Abstract
Multiple myeloma causes major morbidity resulting from osteolytic lesions that can be detected by metastatic bone surveys. Magnetic resonance imaging and positron emission tomography can detect bone marrow focal lesions long before development of osteolytic lesions. Using data from patients enrolled in Total Therapy 3 for newly diagnosed myeloma (n=303), we analyzed associations of these imaging techniques with baseline standard laboratory variables assessed before initiating treatment. Of 270 patients with complete imaging data, 245 also had gene expression profiling data. Osteolytic lesions detected on metastatic bone surveys correlated with focal lesions detected by magnetic resonance imaging and positron emission tomography, although, in two-way comparisons, focal lesion counts based on both magnetic resonance imaging and positron emission tomography tended to be greater than those based on metastatic bone survey. Higher numbers of focal lesions detected by magnetic resonance imaging and positron emission tomography were positively linked to high serum concentrations of C-reactive protein, gene-expression-profiling–defined high risk, and the proliferation molecular subgroup. Positron emission tomography focal lesion maximum standardized unit values were significantly correlated with gene-expression-profiling–defined high risk and higher numbers of focal lesions detected by positron emission tomography. Interestingly, four genes associated with high-risk disease (related to cell cycle and metabolism) were linked to counts of focal lesions detected by magnetic resonance imaging and positron emission tomography. Collectively, our results demonstrate significant associations of all three imaging techniques with tumor burden and, especially, disease aggressiveness captured by gene-expression-profiling–risk designation. (Clinicaltrials.gov identifier: NCT00081939)
Introduction
Multiple myeloma (MM) often presents with bone-related clinical symptoms that result from osteolytic lesions (OLs), associated pathological fractures, or severe osteopenia that often results in spinal compression fractures.1 For decades, X-ray imaging has been the standard tool for evaluating bone and is part of the Durie–Salmon staging system.2 Unfortunately, X-ray-based metastatic bone survey (MBS) does not detect OLs until 70% or more calcium has already been lost.3 Technetium-based bone scintigraphy is insensitive to detecting bone disease in MM because the radiotracer is taken up by osteoblasts, which are severely impaired in number and function in MM,4 so this test is only helpful in patients who have healing pathological fractures.
MM bone disease results from hyperactivation of osteoclasts5 and inhibition of osteoblasts.6 Detailed correlative analyses of MBS-defined OLs and MRI-FLs with gene expression profiling (GEP) analysis of highly purified myeloma plasma cells revealed that myeloma plasma cells release the osteoblast-inactivating molecule DKK1.7 Monoclonal antibody therapy directed against DKK1 was recently developed as a means to alleviate bone disease in breast cancer and MM.8,9
We and others have observed that MRI is much more sensitive for detecting focal lesions (FLs) than MBS detection of OLs, but both methods had prognostic implications. In terms of the median time to response, MRI-defined complete response lagged behind clinical complete response by 18 months.10 Examination of bone marrow samples obtained by computed tomography (CT)-guided fine-needle aspiration demonstrated the presence of subpopulations of low- or non-secretory myeloma cells residing in the lingering FLs defined by MRI (MRI-FLs).10 We have evidence that persistence of MRI-FLs is an important contributor to disease relapse, and MRI-FLs have been linked to earlier progression from asymptomatic MM to symptomatic MM.10,11 Thus, MRI-defined complete response has become an important objective of therapy.12
Another highly sensitive imaging tool in MM diagnostics is fluorodeoxyglucose (FDG)-based positron emission tomography (PET).11,13 We and others have reported that FDG-PET is useful for detecting FDG-avid intramedullary FLs of the entire skeleton, as well as extra-medullary disease (EMD).14,15 Although EMD is present at diagnosis in only a few percent of patients, it is becoming an increasingly vexing problem later in the disease course because MM survival has been extended so markedly.16,17 As in management of malignant lymphoma,18 including Hodgkin's disease,19 early complete suppression of FDG avidity in FLs prior to transplantation intervention has favorable implications for clinical outcomes in MM.20,21
The purpose of this study was to examine, among patients with newly diagnosed MM enrolled in the Total Therapy 3 (TT3) clinical trial,20,21 the mutual correlation between MBS-OL counts and FL counts (defined by MRI and PET), and their associations with baseline prognostic variables.
Design and Methods
The study focused on 270 of 303 patients enrolled in TT3 who had baseline examinations of all three imaging tests performed (MBS, MRI and PET), 245 of whom also had GEP data obtained prior to therapy. The TT3 trial, which has added bortezomib to a multi-agent chemotherapy and tandem autograft-supported high-dose melphalan regimen, had been approved by the Institutional Review Board, and all patients signed a written informed consent for trial participation, in keeping with institutional and international Helsinki Declaration guidelines. Bone marrow sampling for GEP also required a separate written informed consent.
