Abstract
Peroxidasins represent the subfamily 2 of the peroxidase-cyclooxygenase superfamily and are closely related to chordata peroxidases (subfamily 1) and peroxinectins (subfamily 3). They are multidomain proteins containing a heme peroxidase domain with high homology to human lactoperoxidase that mediates one- and two-electron oxidation reactions. Additional domains of the secreted and glycosylated metalloproteins are type C-like immunoglobulin domains, typical leucine-rich repeats, as well as a von Willebrand factor C module. These are typical motifs of extracellular proteins that mediate protein–protein interactions. We have reconstructed the phylogeny of this new family of oxidoreductases and show the presence of four invertebrate clades as well as one vertebrate clade that includes also two different human representatives. The variability of domain assembly in the various clades was analyzed, as was the occurrence of relevant catalytic residues in the peroxidase domain based on the knowledge of catalysis of the mammalian homologues. Finally, the few reports on expression, localization, enzymatic activity, and physiological roles in the model organisms Drosophila melanogaster, Caenorhabditis elegans, and Homo sapiens are critically reviewed. Roles attributed to peroxidasins include antimicrobial defense, extracellular matrix formation, and consolidation at various developmental stages. Many research questions need to be solved in future, including detailed biochemical/physical studies and elucidation of the three dimensional structure of a model peroxidasin as well as the relation and interplay of the domains and the in vivo functions in various organisms including man.
Keywords: Peroxidasin, Peroxidase, Immunoglobulin domain, Leucin-rich repeat domain, von Willebrand factor C
Introduction
Recently, we have reconstructed the phylogenetic relationships of the main evolutionary lines of the mammalian heme-containing peroxidases myelo-peroxidase (MPO), eosinophil peroxidase (EPO), lactoperoxidase (LPO), and thyroid peroxidase (TPO) [1]. Based on their occurrence and the fact that two main enzymatic activities are related to these metalloproteins, the peroxidase-cyclooxygenase superfamily was defined and shown to occur in all kingdoms of life [1]. Seven clearly separated subfamilies were found with chordata heme peroxidases comprising subfamily 1. Its representatives are involved in the innate immune system (MPO, EPO, and LPO) as well as hormone biosynthesis (TPO). Mature vertebrate peroxidases consist of only one (monomeric EPO and LPO) or two (homodimeric MPO and TPO) glycosylated, mainly α-helical domains with one autocatalytically modified heme per domain [2]. Well characterized chordata peroxidases evolved from multidomain enzymes called peroxidasins (subfamily 2) [1]. These peculiar proteins are found in vertebrates and invertebrates and have – in addition to a heme-containing domain of high homology with chordata peroxidases – flanking domains which are known to be important for protein–protein interaction or cell adhesion. These include leucine-rich regions, immunoglobulin-like domains, as well as a von Willebrand factor C (VWC) module.
In 1994, the first peroxidasin was described and found to occur in hemocytes of Drosophila [3]. Hemocytes are migratory cells present in the hemolymph of insects and are involved in (targeted) consolidation of extracellular matrix as well as in intracellular phagocytosis of apoptotic cells or foreign material (e.g., pathogens). In the following years, homologous genes were detected among numerous ecdysozoan [4] and almost all deuterostomian genomes [5] [6], implicating their importance in both invertebrate and vertebrate physiology. The human homologue of this subfamily, first cloned as a shorter mRNA fragment, was originally identified as a p53-responsive gene [7]. Finally, on a protein basis, the first human peroxidasin was detected in human colon cancer cells, but later also in squamous lung carcinoma cells, where it was initially named melanoma gene 50 (MG50) [8]. In the human genome, two different peroxidasins are encoded [1] and were originally designated as vascular peroxidase (VPO) 1 and 2 (VPO 2 is sometimes referred to as cardiac peroxidase), in relation to their high expression in the vascular system or heart [9]. However, both enzymes are multidomain proteins of subfamily 2 of the peroxidase-cyclooxygenase superfamily and thus should be named systematically human peroxidasin 1 (hsPxd01) and human peroxidasin 2 (hsPxd02). In recent years, several papers were published that report expression patterns [9–13], localization [9] [14], enzymatic activities [9], and physiological relevance of peroxidasins [15–18], but still many open questions remain regarding the relation between structure and function and the putative role(s) in the extracellular matrix formation and the innate immune system of invertebrates and vertebrates.
Here, we have reconstructed the molecular evolution of peroxidasins. We have analyzed the multidomain structure of the two main groups, i.e., invertebrate and vertebrate peroxidasins, and discuss the function of the enzymatic domain based on the knowledge about catalysis of human peroxidases [2]. Finally, we critically relate these facts to biochemical and biological features of these proteins.
Results and Discussion
Phylogenetic Reconstruction
Genes encoding peroxidasins are found in vertebrates as well as in invertebrates. For the phylogenetic reconstruction, all available peroxidasin sequences (i.e., 106 in December 2011) were aligned and analyzed by three different methods. Fig. 1 depicts the reconstructed unrooted tree obtained by the neighbor-joining (NJ) method; however, very similar trees were achieved by the minimum-evolution (ME) and maximum-likelihood (ML) methods. A clear separation into five peroxidasin clades is evident, with Clades 1–4 representing invertebrate and Clade 5 vertebrate oxidoreductases (Fig. 1). For the phylogenetic analysis, the full gene length including all domains was used; however, analyses based on only the peroxidase domain gave a very similar evolutionary tree (results not shown1).
Fig. 1.
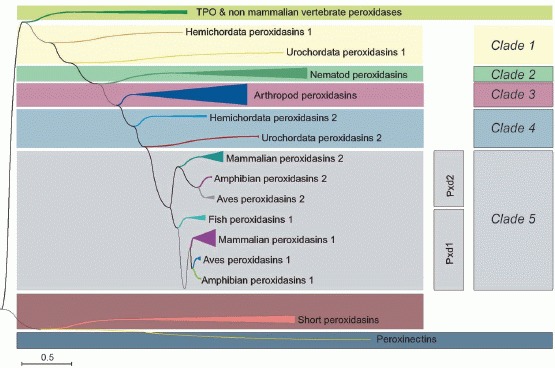
Reconstructed unrooted phylogenetic tree of selected representatives of chordata peroxidases peroxinectins, and the peroxidasin subfamily, obtained by the neighbor joining (NJ) method. For the reconstruction, the complete peroxidasin sequences encoding all domains were used. The abbreviations of the analyzed sequences are identical with those used in the PeroxiBase and are listed in the Table.
Closely related with subfamily 3 (i.e., peroxinectins) of the peroxidase-cyclo-oxygenase superfamily [1], and thus somehow linking subfamilies 2 and 3, are short peroxidasins that lack all characteristic peroxidasin domains except the peroxidase domain. So far (December 2011), only eight genes are known and all occur in nematodes. Since these putative proteins do not show the typical multidomain structure of neither peroxinectins nor peroxidasins, they are not designated as a distinct clade. Echinozoan and ecdysozoan peroxinectins have integrin-binding motifs in addition to the peroxidase domain [1] [18] and, so far, no vertebrate representative was found.
