Abstract
The genetic diversity of HIV-1 represents a major challenge in vaccine development. In this study, we establish a rationale for eliminating HIV-1–infected cells by targeting cellular immune responses against stable human endogenous retroviral (HERV) antigens. HERV DNA sequences in the human genome represent the remnants of ancient infectious retroviruses. We show that the infection of CD4+ T cells with HIV-1 resulted in transcription of the HML-2 lineage of HERV type K [HERV-K(HML-2)] and the expression of Gag and Env proteins. HERV-K(HML-2)–specific CD8+ T cells obtained from HIV-1–infected human subjects responded to HIV-1–infected cells in a Vif-dependent manner in vitro. Consistent with the proposed mode of action, a HERV-K(HML-2)–specific CD8+ T cell clone exhibited comprehensive elimination of cells infected with a panel of globally diverse HIV-1, HIV-2, and SIV isolates in vitro. We identified a second T cell response that exhibited cross-reactivity between homologous HIV-1-Pol and HERV-K(HML-2)-Pol determinants, raising the possibility that homology between HIV-1 and HERVs plays a role in shaping, and perhaps enhancing, the T cell response to HIV-1. This justifies the consideration of HERV-K(HML-2)–specific and cross-reactive T cell responses in the natural control of HIV-1 infection and for exploring HERV-K(HML-2)–targeted HIV-1 vaccines and immunotherapeutics.
Introduction
The genetic diversity of HIV-1 is considerable, with amino acids in Env differing by as much as 20% within a subtype and by more than 35% between subtypes and those in Gag amino acid differing by roughly 8% between clades (1). This poses a major challenge to the development of an effective vaccine by limiting the likelihood that vaccine-elicited immune responses will recognize the diverse strains of HIV-1 to which a vaccinee could be exposed. The almost unparalleled propensity of HIV-1 to mutate in order to evade effective immune pressure is perhaps an even greater barrier to achieving enduring vaccine-mediated protection. A leading hypothesis for the lack of efficacy of the recent phase IIB STEP HIV-1 vaccine trial is that vaccine-elicited T cell responses lacked sufficient breadth to recognize transmitting viral strains or variants that rapidly emerged once an infection was seeded (2). Devising strategies to mitigate the impact of sequence diversity on candidate vaccines is an area of intense research (3). Here we explore what we believe to be a novel approach to circumventing the challenges of HIV-1 diversity and mutability by targeting T cell responses against antigens derived from the HML-2 lineage of type K human endogenous retroviruses [HERV-K(HML-2)] as surrogate markers of HIV-1–infected cells.
Human endogenous retroviruses (HERVs) are the DNA remnants of ancient infectious retroviruses that infected the germ line of our evolutionary ancestors and became fixed in the human population. HERVs, which colonized the human genome in this manner, have expanded through infection or retrotransposition to the point where HERV sequences now comprise 8% of the human genome (4, 5). Of particular relevance to this study is the relatively young and intact HERV-K human mouse mammary tumor virus-like type 2 (HML-2) family, which is present at an estimated 89 proviral copies per haploid genome (6). Some of these HERV-K(HML-2) insertions contain complete open reading frames for viral proteins and, although no replication-competent HERV-K(HML-2) provirus has been identified yet, infectious HERV-K(HML-2) viruses can be reconstituted either from consensus sequences or by complementation among sequences from as few as 3 proviral loci (7–20). Despite this capacity for expression, HERV-K(HML-2) proteins have not been observed in healthy adult tissues but rather have been uniquely associated with disease states such as teratocarcinoma (21–24).
We have previously presented the hypothesis that the manipulation of the host cellular environment by HIV-1 to one which favors retroviral expression and replication may result in the expression of HERV proteins (25). Following from this, we have speculated that such expression could be targeted by HERV-specific T cells, resulting in the specific elimination of HIV-1–infected cells. As HERV antigens are encoded in the human genome, this targeting would occur irrespective of HIV-1 sequence variability and would be exempt from immune escape. The implications of this would be two fold. First, it would validate avenues of study considering a role for HERV-specific T cells in natural control of HIV-1. Second, it would facilitate a new paradigm in the development of HIV-1 vaccines, whereby strategies aimed at eliciting HERV-specific T cell responses could be considered as a means of overcoming the challenge of HIV-1 sequence diversity. Early support for the induction of HERV antigen expression in HIV-1–infected subjects was provided by our observation that T cell responses to a variety of HERV-derived peptides are detectable in HIV-1–infected subjects but not in uninfected controls (25, 26). Supporting the in vivo relevance of these responses, we observed that strong HERV-specific T cell responses were associated with control of HIV-1 replication (25, 27). However, these observations could have resulted from a number of etiologies and are insufficient to either infer HIV-1–induced HERV expression or to validate HERV antigens as novel HIV-1 vaccine targets. In this study, we focus in on the HERV-K(HML-2) family of endogenous retroviruses described above. We present data indicating that HIV-1 infection of primary CD4+ T cells results in the induction of HERV-K(HML-2) RNA and protein expression. Critically, we present the first demonstration to our knowledge that HERV-K(HML-2)–specific CD8+ T cells specifically respond to and eliminate cells infected with diverse isolates of HIV-1, HIV-2, and SIV. This is shown to be dependent upon a specific host-virus interaction, as deletion of the vif gene abrogated recognition. These data provide proof of concept both for considering a role for HERV-K(HML-2)–specific T cell responses in natural control of HIV-1 infection and for pursuing HERV-K(HML-2)–targeted HIV-1 vaccine and immunotherapeutic strategies.
Results
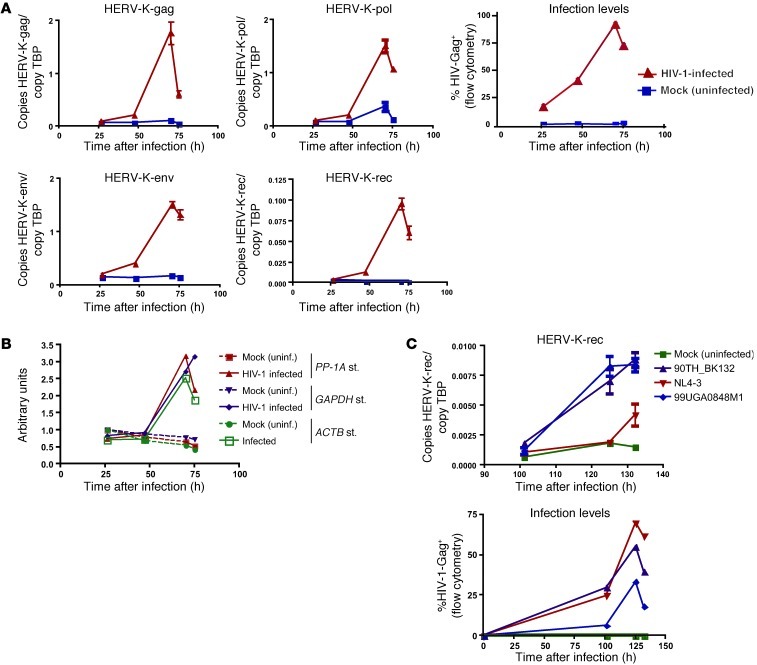
HIV-1 infection results in the accumulation of HERV-K(HML-2) transcripts.
We performed quantitative RT-PCR on RNA from primary CD4+ T cells that had either been infected with the primary isolate of HIV-1 90TH_BK132 or maintained as mock-infected (uninfected) controls. HERV-K(HML-2)-gag, -pol, and -env probes/primers were designed to target uninterrupted viral sequences, while the HERV-K(HML-2)-rec probes/primers span the rec splicing site and therefore only amplify spliced viral RNA. We observed induction of HERV-K(HML-2) RNA as measured by each of these probe/primer combinations, the kinetics of which mirrored HIV-1 replication (Figure 1A). HERV-K(HML-2)-gag, -pol, and -env templates were each detected at similar quantities to one another, and at their peaks exceeded the levels of the TATA-box binding protein (TBP) transcript used to standardize the assay. Spliced HERV-K(HML-2)-rec RNA was present at very low levels in uninfected cells but increased to approximately 10% the level of TBP at the peak of infection (Figure 1A). This pattern of HERV-K(HML-2) induction was maintained when standardization was performed against a variety of housekeeping genes (Figure 1B), and similar results were observed in cells from each of 3 other donors tested. Notably, levels of HERV-K(HML-2) RNA peaked contemporaneously with the peak of HIV-1 infection, as measured by intracellular Gag staining. Thus, the delay of 70 hours between initiation of HIV-1 infection and the peak of HERV-K(HML-2) RNA induction can be primarily attributed the time required to achieve high levels of HIV-1 infection. However, the relatively low levels of HERV-K(HML-2) RNA induction observed at 47 hours may speak to some delay between HIV-1 infection and HERV-K(HML-2) RNA induction. We consistently observed substantially less accumulation of HERV-K(HML-2) transcripts in cells infected with the laboratory-adapted isolate HIV-1-NL4-3, as compared with primary isolates, despite equivalent or greater levels of infection with the former. This is represented by spliced rec quantitation data from a second donor, as shown in Figure 1C, but was observed for each of the HERV-K(HML-2) amplification targets. Thus, the infection of primary CD4+ T cells with primary isolates of HIV-1 results in the accumulation of both spliced and unspliced HERV-K(HML-2) transcripts, with a substantially lesser effect observed upon infection with the laboratory-adapted isolate NL4-3.
Figure 1. HIV-1 infection results in the induction of HERV-K(HML-2) transcripts.
(A) CD4+ T cells were enriched from the PBMCs of HIV-1–uninfected individuals, activated, and then either infected with HIV-1 or maintained as mock-infected (uninfected) controls. Copy numbers of HERV-K(HML-2) and TBP RNA were determined using TaqMan-based qPCR by comparison to linearized plasmid standards. Data from an infection with the HIV-1 isolate 90TH_BK132, depicting the mean number HERV-K(HML-2) copies per copy TBP are shown. Quantitations were performed in triplicate, and error bars represent SEM. Also shown are levels of HIV-1 infection (% HIV-Gag+) at the indicated time points, as measured by flow cytometric staining for CD4 and HIV-Gag. Similar data were observed in cells from each of 4 donors tested and with each of the 2 primary HIV-1 isolates tested (90TH_BK132 and 99UG_A0848M1). (B) RNA from the sample analyzed in A was amplified using the MessageAmp Kit (Ambion). HERV-K(HML-2)-pol RNA was quantified by TaqMan qPCR and standardized to relative quantities of the housekeeping genes PP-1A, GAPDH, or ACTB determined by SYBR qPCR. Values are expressed using the following formula: arbitrary units = (standardized quantity)/(standardized quantity in uninfected 26-hour time point). uninf., uninfected; st., strain. (C) Representative absolute qPCR data depicting copies of spliced HERV-K(HML-2)-rec per copy TBP for cells from a single subject infected with the indicated HIV-1 viruses. Quantitations were performed in triplicate, and error bars represent SEM. Levels of HIV-1 infection corresponding to qPCR sampling points, as measured by flow cytometry are also shown.
