or Taqman Universal PCR Master Mix (Applied Biosystems). Relative mRNA expression levels were quantified with the relative standard curve method according to the manufacturer’s instructions (Applied Biosystems). 18S ribosomal RNA was used for normalization (50). Primer sequences for the analysis of Vhl, Vegf, Dmt1, Tfrc, and Hamp1 have been published elsewhere (30, 32, 35). The following primer sequences were used for mRNA detection: Epo (forward, 5′-TGGTCTACGTAGCCTCACTTCACT-3′; reverse, 5′-TGGAGGCGACATCAATTCCT-3′); Gdf15 (forward, 5′-CAGAGCCGAGAGGACTCGAA-3′; reverse, 5′-CCGGTTGACGCGGAGTAG-3′); Twsg1 (forward, 5′-AGCATGCACTCCTTACAGCA-3′; reverse, 5′-ACAAAGCACTCTGTGCCAGC-3′); Tmprss6 (forward, 5′-ACAGGGTGGCGATGTACGA-3′; reverse, 5′-GCACCCATAGACCGAGGTGAT-3′); furin (forward, 5′-GTGCCTGCTCAGTGCCAG-3′; reverse, 5′-CGCTCGTCCGGAAAAGTT-3′); and human HAMP (forward, 5′-CAGCTGGATGCCCATGTTC-3′; reverse, 5′-AGCCGCAGCAGAAAATGC-3′). Nuclear protein extracts for Western blot analysis were prepared, and Hif-1α and Hif-2α were detected as previously described (32); H-ferritin and β-actin protein levels were analyzed with antibodies from Alpha Diagnostic International and Sigma-Aldrich.
Cell culture.
Hep3B cells were cultured in DMEM supplemented with 10% FBS; recombinant Gdf15 (MyBioSource, LLC) was added to the culture medium to achieve a final concentration of 750 pg/ml.
Statistics.
Data reported represent mean values ± SEM. Statistical analyses were performed with Prism 5.0b software (GraphPad Software) using the unpaired Student’s t test. For consistency, all P values reported were derived from unpaired 1-tailed Student’s t test analysis. P < 0.05 was considered statistically significant.
Study approval.
All procedures involving mice were performed in accordance with NIH guidelines for the use and care of live animals and were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Vanderbilt University.
Supplementary Material
Acknowledgments
This work was supported by the Krick-Brooks chair in Nephrology (to V.H. Haase) and by NIH grants R01-DK081646 and R01-DK080821 (both to V.H. Haase). The authors wish to thank Pinelopi Kapitsinou for critical reading of the manuscript.
Footnotes
Conflict of interest: Volker H. Haase serves on the Scientific Advisory Board of Akebia Therapeutics. Knut Niss was an employee of Pfizer Inc. and is now an employee of EMD Millipore Corp.
Citation for this article: J Clin Invest. 2012;122(12):4635–4644. doi:10.1172/JCI63924.