At the origin of evolution (Clade 1) of multidomain peroxidasins, proteins from hemichordates and urochordates are found, which represent the closest neighbors of chordata peroxidases (Fig. 1). Clearly separated from Clade 1 are nematode peroxidasins (Clade 2), arthropod peroxidasins (Clade 3), and a second group of hemichordate and urochordate proteins (Clade 4; Fig. 2).
Fig. 2.
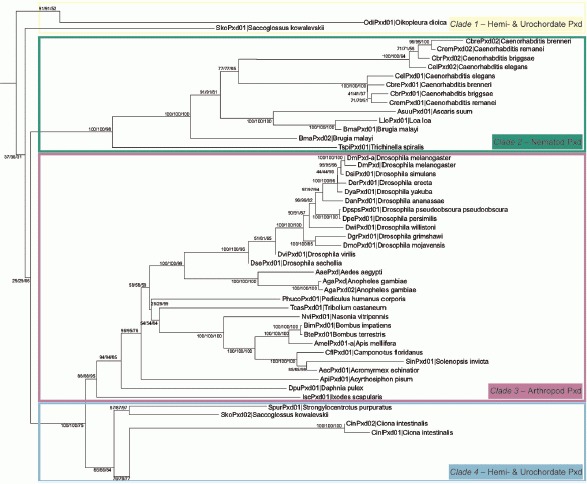
Detail of the reconstructed phylogenetic tree of peroxidasins showing invertebrate peroxidasin evolution (Clades 1-4). Bootstrap values in the nodes were obtained from neighbor-joining (NJ), minimum-evolution (ME), and minimum-likelihood (ML) methods, respectively.
Vertebrate peroxidasins (Clade 5) can be divided into two branches, namely peroxidasins 1 (Pxd1) and peroxidasins 2 (Pxd2), sometimes referred as peroxidasin-like proteins. In each branch, a clear segregation in fish, amphibian, bird, and mammalian proteins is obvious (Fig. 3). In the human genome, two peroxidasins are encoded that share a sequence identity of 63%. Peroxidasin 1 from Homo sapiens (hsPxd01) is located on chromosome 2p25, whereas human peroxidasin 2 (hsPxd02) is located on chromosome 8q11 [9]. A closer look to Fig. 3 depicts the presence of a third human peroxidasin, which is almost identical to hsPxd02 – except for five amino acids – and might be a splicing variant.
Fig. 3.
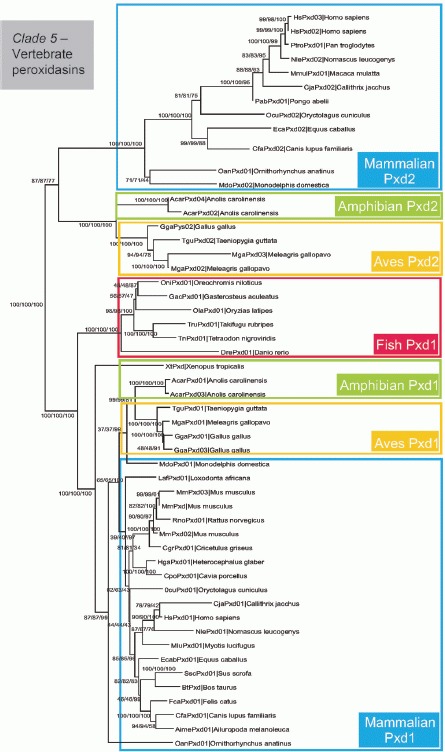
Detail of the reconstructed phylogenetic tree of peroxidasin evolution showing vertebrate peroxidasins (Clade 5). Bootstrap values in the nodes were obtained from NJ, ME, and ML methods, respectively.
Domain Assembly and Architecture
So far, no three-dimensional structure of a peroxidasin is known, but comparative sequence analysis and secondary-structure prediction clearly indicate the presence of four different domains of distinct assembly and architecture. Fig. 4 shows the domain assembly in invertebrate and vertebrate peroxidasins. In the latter, and thus also in both human peroxidasins, the primary transcript has a signal peptide for extracellular secretion followed by five leucine-rich repeats with N-terminal and C-terminal capping motifs, four successive immunoglobulin domains, one heme-binding peroxidase domain, and a C-terminal von Willebrand factor C module (Fig. 4).
Fig. 4.
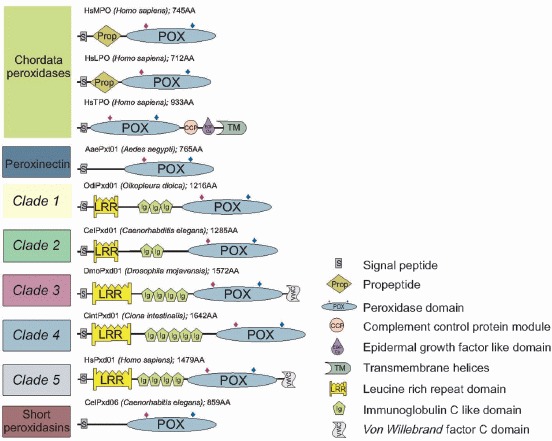
Schematic presentation of subunit assembly of representatives of the chordata heme peroxidases, peroxinectins, short peroxidasins, and the five clades of the peroxidasin subfamily. The domain architectures were adopted from the Pfam-Database at the Sanger Institute (http://www.sanger.ac.uk) and Prosite Database at ExPASy, the SIB Bioinformatics Resource Portal (http://prosite.expasy.org). The rhombuses indicate the position of the distal and proximal histidine in the peroxidase domain.
Clade 1 and nematode peroxidasins (Clade 2) vary in the number of both the leucine-rich repeats as well as immunoglobulin motifs. Additionally, they do not contain a C-terminal von Willebrand factor C, suggesting that this feature is phylogenetically the newest addition to these multidomain proteins. Arthropod peroxidasins (Clade 3) also show variable numbers of leucine-rich repeats and immunoglobulin domains, but the majority of the putative proteins seem to contain already a C-terminal von Willebrand factor C, whereas in the second group of hemichordate and urochordate peroxidases (Clade 4) the von Willebrand factor C is absent.
Up to now, we have no detailed knowledge about the relation between domain assembly and structure and their physiological relevance in peroxidasins. The only enzymatic domain is the heme-containing peroxidase domain and its sequence is critically analyzed below. All the other domains are found in thousands of proteins in various contexts.
Leucine-Rich Repeats
Leucine-rich repeats (LRR) occur in numerous proteins and all of them appear to be involved in protein–protein interactions including cell adhesion and signal transduction, extracellular matrix assembly, platelet aggregation, neuronal development, RNA processing, and immune response [19]. A striking example for the functional variety of LRRs is the adaptive immune system of jawless fishes. They possess variable lymphocyte receptors (VLR) built from proteins that are completely unrelated to immunoglobulins. Interestingly, they are reported to be generated from one or two germ lines of VLR genes by genomic rearrangement of flanking LRR cassettes [20].