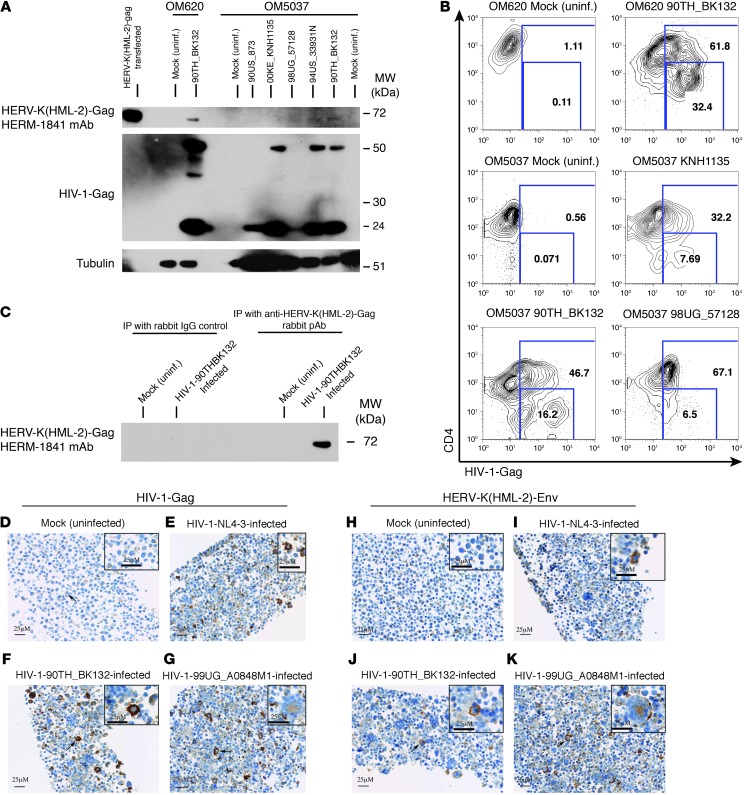
HIV-1 infection results in expression of HERV-K(HML-2)-Gag and -Env proteins.
We examined CD4+ T cells, which had been isolated from HIV-1–uninfected subjects and then either infected with HIV-1 or mock infected in vitro, for the expression of HERV-K(HML-2)-Gag and -Env proteins. The anti–HERV-K-Gag mouse mAb HERM-1841 detected a 72-kDa band in HIV-1–infected cells, which was absent from uninfected controls (Figure 2A). This matched the migration of HERV-K(HML-2)-Gag detected in CD4+ T cells transfected with the codon-optimized HERV-K(HML-2)-Gag mRNA (Figure 2A). The HERV-K(HML-2)-Gag protein was present at low abundance, with the image displayed in Figure 2A representing an overnight exposure for HERV-K(HML-2)-Gag compared with a 5-minute exposure for HIV-1-Gag. This was anticipated by our qPCR data, in which we typically observed HIV-1-gag RNA levels on the order of 2,500 copies per copy TBP compared with less than 2 copies HERV-K(HML-2)-gag per copy TBP. The detection of HERV-K(HML-2)-Gag in these cultures was contingent upon high levels of HIV-1 infection, with a substantial proportion of late-stage–infected cells (CD4 downregulated). Figure 2B displays frequencies of HIV-1–infected (Gag+) cells in a subset of the samples lysed for the Western blot shown in Figure 2A. The cells from subject OM620, in which HERV-K(HML-2)-Gag was clearly detected, were greater than 61% infected, with 32.4% exhibiting late-stage infection. Cells from subject OM5037, with intermediate infections (90TH_BK132, 94US_33931N), gave rise to only faint bands, whereas HERV-K(HML-2)-Gag was not detected in less advanced infections (00KE_KNH1135, 90US_873). The 98UG_57128 infection was unusual in that HERV-K(HML-2)-Gag was detected despite very little HIV-1-Gag detection by Western blot (Figure 2A). By flow cytometry, we observe that these cells were highly infected but that Gag staining was dim, suggesting that the epitope recognized by the anti–HIV-1-Gag mAb Kc57 may be mutated in this virus. We were able to enhance the sensitivity of HERV-K(HML-2)-Gag detection by immunoprecipitating HERV-K(HML-2)-Gag using a custom rabbit anti–HERV-K(HML-2)–capsid polyclonal antibody (pAb) “4890-Gly” and performing Western blotting on the eluate with HERM-1841 (Figure 2C). Thus, HIV-1 infection results in the induction of low levels of HERV-K(HML-2)-Gag.
Figure 2. HIV-1 infection results in the induction of HERV-K(HML-2) protein expression.
CD4+ T cells were either infected with primary isolates of HIV-1 or maintained as mock infection controls. (A) Western blots of a single membrane probed consecutively with the anti–HERV-K(HML-2)-Gag mAb HERM-1841, the anti–HIV-1-Gag mAb Kc57, and an anti-tubulin pAb. Samples represent infections and mock (uninfected) controls for 2 subjects: OM620 and OM5037. (B) Portions of the samples displayed in A were removed prior to lysis and stained with anti-CD4 (y axis) and anti–HIV-1-Gag (x axis). The resulting flow cytometry data are shown. Numbers represent total percentages of HIV-1-Gag+ (top box) and percentages of HIV-1-Gag+CD4dim (bottom box) cells. (C) Immunoprecipitations were performed on lysates from HIV-1–infected or uninfected primary CD4+ T cells using either an anti–HERV-K(HML-2)-Gag rabbit pAb or a rabbit IgG control. Immunoprecipitates were separated by SDS-PAGE and probed with the anti–HERV-K(HML-2)-Gag murine mAb HERM-1841. (D–G) Immunohistochemistry data depicting primary CD4+ T cells stained with the HIV-1-Gag–specific antibody Kc57. (D) Uninfected, (E) HIV-1-NL4-3–infected, (F) HIV-1-90TH_BK132–infected, and (G) 99UG_A0848M1-infected primary CD4+ T cells. (H–K) Immunohistochemistry data from serial sections corresponding to those shown in D through G, stained with the HERV-K(HML-2)-Env–specific antibody HERM-1811-5. In D, F, G, J, and K, arrows emphasize examples of the staining characteristics described in the text. Scale bar: 25 μM.
We have been unable to detect HERV-K(HML-2)-Env protein expression in HIV-1–infected cells by Western blot using the mAb HERM-1811, while detecting expression in lysates of HERV-K(HML-2)-Env mRNA transfected CD4+ T cells (data not shown). We have noted rapid degradation of these positive control samples, suggesting that the transmembrane Env protein may be susceptible to aggregation following cell lysis. As such factors may have impaired our ability to detect low levels of Env expression by Western blot, we also studied HERV-K(HML-2)-Env expression by immunohistochemistry. Figure 2 depicts HIV-1-Gag staining in CD4+ T cells from a single donor, maintained as a mock-infected control (Figure 2D), infected with the laboratory-adapted HIV-1 isolate NL4-3 (Figure 2E), or infected with one of the primary HIV-1 isolates 90TH_BK132 (Figure 2F) or 99UG_A0848M1 (Figure 2G). Figure 2, H–K, depicts HERV-K(HML-2)-Env staining in matched samples using HERM-1811. In mock-infected cells (Figure 2, D and H), a nonspecific punctate cytoplasmic or membrane-associated staining (minimal in distribution and generally mild in intensity) was noted in a subset of mononuclear cells and cellular debris with both antibodies. We observed an induction of HERV-K(HML-2)-Env expression in cells infected with the primary isolates of HIV-1 90TH_BK132 (Figure 2J) or 99UG_A0848M1 (Figure 2K). In these samples, HERV-K(HML-2)-Env expression was induced on the membranes of many large mononuclear and syncytial cells (coarsely granular staining, up to marked intensity) and in the cytoplasm (homogenous staining, moderate intensity) of syncytial cells, mirroring the cell distribution and microscopic characteristics of the staining observed with the HIV-1-Gag antibody in the serial microscopic section. The specificity of the HERV-K(HML-2)-Env staining is supported by its agreement with our qPCR data, as it indicates greater induction of HERV-K(HML-2) in cells infected with primary isolates of HIV-1, as compared with cells infected with HIV-1-NL4-3, despite similar levels of infection (Figure 2, E and I). This demonstrates that HERM-1811 staining does not simply result from nonspecific binding to large mononuclear and syncytial cells formed secondary to viral infection. We have tested HERM-1811 for cross-reactivity with HIV-1-Env and other viral proteins and found none. Thus, HIV-1 infection results in expression of HERV-K(HML-2)-Env.
Characterization of a HERV-K(HML-2)-Env–specific CD8+ T cell clone.
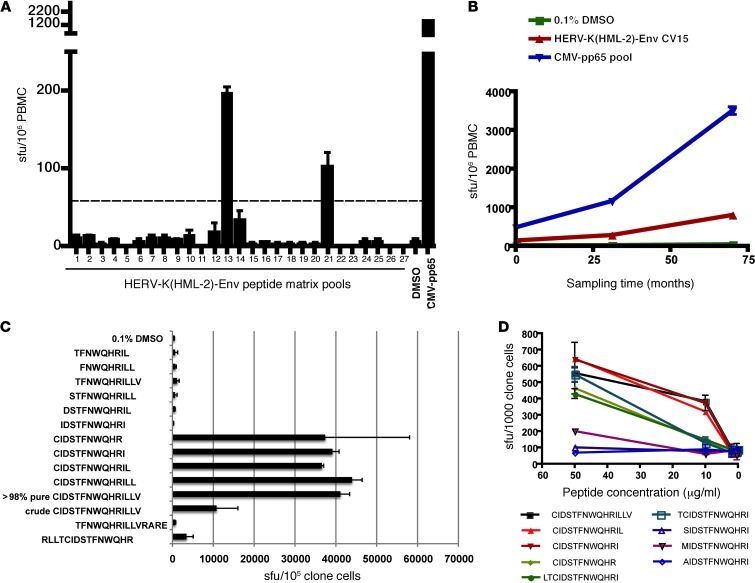
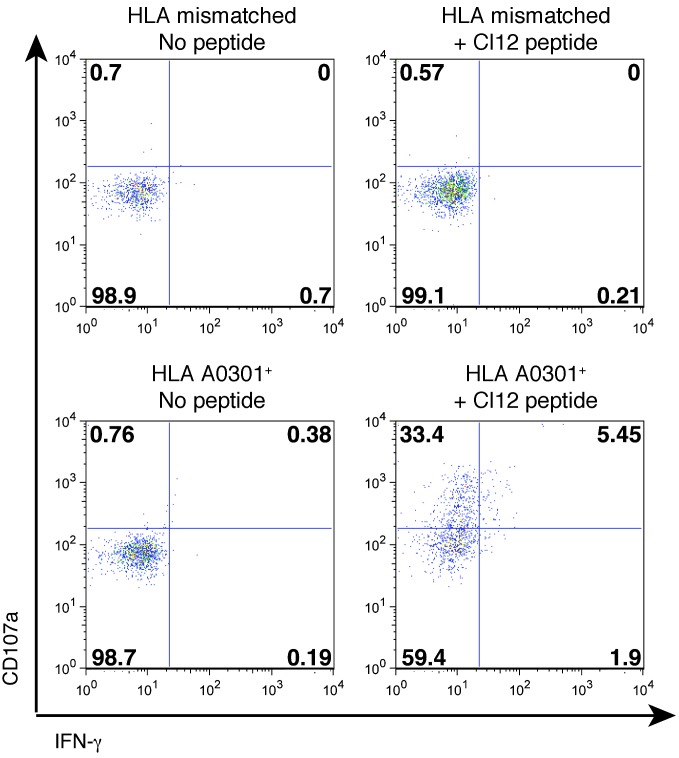
The induction of HERV-K(HML-2) antigen expression in HIV-1–infected cells could serve to stimulate HERV-K(HML-2)–specific CD8+ T cells, resulting in the targeted elimination of infected cells. We studied this using a HERV-K(HML-2)-Env–specific CD8+ T cell response that was identified in the PBMCs of an HIV-1–infected elite controller (subject OM9) by IFN-γ ELISPOT using a peptide matrix mapping methodology with pools of 15-mer peptides overlapping by 11 amino acids. We identified responses to matrix pools 13 and 21, mapping to the peptide CIDSTFNWQHRILLV (Figure 3A). This response was observed at 138 spot-forming units (sfu) per 106 PBMCs at first sampling, approximately 11 years after infection, and increased to 813 sfu per 106 PBMCs over the subsequent 5 years (Figure 3B). HERV-K(HML-2)-Env-CIDSTFNWQHRILLV–specific T cell clones were obtained, and the minimal determinant was established as CIDSTFNWQHR, although the 11-mer CIDSTFNWQHRI elicited a stronger response at 10 μg/ml (Figure 3, C and D). We established that the response was restricted by HLA-A*0301 by testing the abilities of a panel of B lymphoblastoid cell lines (BLCLs) to present to the clone. We present an example of a negative response to a peptide-pulsed BLCLs that was mismatched with subject OM9 on all MHC-I alleles and a positive response to a BLCLs that shared only HLA-A*0301 with OM9 (Figure 4). T cell receptor sequences for this clone are given in Supplemental Table 1 (supplemental material available online with this article; doi: 10.1172/JCI64560DS1), and IMGT/V-QUEST characterizations of these sequences are presented in Supplemental Tables 2 and 3.
