Abstract
The mitotic checkpoint plays an important role in preventing chromosome segregation errors and the production of aneuploid progeny. In this issue, Zhang et al. examine mice and cells lacking the deubiquitinating enzyme USP44. Surprisingly, they find that USP44 prevents chromosome segregation errors through a function independent of its previously identified role in the mitotic checkpoint. Usp44-null animals develop aneuploidy and experience increased rates of tumorigenesis, implicating USP44 as novel tumor suppressor.
Ubiquitination is a reversible posttranslational modification that regulates a broad range of cellular processes. Ubiquitin is a small, 76-amino-acid protein that can be covalently conjugated with the aid of ubiquitin ligases to cellular proteins and to itself to create divergent chains with differing lysine linkages. Substrates marked with a polymer of ubiquitin molecules are often targeted for destruction by the 26S proteasome. Ubiquitin ligases are counterbalanced by deubiquitinases (DUBs) that remove molecules of ubiquitin previously added to target proteins. Despite their importance in controlling protein homeostasis, the substrates and physiological functions of most DUBs remain unknown (1). In this issue, Zhang et al. examine the role of the DUB ubiquitin-specific protease 44 (USP44) in ensuring accurate chromosome segregation and uncover a novel role of this enzyme as a tumor suppressor (2).
Dividing the genome
Each time a cell divides, it must partition its replicated genome to ensure that each new daughter cell receives a single copy of every chromosome. Aneuploidy, or an abnormal chromosome number, arises from errors in chromosome segregation during mitosis and is a remarkably common characteristic of human cancers. More than a decade ago aneuploidy was proposed to promote tumorigenesis, but current evidence suggests that its role is complex, both promoting and inhibiting tumorigenesis, depending on the genetic context in which it is found (3).
Aneuploidy often arises as a consequence of an underlying chromosomal instability (CIN), a condition characterized by frequent and continuing chromosome segregation errors during division (4). A major safeguard to protect against CIN is the mitotic checkpoint (also known as the spindle assembly checkpoint), a surveillance mechanism that operates in every division to prevent chromosome segregation errors and the resultant aneuploidy (5). In mitosis, a bipolar microtubule spindle apparatus forms to segregate chromosomes into the daughter cells. Chromosomes attach to microtubules of the mitotic spindle at a proteinaceous complex known as the kinetochore. Unattached kinetochores release an inhibitory signal that culminates in the creation of a protein complex that binds to CDC20 and prevents its activation of the anaphase-promoting complex (APC), an E3 ubiquitin ligase (Figure 1). Following attachment of all kinetochores to spindle microtubules, the mitotic checkpoint is silenced and APCCDC20 ubiquitinates cyclin B1 and securin, targeting them for proteasomal degradation, leading to anaphase onset with separation of the duplicated sister chromosomes, followed by mitotic exit.