Seven subclasses of LRR motifs differing in length and consensus sequence of the variable segments (typical, RI-like, CC, PS, SDS22-like, bacterial, and TpLRR) have been proposed [21]. In peroxidasins, only typical LRR cassettes are found. This motif exhibits a solenoid structure where each repeat represents a turn that is typically 20–30 amino acids long. It contains a conserved eleven-residue hallmark sequence (LxxLxLxxNxL) corresponding to a β-sheet that forms the concave side of the solenoid [19]. By contrast, the convex side shows a greater variability and may consist of α-helical structures including 310 helices, polyproline II conformation, and β-turns depending on the amino acid composition and length of the variable segment [22]. The conserved leucines as well as other mainly hydrophobic amino acids are arranged to a characteristic tightly packed structure forming the inner core of the solenoid and contribute to the stability of the LRR domain. Many ligand–LRR protein complexes depict the ligand more or less surrounded by the concave surface of the parallel β-sheets of the LRR domain, however, ligand interactions with the convex side have also been observed. Generally, ligand binding does not induce major rearrangements in the LRR structure, since the free and bound forms superimpose [19].
The hydrophobic core of the LRR domains may be protected from opening by N-terminal and C-terminal cysteine-rich flanking regions, which are often found in extracellular LRR proteins including peroxidasins [21]. The N-terminal capping motif (LRRNT) is described as a single β-strand, antiparallel to the main β-sheet, followed by a short LRR of 20 or 21 residues, whereas the C-terminal capping motif (LRRCT) possesses an a-helix covering the hydrophobic core of the last LRR [21]. Fig. 5, a, shows a model of the LRR domain of hsPxd01 that is built from five tandem typical LRRs framed by LRRNT and LRRCT.
Fig. 5.
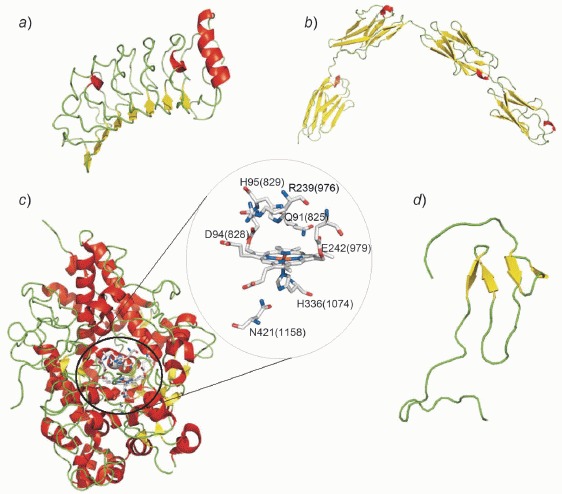
Three-dimensional structures of domains of human peroxidasin 1 (hsPxd01). a) Model of leucine-rich repeat (LRR) domain with N- and C-terminal capping motifs. b) Model of C-like immunoglobulin domain (Ig). c) Overall structure of homologous goat LPO (PDB code 2R5L) with detailed presentation of catalytic active site residues. Numbering corresponds to the mature LPO, whereas numbering in parentheses corresponds to full length hsPxd01, respectively. d) Model of von Willebrand factor C (VWC). All models have been calculated using the ESyPred3D server.
Immunoglobulin Domains
Since peroxidasins in addition contain immunoglobulin (Ig) domains, they are also accounted to the heterogenic immunoglobulin superfamily (IgSF). This major group of proteins differs in tissue distribution, amino acid composition, and biological role, but all representatives possess at least one structurally discrete domain of ca. 100 amino acids exhibiting the Ig-fold [23]. The differentiation of immunoglobulin domains is based on the composition of the two β-sheets that form the sandwich-like fold. The constant domains (C-domains) are comprised of seven strands and the variable domains (V-domains) of eight, nine, or ten strands [24]. The standardized IMGT (International ImMunoGeneTics Information System) discriminates immunoglobulin domains according to following categories: C-domains (including immunoglobulins and T-cell receptors of all jawed vertebrates), C-like domains (proteins other than immunoglobulins or T-cell receptors), V-domains (immunoglobulins and T-cell receptors with V-J-region and V-D-J-region) and V-like domains (proteins other than immunoglobulins or T-cell receptor with V-J-region and V-D-J-region) [25]. According to this systematics, peroxidasins have C-like Ig-domains.
As mentioned before, the Ig-fold is a wide-spread protein motif with various biological functions, although the common feature seems to be cell adhesion and pattern recognition. An example for a structural function of immunoglobulin domains are the Ig-tandems of titin, a myofilament protein and the largest mammalian protein. Von Castelmur et al. [26] determined the crystal structure of a fragment from the skeletal I-band of soleus titin. This structure was also used as a template for tertiary-structure modelling of the Ig-domains of hsPxd01 (Fig. 5, b).
Von Willebrand Factor Type C
The C-terminal von Willebrand factor type C (VWC) module is also referred as chordin-like, cysteine-rich (CR) repeat. It contains ca. 60–80 amino acids and is defined by a consensus sequence of ten cysteines [27]. The name originates from the five types of structural domains comprising the von Willebrand factor (VWF), a multimeric blood glycoprotein that binds and stabilizes clotting factor VIII and mediates platelet adhesion [28]. This motif has been identified in more than 500 extracellular matrix proteins including CCN (cysteine-rich protein 61, connective tissue growth factor proteins, nephroblastoma overexpressed gene), procollagen, thrombospondin, glycosylated mucins, and neuralins with varying copy numbers [29]. Peroxidasins contain only one C-terminal copy, similar to the CCN proteins.
For most VWC modules, the cellular role has still be to investigated [29], however, binding and regulating bone morphogenetic proteins (BMPs) and transforming the tissue growth factor beta (TGF-β) are the two most common functions attributed to the VWC domain [30]. Moreover, the VWC domain might be involved in oligomerization of proteins [28].
The three-dimensional structure of a prototypical chordin-like, cysteine-rich repeat from collagen IIAwas determined by O'Leary et al. [27]. They found that the VWC domain exhibits a two-subdomain architecture connected via a short linker region. A model of the VWC domain of hsPxd01 is shown in Fig. 5, d. The C-terminal subdomain adopts a rather irregular and flexible structure, whereas the N-terminal subdomain of peroxidasin consists of two double stranded antiparallel β-sheets, in contrast to the N-terminal subdomain of collagen IIA that contains one double-stranded and one triple-stranded antiparallel β-sheet. Interestingly, there is a structural similarity between the N-terminal subdomain of the VWC module and the fibronectin type 1 (FN1) domain. Since FN1 domains are only found in vertebrates, in contrast to VWCs, which have been identified in all eukaryotes, an evolutionary relationship between these two domains might be possible [27].