The protocol details have been described previously,20,21 as have imaging parameters for MBS, MRI and PET.10,14
All radiographic studies were evaluated by radiologists who specialize in imaging of MM, each with more than a decade of experience in their respective field. All data were reviewed by at least 2 radiologists independently prior to entry in our database. MBS-derived parameters captured OL number. MRI, which was limited to the axial skeleton (i.e. head, spine and pelvis) identified FL number and bone marrow background intensity, where the presence of diffuse hyper-intense marrow (DHIM) suggested high tumor burden. We define diffuse marrow involvement on short time inversion recovery (STIR) as the marrow signal intensity in relation to adjacent muscles. Usually, we use the muscles of the pelvis and determine the intensity relative to the muscles as hyperintense, isointense or hypointense in relation to adjacent muscles. With PET imaging, we documented FL number and the maximum standardized uptake value (FL-SUV), indicative of FL metabolic activity. In addition, we registered the presence of EMD and SUVdiff, the latter of which reflected the intensity of bone marrow background at sites devoid of FLs (i.e. diffuse involvement by MM) and was typically measured in the lower lumbar spine. We categorize and have standardized diffuse background marrow at our institution as mild (max SUV-diff 0-1.9), moderate (2.0-2.9), or severe (3+) in the lower lumbar spine red marrow. A region of interest is drawn typically at the L5 vertebral body marrow with calculation of maximum SUV based upon lean body mass). In patients without prior treatment, baseline diffuse marrow uptake on FDG-PET should not be greater than 2.0 given there is no active disease in the region of measurement or causative factors, such as degenerative disease, etc. This cut off also coincides with normal liver uptake based upon maxSUV by lean body mass, which is typically not greater than 2.0.22
Cut-off points for imaging parameters were applied as previously reported10,14,15 and included multiple prognostic OL and FL number cut offs, i.e. >0 and >2 for MBS-OL,10 >0 and >7 for MRI-FL,10 and >0 and >3 for PET-FL.14 Odds ratios and corresponding 95% confidence intervals were calculated from 2-by-2 contingency tables for the comparison of dichotomous imaging parameters with dichotomous GEP and standard laboratory variables. For these comparisons, P values were calculated using Fisher's exact test. Given the large scale of this analysis, a more conservative significance level of 0.01 is used for the comparison of baseline prognostic factors and dichotomized imaging parameters. Comparisons between FL and OL counts were performed using Wilcoxon's signed-rank test.
GEP data included designation of molecular subtype according to the following categories: CD-1, CD-2, MS and MF translocations, HY (hyperdiploidy), LB (low bone disease), and PR (proliferation).23 In addition, we determined the status of TP53 deletion (delTP53),24 prognostic risk linked to progression-free and overall survival (based on 70-gene and 80-gene models),25,26 Proliferation Index (PI), and Centrosome Index (CI).27 We also examined whether bone-related genes and those constituting the 70-gene risk model