Figure 3. Identification of a HERV-K(HML-2)-Env–specific CD8+ T cell response and fine mapping of the T cell determinant.
(A and B) IFN-γ ELISPOT was performed on PBMCs from the HIV-1–infected elite controller OM9. Results depicting mean sfu per 106 PBMCs, with error bars representing standard deviation, are shown. (A) All tests were performed in duplicate. The dashed line represents the threshold for a positive response as defined by meeting the criteria of >3x background and >50 sfu per million PBMCs after background subtraction. (B) All tests were performed in quadruplicate. (C and D) IFN-γ ELISPOT was performed on a HERV-K(HML-2)-Env–specific CD8+ T cell clone from OM9. (C) The specificity of the HERV-K(HML-2)-Env–specific clone was confirmed using the original CIDSTFNWQHRILLV peptide (crude) and a newly synthesized batch of the same peptide (>98% pure) and fine mapped using a panel of truncated peptides. IFN-γ ELISPOT data depicting mean spots per 106 clone cells (tested in duplicate), with error bars representing standard deviation, are shown. (D) The reactivity of the HERV-K(HML-2)-Env–specific T cell clone to serial dilutions of the indicated peptides was tested in the presence of autologous BLCLs. Tests were performed in triplicate, and error bars represent SEM.
Figure 4. MHC-I restriction of a HERV-K(HML-2)-Env–specific T cell clone.
The MHC-I restriction of the HERV-K(HML-2)-Env–specific T cell clone was determined by testing its responsiveness to a panel of peptide-pulsed BLCLs. Shown are flow cytometry data depicting an example of a lack of recognition of a peptide-pulsed BLCL that did not share any MHC-I alleles with OM9 (top) and recognition of a peptide-pulsed BLCL that was matched on only HLA-A*0301 (bottom). CD107a staining (degranulation) and IFN-γ production were assessed as measures of responsiveness. Numbers represent percentages of cells in each quadrant.
HERV-K(HML-2)-Env–specific T cells recognize HIV-1–infected cells.
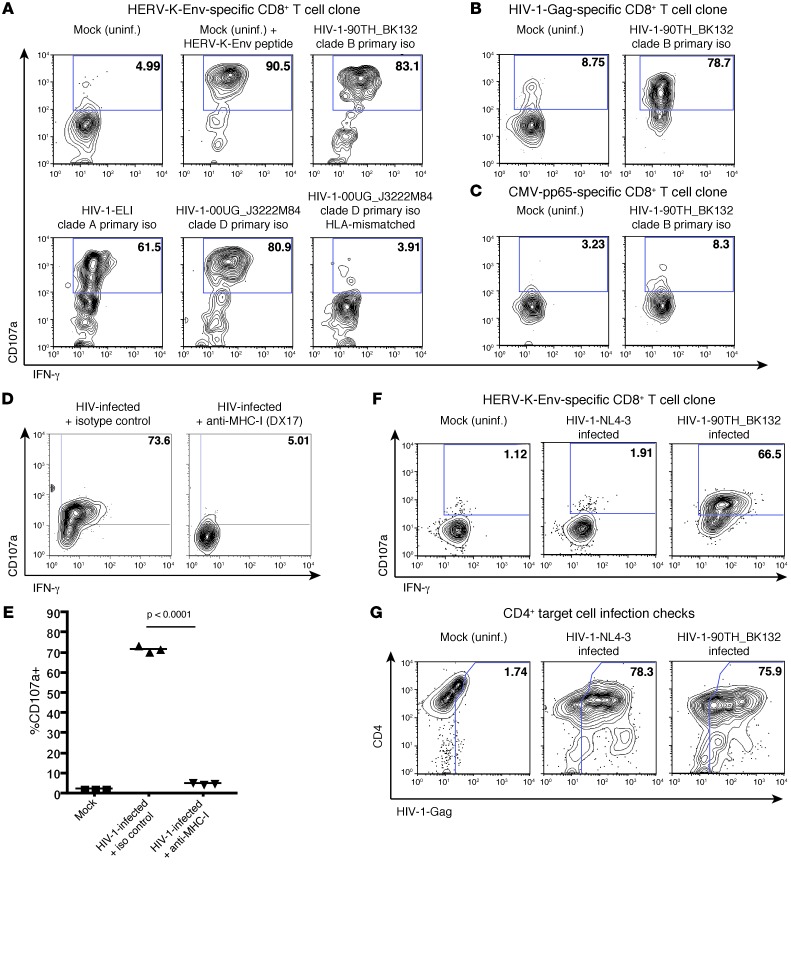
The HERV-K(HML-2)-Env–specific CD8+ T cell clone was cocultured with autologous CD4+ T cells that had either been infected with 1 out of 3 diverse isolates of HIV-1 or maintained as an uninfected (mock) control. We observed induction of IFN-γ production and degranulation (CD107a) in clone cells that had been cocultured with cells infected with each of the HIV-1 isolates (Figure 5A). These responses were contrasted by a lack of recognition of uninfected autologous target cells, as well as of HIV-1–infected HLA-mismatched target cells, and were of similar magnitude to the response observed to uninfected cells pulsed with the cognate HERV-K(HML-2)-Env peptide (Figure 5A). In parallel, we observed recognition of HIV-1–infected targets by the HIV-1-Gag–specific clone (Figure 5B) and a lack of recognition with the CMV-pp65–specific clone (Figure 5C). The recognition of HIV-1–infected cells by the HERV-K(HML-2)-Env–specific CD8+ clone was blocked by pretreatment with anti–MHC-I antibody (Figure 5, D and E). In agreement with our qPCR and immunohistochemistry data, we observed little recognition of cells infected with the laboratory-adapted isolate by the HERV-K(HML-2)–specific clone despite high levels of infection (Figure 5, F and G). Thus, the induction of HERV-K(HML-2) antigen expression by primary isolates of HIV-1 results in the activation of HERV-K(HML-2)-Env–specific T cells.
Figure 5. HERV-K(HML-2)-Env–specific CD8+ T cells respond to cells infected with diverse primary isolates of HIV-1.
(A–C) Flow cytometry data, gated on CD8+ clone cells, depicting CD107a staining (degranulation) (y axis) by IFN-γ production (x axis) for (A) HERV-K(HML-2)-Env–specific, (B) HIV-1-Gag–specific, or (C) CMV-pp65–specific CD8+ T cells cocultured with autologous CD4+ T cells that had either been infected with the indicated primary isolates (iso) of HIV-1 or maintained as uninfected (mock) controls. In the top middle panel of A, uninfected cells pulsed with the HERV-K-Env–derived CIDSTFNWQHRI peptide are shown as a confirmation of clone specificity. A lack of recognition of HIV-1-00UG_J3222M84–infected allogeneic CD4+ T cells (MHC-I mismatched) by the HERV-K(HML-2)–specific T cell clone (bottom right panel) is also shown in A. (D and E) CD4+ T cells from subject OM9 were either infected with the HIV-1 primary isolate 90TH_BK132 or maintained as a mock infection control. These target cells were incubated with 10 μg/ml of either the anti–MHC-I antibody DX17 or an isotype control and then cocultured with the HERV-K(HML-2)-Env–specific T cell clone. (D) Representative flow cytometry plots depicting CD107a staining (degranulation) by IFN-γ are shown. (E) Summary data from 3 independent replicates are shown. Symbols represent individual replicates, and horizontal bars indicate the means. (F) Data shown are analogous to those presented in A and compare responsiveness of the HERV-K(HML-2)-Env–specific clone to that of cells infected with the laboratory-adapted isolate of HIV-1-NL4-3 with responsiveness to the primary isolate 90TH_BK132. (G) Flow cytometry plots for CD4+ targets corresponding to F, representing the level of HIV-1 infection (HIV-1-Gag [x axis], CD4 [y axis]). Numbers represent percentages of (A–D and F) CD107a+ and (G) HIV-1-Gag+ cells.
The HERV-K(HML-2)-Env–specific CD8+ T cell clone has been screened for responsiveness to autologous CD4+ T cells pulsed with HIV-1 clade B consensus Gag, Pol, Env, Vif, Vpr, Rev, and Vpu master pools and matrix mapping pools (obtained from NIH AIDS Research and Reference Reagent Program peptide sets) and showed no responsiveness (data not shown). The NCBI database was queried for HIV-1 sequences with similarity to the HERV-K(HML-2) determinant CIDSTFNWQHRILLV using the protein BLAST search algorithm. The only HIV-1 peptide identified with an appreciable degree of similarity to the HERV-K(HML-2)-Env peptide was the HIV-1-Env–derived peptide CNDSTFNGTGPCSNV (shares 7 out of 15 amino acids). We synthesized this peptide and determined that the HERV-K(HML-2)-Env–specific T cell clone was unresponsive to it (data not shown). We further searched the NCBI database for HIV sequences from alternative reading frames with similarity to the HERV-K(HML-2)-Env–derived peptide by performing a TBLASTN query (searches translated nucleotide databases with a protein query) and found none of significance (expect threshold = 100).
Induction of HERV-K(HML-2)-Env antigen expression requires HIV-1 integration and is dependent upon Vif.
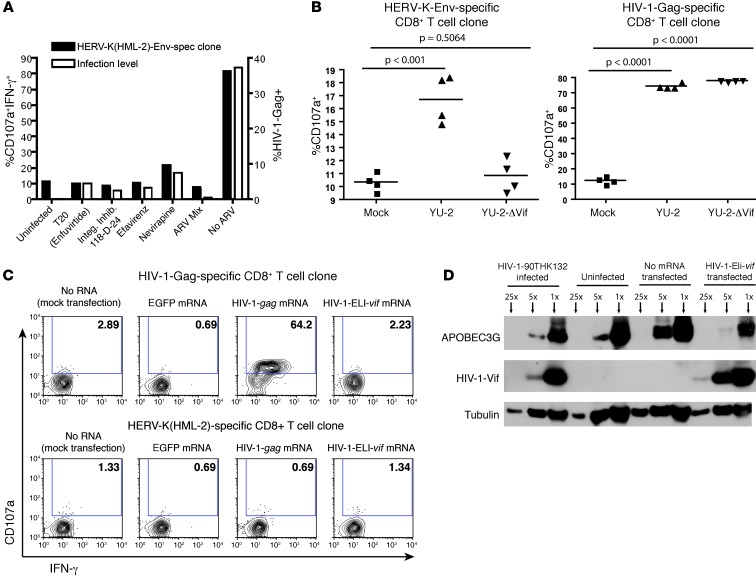
To begin to elucidate the mechanism of HERV-K(HML-2) induction, we tested the effect of blocking HIV-1 entry, reverse transcription, or integration with antiretroviral drugs on recognition by the HERV-K(HML-2)-Env–specific T cell clone. T cell activity was assessed 16 hours after a high-titer infection to ensure that any observed effect could be attributed to blocking the specific replication phase of the inoculum virus. Each of the drugs tested effectively abolished recognition of target cells (Figure 6A). Thus, completion of the infection cycle and integration of the HIV-1 provirus is necessary to induce HERV-K(HML-2) expression. We then tested the ability of the HERV-K(HML-2)-Env–specific T cell clone to respond to CD4+ T cells infected with WT and Vif-deleted (ΔVif) variants of the primary isolate clone YU-2-ΔVif. We magnetofected target cells with high levels of YU-2 and YU-2-ΔVif and tested recognition 16 hours after infection to ensure that only a single round of infection had occurred. At this time point, we observed high-level infections with both viruses, with greater than 40% of cells staining Gag+ (data not shown). The HERV-K(HML-2)-Env–specific T cell clone recognized WT virus but failed to recognize cells infected with YU-2-ΔVif (Figure 6B). In parallel, an HIV-1-Gag–specific CD8+ T cell clone responded to cells infected with either YU-2 or YU-2-ΔVif (Figure 6B). Finally, we tested the ability of the HERV-K(HML-2)-Env–specific T cell clone to respond to CD4+ T cells that had been transfected with codon-optimized vif mRNA. Despite Vif expression levels and corresponding APOBEC3G knockdowns, which were equivalent or greater than those observed in HIV-1–infected cells, we observed a lack of recognition (Figure 6, C and D). Thus, the expression of HIV-1-Vif is necessary, but not sufficient, for the induction of HERV-K(HML-2) expression. The mechanism by which Vif contributes to the induction of HERV-K(HML-2) protein expression is presently unknown.