References
- 1.Haase VH. Hypoxic regulation of erythropoiesis and iron metabolism. Am J Physiol Renal Physiol. 2010;299(1):F1–F13. doi: 10.1152/ajprenal.00174.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112(2):219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jelkmann W. Erythropoietin after a century of research: younger than ever. Eur J Haematol. 2007;78(3):183–205. doi: 10.1111/j.1600-0609.2007.00818.x. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Ang SO, et al. Disruption of oxygen homeostasis underlies congenital Chuvash polycythemia. Nat Genet. 2002;32(4):614–621. doi: 10.1038/ng1019. [DOI] [PubMed] [Google Scholar]
- 6.Gordeuk VR, et al. Chuvash polycythemia VHLR200W mutation is associated with down-regulation of hepcidin expression. Blood. 2011;118(19):5278–5282. doi: 10.1182/blood-2011-03-345512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicolas G, et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110(7):1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peyssonnaux C, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J Clin Invest. 2007;117(7):1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volke M, et al. Evidence for a lack of a direct transcriptional suppression of the iron regulatory peptide hepcidin by hypoxia-inducible factors. PLoS One. 2009;4(11):e7875. doi: 10.1371/journal.pone.0007875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braliou GG, Verga Falzacappa MV, Chachami G, Casanovas G, Muckenthaler MU, Simos G. 2-Oxoglutarate-dependent oxygenases control hepcidin gene expression. J Hepatol. 2008;48(5):801–810. doi: 10.1016/j.jhep.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 11.Gao J, Chen J, Kramer M, Tsukamoto H, Zhang AS, Enns CA. Interaction of the hereditary hemochromatosis protein HFE with transferrin receptor 2 is required for transferrin-induced hepcidin expression. Cell Metab. 2009;9(3):217–227. doi: 10.1016/j.cmet.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemeth E, Ganz T. The role of hepcidin in iron metabolism. Acta Haematol. 2009;122(2–3):78–86. doi: 10.1159/000243791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babitt JL, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38(5):531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 14.Andriopoulos B, Jr, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41(4):482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41(4):478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 16.Silvestri L, Pagani A, Camaschella C. Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron homeostasis. Blood. 2008;111(2):924–931. doi: 10.1182/blood-2007-07-100677. [DOI] [PubMed] [Google Scholar]
- 17.McMahon S, Grondin F, McDonald PP, Richard DE, Dubois CM. Hypoxia-enhanced expression of the proprotein convertase furin is mediated by hypoxia-inducible factor-1: impact on the bioactivation of proproteins. J Biol Chem. 2005;280(8):6561–6569. doi: 10.1074/jbc.M413248200. [DOI] [PubMed] [Google Scholar]
- 18.Du X, et al. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320(5879):1088–1092. doi: 10.1126/science.1157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakhal S, Schodel J, Townsend AR, Pugh CW, Ratcliffe PJ, Mole DR. Regulation of type II transmembrane serine proteinase TMPRSS6 by hypoxia-inducible factors: new link between hypoxia signaling and iron homeostasis. J Biol Chem. 2010;286(6):4090–4097. doi: 10.1074/jbc.M110.173096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8(6):502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto JP, et al. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPalpha. Blood. 2008;111(12):5727–5733. doi: 10.1182/blood-2007-08-106195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pak M, Lopez MA, Gabayan V, Ganz T, Rivera S. Suppression of hepcidin during anemia requires erythropoietic activity. Blood. 2006;108(12):3730–3735. doi: 10.1182/blood-2006-06-028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakhal S, et al. Regulation of growth differentiation factor 15 expression by intracellular iron. Blood. 2009;113(7):1555–1563. doi: 10.1182/blood-2008-07-170431. [DOI] [PubMed] [Google Scholar]
- 24.Tanno T, et al. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 2007;13(9):1096–1101. doi: 10.1038/nm1629. [DOI] [PubMed] [Google Scholar]
- 25.Tamary H, et al. Elevated growth differentiation factor 15 expression in patients with congenital dyserythropoietic anemia type I. Blood. 2008;112(13):5241–5244. doi: 10.1182/blood-2008-06-165738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez JM, et al. Growth differentiation factor 15 production is necessary for normal erythroid differentiation and is increased in refractory anaemia with ring-sideroblasts. Br J Haematol. 2009;144(2):251–262. doi: 10.1111/j.1365-2141.2008.07441.x. [DOI] [PubMed] [Google Scholar]
- 27.Finkenstedt A, et al. Regulation of iron metabolism through GDF15 and hepcidin in pyruvate kinase deficiency. Br J Haematol. 2009;144(5):789–793. doi: 10.1111/j.1365-2141.2008.07535.x. [DOI] [PubMed] [Google Scholar]