The Enzymatic Heme Domain
Subfamilies 1–3 of the peroxidase-cyclooxygenase superfamily contain one enzymatic peroxidase domain [1]. Multiple sequence alignment reveals highly conserved regions (Figs. 6 and 7). They correspond to functionally and structurally essential motifs, as known from mammalian peroxidases. The secondary structure of mammalian peroxidases as well as of the peroxidase domain of peroxidasins is predominantly a-helical with a central heme-containing core composed of five helices (Fig. 5, c).
Fig. 6.
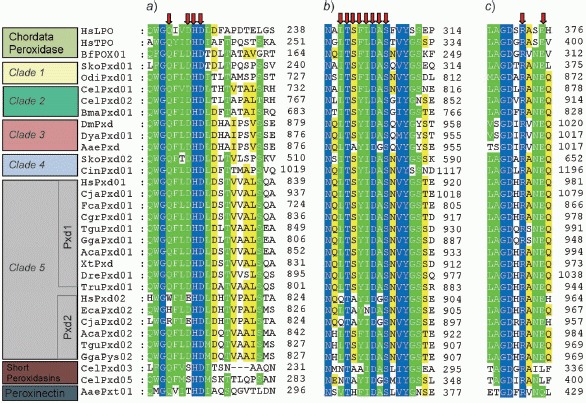
Selected parts of the multiple sequence alignment of 32 sequences from members of the peroxidasin subfamily as well as representatives of chordata peroxidases, peroxinectins, and short peroxidasins. Catalytically important residues of the distal heme cavity are highlighted by arrows. a) Area around the essential distal Asp-His-Asp as well as Gln (compare with Fig. 5, c). b) Sequence area around the known Ca2+-binding site in mammalian peroxidases. c) Area around the catalytic distal Arg and Glu, the latter being involved in heme to protein ester linkage in mammalian peroxidases.
Fig. 7.
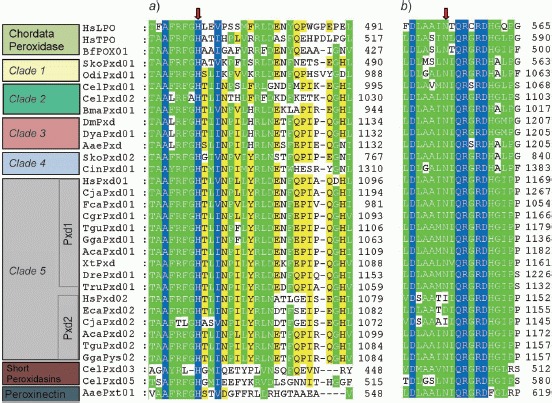
Selected parts of the multiple sequence alignment of 32 sequences from members of the peroxidasin subfamily as well as representatives of chordata peroxidases, peroxinectins, and short peroxidasins. Catalytically important residues of the proximal heme cavity are depicted. Sequence areas around a) the proximal histidine and b) its H-bonding partner asparagine are shown.
Both the distal and proximal histidines in the heme cavity as well as the corresponding H-bonding partners are located within α-helices. Essential distal residues in mammalian peroxidases are Gln91, His95, and Arg239 (Fig. 5, c; numbering corresponds to mature lactoperoxidase and numbering in parentheses corresponds to hsPxd01). The peroxidase-typical distal pair His-Arg is found in all heme peroxidases from both superfamilies (peroxidase-cyclooxygenase and peroxidase-catalase super-family) [1] and is also fully conserved in all peroxidasins (Fig. 6). This pair is important in the heterolytic cleavage of H2O2 [2]. In mammalian peroxidases, Gln91 (Fig. 5, c) is involved in the maintenance of the distal H-bond network as well as halide binding [2]. This essential glutamine, which is part of a conserved motif ‘W/F-G-Q-F’, is present in almost all analyzed peroxidasin sequences, except short peroxidasins and group Pxd2 of Clade 5 enzymes including hsPxd02 (Fig. 6).
The second adjacent conserved motif in mammalian peroxidases, which is also fully conserved in peroxidasins and includes the distal His95, is ‘Asp-His-Asp’. Asp94 is known to be involved in ester-bond formation with the OHCH2 group at C(5) of pyrrole ring C of the heme (Fig. 5, c), and it is found in all peroxidasins, except group Pxd2 of Clade 5 and short peroxidasins. Regarding Asp96, its role as a ligand of Ca2+ is well established. The role of the distal Ca2+-binding site could be to stabilize both the distal heme cavity architecture as well as to mediate the assembly of the mature peroxidases [2]. In both lactoperoxidase and myeloperoxidase, the Ca2+-binding site has a typical pentagonal bipyramidal coordination geometry [2] and the cation is liganded by several highly conserved residues. One is the already mentioned Asp96 and the other ligands are part of a loop that consists of eight residues, i.e., ‘Leu-Thr-Ser-Phe-Val-Asp-Ala-Ser’ (amino acid number 333–340 of the mature LPO), and is found in all chordata peroxidases. This motif is also present in all peroxidasins, although some mutations, especially in short peroxidasins and group Pxd2 of Clade 5, are observable.
All mammalian peroxidases have their prosthetic group covalently bound to the protein and, besides Asp94 mentioned above, a conserved Glu242 forms the second ester bond. Except short peroxidasins, all other members of this protein family have this glutamate residue fully conserved (Fig. 6).
Structurally and functionally important motifs on the proximal side of mammalian peroxidases include the proximal histidine (His336) and its H-bonding partner asparagine (Asn421; Fig. 5, c). Both residues govern the heme-iron reactivity by controlling the electron density at the metal [2]. Inspection of Fig. 7 again demonstrates that Clades 1–4 and group Pxd1 of Clade 5 are chordata peroxidase-like, whereas in group Pxd2 of vertebrate peroxidasins variable amino acids (Ile, Asp, Asn) are found at the corresponding position of Asn421.
Summing up, sequence analysis of the enzymatic domain of peroxidasins suggests a similar function as in vertebrate peroxidases. All catalytically relevant residues (with exception of the MPO-specific methionine that is responsible for the third covalent linkage between the autocatalytically modified heme and the protein) are also found in the peroxidase domain of most of the peroxidasins. This suggests similar enzymatic features, including catalysis of one- and two-electron oxidation reactions, and some preliminary biochemical data support this observation [9] [14], although the reported activities (e.g., tyrosine and halide oxidation) seem to be significantly lower compared to mammalian peroxidases. Only short peroxidasins and group Pxd2 of vertebrate peroxidases (including hsPxd02) show alternative amino acids at structurally and functionally important positions, and it is not clear at the moment whether these enzymes are functionally comparable with peroxidases.
Physiological Roles
So far, only a handful of studies have been published that might give an indication of the in vivo function of peroxidasins. Focusing on the enzymatic domain only, one could speculate about similar biological roles as reported for the closely related and homologous chordata peroxidases including MPO, EPO, and LPO. The latter are involved in innate immune-defense reactions against invading pathogens and are stored in high concentration in granules of neutrophilic (MPO) or eosinophilic leukocytes (EPO) that are recruited to sites of pathogen invasion and inflammation [31] [32], whereas secreted lactoperoxidase acts as extracellular bactericidal agent in exocrine secretions [33]. These peroxidases catalyze the production of hypohalous acids or hypothiocyanate from H2O2 and (pseudo-) halide anions, but also act as efficient one-electron oxidants [2].