Figure 6. Induction of HERV-K(HML-2)-Env antigen expression requires entry, reverse transcription, and integration of HIV-1 and is Vif dependent.
(A) Primary CD4+ T cells from subject OM9 were treated with 1 μM each of efavirenz, nevirapine, integrase inhibitor 118-D-24, with 10 μg/ml of T20, or with a combination of each of these drugs (ARV Mix) throughout a 16-hour exposure to HIV-1 90TH_BK132 and then cocultured with the HERV-K(HML-2)-Env–specific T cell clone. Flow cytometry data comparing the frequencies of CD107a-IFN-γ dual-positive clone cell populations in response to these target cells are shown. Levels of HIV-1 infection were assessed by intracellular staining for Gag. (B) Activated CD4+ T cells from subject OM9 were infected with HIV-1 YU-2 or YU-2ΔVif for 16 hours or were maintained as mock infection controls and then cocultured with the indicated T cell clones. Summary data from 4 independent replicates are shown. Symbols represent individual replicates, and horizontal bars indicate the means. P values were calculated using Student’s t test. (C and D) Activated CD4+ T cells from subject OM9 were transfected with mRNA encoding EGFP, HIV-1-gag, and HIV-1-vif (from primary isolate ELI) or mock transfected and then cocultured with the indicated T cell clones. (C) Shown are flow cytometry data from a representative experiment, gated on CD8+ clone cells. Numbers represent percentages of CD107a+ cells. (D) Western blot depicting levels of APOBEC3G and HIV-1-Vif expression compared with a tubulin loading control in CD4+ T cells that had been infected with HIV-1-90TH_BK132, maintained as an uninfected control, mock electroporated (no RNA), or electroporated with vif mRNA. Samples were run at 1:1 (×1), 1:5 (×5), and 1:25 (×25) dilutions.
Elimination of cells infected by diverse isolates of HIV-1, HIV-2, and SIV by HERV-K(HML-2)-Env–specific T cells.
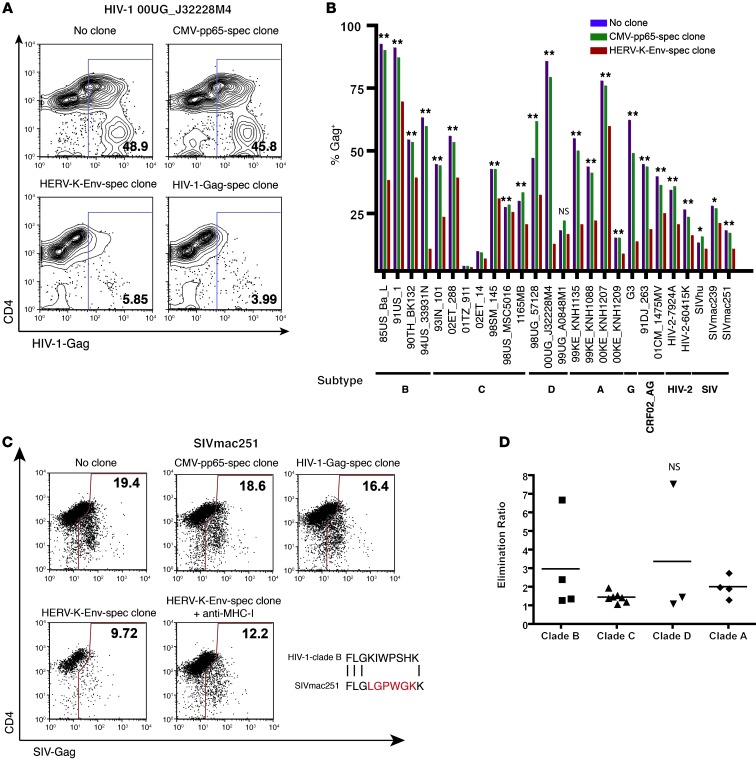
We tested the ability of the HERV-K(HML-2)-Env–specific clone to eliminate HIV-1–infected cells. Primary CD4+ T cells were infected with the clade D primary isolate of HIV-1 00UG_J32228M4. When the frequency of infected cells reached greater than 20% Gag+ by flow cytometry these targets were split and cultured for 24 hours with either the CMV-pp65–, HERV-K(HML-2)–, or HIV-1-Gag–specific clones at a ratio of 1 clone to 10 targets or maintained as no-clone controls. Frequencies of infected targets increased over this period in the no-clone and CMV-pp65–specific clone conditions to greater than 45% Gag+. In contrast, we observed dramatic reductions in frequencies of infected targets upon coculture with either the HERV-K(HML-2)-Env–specific (5.85% Gag+) or the HIV-1-Gag–specific (3.99% Gag+) clones (Figure 7A). This reduction in the baseline frequency of infected cells, observed over a 24-hour period, is consistent with the direct elimination of infected targets by HERV-K(HML-2)-Env–specific T cells. Thus, HERV-K(HML-2)–specific CD8+ T cells eliminate HIV-1–infected cells, resulting in the suppression of viral replication. The preferential elimination of infected cells, compared with that of bystander cells, supports the hypothesis that the HERV-K(HML-2)-Env expression induced in HIV-1–infected cell cultures is restricted to infected cells.
Figure 7. HERV-K(HML-2)-Env–specific CD8+ T cells eliminate cells infected with diverse isolates of HIV-1 and suppress viral replication.
(A and B) Primary CD4+ T cells from subject OM9 were infected with a panel of HIV-1, HIV-2, or SIV isolates for 24 to 72 hours. The indicated T cell clones were then added at an effector/target ratio of 1:10 and cocultured for 24 hours. Levels of infection were measured by flow cytometry. (A) Representative data depicting elimination of cells infected with the clade D primary isolate 00UG_J32228M4. Numbers represent percentages of HIV-1-Gag+ cells. (B) Summary data for the panel of isolates. Statistical significance was calculated by probability binning using Flowjo, comparing target cells cocultured with the HERV-K(HML-2)-Env–specific clone with those cultured in the absence of clone. *P < 0.05 [T(X)>3.0], **P < 0.01 [T(X)>4.0]. (C) Representative data from a SIVmac251 elimination assay. The experimental setup and data presentation are analogous to A but include some additional controls. An HIV-1-Gag–specific CD8– T cell clone is shown to fail to eliminate cells infected with SIVmac251. The subtype B consensus sequence of the T cell determinant for this HIV-1-Gag–specific T clone is given below and is aligned with the corresponding sequence from SIVmac251-Gag. Amino acid mismatches are highlighted in red. The bottom left panel demonstrates the effect of blocking antigen presentation using 10 μg/ml anti–MHC-I antibody on elimination. Numbers represent percentages of HIV-1-Gag+ cells. (D) Using the data presented in B, we calculated an elimination ratio as follows: = percentage infected with no clone/percentage infected with HERV-K(HML-2)-Env–specific clone. Symbols represent individual replicates, and horizontal bars indicate the means. There was no statistically significant difference between any of the 2 groups by Student’s t test.
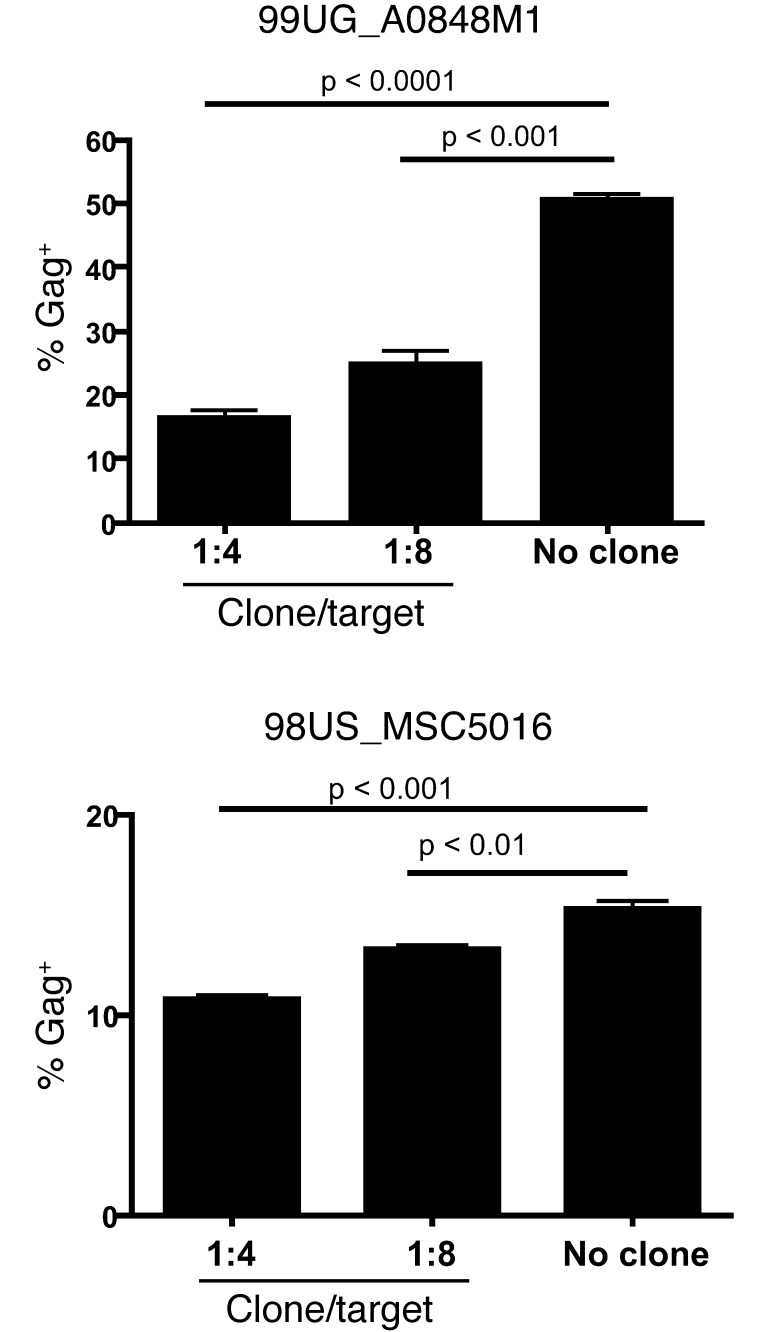
Since the HERV-K(HML-2)-Env–specific clone recognizes a stable host-genome-encoded antigen, we reasoned that it should eliminate HIV-1–infected cells irrespective of HIV-1 sequence variability. To test this, we obtained a panel of 21 diverse HIV-1 primary isolates representing clades A–D and G and circulating recombinant forms (Supplemental Table 4 and ref. 28). CD4+ T cells from subject OM9 were infected individually with viruses from this HIV-1 panel as well as with HIV-2 and SIVmac isolates. Target cells were then either cocultured with HERV-K(HML-2)-Env– or CMV-pp65–specific CD8+ T cell clones at a ratio of 1 clone to 10 targets or maintained as no clone controls. The HERV-K(HML-2)-Env–specific clone exhibited elimination of cells infected with diverse isolates, HIV-1, HIV-2, and SIV viruses, tested, with statistically significant reductions in cells infected with each isolate, except for 99UG_A0848M1 over these 24-hour cocultures (Figure 7B). In contrast, an HIV-1-Gag–specific clone, tested in parallel, failed to eliminate cells infected with HIV-2 and SIV viruses (Figure 7C and data not shown). This corresponds with mismatches between the HIV-1-Gag T cell determinant recognized by the clone and the corresponding HIV-2 and SIVmac239 sequences (SIVmac239 is a cloned virus derived from the SIVmac251 swarm) (Figure 7C). Additional data demonstrating the elimination of SIV-infected target cells are presented in Supplemental Figures 1 and 2. Thus, consistent with the proposed mode of action, the HERV-K(HML-2)-Env–specific T cell clone eliminated cells infected with diverse primate lentiviruses. We did, however, observe some heterogeneity in the ability of the HERV-K(HML-2)-Env–specific clone to eliminate cells infected with different isolates of HIV-1. On one end of the spectrum, cells infected with the clade B virus 94US_33931N or the clade D virus 00UG_J32228M4 were eliminated very effectively, while, on the other end, we observed only a small degree of elimination of cells infected with the clade C virus 98US_MSC5016. We calculated an elimination ratio [percentage infected with no clone/percentage infected with HERV-K(HML-2)-Env–specific clone] and grouped viruses by clade (Figure 7D) but did not observe any significant differences in the extent of elimination of cells infected with viruses from any 2 clades. We did not observe a statistically significant reduction in cells infected with HIV-1 99UG_A0848M1 in our initial experiment. We posited that this was an experimental artifact, rather than a true inability of the HERV-K(HML-2)-Env–specific clone to kill cells infected with this isolate. In our initial experiment, the infection with 99UG_A0848M1 was particularly potent, such that the majority of cells were immediately infected by our synchronized magnetofection protocol. Thus, at the initiation of coculture with clone, these cells had already been infected for 24 hours. Over the subsequent 24-hour coculture period, the majority of these infected cells died whether or not they were cocultured with clone, and thus we were unable to distinguish a significant effect of clone killing. When we repeated this experiment, tailoring the kinetics of the assay to this viral stock, we observed clear, dose-dependent, elimination of 99UG_A0848M1-infected cells by the HERV-K(HML-2)-Env–specific clone (Figure 8). In parallel, we repeated the elimination assay with 98US_MSC5016 and observed that the relatively modest, though statistically significant, elimination displayed in Figure 7B was reproducible (Figure 8). Thus, while there is some degree of heterogeneity in the extent to which different isolates were targeted by the HERV-K(HML-2)-Env–specific T cells, we observed a significant reduction in infected cells with all isolates tested, even at the effector/target ratio of 1:10 over a relatively short period of coculture.