were individually linked to imaging variables. For this purpose, Wilcoxon's rank sum test was applied to each pairwise comparison of dichotomized imaging parameters with the log base 2 expression levels for the provided bone-related genes and the GEP-70 probes. A Bonferroni adjusted significance level of 0.0001 was deemed sufficient to maintain an overall error rate of approximately 0.05 for each set of bone- or risk-associated probes.28 Microarray data used in this study have been deposited in the NIH Gene Expression Omnibus under accession number GSE2658.
Results
Patients' characteristics
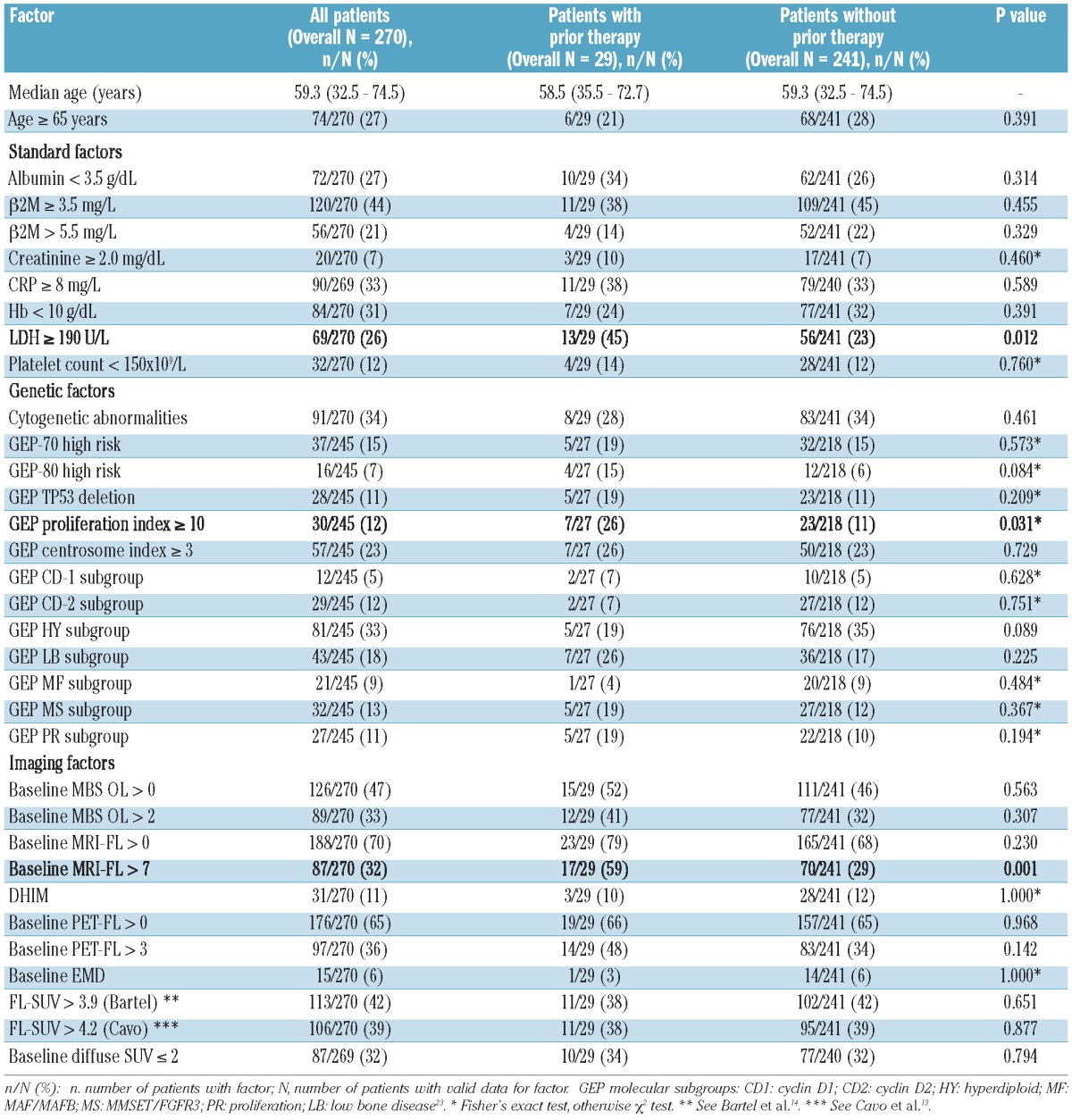
Patients' baseline features are presented in Table 1. Median age was 59 years; 27% were 65 years or older. Elevated serum levels of beta-2-microglobulin (β2M) of 3.5 mg/L and over and of more than 5.5 mg/L were observed in 44% and 21% of patients, respectively. Serum albumin levels were low (<3.5 g/dL) in 27% of patients. Metaphase-based cytogenetic abnormalities (CA) were documented in 34% of patients. GEP data, available in 245 patients, revealed high-risk designation in 15% and 7%, respectively, according to 70-gene and 80-gene risk models. The molecular subgroup distribution was typical of patterns observed in our patient population. MBS-OLs were noted in 47% of patients; 33% had over 2 MBS-OLs. MRI-FLs over 0 and more than 7 were present in 70% and 32% of patients, respectively, with 11% of patients displaying DHIM. PET-FLs over 0 and over 3 were present in 65% and 36%, respectively, with FL-SUV over 3.914 and over 4.215 present in 42% and 39% of patients, respectively. EMD was detected in 6% of patients and SUVdiff of 2 or below in 32%. Among 32 individual variables examined (Table 1), only three showed differences among patients with and without prior therapy. The differences related to lactate dehydrogenase (LDH) and GEP PI were only marginally significant; however, patients with prior therapy clearly showed greater numbers of MRI-FLs. In spite of this, the high degree of similarity between other imaging parameters justified the inclusion of all 270 patients in further analyses.
Table 1.
Patients' characteristics.
Relationship between imaging variables
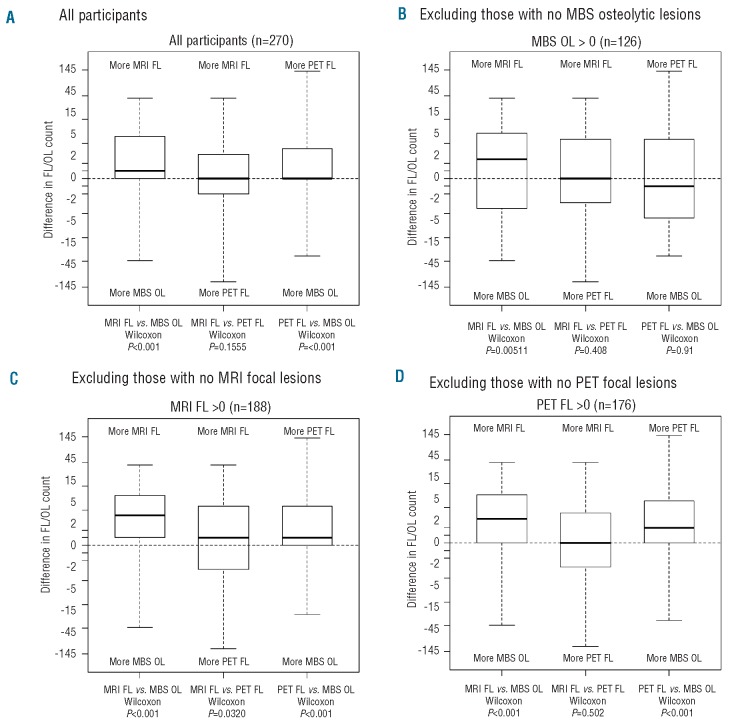
Figure 1 provides a visual comparison of the differences in FL and OL counts for the three imaging methods, along with P values from the Wilcoxon's signed-rank test for each comparison. Briefly, the Wilcoxon's test examines whether the FL/OL counts from one imaging method tend to be larger in magnitude or more frequently greater than counts from another imaging method. In Figure 1A, we present the comparisons for all 270 participants. No differences were seen between the MRI and PET methods. This is highlighted in the box-and-whisker diagram for this comparison, which shows an even distribution of patients with more MRI-FLs versus more PET-FLs (i.e. the differences are symmetric around zero). However, both MRI and PET methods detected more FLs than the OLs detected by MBS. This was more clearly seen for MRI-FLs, as the median difference was shifted away from MBS-OLs (P<0.001), and was more subtle for PET-FLs, where the median difference was zero but the distribution highly skewed away from MBS-OLs (P<0.001).