Another physiological role of chordata peroxidases is cell adhesion and formation of extracellular matrix (ECM). It has been demonstrated that the extracellular oxidants generated by cationic mammalian peroxidases (e.g., MPO) can have both damaging and protective effects on the ECM, including inactivation of metalloproteinases [34] [35] and formation of dityrosine crosslinks [36].
Among peroxidase-domain containing proteins of the same superfamily, also peroxinectins (subfamily 3 of the peroxidase-cyclooxygenase superfamily) might give some indications about the functionality of peroxidasins, although from this subfamily (that has no vertebrate representative) only very few reports can be found in the literature. These metalloenzymes seem to be involved in the promotion of cell–ECM adhesion via their integrin-binding motifs [37]. Interestingly, also MPO has been shown to promote integrin-mediated adhesion of neutrophils [38].
Regarding the overall structure and size as well as domain assembly, peroxidasins are the most complex representatives of peroxidase-domain-containing enzymes of the peroxidase-cyclooxygenase superfamily. Sequence analysis clearly suggests an enzymatic functionality similar to LPO and EPO (except in short peroxidasins and group Pxd2 of Clade 5). The motifs typical of extracellular proteins (Igs, LRRs, and VWC) might contribute to association with ECM. Both the roles in innate immune defense and extracellular matrix consolidation are often addressed in various contexts in the literature.
As already mentioned, the first peroxidasin was identified in 1994 in Drosophila melanogaster (Clade 3 peroxidasins; Fig. 2) and has shown to be associated with the function of insect hemocytes and plasmatocytes [3] [7]. Similar to neutrophils, hemocytes are migratory cells that phagocytose foreign and dead cells and deposit ECM. Peroxidasin expression in Drosophila is widely used as a molecular marker for the early hemocyte lineage [39], and hemocytes are crucial for a morphogenetic event known as condensation of the ventral nerve cord (VNC). If hemocyte migration is blocked, deposition of ECM components, including peroxidasin and type IV collagen, does not occur, leading to failure of ventral nerve cord condensation [40]. Drosophila peroxidasin was reported to be a homotrimer that catalyzes H2O2-mediated tyrosine and iodide oxidation [3].
The second invertebrate model organism in which the in vivo role of peroxidasin was studied is Caenorhabditis elegans [15]. Two orthologs are found in C. elegans (CelPxd01 and CelPxd02; Fig. 2) with apparently antagonistic functions. CelPxd02 was identified upon screening for mutants defective in embryonic worm development. It was found to be essential for specific stages of morphogenesis and epidermal muscle attachment as well as postembryonically for basement membrane integrity [15]. The peroxidase activity (which – based on sequence analysis – should be LPO-like) was shown to be responsible for these developmental roles. In adult worms, loss of CelPxd02 promotes regrowth of axons after injury, providing evidence that C. elegans extracellular matrix can play an inhibitory role in axon regeneration. By contrast, loss of CelPxd01 did not cause developmental effects. Moreover, CelPxd02 mutant phenotypes were suppressed by loss of function of CelPxd01 and exacerbated by its overexpression, suggesting antagonistic roles.
The first study on a (non-mammalian) vertebrate peroxidasin was performed on group 1 peroxidasin (Fig. 3) from Xenopus tropicalis in 2005 [10]. This representative is expressed in several distinct tissues during early development, including the neural tube and the tail-forming region, and has also a role in modifying extracellular matrix components necessary in morphogenesis, again suggesting a role of the peroxidase domain in catalyzing one-electron oxidation reactions.
Investigations on human peroxidasins started in 1999 [7]. It was demonstrated that besides the full-length mRNA of hsPxd01, also a shorter 4.5 kb mRNA version occurs in human colon EB1 cancer cells undergoing p53-dependent apoptosis. Characterization of the smaller mRNA showed that the N-terminal part of hsPxd01 including the signal peptide was not present, whereas the C-terminal portion including the peroxidase domain was present. The authors hypothesized that the protein missing a signal sequence would accumulate in the cytoplasm, and since the peroxidase domain is intact, it could potentially increase the cellular production of reactive oxygen species (ROS), which are powerful inducers of apoptosis by inactivation of the p53 tumor suppressor protein [7]. Based on the knowledge of catalysis of homologous mammalian peroxidases [2], the effect on p53 could be mediated by hypohalous acids or hypothiocyanate released from peroxidasin rather than by activated oxygen species, since peroxidases consume H2O2 and do not release O2•− at reasonable rates.
Finally, the cloning of human and mice peroxidasins and initial characterization of hsPxd01 were reported [9]. Heme-containing hsPxd01was recombinantly expressed in HEK cells, and the prosthetic group of hsPxd01 was shown to be covalently attached to the protein via two ester bonds very similar to LPO or EPO [9]. The protein was shown to exhibit both peroxidase activity (tetramethylbenzidine oxidation) as well as halogenation activity (two-electron oxidation of Cl−). However, the reported activities were very low compared to mammalian peroxidases. The finding of chlorination activity at neutral pH, which so far was only attributed to MPO, is peculiar, since hsPxd01 cannot form the MPO-typical (e− withdrawing) sulfonium-ion linkage and thus this reactivity must be addressed in future biochemical studies.
Based on its halogenation activity and localization, the role of hsPxd01 in low-density lipoprotein oxidation and endothelial cell apoptosis was investigated [17]. It was demonstrated that expression of hsPxd01 in endothelial cells correlates with LDL oxidation in a time- and concentration-dependent manner. Activity of hsPxd01 was shown to be dependent on NADPH-oxidase activity, which initially forms O2•− that dismutates to H2O2 necessary to initiate both peroxidase and halogenation activity.
Peterfi et al. [14] demonstrated the secretion of human hsPxd01 as well as the formation of peroxidasin-containing, fibril-like structures by differentiated myofibroblasts. Since myofibroblasts appear during wound healing, it is tempting to speculate that the peroxidase domain functions in an antimicrobial manner, whereas the other domains could participate in the ECM formation. Stabilization of the ECM could also be achieved by the enzymatic formation of dityrosine crosslinks.
In another study [18], homozygous mutations in the gene encoding hsPxd01were associated with a spectrum of ocular anterior segment dysgenesis phenotypes. Although the precise effect of these mutations is unknown, the pathogenic impact is obvious. Finally, it was demonstrated that hsPxd01 is also expressed in various cancer cells, including breast cancer, melanoma, colon cancer, and metastatic gilomas [8] [11] [12]. Hence, hsPxd01 seems to be a signature gene of heme oxygenase-1 (HO-1) [12], an enzyme associated with tumor angiogenesis. A loss of the adhesion promoting effect of HO-1 in hsPxd01-silenced cells was observed. This suggests also a role of hsPxd01 in cell adhesion and invasion as well as in the transition of benign tumors to invasive and malignant cancers [12].