Figure 8. Confirmation of elimination of select viral isolates by HERV-K(HML-2)-Env–specific T cells.
Shown are data from an additional elimination assay, testing the ability of the HERV-K(HML-2)-Env–specific T cell clone to eliminate cells infected with 99UG_A0848M1 and 98US_MSC5016. Each virus was tested in triplicate, and error bars represent standard deviation. P values were calculated by Student’s t test.
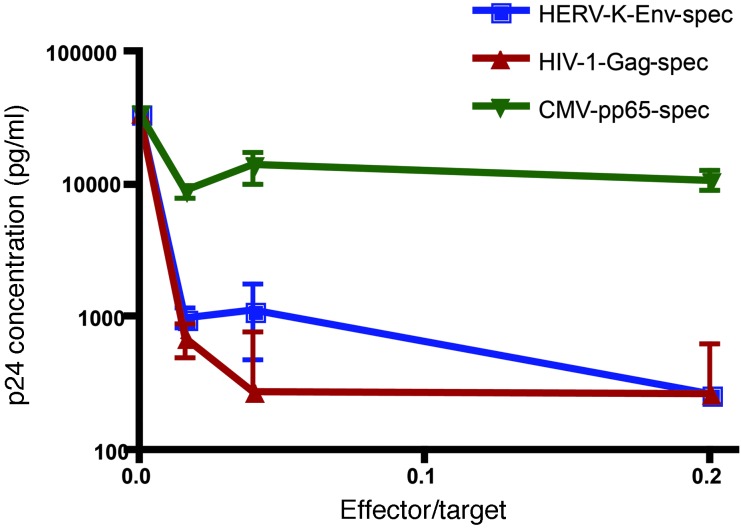
HERV-K(HML-2)-Env–specific T cells recognize low levels of HIV-1 infection and eliminate infected cells rapidly enough to suppress viral outgrowth.
The elimination assays described above test the abilities of T cell clones to eliminate cells infected with fairly high levels of virus. While this represents a stringent test of CTL function, we were also interested in determining whether HERV-K(HML-2)-Env–specific T cells could target low levels of HIV-1 infection rapidly enough to suppress viral outgrowth. In order to test this, HERV-K(HML-2)-Env–, CMV-pp65–, and HIV-1-Gag–specific CD8+ T cells were cocultured with autologous CD4+ T cells that had been infected with a low MOI of 0.01 of HIV-1-91_US4 (Figure 9). Nine days after infection, we observed potent suppression of viral replication (p24 production) by both the HIV-1-Gag– and the HERV-K(HML-2)-Env–specific clones and a lack of suppression by the CMV-pp65–specific clone. Thus, although high levels of HIV-1 infection were required to detect HERV-K(HML-2) expression by Western blot, HERV-K(HML-2)-Env–specific T cells are able to effectively recognize an infection seeded at low levels and to inhibit viral outgrowth.
Figure 9. HERV-K(HML-2)-Env–specific T cells suppress HIV-1 replication.
Primary CD4+ T cells from subject OM9 were activated and then infected with the HIV-1 primary isolate 91_US4. These target cells were cocultured with the HERV-K(HML-2)-Env, HIV-1-Gag, or CMV-pp65 clones at the indicated effector/target ratios for 9 days. Concentrations of p24 in supernatants were determined by ELISA. Shown are results from a representative experiment depicting mean p24 concentration calculated from 5 replicates, with error bars representing SEM.
An additional line of evidence supporting that HERV-K(HML-2)-Env–specific T cells can recognize low levels of HIV-1 infection comes from the antiretroviral suppression assay depicted in Figure 6. While nevirapine substantially reduced levels of HIV-1 infection in this assay, some HIV-1-Gag expression was observed. The low level of infection observed in these nevirapine-treated cells was associated with detectable recognition by the HERV-K(HML-2)–specific T cell clone. Notably, these recognition assays were performed just 16 hours after infection, demonstrating that the induction of HERV-K(HML-2) antigen expression does not require protracted time lines. A second repeat of this experiment, showing the low levels of intracellular Gag expression detected in nevirapine-treated cells, is presented in the Supplemental Results and Supplemental Figure 1.
Cross-reactive HIV-1/HERV-K(HML-2)–specific T cells exist and recognize cells infected with a broader range of lentiviruses than that predicted by epitope sequence.
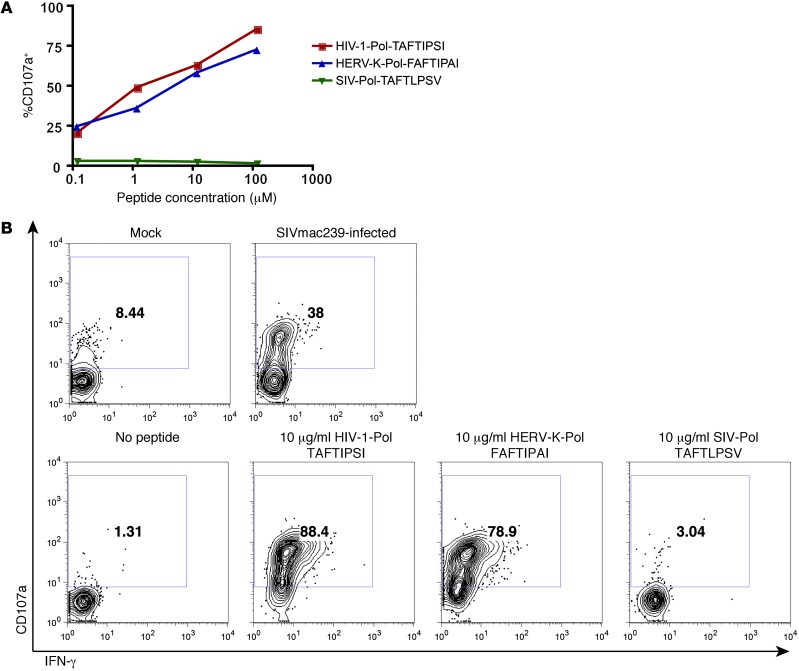
HERV-K(HML-2)-Env– and Gag–specific T cell responses detectable by 15-mer peptides are rare (29). Hence, to generate additional HERV-K(HML-2)–specific T cell clones we screened subjects using optimal HERV-K(HML-2) determinants identified in our previous studies (25–27). We generated a CD8+ T cell clone specific for the HERV-K(HML-2)-Pol epitope FAFTIPAI from an individual with early (<1 year) HIV-1 infection and determined this response to be HLA-B*51 restricted. We had initially selected this HERV-K(HML-2) epitope based on its similarly to the known HLA-B*51–restricted HIV-1-Pol epitope, TAFTIPSI. Thus, in characterizing this clone, we tested it for cross-reactivity between the HERV-K(HML-2)-Pol and HIV-1-Pol epitope sequences. We observed that the clone was cross-reactive and that it exhibited very similar functional avidities for these 2 determinants (Figure 10A). In contrast, it did not recognize the SIVmac239-derived sequence, TAFTLPSV. The direct recognition of an HIV-1–derived determinant posed a challenge to testing whether this clone could recognize HERV-K(HML-2)-Pol expression induced in HIV-1–infected cells. Represented within our diverse clade of HIV-1 isolates were the variant sequences TAFTIPST and TAFTIPSL; however, this T cell clone exhibited direct recognition of cells pulsed with these peptides as well (data not shown). This clone, however, failed to recognize cells pulsed with even up to 100 μg/ml of peptide corresponding to the homologous SIVmac239 sequence TAFTLPSV. We therefore tested the clone for recognition of HLA-B*51+CD4+ T cells that had either been infected with SIVmac239 or maintained as mock-infected controls and observed robust recognition of SIVmac239-infected cells (Figure 10B). Simultaneously, we reconfirmed the responsiveness of the clone to autologous BLCLs pulsed with HIV-1-Pol TAFTIPSI and HERV-K(HML-2)-Pol FAFTIPAI and the lack of a response to SIVmac239-Pol-TAFTLPSV (Figure 10B). We interpret these data as indicating that this T cell clone recognizes HERV-K(HML-2)-Pol expressed upon lentiviral infection of primary CD4+ T cells. By virtue of this indirect recognition, this HERV-K(HML-2)-Pol/HIV-1-Pol cross-reactive CD8+ T cell clone recognizes cells infected with a broader range of viruses than would have been anticipated by testing direct recognition of variant HIV-1/SIV peptides.
Figure 10. Recognition of SIVmac239-infected cells by a HERV-K(HML-2)-Pol/HIV-1-Pol cross-reactive T cell clone.
A HERV-K(HML-2)-Pol–specific CD8+ T cell clone was isolated from an individual with early (<1 year) HIV-1 infection. (A) This clone was cultured with an autologous BLCL pulsed with the indicated concentrations of the HIV-1-Pol peptide TAFTIPSI, the HERV-K(HML-2)-Pol peptide FAFTIPAI, or the SIVmac239-Pol peptide TAFTLPSV in the presence of PE-conjugated anti-CD107a antibody and brefeldin A. Shown are flow cytometry data depicting the percentage of CD107a+ (degranulating) cells, gated on viable CD8+ clone cells. (B) The HERV-K(HML-2)-Pol–specific CD8+ T cell clone was cultured with HLA-B*51+CD4+ T cells that had been infected with SIVmac239 or maintained as mock-infected controls (top row) or infected with autologous BLCLs pulsed with the indicated peptides (bottom row). Shown are flow cytometry data, gated on CD8+ clone cells and displaying IFN-γ (x axis) by CD107a (y axis). Numbers represent percentages of CD107a+ cells.