Figure 1.
Comparisons of number of focal lesions (MRI-FLs and PET-FLs) and osteolytic lesions (MBS-OLs). For each patient, we calculated the difference in the total number of FLs and OLs detected with each method. The distributions of the differences for each pairwise comparison are presented as box-and-whisker plots in the figures below. The lower and upper edges of the box correspond to the first and third quartiles, respectively. The thicker bars in the middle represent the median, with the whiskers extending to the minimum and maximum values. P values from the Wilcoxon signed-rank test are given for each comparison. (A) Among all 270 patients with complete imaging information, no difference was noted between FLs detected by MRI and FLs detected by PET (middle box-whisker), whereas both MRI (left) and PET (right) detected more FLs than the number of OLs observed on MBS. (B) When limited to the 126 patients with at least one MBS-OL, MRI-FL was higher than MBS-OL (left) while no differences were noted between MRI-FL and PET-FL (middle) and between PET-FL and MBS-OL (right). (C) When restricted to the 188 subjects with at least one MRI-FL, data consistent with that noted in Figure 1A were observed. (D) This also applied to the 176 individuals with at least 1 PET-FL.
Since these differences may have been magnified by the large proportion of patients with no MBS-OLs compared to the numbers of patients with no MRI- and PET-FLs (53% compared to 30% and 35%, respectively), we excluded patients with no MBS-OLs and examined only those patients with at least one MBS-OL (n=126; Figure 1B). For this subset, the comparisons are intentionally biased towards more MBS-OLs. However, MRI-FL counts still tended to be higher than MBS-OL counts (P=0.005), suggesting the ability of MRI to detect more FLs is not limited to those with no measurable MBS-OLs. No significant differences were observed when PET-FLs were compared to MRI-FLs or MBS-OLs. For completeness, we also examined the subsets of patients with at least one MRI-FL (n=188; Figure 1C) and at least one PET-FL (n=176; Figure 1D). The results were consistent with those observed in Figure 1A. Together, these results demonstrate the superior ability of both MRI and PET to detect FLs compared with the ability of MBS to detect OLs.
Log odds ratios for association of baseline prognostic variables with imaging parameters
Cut-off points for MBS-OLs (>0 and >2) were examined for associations with other imaging parameters, GEP variables, and standard prognostic variables (Figure 2A, left panel). Both cut-off points for MBS-OLs positively correlated with the cut-off points for the other two imaging methods: MRI-FL over 0 and over 7 and PET-FL over 0 and over 3. Among GEP variables, high risk (70-gene model), PR subtype, and CI, all reflecting disease aggressiveness, were significantly linked to more than 2 MBS-OLs. DelTP53 was neutral relative to MBS-OLs. Among standard laboratory prognostic variables, high serum levels of β2M (>5.5 mg/L) were associated with more than 2 MBS-OLs.
Figure 2.
Log odds ratios measuring the association of baseline prognostic factors with imaging parameters at baseline. DHIM comparisons were limited to the subset of participants with no detected MRI-FLs (n=82 for standard laboratory and imaging variables and n=74 for GEP variables). (A) MBS and MRI. Left: both MBS-OL cut-off points were correlated with both cut-off points for MRI-FL and PET-FL. Among GEP variables, high risk (70-gene model), PR subtype, and CI, all reflecting disease aggressiveness, were significantly linked to more than 2 MBS-OLs. DelTP53 was neutral relative to MBS-OLs. Among standard laboratory prognostic variables, high serum levels of β2M (>5.5 mg/L) were associated with more than 2 MBS-OLs. Middle: for both MRI-FL cut-off points, there were strong positive correlations with MBS-OLs and PET-FLs, but not with PET FL-SUV, EMD, or SUVdiff. Among GEP features, PI and CI positively correlated with MRI-FL >7. Among GEP-defined molecular subgroups, the PR subgroup positively correlated with MRI-FLs. Both MRI-FL cut-off points correlated with high-risk scores from the 70-gene model, but only the higher cut-off point (>7) was linked to high-risk scores from the 80-gene model. Among standard laboratory prognostic variables, CRP had significant positive correlations to MRI-FL >7. Right: for the subset with no MRI-FL, DHIM was associated with increased LDH and β2M. The comparison of GEP-80 risk groups was not possible because of the small number of GEP-80 high-risk subjects. Some parameters could not be estimated due to small sample sizes or association by definition (for example, MRI-FL >7 vs. MRI-FL >0). (B) PET. Left: PET-based FL number was examined for correlations with other imaging parameters, standard laboratory prognostic variables, and GEP variables. At both cut-off points (>0 and >3), PET-FLs had highly significant positive correlations with MBS-OLs and MRI-FLs. GEP-derived variables that were significantly positively associated with PET-FL included high-risk myeloma defined by the 70-gene model as well as PI and CI. Among GEP-defined molecular subgroups, the PR subgroup showed positive correlation and the LB subgroup negative correlation to PET-FLs. Among standard laboratory prognostic variables, elevated CRP and LDH were seen more often with more PET-FLs. Middle: PET FL-SUV was highly associated with more than 3 PET-FLs, high-risk disease (70-gene model), and SUVdiff. Right: PET-EMD was linked to high β2M. Comparison of the CD-1 sub-group versus all other GEP subgroups was not possible because of the small number of CD-1 subjects.