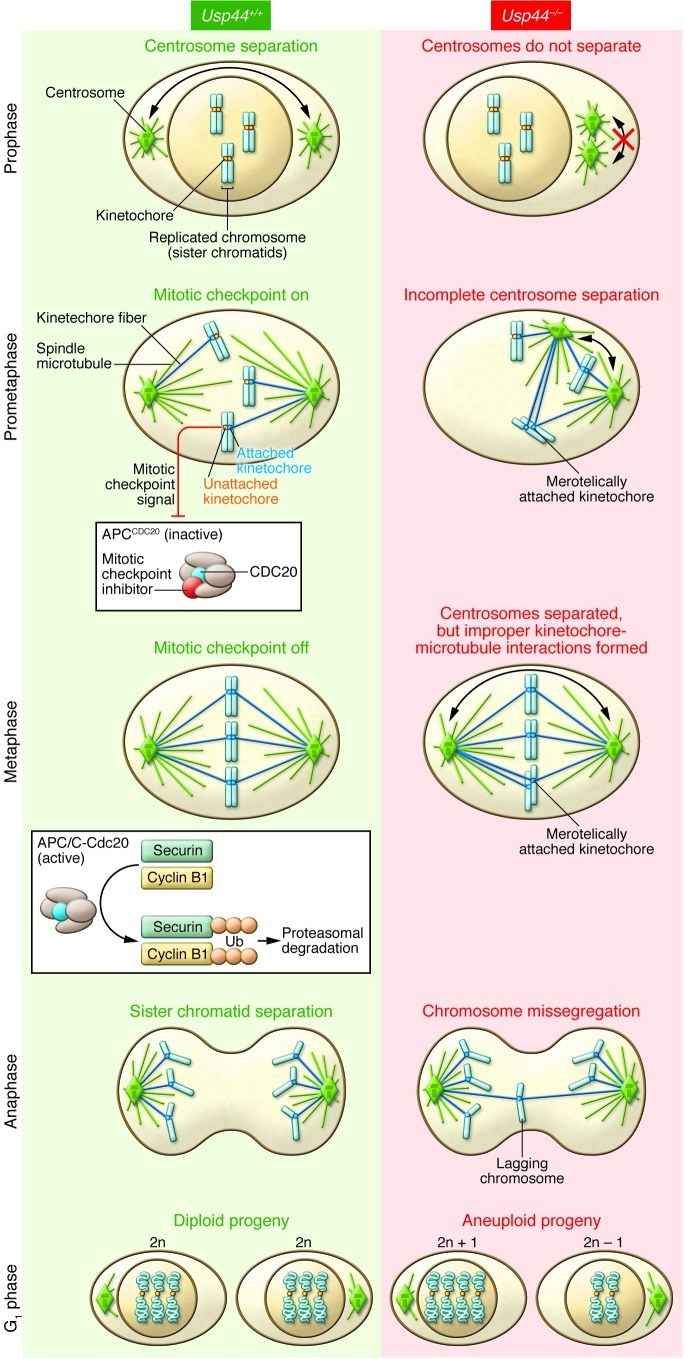
Figure 1. Loss of USP44 leads to errors in chromosome segregation.
Usp44+/+ cells: Centrosomes separate in prophase to instruct the formation of a bipolar microtubule spindle apparatus. Chromosomes attach to microtubules of the mitotic spindle at their kinetochores. Unattached kinetochores create a diffusible mitotic checkpoint signal that culminates in the inhibition of the E3 ligase activity of APCCDC20. At metaphase when all kinetochores are correctly attached to microtubules of the spindle, the mitotic checkpoint is turned off and APCCDC20 ubiquitinates securin and cyclin B1 to target them for destruction by the 26S proteasome. Securin destruction promotes sister chromatid disjunction, while cyclin B1 destruction promotes mitotic exit. Usp44–/– cells: Incomplete centrosome separation prior to nuclear envelope breakdown creates a spindle geometry that predisposes to the formation of improper kinetochore-microtubule interactions. The close positioning of the two centrosomes leads to an increase in the formation of merotelic kinetochore attachments (where a single kinetochore attaches to two different centrosomes) (14). These attachments persist into anaphase, resulting in lagging anaphase chromosomes and chromosome missegregation.
Breaking the chains: USP44’s role in regulating mitosis
An shRNA screen for components of the ubiquitin pathway that are involved in mitotic checkpoint signaling uncovered the DUB USP44 (6). USP44-depleted cells underwent premature entry into anaphase in unperturbed cell cycles and were unable to sustain activation of the mitotic checkpoint in the presence of spindle toxins. This points to a role of USP44 in inhibiting or antagonizing the activity of APCCDC20. The simplest model is that USP44 deubiquitinates APCCDC20 substrates, such as securin and cyclin B1. However, further analysis has suggested a more direct role of USP44 in the mitotic checkpoint: Since polyubiquitination of CDC20 is thought to be required to activate checkpoint-inhibited APCCDC20, the conventional view is that USP44 restrains APCCDC20 activity by stripping ubiquitin molecules from CDC20 to maintain inhibition by the mitotic checkpoint (6). Aspects of this model have remained controversial, however (7, 8), and the precise mechanism(s) by which USP44 functions in the mitotic checkpoint was imprecisely defined.