Summing up, peroxidasins are multidomain heme peroxidases with a substrate spectrum similar to mammalian peroxidases. Based on sequence analysis and reported biochemical data, they perform one-electron oxidation reactions including tyrosine oxidation as well as two-electron oxidation reactions of (pseudo-) halides. Hypochlorous acid formation is questionable and must be addressed in future investigations. In any case, physiological studies have shown that the enzymatic domain is essential for the in vivo activity and might contribute to extracellular matrix consolidation as well as antimicrobial defense. The additional domains (Igs, LRRs, and VWC) are typical for extracellular proteins and may help in targeting the peroxidase to its site of action. However, a distinct function of the individual domains and their relation to the reported in vivo roles has not been demonstrated so far. Clearly, more detailed biochemical and biophysical studies as well as the elucidation of the three-dimensional structure of peroxidasins are necessary. These studies must include invertebrate and both groups of vertebrate peroxidasins, since – as sequence analysis suggests – they most probably differ in catalysis and, in consequence, in function.
This project was supported by the Austrian Science Fund and the doctoral program BioToP – Biomolecular Technology of Proteins (FWF W1224).
Experimental Part
Data Mining
All currently (December 2011) available 106 peroxidasin sequences were collected from public databases (Uniprot, NCBI, PeroxiBase, Ensembl). Selected representatives of non-mammalian vertebrate peroxidase (BbePOX01 Branchiostoma floridae, BbePOX01 Branchiostoma belcheri, HrPOX Halocynthia roretzi), HsTPO Human thyroid peroxidase, and Aedes aegypti peroxinectin (AaePxt01) sequences were retrieved from the PeroxiBase database [41] and used as an outgroup. Sequence data are given in the Table.
Multiple Sequence Alignment
A multiple sequence alignment of the 112 enzymes was constructed with ClustalW [42] with following parameters: for pairwise alignment, gap opening penalty 9 and gap extension penalty 0.1; for multiple alignment, gap opening penalty 8 and gap extension penalty 0.2. Gonnet protein weight matrix was used and the gap separation distance was set to 4. The delay divergent cut off was defined with 25%. The output was displayed with Genedoc [43].
Phylogeny Reconstruction
Distance Method. The multiple sequence alignment of the 112 selected enzymes was subjected to the neighbor-joining (NJ) method of the MEGA5 package [44] with the Jones–Taylor–Thornton (JTT) model of amino acid substitution, pairwise deletion of gaps, and homogenous pattern among lineages. The test of phylogeny was conducted with the bootstrap method, by setting the bootstrap replications to 1000. The optimized γ parameter for this set was determined as γ = 1.75.
Table Abbreviations of the 106 Peroxidasin Sequences Used in This Study
| Name | Entry IDa) | Organism | Lenght (aa) |
|---|---|---|---|
| AecPxd01 | 10036 | Acromyrmex echinatior | 1305 |
| ApiPxd01 | 7638 | Acyrthosiphon pisum | 1669 |
| AaePxd | 3368 | Aedes aegypti | 1528 |
| AcarPxd01 | - | Anolis carolinensis | 1482 |
| AcarPxd02 | - | Anolis carolinensis | 1422 |
| AcarPxd03 | - | Anolis carolinensis | 1436 |
| AcarPxd04 | - | Anolis carolinensis | 1374 |
| AgaPxd | 4043 | Anopheles gambiae | 1559 |
| AgaPxd02 | 7639 | Anopheles gambiae | 1514 |
| AimePxd01 | 7610 | Ailuropoda melanoleuca | 1466 |
| AmelPxd01-a | 3671 | Apis mellifera | 1293 |
| AsuuPxd01 | - | Ascaris suum | 1259 |
| BmalPxd02 | 7644 | Brugia malayi | 999 |
| BmalPxd01 | 7642 | Brugia malayi | 1149 |
| BimPxd01 | - | Bombus impatiens | 1290 |
| BtePxd01 | - | Bombus terrestris | 1290 |
| BtPxd | 3356 | Bos taurus | 1475 |
| CbrePxd01 | Caenorhabditis brenneri | 1280 | |
| CbrePxd02 | - | Caenorhabditis brenneri | 1382 |
| CbrPxd01 | 4289 | Caenorhabditis briggsae | 1288 |
| CbrPxd02 | 4288 | Caenorhabditis briggsae | 1355 |
| CbrPxd03b) | 4287 | Caenorhabditis briggsae | 658 |
| CbrPxd04b) | 4286 | Caenorhabditis briggsae | 728 |
| CbrPxd05b) | 4285 | Caenorhabditis briggsae | 724 |
| CbrPxd06b) | 4284 | Caenorhabditis briggsae | 864 |
| CelPxd01 | 3359 | Caenorhabditis elegans | 1285 |
| CelPxd02 | 4103 | Caenorhabditis elegans | 1328 |
| CelPxd03b) | 4139 | Caenorhabditis elegans | 655 |
| CelPxd04b) | 4142 | Caenorhabditis elegans | 729 |
| CelPxd05b) | 4141 | Caenorhabditis elegans | 724 |
| CelPxd06b) | 4140 | Caenorhabditis elegans | 859 |
| CremPxd01 | - | Caenorhabditis remanei | 1317 |