Cut-off points for MRI-FLs (>0 and >7) were examined for their associations with other imaging parameters, GEP variables, and standard laboratory prognostic variables (Figure 2A, middle panel). For both cut-off points, there were strong positive correlations with MBS-OLs and PET-FLs, but not with PET FL-SUV, EMD, or SUVdiff. Among GEP features, both PI and CI positively correlated with the MRI-FL cut-off point over 7. Among GEP-defined molecular subgroups, the PR subgroup positively correlated with MRI-FL counts. Both MRI-FL cut-off points correlated with high-risk scores from the 70-gene model, but only the higher cut-off point (>7) was linked to high-risk scores from the 80-gene model. Among standard laboratory prognostic variables, CRP had significant positive correlations to the over 7 cut-off point. For the subset with no MRI-FLs, DHIM was associated with increased LDH and β2M (Figure 2A, right panel).
In the case of PET variables, PET-FL at both cut-off points (>0 and >3) had highly significant positive correlations with MBS-OLs and MRI-FLs (Figure 2B, left panel). GEP-derived variables that were significantly positively associated with PET-FL counts included high-risk designation defined by the 70-gene model as well as PI and CI. Among GEP-defined molecular subgroups, the PR subgroup showed positive correlation and the LB subgroup negative correlation to PET-FLs. Among standard laboratory prognostic variables, elevated CRP and LDH were seen more often with more PET-FLs. However, while highly suggestive, the greater incidence of CA coinciding with PET-FL was not statistically significant. FL-SUV was highly associated with more than 3 PET-FLs, high-risk disease (70-gene model), and SUVdiff (Figure 2B, middle panel). PET-EMD was linked to high β2M (Figure 2B, right panel). Online Supplementary Table S1 provides odds ratios and P values for the data presented in Figure 2.
We next examined bone-related genes and those constituting the 70-gene risk model (Online Supplementary Table S2) for their individual associations with the imaging parameters. Online Supplementary Figure S1 graphically depicts the P values for the comparisons of the dichotomized imaging parameters with bone-related genes, including factors directly affecting osteoblastogenesis (PTHLH and genes associated with Wnt signaling, including DKK1, FRZB, SRFP2, WNT10A, and WNT10B, and the gene family of LRP receptors), in addition to genes known to affect osteoclastogenesis (CCL3, CST6, TNFSF11, TNFRSF11B, and IL6). While no significant comparisons were observed for the dichotomized MBS or MRI subgroups, the median expression levels of 208433_at, an LRP8 probe, were significantly different for subjects with SUVdiff≤2. Expression levels of 205282_at, another LRP8 probe, were highly suggestive of association with PET-FLs over 0 and over 3, but did not exceed the significance level adjusted for the multiple comparisons (Online Supplementary Table S3).
Given the strong correlation of imaging parameters to 70-gene model-defined high risk (Figure 2), we also compared the expression levels of the individual GEP-70 probes with the dichotomized imaging parameters. Online Supplementary Figure S2 graphically depicts and Online Supplementary Table S4 indicates the P values from this set of comparisons. No significant correlations were observed for either MBS-OL cut-off point. GEP-70 probes associated with the MRI-FL over 0 cut-off point were for the following genes: C6orf173 (226936_at), STK6 (204092_s_at), and TRIP13 (204033_at). For the MRI-FL over 7 cut-off point, comparisons for ENO1 (201231_s_at) and TRIP13 (204033_at) exceeded the prescribed significance level. TRIP13 (204033_at) was also highly correlated with the PET-FL cut-off points (>0 and >3). The comparison of FABP5 (202345_s_at) for the PET-FL cut-off point over 0 was highly suggestive of an association, but statistical significance was only reached for the PET-FL cut-off point over 3. ENO1 (alpha-enolase) and FABP5 (fatty acid–binding protein 5) are implicated in glycolysis29 and cellular lipid transport,30 respectively, while STK631 and TRIP1332 are involved in cell cycle and control of DNA replication.
Discussion
The patient population evaluated here is representative of those patients with MM who, according to CRAB criteria (creatinine level, renal failure, anemia, bone disease),33 require therapy; however, this population differs in age distribution from most reported transplant trials, with a median age of 59.3 years (range 32.5-74.5 years), including 27% who were 65 years and older. International Staging System (ISS) distributions34 were also similar to patients enrolled in large clinical trials by the Intergroupe Francophone de Myelome35 and by Spanish,36 Italian,37 and the joint Dutch and German trials.38
In evaluating similarities and differences between the three imaging techniques, we noted a tendency towards higher counts for MRI-FLs and PET-FLs compared to MBS-OLs. These data validate the use of MRI and PET in early surveillance of FLs, so patients and their physicians will be able to identify potential problems. Regarding standard laboratory parameters, β2M, reflecting tumor burden and renal function,39 was linked to MBS-OLs, MRI-DHIM and PET-EMD, while CRP, affected by interleukin-6 activity,40 correlated with the MRI-FL cut-off point over 7 and PET-FLs. In the case of LDH as a feature of tumor aggressiveness41 and anaerobic glycolysis,42 significant association was present only for MRI-DHIM. While the odds ratios suggested high LDH was associated with increased MBS-OLs, PET-FLs, and PET-EMD, these comparisons were not statistically significant. Additionally, the presence of CA may be linked to PET-FL, but the associated odds ratio of only 1.79 was not statistically significant. In comparison, we estimate the odds ratio for GEP-70 high-risk MM to be 4.34.