- 28.Tanno T, Noel P, Miller JL. Growth differentiation factor 15 in erythroid health and disease. Curr Opin Hematol. 2010;17(3):184–190. doi: 10.1097/MOH.0b013e328337b52f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1(1):113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapitsinou PP, et al. Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. Blood. 2010;116(16):3039–3048. doi: 10.1182/blood-2010-02-270322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rankin EB, et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol. 2009;29(16):4527–4538. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rankin EB, et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117(4):1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minamishima YA, Kaelin WG., Jr Reactivation of hepatic EPO synthesis in mice after PHD loss. Science. 2010;329(5990):407. doi: 10.1126/science.1192811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeigler BM, Vajdos J, Qin W, Loverro L, Niss K. A mouse model for an erythropoietin-deficiency anemia. Dis Model Mech. 2011;3(11–12):763–772. doi: 10.1242/dmm.004788. [DOI] [PubMed] [Google Scholar]
- 35.Gardenghi S, et al. Ineffective erythropoiesis in beta-thalassemia is characterized by increased iron absorption mediated by down-regulation of hepcidin and up-regulation of ferroportin. Blood. 2007;109(11):5027–5035. doi: 10.1182/blood-2006-09-048868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanno T, et al. Identification of TWSG1 as a second novel erythroid regulator of hepcidin expression in murine and human cells. Blood. 2009;114(1):181–186. doi: 10.1182/blood-2008-12-195503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piperno A, et al. Modulation of hepcidin production during hypoxia-induced erythropoiesis in humans in vivo: data from the HIGHCARE project. Blood. 2011;117(10):2953–2959. doi: 10.1182/blood-2010-08-299859. [DOI] [PubMed] [Google Scholar]
- 38.Hickey MM, Lam JC, Bezman NA, Rathmell WK, Simon MC. von Hippel-Lindau mutation in mice recapitulates Chuvash polycythemia via hypoxia-inducible factor-2alpha signaling and splenic erythropoiesis. J Clin Invest. 2007;12(117):3879–3889. doi: 10.1172/JCI32614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117(23):e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mastrogiannaki M, et al. Hepatic HIF-2 down-regulates hepcidin expression in mice through epo-mediated increase in erythropoiesis. Haematologica. 2012;97(6):827–834. doi: 10.3324/haematol.2011.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang AS, et al. Suppression of hepatic hepcidin expression in response to acute iron deprivation is associated with an increase of matriptase-2 protein. Blood. 2011;117(5):1687–1699. doi: 10.1182/blood-2010-06-287292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos E, et al. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology. 2011;53(4):1333–1341. doi: 10.1002/hep.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alberghini A, Recalcati S, Tacchini L, Santambrogio P, Campanella A, Cairo G. Loss of the von Hippel Lindau tumor suppressor disrupts iron homeostasis in renal carcinoma cells. J Biol Chem. 2005;280(34):30120–30128. doi: 10.1074/jbc.M500971200. [DOI] [PubMed] [Google Scholar]
- 44.Talbot NP, et al. Regulation of hepcidin expression at high altitude. Blood. 2012;119(3):857–860. doi: 10.1182/blood-2011-03-341776. [DOI] [PubMed] [Google Scholar]
- 45.Ginzburg Y, Rivella S. beta-thalassemia: a model for elucidating the dynamic regulation of ineffective erythropoiesis and iron metabolism. Blood. 2011;118(16):4321–4330. doi: 10.1182/blood-2011-03-283614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashby DR, et al. Erythropoietin administration in humans causes a marked and prolonged reduction in circulating hepcidin. Haematologica. 2009;95(3):505–508. doi: 10.3324/haematol.2009.013136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanno T, Rabel A, Lee YT, Yau YY, Leitman SF, Miller JL. Expression of growth differentiation factor 15 is not elevated in individuals with iron deficiency secondary to volunteer blood donation. Transfusion. 2010;50(7):1532–1535. doi: 10.1111/j.1537-2995.2010.02601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Pop R, Sadegh C, Brugnara C, Haase VH, Socolovsky M. Suppression of Fas-FasL coexpression by erythropoietin mediates erythroblast expansion during the erythropoietic stress response in vivo. Blood. 2006;108(1):123–133. doi: 10.1182/blood-2005-11-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19(15):4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rankin EB, Higgins DF, Walisser JA, Johnson RS, Bradfield CA, Haase VH. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol Cell Biol. 2005;25(8):3163–3172. doi: 10.1128/MCB.25.8.3163-3172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.