Recognition of HIV-1–infected cells by additional HERV-K(HML-2)-Gag– and -Env–specific cell lines.
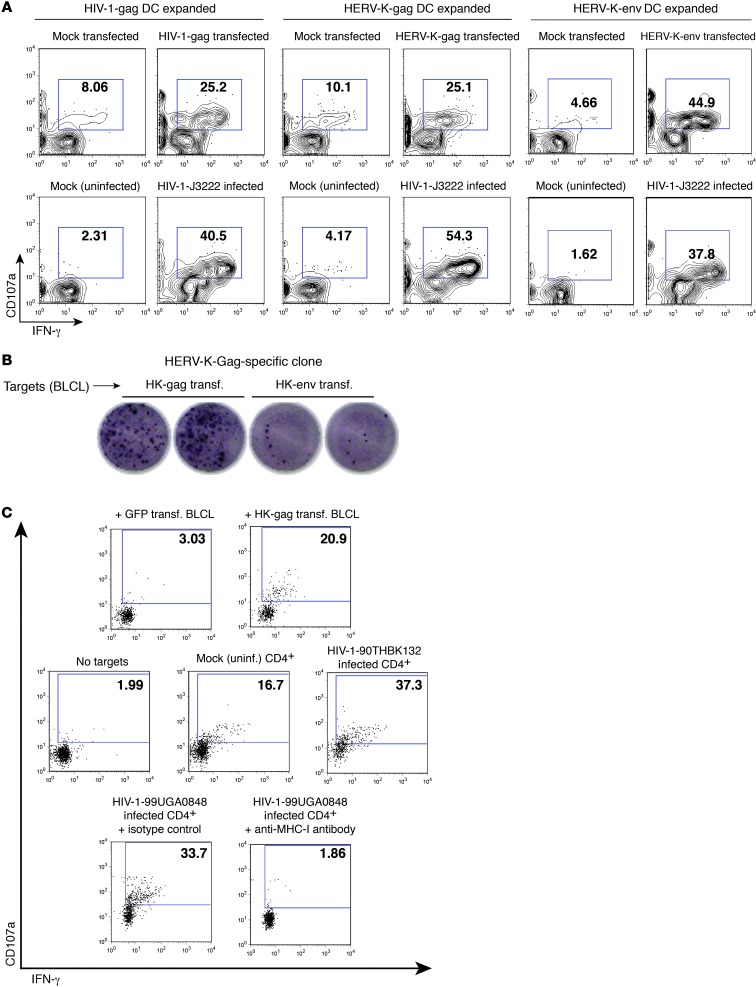
We have drawn from responses detected by an alternative method to corroborate our results. PBMCs were cocultured with autologous mature DCs that had been transfected with HERV-K(HML-2) antigen-encoding mRNA. Using this approach, we have detected HERV-K(HML-2)-Gag and -Env–specific CD8+ T cell responses in a subset of HIV-1–infected subjects but not in HIV-1–uninfected controls (unpublished data). In this study, we confirmed the specificities of HIV-1-Gag–, HERV-K(HML-2)-Gag–, and HERV-K(HML-2)-Env–expanded CD8+ T cell lines, obtained from an HIV-1–infected individual, by testing their responsiveness to autologous CD4+ T cells, which had been transfected with mRNA encoding the corresponding antigen, mock transfected (no RNA), infected with HIV-1, or mock infected (Figure 11A). We observed recognition of both antigen-transfected and HIV-1–infected CD4+ T cells by the HIV-1-Gag–, HERV-K-Gag–, and HERV-K-Env–expanded CD8+ T cell lines (Figure 11A). Clones were obtained by limiting dilution and screened for antigen specificity using mRNA transfected autologous BLCLs. We succeeded in identifying a HERV-K(HML-2)-Gag–specific CD8+ T cell clone from subject OM2 (Figure 11B) but failed to isolate additional HERV-K(HML-2)-Env–specific clones. This HERV-K(HML-2)-Gag–specific clone was tested for recognition of HIV-1–infected or mock-infected autologous CD4+ T cells. We observed some low-level outgrowth of autologous HIV-1 in the mock-infected controls (data not shown) and, likely as a result of this, observed some background recognition of mock-infected targets (as compared with no targets) (Figure 11C). Despite this, we were able to observe clear recognition of autologous CD4+ T cells infected with high levels of HIV-1. This recognition was blocked by pretreatment with an anti–MHC-I antibody (Figure 11C). Notably, there is very little homology between HERV-K(HML-2)-Gag and HIV-1-Gag. There are 2 regions of microhomology in the capsids of the 2 proteins with potential epitopes sharing 6 out of 9 and 5 out of 9 amino acids. Cross-reactivity between HERV-K(HML-2)-Gag and HIV-1-Gag is therefore a very unlikely etiology for these observations. Thus, the recognition of HIV-1–infected cells by HERV-K(HML-2)–specific CD8+ T cells has been confirmed for clones from 2 separate subjects targeted against distinct HERV-K(HML-2) antigens.
Figure 11. Recognition of HIV-1–infected cells by additional HERV-K(HML-2)–specific lines and clones.
HIV-1-Gag–, HERV-K-Gag–, and HERV-K-Env–specific CD8+ T cell lines were established by a 9-day coculture of PBMCs with autologous mature DCs that had been transfected with codon-optimized mRNA encoding the corresponding antigen. (A) Expanded cell lines were cocultured with autologous CD4+ T cells that had been transfected with mRNA or maintained as a mock transfection control or had been infected with the primary isolate of HIV-1 J3222 or maintained as a mock-infected control. Shown are flow cytometry data, gated on the CD8+ lymphocyte population. (B) DC-expanded cell lines were plated at limiting dilution on irradiated feeder cells, then screened for antigen specificity by coculture with autologous BLCLs that had been transfected with the indicated mRNAs. Shown are IFN-γ ELISPOT data for 1 clone derived from the HERV-K(HML-2)-Gag expansion, exhibiting recognition of gag transfected BLCLs. (C) The HERV-K(HML-2)-Gag–specific CD8+ T cell clone from B was reexpanded. Shown are flow cytometry data, gated on CD8+ lymphocytes, for HERV-K(HML-2)–specific T cell clones cocultured for 6 hours with autologous BLCLs that had been transfected with GFP (negative control) or HERV-K(HML-2)-Gag encoding mRNA (top panel); no targets or autologous CD4+ T cells that had been infected with HIV-1-90TH_BK132 or mock infected (middle panel); or autologous HIV-1-99UG_A0848M1–infected CD4+ T cells in the presence of either an anti–MHC-I blocking antibody or an isotype control (bottom panel). Panels in C present results from 3 separate experiments performed during the same week, with the same expansion of clones. (A and C) Numbers represent percentages of CD107a+ cells.
Discussion
This study demonstrates the first examples to our knowledge of specific recognition and elimination of HIV-1–infected cells by HERV-K(HML-2)–specific T cells. To our knowledge, these are also the first demonstrations of such activity for any non–HIV-1–specific T cell. This bears important considerations for understanding the T cell response to HIV-1 infection by demonstrating that even a comprehensive evaluation of HIV-1–specific T cells may not capture all T cell responses directed against HIV-1–infected cells. We have previously reported that strong HERV-specific T cell responses (considering peptides from diverse HERV families in a broad sense) are associated with natural control of HIV-1 viremia (27). This study provides a potential mechanism underlying this observation. However, it certainly cannot be assumed based on our data that the expression of antigens derived from HERV families other than HERV-K(HML-2) are induced in HIV-1–infected cells. Thus, our data provide rationale for moving forward with studies that focus in on evaluating any associations between HERV-K(HML-2)–specific T cell responses and control of HIV-1 viremia. Such studies should consider that methods that are commonly used to detect exogenous virus–specific T cell responses, such as ELISPOT assays with pools of 15-mer peptides, may not be optimal for detecting HERV-K(HML-2)–specific T cell responses. Although the HERV-K(HML-2)-Env–specific CD8+ T cell response featured in this study was detected by ELISPOT peptide-matrix mapping, subsequent titration experiments demonstrated that peptide concentrations were barely above the threshold required to elicit a response. In the case of the HERV-K(HML-2)-Gag–specific T cell clone isolated from subject OM2, although the clone specifically responds to autologous BLCLs and CD4+ cells transfected with HERV-K(HML-2)-Gag mRNA, we have been unable to map a T cell determinant using 15-mer peptides. The etiology of these unusual features is unclear but may be related to clonal deletion of higher-avidity HERV-K(HML-2)–specific T cells. In this regard, it is interesting to note that when we have vaccinated mice against murine L1 retrotransposable element antigens, we have observed that robust T cell responses were detectable upon screening with overlapping 9-mer peptides, while, in parallel, a complete lack of responses was observed with overlapping 15-mer peptides (30). Comprehensive evaluation of HERV-K(HML-2)–specific T cell responses may therefore require the application of more sensitive methods, such as DC-mediated expansions.
Our identification of a cross-reactive HERV-K(HML-2)/HIV-1-Pol–specific CD8+ T cell response raises the intriguing possibility that the endogenous retroviral antigens within our genome may play a role in shaping the immune response to HIV-1. A recent study has provided evidence that, in mice, an endogenous retroviral antigen with homology to Friend murine leukemia virus (F-MLV) drives negative selection of the naive CD4+ T cell repertoire available to respond to a specific F-MLV-Env determinant (31). The result of this is the deletion of low-avidity cross-reactive T cells and the promotion of a higher avidity to CD4+ T cell responses to retroviral infection. Future studies will aim to determine whether the HERV-K(HML-2)/HIV-1 cross-reactivity observed here is an isolated case or whether it is shared by multiple subjects and across additional pairs of T cell determinants. While one might anticipate that cross-reactivity between HIV-1 and HERV-K(HML-2) may lead to impairment of HIV-1–specific T cell responses through tolerization, the F-MLV scenario highlighted above illustrates how this may also serve to enhance HIV-1–specific responses. At present, what can be said conclusively is that any tolerization to the HERV-K(HML-2)-Pol–reactive T cell clones was incomplete and did not preclude the elicitation of a cross-reactive HIV-1-Pol–specific T cell response in this subject. Whether or not there are any autoimmune implications for these HERV-K(HML-2)-Pol–specific T cell clones depends upon whether HERV-K(HML-2)-Pol protein is expressed in healthy tissues, which has not been demonstrated. The identification of a HERV-K(HML-2) cross-reactive T cell response also presents another dimension that may be important in understanding protective HIV-1–specific T cell responses, whereby the effectiveness of HIV-1–specific CTL responses may be augmented by their reactivity to the HERV-K(HML-2) antigens that also mark infected cells. In the case of the specific clone identified in this study, a lack of appreciation of the HERV-K(HML-2) reactivity would likely have led to an underestimation of the breadth of reactivity of this clone, which effectively responded to SIVmac239-infected CD4+ T cells, despite failing to directly recognize the SIVmac239-encoded T cell determinant. It is intriguing to note that HIV-1-Pol-TAFTIPSI is 1 out of 3 dominant responses restricted by the “protective” HLA-B51 allele, and further study is needed in order determine whether HERV-K(HML-2) cross-reactivity may play any role in this protection.