The strongest correlations with imaging parameters were seen for GEP-derived variables, such as high-risk designation (70-gene model)25 and PR subtype,23 significantly linked to most imaging variables, including MBS-OLs, MRI-FLs, PET-FLs, and FL-SUV (only for high-risk disease). PI and CI showed links to MRI-FLs and PET-FLs. LB subtype was inversely linked to PET-FLs, and, although not statistically significant, showed a similar trend with MRI-FL. The absence of correlation with MRI-FLs may relate to the greater sensitivity of PET for FL detection. As a metabolic rather than anatomical measurement, PET may reflect gene expression differences better than MRI. MS and MF subtypes, often considered prognostically unfavorable,23 displayed no link to imaging parameters, which was also the case with the CD-1 and CD-2 subtypes and GEP-derived delTP53.24
Interestingly, the proliferation index (PI), PR group, and metabolism did not correlate with DHIM and SUV-diff. This can be explained by metabolism being measured in the serum, reflecting a systemic parameter; the PI, and especially the PR group, represent a balance of expression of select groups of genes and include patients from the other GEP-defined groups
The availability of GEP data on more than 56,000 gene probes led us to examine which genes, among bone-relevant genes and those associated with high-risk disease, were linked to imaging parameters. While many of the suspected candidates were highly suggestive of an association with some imaging variable (e.g. DKK1 with MBS-OL >0, P<0.0056; MBS-OL >2, P<0.0036; MRI-FL >0, P<0.0069), only LRP8 had statistically significant correlations once adjustment was made for multiple comparisons. On the other hand, several of the genes constituting the 70-gene risk model, itself linked to imaging features, were found to be highly associated with MRI-FL and PET-FL counts, such as STK6, ENO1, TRIP13, and FABP5.
STK6, also known as aurora kinase A, is recruited into the centrosome early in G2 and has been implicated in the activation of CDK1/cyclin B on the centrosome cell cycle transit from G2 through to cytokinesis.31 The role of STK6 in MM growth has been recently evaluated, and several STK6 inhibitors have been successfully tested against myeloma cells in pre-clinical studies.43,44 TRIP13 is a AAA+ ATPase family member associated with and required for completion of proper meiosis,32 and is involved in promotion of early steps of the double-strand breaks repair process upstream of the assembly of RAD51 complexes.45 Increased homologous recombination activity and elevated expression of HR-related genes, including RAD51, has been suggested to mediate DNA instability and progression of MM.46 FABP5 is expressed in various malignancies; it is a major target of the proto-oncogene c-MYC47 and is involved in resistance of solid tumor cells to retinoic acid treatment.48 ENO1, also known as a c-MYC promoter-binding protein, is a metalloenzyme metabolic enzyme involved in the synthesis of pyruvate in the anaerobic glycolysis pathway.49 In the cytoplasm, ENO1 was found to be associated with PCNA,50 a proliferating cell nuclear antigen that is up-regulated in the PR MM subtype.23 ENO1 is also expressed on the cell surface and participates in plasminogen receptor-promoting plasmin activation, extracellular matrix degradation, and tumor metastasis.29 Furthermore, ENO1 can be translated into the c-myc promoter-binding protein (MBP-1), a nuclear protein that binds to the c-MYC P2 promoter and acts as a c-MYC transcription repressor. ENO1 is over-expressed in various tumors and is associated with poor outcome.49 In myeloma cell lines, ENO1 has been identified as an IL-6 target gene.51
Taken together, the unique association between MRI-FLs and PET-FLs and certain significant genes associated with cell cycle, DNA replication, or repair of DNA double-strand breaks (e.g. TRIP13, STK6) is in accordance with the well-recognized chromosomal instability features in MM. The correlation of the two metabolism-associated genes, ENO1 and FABP5, with MRI-FLs and PET-FLs further implicates hyper-metabolic MM with adverse clinical parameters and poor outcome.
Our work represents an excellent example of clarifying how clinical disease manifestation, i.e. MM bone disease, is linked to and explained by tumor-cell molecular genetic features. We postulate that GEP of myeloma cells can also reveal unique signatures linked to other variables, such as anemia, immune paralysis, and renal failure, similar to LDH and CA. Recognizing the critical role of the bone marrow environment for MM progression and disease manifestation, work is currently in progress to examine GEP signatures of whole bone marrow biopsies.
Acknowledgments
The authors would like to thank Peggy Brenner, Office of Grants and Scientific Publications, University of Arkansas for Medical Sciences, for editorial assistance.
Funding: This work was supported by a grant from the National Cancer Institute (CA 55819).