| CremPxd02 | - | Caenorhabditis remanei | 1360 |
| CflPxd01 | 7646 | Camponotus floridanus | 1303 |
| CfaPxd01 | 10035 | Canis lupus familiaris | 1588 |
| CfaPxd02 | 3358 | Canis lupus familiaris | 1429 |
| CjaPxd01 | 7609 | Callithrix jacchus | 1577 |
| CjaPxd02 | 7617 | Callithrix jacchus | 1456 |
| CpoPxd01 | - | Cavia porcellus | 1479 |
| CgrPxd01 | - | Cricetulus griseus | 1475 |
| CintPxd01 | 7647 | Ciona intestinalis | 1642 |
| CintPxd02 | - | Ciona intestinalis | 1279 |
| DrPxd01 | 7648 | Danio rerio | 1530 |
| DpulPxd01 | - | Daphnia pulex | 1262 |
| DanPxd01 | 7649 | Drosophila ananassae | 1531 |
| DerPxd01 | 7650 | Drosophila erecta | 1526 |
| DgrPxd01 | 7651 | Drosophila grimshawi | 1534 |
| DmPxd | 3370 | Drosophila melanogaster | 1535 |
| DmPxd-a | 3369 | Drosophila melanogaster | 1527 |
| DmoPxd01 | 7652 | Drosophila mojavensis | 1572 |
| DpePxd01 | 7653 | Drosophila persimilis | 1534 |
| DpspsPxd01 | 7654 | Drosophila pseudoobscura pseudoobscura | 1529 |
| DsePxd01 | 7655 | Drosophila sechellia | 880 |
| DsiPxd01 | 7656 | Drosophila simulans | 1528 |
| DviPxd01 | 7657 | Drosophila virilis | 892 |
| DwiPxd01 | 7658 | Drosophila willistoni | 1540 |
| DyaPxd01 | 7659 | Drosophila yakuba | 1528 |
| EcabPxd01 | 5825 | Equus caballus | 1431 |
| EcabPxd02 | 7625 | Equus caballus | 1466 |
| FcaPxd01 | 7692 | Felis catus | 1293 |
| GgaPxd01 | 4049 | Gallus gallus | 1456 |
| GgaPxd02 | 7673 | Gallus gallus | 1338 |
| GacPxd01 | 7682 | Gasterosteus aculeatus | 1440 |
| HglPxd01 | - | Heterocephalus glaber | 1412 |
| HsPxd01 | 3355 | Homo sapiens | 1479 |
| HsPxd02 | 5398 | Homo sapiens | 1463 |
| HsPxd03 | 5827 | Homo sapiens | 1463 |
| IsPxd01 | 7663 | Ixodes scapularis | 1111 |
| LloPxd01 | - | Loa loa | 1236 |
| LafPxd01 | - | Loxodonta africana | 1475 |
| MmulPxd01 | 7616 | Macaca mulatta | 1413 |
| MgaPxd01 | - | Meleagris gallopavo | 1459 |
| MgaPxd02 | - | Meleagris gallopavo | 1342 |
| MgaPxd03 | - | Meleagris gallopavo | 1374 |
| MdoPxd01 | 5814 | Monodelphis domestica | 1627 |
| MdoPxd02 | 5830 | Monodelphis domestica | 1542 |
| MmPxd01 | 3357 | Mus musculus | 1475 |
| MmPxd02 | 7680 | Mus musculus | 1431 |
| MmPxd03 | 7681 | Mus musculus | 1379 |
| MluPxd01 | - | Myotis lucifugus | 1133 |
| NviPxd01 | 7676 | Nasonia vitripennis | 1240 |
| NlePxd01 | - | Nomascus leucogenys | 1747 |
| NlePxd02 | - | Nomascus leucogenys | 1463 |
| OdiPxd01 | - | Oikopleura dioica | 1216 |
| OanPxd01 | 5826 | Ornithorhynchus anatinus | 1469 |
| OanPxd02 | 7674 | Ornithorhynchus anatinus | 1218 |
| OniPxd01 | - | Oreochromis niloticus | 1462 |
| OcuPxd01 | 7637 | Oryctolagus cuniculus | 1411 |
| OcuPxd02 | 7624 | Oryctolagus cuniculus | 1467 |
| OlaPxd01 | 7686 | Oryzias latipes | 1483 |
| PtroPxd01 | 5828 | Pan troglodytes | 1463 |
| PhucoPxd01 | 7677 | Pediculus humanus corporis | 1266 |
| PabePxd01 | 7623 | Pongo abelii | 1300 |
| RnoPxd01 | 7664 | Rattus norvegicus | 1475 |
| SkoPxd01 | 7615 | Saccoglossus kowalevskii | 794 |
| SkoPxd02 | 7622 | Saccoglossus kowalevskii | 1055 |
| SinPxd01 | - | Solenopsis invicta | 1299 |
| SpurPxd01 | 4101 | Strongylocentrotus purpuratus | 1421 |
| SscPxd01 | - | Sus scrofa | 1479 |
| TguPxd01 | 7678 | Taeniopygia guttata | 1490 |
| TguPxd02 | 7679 | Taeniopygia guttata | 1475 |
| TspiPxdOl | - | Trichinella spiralis | 1276 |
| TrubPxd01 | 7685 | Takifugu rubripes | 1434 |
| TnPxd01 | 7618 | Tetraodon nigroviridis | 1379 |
| TcasPxd01 | 4102 | Tribolium castaneum | 1388 |
| XtPxd | 4051 | Xenopus tropicalis | 1460 |
Entry ID in the PeroxiBase (http://peroxibase.toulouse.inra.fr/).
Member of the short peroxidasins.
Minimum-Evolution (ME) Method
The phylogeny was also reconstructed using the ME method of the MEGA5 package [44] with following options: JTT model; bootstrap replications 1000; γ set to 1.75; close-neighbor interchange (CNI) was chosen as heuristic model and search level 2; the initial tree was obtained by NJ; homogenous pattern among lineages and pairwise deletion for gaps/missing data treatment.
Maximum-Likelihood (ML) Trees
Additionally, the ML trees were inferred with the MEGA5 package [44] by using the JTT model, the γ parameter set to 3, and with 100 bootstrap replications; CNI and all sites were used for gaps/missing data treatment.
Protein secondary and tertiary structure prediction of the various domains of peroxidasin was performed with PSIPRED [45] and EsyPred3D [46].
Footnotes
A supplementary figure showing the unrooted phylogenetic tree reconstructed with only the catalytic peroxidase domain is available upon request from the author.