The unprecedented breadth of elimination achieved by HERV-K(HML-2)–specific T cell responses against diverse primary isolates of HIV-1, both implied by the proposed mode of action and observed in this study, comprises an enticing advantage over HIV-1–specific T cell responses, which could be exploited in the development of HERV-K(HML-2)–targeted vaccines. As is the case for any CD8+ T cell–based vaccine, it might be unlikely that this strategy would afford sterilizing protection from infection. However, a prophylactically given vaccine that elicits mucosal HERV-K(HML-2)–specific CD8+ T cell responses may serve to prevent viral dissemination and thus achieve long-term control of viremia, as has been observed in rhesus macaques with robust vaccine-elicited SIV-specific CD8+ T cell responses (32). The elicitation of HERV-K(HML-2)–specific CD8+ T cell responses by a therapeutic vaccine may also be of value in the setting of chronic infection. In moving toward developing this vaccine strategy, it is important to consider whether immune tolerance to HERV-K(HML-2) antigens is likely to exist and, if so, whether breaking such tolerance may carry the risk of inducing autoimmunity. While HERV-K(HML-2) RNA appears to be readily detected in all tissues, including the thymus (33), we are of the position that the expression of HERV-K(HML-2)-Gag and -Env protein has not been convincingly demonstrated in any healthy adult human tissue, despite extensive screening (30). The fact that we are not able to more readily detect T cell responses to these antigens in HIV-1–infected individuals using 15-mers may be indicative of tolerance (29). Alternatively, however, since our data indicate very low levels of HERV-K(HML-2) antigen expression as compared with HIV-1 antigen expression, and since the expression of the two would be tightly associated, it is possible that T cells specific for the former are simply not able to effectively compete for priming by DCs. This scenario would provide a clear opportunity for intervention, as vaccination with HERV-K(HML-2) antigens alone would have the potential to elicit effective anti–HIV-1 T cell responses, which were not elicited by natural infection. In moving forward with HERV-K(HML-2)–targeted vaccine strategies, we envision that, after extensive additional nonhuman primate experiments, initial testing in humans would occur in the context of HIV-1–infected subjects to determine whether therapeutic vaccination could elicit T cell responses capable of controlling HIV-1 replication in the absence of antiretroviral drugs. If a favorable safety profile is established in this setting, consideration could then be made for applying the strategy in prophylactic vaccines to prevent dissemination of initial HIV-1 infection and thus the establishment of chronic viremia.
In additional to these immunological implications, this study, in a more general sense, provides evidence for biologically relevant interactions between HIV-1 and HERV-K(HML-2). A precedent exists in mice for the mobilization of endogenous retroviruses by exogenous retroviruses, where infection with exogenous ectopic MuLV has been shown to result in the replication of defective endogenous polytopic retroviruses (34). This has led to the suggestion that periods of replication of endogenous retroviruses in the mouse have been initiated by exogenous retroviruses capable of circumventing restriction factors. Although we do not provide any evidence to support that HIV-1 infection may drive actual replication of HERV-K(HML-2), we demonstrate that the HIV-1 viral protein Vif is in part responsible for the induction of HERV-K(HML-2) antigen expression in HIV-1–infected cells, although additional factors appear to be required. One potential mode by which Vif could contribute to the derepression of HERV-K(HML-2) expression is through the degradation of APOBEC3 proteins, which are known to restrict the replication of reconstituted HERV-K(HML-2) viruses (35). It is unclear, however, if or how reverse transcription of HERV-K(HML-2), the stage at which APOBEC3 proteins exert their mutagenic effects, would be necessary for the induction of HERV-K(HML-2) protein expression. One speculative possibility is that APOBEC3 proteins may suppress HERV-K(HML-2) expression in a deamination-independent manner, for example, by binding to HERV-K(HML-2) transcripts in the cytoplasm and sequestering them away from the translational machinery, analogous to the reported sequestration of Alu retrotransposon RNA by APOBEC3G (36). Further study is required to determine the precise mechanism by which Vif contributes to the induction of HERV-K(HML-2) antigen expression as well as to determine which additional viral factors may be required to remove, as of yet, unknown barriers to expression. Delineation of the mechanisms through which HIV-1 interacts with HERV-K(HML-2) is likely to shed light on additional restriction factors with activity against both endogenous retroviruses and contemporary exogenous retroviruses.
In this study, we present multiple lines of evidence indicating that infection with the laboratory-adapted isolate of HIV-1-NL4-3 results in a lesser induction of HERV-K(HML-2) expression than diverse primary isolates of HIV-1. We also observed some heterogeneity in the ability of the HERV-K(HML-2)-Env–specific T cell clone to eliminate diverse isolates of HIV-1. We propose that this reflects differential potencies of viruses in some function required for the induction of HERV-K(HML-2) expression. The nature of this function is presently unclear; however, it is intriguing, in light of the demonstration that APOBEC3F is a much more potent inhibitor of a reconstituted HERV-K(HML-2) virus than APOBEC3G (20), to note that NL4-3-Vif, while effective in degrading APOBEC3G, does not effectively degrade other APOBEC3 proteins, including APOBEC3F (37). Critically, while it appears that some primary isolates of HIV-1 may induce HERV-K(HML-2) expression to a lesser degree than others, none were exempt from elimination. It is plausible that, while the functionality required for the induction of HERV-K(HML-2) expression is conserved among primary isolates, it may be diminished by the passaging of virus on cell lines (laboratory adaptation). Further study is clearly required in this area, including the testing of additional laboratory-adapted isolates; however, it is advisable, at present, that studies related to the interaction between HIV-1 and HERV-K(HML-2) should focus on primary isolate HIV-1 viruses that have been cultured on primary CD4+ T cells.
In conclusion, our study of the interface among endogenous retroviruses, exogenous retrovirus, and the immune system has revealed a mechanism by which the human immune system can target HIV-1–infected cells. This suggests a potential means of dealing with the obstinate challenge of HIV-1 sequence diversity. While we have focused on this potential utility for HERV-K(HML-2)–specific T cell responses, we also feel that our study sets a precedent for more generally considering the ancient interaction between the our immune systems and the diverse “self viral antigens” encoded by our genomes.
Methods
Viruses.
All viruses, with the exceptions of YU-2, YU-2ΔVif, and NL4-3, were obtained as viral stocks from the NIH AIDS Research and Reference Reagent Program (Supplemental Table 4) and amplified on activated primary CD4+ T cells from an HIV-1–uninfected donor. A plasmid encoding full-length NL4-3 was also obtained from the NIH AIDS Research and Reference Reagent Program. Plasmids encoding YU-2 and YU-2ΔVif were a gift from Michael Malim (King’s College, London, United Kingdom) and have been previously described (1). These plasmids were transfected into 293T cells using Fugene 6 (Roche) to generate viral stocks. All viruses were purified by centrifugation through a 20% sucrose cushion.
HIV-1, HIV-2, and SIV infections.
CD4+ T cells were enriched by negative selection (Easysep, Stemcell), activated for 24 to 72 hours using 1 μg/ml each of antibodies to CD3 (OKT-3, ebioscience), CD28 (CD28.2, ebioscience), and 1 μg/ml SEB (Sigma-Aldrich) in RPMI with 10% FBS (Gibco) and 50 U/ml IL-2 (Hoffmann-La Roche Inc.). Infections were generally performed by a magnetofection process that has been previously described (2). Recognition and elimination of infected cells was also observed when standard infections were performed. Infections were monitored by flow cytometry after surface staining cells with anti–CD4-APC (BD) and intracellular staining for HIV-Gag (Kc57-PE, Beckman Coulter).
Quantitative PCR.
Primers and probes sequences are available in Supplemental Table 5. RNA was isolated using RNeasy Plus (Qiagen) and treated with DNAse (Turbo-DNA-free, Ambion). Real-time PCRs were performed using the TaqMan One-Step RT-PCR Master Mix Kit with the ABI Prism 7900HT instrument (Applied Biosystems). Standard curves were generated using 10-fold serial dilutions of linearized plasmid standards containing the target sequence (109 – 103 copies). Reaction conditions were as follows: 48°C for 15 minutes, 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds and 58°C for 1 minute).
Western blotting.
Cells were lysed in RIPA buffer plus protease inhibitor cocktail (Roche) at concentrations of 1 × 107 to 5 × 107 cells per ml. Proteins were separated by reducing SDS-PAGE under reducing conditions (NuPage, Invitrogen) and transferred to PVDF membranes. Membranes were blocked with 5% milk in PBS plus 0.2% TWEEN-20 and probed with 0.2 μg/ml of anti–HERV-K-Gag (HERM-1841, Austral Biologicals). Membranes were probed with HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories Inc.) and detected using ECL substrate (GE Healthcare).
Generation of HERV-K(HML-2) antibody 4890-Gly.
HERV-K(HML-2) capsid (consensus residues 282–554) was produced in E. coli and purified by Ni affinity. Rabbits were immunized with six 200 μg doses of capsid in complete or incomplete Freund’s adjuvant. Specific antibody was affinity purified from whole serum using capsid linked to Thiopropyl 6B Sepharose (GE Healthcare) and eluted with TEA or glycine.
Immunoprecipitation.
Immunoprecipitation was performed using the Dynabeads Protein G Kit (Invitrogen) following the manufacturer’s instructions.
Immunohistochemistry.
4-μm sections of paraffin-embedded cell pellet were mounted on Superfrost Plus slides (Fisher). Immunohistochemistry was performed using a Ventana XT autostainer (Roche Diagnostics) with heat pretreatment (mCC1). Anti–HERV-K-Env (Austral Biologicals, HERM-1811-5) was used at a concentration of 2.5 μg/ml. A hematoxylin counterstain was used to contrast the brown positive stain of the immunohistochemistry.
IFN-γ ELISPOT.
ELISPOT assays were performed as previously described (25). HIV peptides were obtained from the NIH AIDS Research and Reference Reagent Program.
T cell cloning.
PBMCs were enriched for antigen-specific cells using the IFN-γ Secretion Detection and Enrichment Kit (Miltenyi Biotec) and cloned via 2 rounds of limiting dilution, as has been previously described (38). T cells were maintained in IL-2, obtained from Maurice Gately (Hoffmann-La Roche Inc.) via the NIH AIDS Research and Reference Reagent Program.
Recognition assays.
T cell clones were cocultured with target cells, anti–CD107a-PE antibody (Biolegend), and 1 μg/ml Brefeldin A (BD) for 6 to 12 hours. Cells were stained with anti–CD8-FITC (Biolegend) and anti–IFN-γ-APC (BD Biosciences).
Antiretroviral suppression recognition assays.
CD4+ cells were treated with 1 μM each of efavirenz, nevirapine, integrase inhibitor 118-D-24, with 10 μg/ml of T20, or with a combination throughout a 16-hour exposure to the HIV-1 primary isolate 90TH_BK132. All antiretroviral drugs were obtained from the NIH AIDS Research and Reference Reagent Program. These target cells were then used in recognition assays with the HERV-K(HML-2)-Env–specific CD8+ T cell clone.
Transfection of primary CD4+ T cells with HIV-1-vif.
Codon-optimized vif from the primary HIV-1 isolate ELI was synthesized by Genscript (genbank accession no. GU945071.1) and cloned into pGEM4Z/GFP/A64, which was provided as a gift by Eli Gilboa (University of Miami, Miami, Florida, USA) (39). Linearized pGEM4Z/ELI-vif/A64 was used as template for in vitro transcription (mMessage mMachine T7 Kit, Ambion). mRNA was treated with DNAse (TURBO DNA-free, Ambion) and isolated using Megaclear (Ambion). Primary CD4+ T cells were transfected with 2 μg mRNA using the Human T cell Nucleofector Kit (Amaxa). Transfection efficiency, assessed by GFP expression, was consistently found to be greater than 80%. Expression of Vif was confirmed by Western blot probing with 1:1,000 Vif antiserum (NIH AIDS Research and Reference Reagent Program, catalog no. 2221).
Elimination assays.
T cell clones were cocultured with infected or uninfected (mock) autologous CD4+ T target cells at a ratio of 1 clone per 10 target cells for 24 hours. Cells were stained with anti–CD8-FITC and anti–CD4-APC (Biolegend), permeabilized, and then stained with anti–HIV-Gag-PE (Kc57-RD1, Beckman Coulter). Cells were fixed in 5% formalin and analyzed on a FACSCalibur instrument. Data analysis was performed using Flowjo (Treestar).