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures: Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Roodman GD. Mechanisms of bone metastasis. N Engl J Med. 2004;350(16):1655-64 [DOI] [PubMed] [Google Scholar]
- 2.Durie BG, Salmon SE. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer. 1975;36(3):842-54 [DOI] [PubMed] [Google Scholar]
- 3.Edelstyn GA, Gillespie PJ, Grebbell FS. The radiological demonstration of osseous metastases. Experimental observations. Clin Radiol. 1967;18(2):158-62 [DOI] [PubMed] [Google Scholar]
- 4.Wahner HW, Kyle RA, Beabout JW. Scintigraphic evaluation of the skeleton in multiple myeloma. Mayo Clin Proc. 1980;55(12):739-46 [PubMed] [Google Scholar]
- 5.Mundy GR, Raisz LG, Cooper RA, Schechter GP, Salmon SE. Evidence for the secretion of an osteoclast stimulating factor in myeloma. N Engl J Med. 1974;291(20):1041-6 [DOI] [PubMed] [Google Scholar]
- 6.Bataille R, Chappard D, Marcelli C, Dessauw P, Baldet P, Sany J, et al. Recruitment of new osteoblasts and osteoclasts is the earliest critical event in the pathogenesis of human multiple myeloma. J Clin Invest. 1991;88(1):62-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349(26):2483-94 [DOI] [PubMed] [Google Scholar]
- 8.Yaccoby S, Ling W, Zhan F, Walker R, Barlogie B, Shaughnessy JD., JrAntibody-based inhibition of DKK1 suppresses tumor-induced bone resorption and multiple myeloma growth in vivo. Blood. 2007;109(5):2106-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulciniti M, Tassone P, Hideshima T, Vallet S, Nanjappa P, Ettenberg SA, et al. Anti-DKK1 mAb (BHQ880) as a potential therapeutic agent for multiple myeloma. Blood. 2009;114(2):371-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker R, Barlogie B, Haessler J, Tricot G, Anaissie E, Shaughnessy JD, Jr, et al. Magnetic resonance imaging in multiple myeloma: diagnostic and clinical implications. J Clin Oncol. 2007;25(9):1121-8 [DOI] [PubMed] [Google Scholar]
- 11.Angtuaco EJ, Fassas AB, Walker R, Sethi R, Barlogie B. Multiple myeloma: clinical review and diagnostic imaging. Radiology. 2004;231(1):11-23 [DOI] [PubMed] [Google Scholar]
- 12.Barlogie B, Crowley J. Could CR mean cure? Blood. 2011;118(3):483. [DOI] [PubMed] [Google Scholar]
- 13.Zamagni E, Patriarca F, Nanni C, Zannetti B, Englaro E, Pezzi A, et al. Prognostic relevance of 18-F FDG PET/CT in newly diagnosed multiple myeloma patients treated with upfront autologous transplantation. Blood. 2011;118(23):5989-95 [DOI] [PubMed] [Google Scholar]
- 14.Bartel TB, Haessler J, Brown TL, Shaughnessy JD, Jr, van Rhee F, Anaissie E, et al. F18-fluorodeoxyglucose positron emission tomography in the context of other imaging techniques and prognostic factors in multiple myeloma. Blood. 2009;114(10):2068-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamagni E, Nanni C, Patriarca F, Englaro E, Castellucci P, Geatti O, et al. A prospective comparison of 18F-fluorodeoxyglucose positron emission tomography-computed tomography, magnetic resonance imaging and whole-body planar radiographs in the assessment of bone disease in newly diagnosed multiple myeloma. Haematologica. 2007;92(1):50-5 [DOI] [PubMed] [Google Scholar]
- 16.Bladé J, Fernández de Larrea C, Rosiñol L, Cibeira MT, Jiménez R, Powles R. Soft-tissue plasmacytomas in multiple myeloma: incidence, mechanisms of extramedullary spread, and treatment approach. J Clin Oncol. 2011;29(28):3805-12 [DOI] [PubMed] [Google Scholar]
- 17.Varettoni M, Corso A, Pica G, Mangiacavalli S, Pascutto C, Lazzarino M. Incidence, presenting features and outcome of extramedullary disease in multiple myeloma: a longitudinal study on 1003 consecutive patients. Ann Oncol. 2010;21(2):325-30 [DOI] [PubMed] [Google Scholar]
- 18.Spaepen K, Stroobants S, Dupont P, Van SS, Thomas J, Vandenberghe P, et al. Prognostic value of positron emission tomography (PET) with fluorine-18 fluorodeoxyglucose ([18F]FDG) after first-line chemotherapy in non-Hodgkin's lymphoma: is [18F]FDG-PET a valid alternative to conventional diagnostic methods? J Clin Oncol. 2001;19(2):414-9 [DOI] [PubMed] [Google Scholar]
- 19.Juweid ME. Utility of positron emission tomography (PET) scanning in managing patients with Hodgkin lymphoma. Hematology Am Soc Hematol Educ Program. 2006:259-1 [DOI] [PubMed] [Google Scholar]
- 20.van Rhee F, Szymonifka J, Anaissie E, Nair B, Waheed S, Alsayed Y, et al. Total Therapy 3 for multiple myeloma: prognostic implications of cumulative dosing and premature discontinuation of VTD maintenance components, bortezomib, thalidomide, and dexamethasone, relevant to all phases of therapy. Blood. 2010;116(8):1220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair B, van Rhee F, Shaughnessy JD, Jr, Anaissie E, Szymonifka J, Hoering A, et al. Superior results of Total Therapy 3 (2003-33) in gene expression profiling-defined low-risk multiple myeloma confirmed in subsequent trial 2006-66 with VRD maintenance. Blood. 2010;115(21):4168-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paquet N, Albert A, Foidart J, Hustinx R. Within-patient variability of (18)F-FDG: standardized uptake values in normal tissues. J Nucl Med. 2004;45(5):784-8 [PubMed] [Google Scholar]