In this issue, Zhang et al. explore the physiological function of USP44 by creating a mouse model possessing a null allele of the gene (2). Surprisingly, given that mitotic abnormalities are observed in cultured cells depleted of USP44 (6), Zhang et al. found that Usp44–/– mice were born at normal frequencies and displayed no apparent defects (2). Since proper functioning of the mitotic checkpoint is required for viability in mammals, this demonstrates that USP44 is not an indispensable component of this checkpoint in mammals (9).
MEFs derived from Usp44–/– animals did, however, exhibit a subtle defect in mitotic checkpoint signaling: Usp44–/– MEFs displayed a reduced duration of mitotic arrest in the presence of spindle toxins and progressed through unchallenged divisions more rapidly. Zhang et al. found that MEFs lacking USP44 also exhibit an increase in the rate of chromosome segregation errors and whole chromosome aneuploidy, but notably show no evidence of structural alterations in chromosomes (2). The simple explanation is that the mitotic checkpoint defect is responsible for the chromosome segregation errors in Usp44–/– cells. This simple interpretation, however, is wrong: further experiments demonstrated that delaying mitotic progression with low doses of an APC inhibitor reduced chromosome segregation errors in cells with a bona fide mitotic checkpoint defect (cells heterozygous for the checkpoint component Mad2; ref. 10), but failed to prevent segregation errors in cells lacking USP44. This finding suggests that the subtle checkpoint defect observed in Usp44–/– cells is not responsible for the CIN in cells lacking the enzyme.
A role in the mitotic checkpoint and beyond
If a mitotic checkpoint defect is not the cause, then what is the mechanism by which loss of USP44 leads to an increased rate of chromosome segregation errors and aneuploidy? The first clue came from the analysis of how cells lacking USP44 missegregated their chromosomes. By far the most frequent mitotic error in Usp44–/– MEFs was an increased frequency of lagging anaphase chromosomes. Lagging chromosomes are caused by the aberrant attachment of a single kinetochore to spindle microtubules emanating from more than one spindle pole. This configuration, known as a merotelic attachment, causes the affected chromosome to be pulled by microtubules anchored to opposite spindle poles, causing them to “lag” in the spindle midzone while the correctly attached chromosomes move poleward during anaphase (11). Several mitotic abnormalities have been shown to increase the rate of formation of merotelic attachments, including centrosome amplification (12, 13) and the incomplete separation of centrosomes prior to nuclear envelope breakdown (NEBD) (14). Consistent with the latter possibility, Usp44–/– MEFs display abnormal spindle geometries and reduced centrosome separation at NEBD. Live-cell imaging revealed a strong correlation between the presence of an initial spindle defect and subsequent anaphase chromosome segregation errors. The current evidence, therefore, suggests that chromosome segregation errors in Usp44–/– cells do not arise from mitotic checkpoint abnormalities as expected, but rather stem from incomplete centrosome separation that predisposes cells to the development of increased numbers of merotelic attachments (Figure 1).
An obvious question raised by these findings is, how does USP44 function to promote centrosome separation? Insight into the mechanism came from the identification of a previously overlooked pool of USP44 that is localized to the centrosome during interphase. Zhang and colleagues identified a domain in USP44 that is required for direct interaction with the centrosome component centrin and recruitment of USP44 to the centrosome. To address the physiological function of the centrin-USP44 complex in chromosome segregation, the authors analyzed the ability of USP44 mutants to rescue the mitotic defects of Usp44–/– cells. As expected, expression of exogenous wild-type USP44 rescued the spindle defects and lagging anaphase chromosomes in Usp44–/– MEFs; however, complementation with a point mutant of USP44 unable to bind centrin or a catalytically inactive USP44 mutant failed to mitigate either defect. Therefore, the deubiquitinating activity and an ability to interact with centrin are both critical for USP44’s function in ensuring accurate chromosome segregation. Interestingly, a proportion of centrin also localizes to the nucleus of cells, where it forms a complex required for nucleotide excision repair following DNA damage (15). It will be interesting to establish whether USP44 is a binding partner of the nuclear pool of centrin and whether this interaction is required for centrin’s function in nucleotide excision repair.