References
- 1.Zamocky M, Jakopitsch C, Furtmüller PG, Dunand C, Obinger C. Proteins: Struct., Funct., Bioinf. 2008;72:589. doi: 10.1002/prot.21950. [DOI] [PubMed] [Google Scholar]
- 2.Furtmüller PG, Zederbauer M, Jantschko W, Helm J, Bogner M, Jakopitsch C, Obinger C. Arch. Biochem. Biophys. 2006;445:199. doi: 10.1016/j.abb.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Nelson RE, Fessler LI, Takagi Y, Blumberg B, Keene DR, Olson PF, Parker C, Fessler JH. EMBO J. 1994;13:3438. doi: 10.1002/j.1460-2075.1994.tb06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott AL, Ghedin E. Parasitol. Int. 2009;58:6. doi: 10.1016/j.parint.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okazaki N, Kikuno R, Ohara I, Inamoto S, Aizawa H, Yuasa S, Nakajima D, Nagase T, Ohara O, Koga H. DNA Res. 2003;10:35. doi: 10.1093/dnares/10.1.35. [DOI] [PubMed] [Google Scholar]
- 6.Team MammalianGeneCollection(MGC)Program. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16899. [Google Scholar]
- 7.Horikoshi N, Cong J, Kley N, Shenk T. Biochem. Biophys. Res. Commun. 1999;261:864. doi: 10.1006/bbrc.1999.1123. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell MS, Kan-Mitchell J, Minev B, Edman C, Deans RJ. Cancer Res. 2000;60:6448. [PubMed] [Google Scholar]
- 9.Cheng G, Salerno JC, Cao Z, Pagano PJ, Lambeth J. Free Radical Biol. Med. 2008;45:1682. doi: 10.1016/j.freeradbiomed.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tindall AJ, Pownall ME, Morris ID, Isaacs HV. Dev. Dyn. 2005;232:377. doi: 10.1002/dvdy.20226. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Carson-Walter EB, Cooper A, Winans BN, Johnson MD, Walter KA. J. Neurooncol. 2010;99:13. doi: 10.1007/s11060-009-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tauber S, Jais A, Jeitler M, Haider S, Husa J, Lindroos J, Knöfler M, Mayerhofer M, Pehamberger H, Wagner O, Bilban M. Mol. Cancer. 2010;9:200. doi: 10.1186/1476-4598-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homma S, Shimada T, Hikake T, Yaginuma H. Gene Expr. Patterns. 2009;9:1. doi: 10.1016/j.gep.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Peterfi Z, Donko A, Orient A, Sum A, Prokai A, Molnar B, Vereb Z, Rajnavölgyi E, Kovacs KJ, Müller V, Szabo AJ, Geiszt M. Am. J. Pathol. 2009;175:725. doi: 10.2353/ajpath.2009.080693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotenstein JR, Swale RE, Fukuda T, Wu Z, Giurumescu CA, Goncharov A, Jin Y, Chisholm AD. Development. 2010;137:3603. doi: 10.1242/dev.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi R, Hu C, Yuan Q, Yang T, Peng J, Li Y, Bai Y, Cao Z, Cheng G, Zhang G. Cardiovasc. Res. 2011;91:27. doi: 10.1093/cvr/cvr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai Y-P, Hu C-P, Yuan Q, Peng J, Shi R-Z, Yang T-L, Cao Z-H, Li Y-J, Cheng G, Zhang G-G. Free Radical Biol. Med. 2011;51:1492. doi: 10.1016/j.freeradbiomed.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan K, Rudkin A, Parry DA, Burdon KP, McKibbin M, Logan CV, Abdelhamed ZI, Muecke JS, Fernandez-Fuentes N, Laurie KJ, Shires M, Fogarty R, Carr IM, Poulter JA, Morgan JE, Mohamed MD, Jafri H, Raashid Y, Meng N, Piseth H, Toomes C, Casson RJ, Taylor GR, Hammerton M, Sheridan E, Johnson CA, Inglehearn CF, Craig JE, Ali M. Am. J. Hum. Genet. 2011;89:464. doi: 10.1016/j.ajhg.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bella J, Hindle KL, McEwan PA, Lovell SC. Cell. Mol. Life Sci. 2008;65:2307. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HM, Oh SC, Lim KJ, Kasamatsu J, Heo JY, Park BS, Lee H, Yoo OJ, Kasahara M, Lee J-O. J. Biol. Chem. 2007;282:6726. doi: 10.1074/jbc.M608471200. [DOI] [PubMed] [Google Scholar]
- 21.Kobe B, Kajava AV. Curr. Opin. Struct. Biol. 2001;11:725. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 22.Enkhbayar P, Kamiya M, Osaki M, Matsumoto T, Matsushima N. Proteins: Struct., Funct., Bioinf. 2004;54:394. doi: 10.1002/prot.10605. [DOI] [PubMed] [Google Scholar]
- 23.Halaby DM, Mornon JP. J. Mol. Evol. 1998;46:389. doi: 10.1007/pl00006318. [DOI] [PubMed] [Google Scholar]
- 24.Barclay AN. Semin. Immunol. 2003;15:215. doi: 10.1016/s1044-5323(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 25.Kaas Q, Ehrenmann F, Lefranc MP. Brief. Funct. Genomics Proteomics. 2007;6:253. doi: 10.1093/bfgp/elm032. [DOI] [PubMed] [Google Scholar]
- 26.von Castelmur E, Marino M, Svergun DI, Kreplak L, Ucurum-Fotiadis Z, Konarev PV, Urzhumtsev A, Labeit D, Labeit S, Mayans O. Proc. Natl. Acad. Sci. U. S.A. 2008;105:1186. doi: 10.1073/pnas.0707163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Leary JM, Hamilton JM, Deane CM, Valeyev NV, Sandell LJ, Downing AK. J. Biol. Chem. 2004;279:53857. doi: 10.1074/jbc.M409225200. [DOI] [PubMed] [Google Scholar]
- 28.Sadler JE. J. Thromb. Haemostasis. 2009;7 S1, 24. [Google Scholar]
- 29.Holbourn KP, Perbal B, Ravi Acharya K. J. Cell. Commun. Signal. 2009;3:25. doi: 10.1007/s12079-009-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J-L, Qiu L-Y, Kotzsch A, Weidauer S, Patterson L, Hammerschmidt M, Sebald W, Mueller TD. Dev. Cell. 2008;14:739. doi: 10.1016/j.devcel.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Hansson M, Olsson L, Nauseef WM. Arch. Biochem. Biophys. 2006;445:214. doi: 10.1016/j.abb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Slungaard A. Arch. Biochem. Biophys. 2006;445:256. doi: 10.1016/j.abb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Ihalin R, Loimaranta V, Tenuovo J. Arch. Biochem. Biophys. 2006;445:261. doi: 10.1016/j.abb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Rees MD, Kennet EC, Whitelock JM, Davies MJ. Free Radical Biol. Med. 2008;44:1973. doi: 10.1016/j.freeradbiomed.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Rosen H, Madtes DK, Shao B, Marten TR, Heinecke JW, Fu X. J. Biol. Chem. 2007;282:31826. doi: 10.1074/jbc.M704894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinecke JW, Li W, 3rd HLDaehnke,, Goldstein JA. J. Biol. Chem. 1993;268:4069. [PubMed] [Google Scholar]
- 37.Johansson MW, Lind MI, Holmblad T, Thornqvist PO, Soderhall K. Biochem. Biophys. Res. Commun. 1995;216:1079. doi: 10.1006/bbrc.1995.2731. [DOI] [PubMed] [Google Scholar]
- 38.Johansson MW, Patarroyo M, Oberg F, Siegbahn A, Nilsson K. J. Cell Sci. 1997;110:1133. doi: 10.1242/jcs.110.9.1133. [DOI] [PubMed] [Google Scholar]
- 39.Stofanko M, Kwon SY, Badenhorst P. Genetics. 2008;180:253. doi: 10.1534/genetics.108.089094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olofsson B, Page DT. Dev. Biol. 2005;279:233. doi: 10.1016/j.ydbio.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 41.Passardi F, Theiler G, Zamocky M, Cosio C, Rouhier N, Teixera F, Margis-Pinheiro M, Ioannidis V, Penel C, Falquet L, Dunand C. Phytochemistry. 2007;68:1605. doi: 10.1016/j.phytochem.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Bioinformatics. 2007;23:2947. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Nicholas KB, Nicholas HB. Genedoc, Tool for Editing and Annotating Multiple Sequence Alignments, 1997, distributed by the author.
- 44.Tamura K, Dudley J, Nei M, Kumar S. Mol. Biol. Evol. 2007;24:1596. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 45.McGuffin LJ, Bryson K, Jones DT. Bioinformatics. 2000;16:404. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 46.Lambert C, Léonard N, Bolle XDe, Depiereux E. Bioinformatics. 2002;18:1250. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]