- 23.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaughnessy JD, Zhou Y, Haessler J, van Rhee F, Anaissie E, Nair B, et al. TP53 deletion is not an adverse feature in multiple myeloma treated with total therapy 3. Br J Haematol. 2009;147(3):347-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276-84 [DOI] [PubMed] [Google Scholar]
- 26.Shaughnessy JD, Jr, Qu P, Usmani S, Heuck CJ, Zhang Q, Zhou Y, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118(13):3512-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chng WJ, Ahmann GJ, Henderson K, Santana-Davila R, Greipp PR, Gertz MA, et al. Clinical implication of centrosome amplification in plasma cell neoplasm. Blood. 2006;107(9):3669-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuehl RO. Design of Experiments: Statistical Principles of Research Design and Analysis, 2nd ed., pp. 469-72, Brooks/Cole, Pacific Grove, California: 2000 [Google Scholar]
- 29.Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci. 2001;58(7):902-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haunerland NH, Spener F. Fatty acid-binding proteins--insights from genetic manipulations. Prog Lipid Res. 2004;43(4):328-49 [DOI] [PubMed] [Google Scholar]
- 31.Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, et al. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114(5):585-98 [DOI] [PubMed] [Google Scholar]
- 32.San-Segundo PA, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97(3):313-24 [DOI] [PubMed] [Google Scholar]
- 33.Kyle RA, Rajkumar SV. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia. 2009;23(1):3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Blade J, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23(15):3412-20 [DOI] [PubMed] [Google Scholar]
- 35.Moreau P, Garban F, Attal M, Michallet M, Marit G, Hulin C, et al. Long-term follow-up results of IFM99-03 and IFM99-04 trials comparing nonmyeloablative allotransplantation with autologous transplantation in high-risk de novo multiple myeloma. Blood. 2008;112(9):3914-5 [DOI] [PubMed] [Google Scholar]
- 36.Rosinol L, Perez-Simon JA, Sureda A, de la Rubia J, de Arriba F, Lahuerta JJ, et al. A prospective PETHEMA study of tandem autologous transplantation versus autograft followed by reduced-intensity conditioning allogeneic transplantation in newly diagnosed multiple myeloma. Blood. 2008;112(9):3591-3 [DOI] [PubMed] [Google Scholar]
- 37.Cavo M, Tacchetti P, Patriarca F, Petrucci MT, Pantani L, Galli M, et al. Bortezomib with thalidomide plus dexamethasone compared with thalidomide plus dexamethasone as induction therapy before, and consolidation therapy after, double autologous stem-cell transplantation in newly diagnosed multiple myeloma: a randomised phase 3 study. Lancet. 2010;376(9758):2075-85 [DOI] [PubMed] [Google Scholar]
- 38.Goldschmidt H, Sonneveld P, Cremer FW, van der Holt B, Westveer P, Breitkreutz I, et al. Joint HOVON-50/GMMG-HD3 randomized trial on the effect of thalidomide as part of a high-dose therapy regimen and as maintenance treatment for newly diagnosed myeloma patients. Ann Hematol. 2003;82(10):654-9 [DOI] [PubMed] [Google Scholar]
- 39.Alexanian R, Barlogie B, Fritsche H. Beta 2 microglobulin in multiple myeloma. Am J Hematol. 1985;20(4):345-51 [DOI] [PubMed] [Google Scholar]
- 40.Tienhaara A, Pulkki K, Mattila K, Irjala K, Pelliniemi TT. Serum immunoreactive interleukin-6 and C-reactive protein levels in patients with multiple myeloma at diagnosis. Br J Haematol. 1994;86(2):391-3 [DOI] [PubMed] [Google Scholar]
- 41.Barlogie B, Smallwood L, Smith T, Alexanian R. High serum levels of lactic dehydrogenase identify a high-grade lymphoma-like myeloma. Ann Intern Med. 1989;110(7):521-5 [DOI] [PubMed] [Google Scholar]
- 42.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107(5):2037-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Y, Reiman T, Li W, Maxwell CA, Sen S, Pilarski L, et al. Targeting aurora kinases as therapy in multiple myeloma. Blood. 2007;109(9):3915-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorgun G, Calabrese E, Hideshima T, Ecsedy J, Perrone G, Mani M, et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115(25):5202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li XC, Schimenti JC. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 2007;3(8):e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shammas MA, Shmookler Reis RJ, Koley H, Batchu RB, Li C, Munshi NC. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma. Blood. 2009;113(10):2290-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97(7):3260-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S, Pramanik D, Mukherjee R, Campbell NR, Elumalai S, de Wilde RF, et al. Molecular determinants of retinoic Acid sensitivity in pancreatic cancer. Clin Cancer Res. 2012;18(1):280-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capello M, Ferri-Borgogno S, Cappello P, Novelli F. alpha-Enolase: a promising therapeutic and diagnostic tumor target. FEBS J. 2011;278(7):1064-74 [DOI] [PubMed] [Google Scholar]
- 50.Naryzhny SN, Lee H. Proliferating cell nuclear antigen in the cytoplasm interacts with components of glycolysis and cancer. FEBS Lett. 2010;584(20):4292-8 [DOI] [PubMed] [Google Scholar]
- 51.Wen XY, Stewart AK, Sooknanan RR, Henderson G, Hawley TS, Reimold AM, et al. Identification of c-myc promoter-binding protein and X-box binding protein 1 as interleukin-6 target genes in human multiple myeloma cells. Int J Oncol. 1999;15(1):173-8 [DOI] [PubMed] [Google Scholar]