USP44 DUBbed a tumor suppressor
Increased levels of aneuploidy were observed in the spleens of Usp44–/– animals, prompting an analysis of whether animals lacking the DUB were tumor-prone. Indeed, Zhang et al. found that aged Usp44–/– mice had an approximately five-fold-increased incidence of spontaneously arising tumors, thus identifying USP44 as a novel tumor suppressor (2). Adenomas of the lung were the most common tumors identified, with an approximately nine-fold increase in Usp44–/– animals relative to controls. Reduced expression of USP44 was also observed in human lung adenocarcinoma, and patients with low levels of the enzyme had a significantly reduced overall survival.
While it is tempting to conclude that the aneuploidy induced by loss of USP44 is responsible for promoting tumor formation, current evidence from other mouse models of CIN have shown that the degree of aneuploidy in cells and tissue is a poor predictor of tumor susceptibility in mice (9). Indeed, there are several mouse CIN models without an increase in spontaneous tumor formation that exhibit similar or greater levels of aneuploidy compared with that observed in Usp44–/– animals (9). Thus, it is plausible that loss of USP44 causes defects in addition to mitotic chromosome segregation errors that predispose animals to tumor development. For instance, USP44 has been shown to prevent embryonic stem cell differentiation by negatively regulating histone H2B monoubiquitination (16). USP44 is likely to have numerous targets, and a complete inventory of all of the enzyme’s substrates will aid our future understanding of its tumor suppressor function.
Future directions
The work by Zhang and colleagues (2) provides new insights into the physiological function of USP44 and the impact that loss of this DUB has on mitosis and cancer susceptibility. These discoveries raise a slew of unanswered questions, including: How does the USP44-centrin complex promote centrosome separation prior to NEBD? What role does USP44’s enzymatic activity play in this process? What are the substrates of USP44, and how is the substrate specificity or function of the enzyme altered by post-translational modifications, interacting proteins, and subcellular localization? What are the critical functions of USP44 that are required for tumor suppression?
Finally, although aneuploidy is commonly associated with cancer, a disease of unremitting cell growth, yeast and mouse cells that are aneuploid for a single chromosome exhibit a proteotoxic stress that reduces their growth rate in culture (17, 18). In the context of tumors, aneuploidy is likely to occur alongside additional “aneuploidy-tolerating” mutations. A recent genetic screen for mutations that improve the proliferative potential of aneuploid yeast cells identified mutations in the deubiquitinating enzyme UBP6, demonstrating the importance of the ubiquitin-proteasome pathway in suppressing the growth of aneuploid cells (19). An interesting question for future study is whether loss of USP44 facilitates the proliferation of aneuploid cells. Identifying the lesions that collaborate with aneuploidy to promote tumor formation is one key area for future research.
Acknowledgments
We thank Quan Zhu for helpful comments on this preview. We apologize to all whose work was not cited because of space restrictions. This work was supported by a grant (GM29513) from the NIH to D.W. Cleveland, who receives salary support from the Ludwig Institute for Cancer Research. A.J. Holland is supported by a Leukemia and Lymphoma Society special fellowship.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(12):4325–4328. doi:10.1172/JCI66420.
See the related article beginning on page 4362.
References
- 1.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J Clin Invest. 2012;122(12):4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13(6):501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 5.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 6.Stegmeier F, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446(7138):876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nature Cell Biol. 2008;10(12):1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell. 2011;44(5):710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409(6818):355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 11.Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786(1):32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4(8):e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23(3):401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araki M, et al. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem. 2001;276(22):18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs G, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46(5):662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 18.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322(5902):703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143(1):71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]