Suppression assays.
CD4+ T target cells were infected with 0.01 MOI of HIV-1 and plated at 50,000 cells per well in a 96-well plate. Clones were added at indicated effector/target ratios (see Results) and cocultured for 9 to 16 days. Supernatants were harvested and assayed for p24 by ELISA (p24 Antigen Detection Kit, NCI Frederick).
DC expansions.
Monocytes were enriched from PBMCs using the Monocyte Isolation Kit II (Miltenyi Biotec) and cultured at 1 × 106 cells per ml in DC media (CellGenix; plus 2 mM l-glutamine plus 800 U/ml GM-CSF [R&D Systems] plus 1,000 U/ml IL-4 [R&D Systems] plus 100 nM nevirapine). Codon-optimized HERV-K(HML-2)-gag and -env were provided by Pfizer Inc. mRNA was produced and transfections were performed as for HIV-1-vif (see above). Protein expression was confirmed by Western blot, and DCs were matured with 10 ng/ml TNF-α plus 10 ng/ml IL-1β (all from R&D Systems) plus 1 μg/ml prostaglandin E2 (Sigma-Aldrich). DCs were combined with autologous PBMCs at a ratio of 1:30 and cocultured for 7 days. Positive responses were identified by proliferation and upregulation of CD25. To confirm antigen specificity of expanded cell lines, CD4+ cells were activated with anti-CD3/CD28 (as above) and transfected with mRNA as with monocytes but using the activated T cell program on an Amaxa 4D nucleofector as per the manufacturer’s specifications. Expanded cell lines were cocultured with these transfected target cells in the presence of anti–CD107a-PE and Brefeldin A for 6 hours.
T cell receptor sequencing.
RNA was isolated from HERV-K(HML-2)-Env–specific clone cells using the RNeasy Kit (Qiagen). cDNA was synthesized using the SMARTer RACE cDNA Amplification Kit (Clonetech). 5′-RACE was performed using a gene-specific primer (GSP) on the 3′ end and the adapter binding sequence on the 5′ end added by the kit. For α chain amplification, a single 3′ GSP was used: α, 5′CAGCAGTGTTTGGCAGCTCT3′. For β chain amplification, a set of 2 GSP 3′ primers was used to target the 2 different possible constant regions: β 1, 5′AGAAGCGCTGGCAAAAGAAG, and β 2, 5′CAGGAGAATCCTGGGTGAGG. Amplification was performed by touchdown PCR, with the following cycling conditions: (a) 5 cycles of 94°C for 30 seconds and 72°C for 3 minutes; (b) 5 cycles of 94°C for 30 seconds, 70°C for 30 seconds, and 72°C for 3 minutes; and (c) 30 cycles of 94°C for 30 seconds, 68°C for 30 seconds, and 72°C for 3 minutes. Nested PCR was performed using a nested GSP on the 5′ end and a nested primer for the adapter sequence (provided by the Clonetech kit). As before, one primer was used for α, α internal, 5′TGTCAGGCAGTGACAAGCAG, while 2 primers were used for β, β 1 internal, 5′TGGGATGCAGAGAGGTGAGA, and β 2 internal, 5′AACCAGGCCCAACACACAAT. A product of the expected size (1 kb) was excised from an agarose gel and cloned using the Invitrogen pCR 4-TOPO Vector TA Cloning Kit. Resulting colonies were sequenced using TOPO vector primers, M13 forward and M13 reverse, using an ABI 3730 instrument. V/J cassettes were determined using the IMGT/V-QUEST bioinformatic tool ( http://www.imgt.org/IMGT_vquest/share/textes/).
Reagents.
The following reagents were obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID: human rIL-2 (Maurice Gately, Hoffmann-La Roche Inc.), pNL4-3, primary isolate HIV-1 viruses (Supplemental Table 4), integrase inhibitor (118-D-24), nevirapine, efavirenz, and peptide sets representing HIV-1 (Vif, Vpr, Vpu, Tat, Rev, Nef, Pol, Gag, and Env).
Statistics.
For the comparisons of recognition of cells infected with HIV-1 YU-2 or YU-2DVif or mock infected in Figure 6B, P values were calculated by Student’s t test. Statistical significance of viral elimination assays presented in Figure 7B was calculated by probability binning using the algorithm included in the Flowjo software package. For the comparison of viral elimination data across different clades presented in Figure 7D, as well as the viral elimination data presented in Figure 8, P values were calculated by 2-tailed Student’s t tests. For each of the statistical tests described above, a P value of less than 0.05 was considered to be significant. With the exception of probability binning, all statistical tests were performed using Prism GraphPad software.
Study approval.
This study was approved by the University of Toronto and UCSF IRBs. Antisera production in rabbits was approved by Covance Research Products’ Denver Site Institutional Animal Care and Use Committee – OLAW assurance no. A3580-01, USDA research registration no. 35-R-0030, and adhered to IACUC guidelines. Informed written consent was received for human samples through an IRB-approved protocol at St. Michael’s Hospital.
Supplementary Material
Acknowledgments
Funding for this work was provided by Pfizer through a sponsored research agreement. This work was also supported by funds from the NIH (AI076059 and AI084113). We thank Tania Watts for useful discussion. We thank Michael Malim for providing the pYU-2 and pYU-2ΔVif plasmids and Eli Gilboa for provision of the pGEM/4Z/A64 plasmid. R.B. Jones and M.A. Ostrowski gratefully acknowledges salary support from the Ontario HIV Treatment Network. Biosafety level 3 laboratory space was provided by the Canadian Foundation for HIV Research in partnership with the Ontario Innovation Trust and the Canadian Foundation for Innovation.
Footnotes
Conflict of interest: Neil C. Sheppard, Lianchun Chen, Peter T. Loudon, Sophie Muscat-King, Ingrid M. Priumboom-Brees, David A. Goodwin, and Melanie Rieger declare a potential conflict of interest as employees of Pfizer. R. Brad Jones declares that he owns stock of Sangamo Biosciences worth $10,000 or more. Ingrid M. Priumboom-Brees declares that she owns stock of Pfizer and Novartis worth $10,000 or more. Peter T. Loudon, Lianchun Chen, and Neil C. Sheppard declare that they each own stocks of Pfizer Inc. worth $10,000 or more. R. Brad Jones, Keith E. Garrison, Douglas F. Nixon, and Mario A. Ostrowski are listed as inventors on a patent application related to this work.
Citation for this article: J Clin Invest. 2012;122(12):4473–4489. doi:10.1172/JCI64560.
References
- 1.Gaschen B, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296(5577):2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 2.Rolland M, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med. 2011;17(3):366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korber BT, Letvin NL, Haynes BF. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J Virol. 2009;83(17):8300–8314. doi: 10.1128/JVI.00114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12(10):1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol. 1999;9(16):861–868. doi: 10.1016/S0960-9822(99)80390-X. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout B, Jebbink M, Zsiros J. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J Virol. 1999;73(3):2365–2375. doi: 10.1128/jvi.73.3.2365-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewannieux M, Blaise S, Heidmann T. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol. 2005;79(24):15573–15577. doi: 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris JM, Haynes RH, McIntosh EM. A consensus sequence for a functional human endogenous retrovirus K (HERV-K) dUTPase. Biochem Cell Biol. 1997;75(2):143–151. doi: 10.1139/o97-027. [DOI] [PubMed] [Google Scholar]
- 11.Kitamura Y, Ayukawa T, Ishikawa T, Kanda T, Yoshiike K. Human endogenous retrovirus K10 encodes a functional integrase. J Virol. 1996;70(5):3302–3306. doi: 10.1128/jvi.70.5.3302-3306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magin C, Lower R, Lower J. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J Virol. 1999;73(11):9496–9507. doi: 10.1128/jvi.73.11.9496-9507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21(3):257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 14.Mueller-Lantzsch N, et al. Human endogenous retroviral element K10 (HERV-K10) encodes a full-length gag homologous 73-kDa protein and a functional protease. AIDS Res Hum Retroviruses. 1993;9(4):343–350. doi: 10.1089/aid.1993.9.343. [DOI] [PubMed] [Google Scholar]
- 15.Reus K, Mayer J, Sauter M, Scherer D, Muller-Lantzsch N, Meese E. Genomic organization of the human endogenous retrovirus HERV-K(HML-2.HOM) (ERVK6) on chromosome 7. Genomics. 2001;72(3):314–320. doi: 10.1006/geno.2000.6488. [DOI] [PubMed] [Google Scholar]
- 16.Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74(8):3715–3730. doi: 10.1128/JVI.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11(19):1531–1535. doi: 10.1016/S0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Bogerd HP, Peng S, Wiegand H, Truant R, Cullen BR. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc Natl Acad Sci U S A. 1999;96(23):13404–13408. doi: 10.1073/pnas.96.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dewannieux M, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16(12):1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3(1):e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boller K, et al. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196(1):349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 22.Herbst H, Sauter M, Mueller-Lantzsch N. Expression of human endogenous retrovirus K elements in germ cell and trophoblastic tumors. Am J Pathol. 1996;149(5):1727–1735. [PMC free article] [PubMed] [Google Scholar]
- 23.Muster T, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63(24):8735–8741. [PubMed] [Google Scholar]
- 24.Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. Endogenous retroviruses and cancer. Cell Mol Life Sci. 2008;65(21):3366–3382. doi: 10.1007/s00018-008-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrison KE, et al. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 2007;3(11):e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tandon R, et al. Identification of human endogenous retrovirus-specific T cell responses in vertically HIV-1-infected subjects. J Virol. 2011;85(21):11526–11531. doi: 10.1128/JVI.05418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sengupta D, et al. Strong human endogenous retrovirus (HERV)-specific T cell responses are associated with control of HIV-1 in chronic infection. J Virol. 2011;85(14):6977–6985. doi: 10.1128/JVI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown BK, et al. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J Virol. 2005;79(10):6089–6101. doi: 10.1128/JVI.79.10.6089-6101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones RB, et al. Human endogenous retrovirus K(HML-2) Gag- and Env-specific T-cell responses are infrequently detected in HIV-1-infected subjects using standard peptide matrix-based screening. Clin Vaccine Immunol. 2012;19(2):288–292. doi: 10.1128/CVI.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacha JB, et al. Vaccination with cancer- and HIV infection-associated endogenous retrotransposable elements is safe and immunogenic. J Immunol. 2012;189(3):1467–1479. doi: 10.4049/jimmunol.1200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young GR, Ploquin MJ, Eksmond U, Wadwa M, Stoye JP, Kassiotis G. Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathog. 2012;8(5):e1002709. doi: 10.1371/journal.ppat.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473(7348):523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meylan F, et al. Negative thymocyte selection to HERV-K18 superantigens in humans. Blood. 2005;105(11):4377–4382. doi: 10.1182/blood-2004-07-2596. [DOI] [PubMed] [Google Scholar]
- 34.Evans LH, et al. Mobilization of endogenous retroviruses in mice after infection with an exogenous retrovirus. J Virol. 2009;83(6):2429–2435. doi: 10.1128/JVI.01926-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YN, Malim MH, Bieniasz PD. Hypermutation of an ancient human retrovirus by APOBEC3G. J Virol. 2008;82(17):8762–8770. doi: 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu YL, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103(42):15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marin M, Golem S, Rose KM, Kozak SL, Kabat D. Human immunodeficiency virus type 1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J Virol. 2008;82(2):987–998. doi: 10.1128/JVI.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones RB, et al. Human immunodeficiency virus type 1 escapes from interleukin-2-producing CD4+ T-cell responses without high-frequency fixation of mutations. J Virol. 2009;83(17):8722–8732. doi: 10.1128/JVI.00433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boczkowski D, Nair SK, Nam JH, Lyerly HK, Gilboa E. Induction of tumor immunity and cytotoxic T lymphocyte responses using dendritic cells transfected with messenger RNA amplified from tumor cells. Cancer Res. 2000;60(4):1028–1034. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.