Abstract
Mutation discovery technologies have enabled the development of reverse genetics for many plant species and allowed sophisticated evaluation of the consequences of mutagenesis. Such methods are relatively straightforward for seed-propagated plants. To develop a platform suitable for vegetatively propagated species, we treated isolated banana shoot apical meristems with the chemical mutagen ethyl methanesulphonate, recovered plantlets and screened for induced mutations. A high density of GC-AT transition mutations were recovered, similar to that reported in seed-propagated polyploids. Through analysis of the inheritance of mutations, we observed that genotypically heterogeneous stem cells resulting from mutagenic treatment are rapidly sorted to fix a single genotype in the meristem. Further, mutant genotypes are stably inherited in subsequent generations. Evaluation of natural nucleotide variation showed the accumulation of potentially deleterious heterozygous alleles, suggesting that mutation induction may uncover recessive traits. This work therefore provides genotypic insights into the fate of totipotent cells after mutagenesis and suggests rapid approaches for mutation-based functional genomics and improvement of vegetatively propagated crops.
Keywords: Musa, induced mutations, TILLING, enzymatic mismatch cleavage, chimerism, ethyl methanesulphonate
Introduction
Asexual or vegetative propagation is common to many plant species owing to pools of totipotent stem cells maintained by plants (Murashig, 1974; Priestley, 1929). Examples include apple, banana, cassava, citrus, coffee, grapevine, hops and potato and thus represent major economic and food security crops. Vegetatively propagated plants can be further defined as either facultative, whereby sexual or meiotic propagation is possible but sometimes inefficient, or obligate where mitotic propagation is the only means available (van Harten, 1998). Prolonged or continual mitotic propagation can alter the genetic make-up of organisms. Lack of meiosis, recombination and independent assortment provide a means of accumulating spontaneous deleterious mutations that cannot be expunged efficiently from a population, a phenomenon known as Muller’s ratchet (Muller, 1932). This has led to models for the preponderance of sexual propagation in extant species (Felsenstein, 1974; Muller, 1964). For example, lack of meiotic propagation means that new genotypic combinations cannot be easily created. This has major implications for crop improvement as crossing is a key feature of most plant improvement schemes.
One strategy for inducing novel genetic variation in both sexually and asexually propagated species is mutagenesis. Since the pioneering work of Muller and Stadler, mutagenesis has been established as an efficient tool for forward genetics, plant breeding and more recently reverse genetic approaches (Ahloowalia et al., 2004; Henikoff et al., 2004; Muller, 1927; Stadler, 1928). In the past decade, high-throughput mutation discovery methods have been developed that allow measurements of densities and spectra of induced mutations without the need for an expressed phenotype. Thus, a more accurate assessment of the total effect of mutagenic treatment can be made. To date, the largest data sets come from chemical mutagenesis of plants and animals developed for TILLING (Targeting Induced Local Lesions IN Genomes) screens. TILLING is a strategy that combines random mutagenesis with mutation discovery for a reverse genetic approach that enables targeted recovery of a spectrum of point mutations in any gene in a genome (Kurowska et al., 2011; McCallum et al., 2000). Widely applicable, TILLING has been established for a range of plant and animal species. In crops, it can be a nontransgenic alternative to create novel genetic diversity and has been used for the improvement of species such as wheat and potato, where altering starch quality is of commercial importance (Muth et al., 2008; Slade et al., 2005). The most commonly reported mutagen, ethyl methanesulphonate (EMS), causes primarily GC-AT base pair transition mutations in many species including Arabidopsis thaliana, Triticum durum (tetraploid wheat), Triticum aestivum (hexaploid wheat), Zea mays and Caenorhabditis elegans (Gilchrist et al., 2006; Kurowska et al., 2011). In other species, such as Drosophila melanogaster, Hordeum vulgare and Oryza sativa, over 25% non-GC-AT changes have been reported (Caldwell et al., 2004; Cooper et al., 2008; Till et al., 2007). This divergence from transition changes may be an artefact intrinsic to the way populations were produced or maintained, or may represent differences in mismatch repair unique to particular species or genotypes. Regardless of species or genotype, nearly all reported mutations have been single nucleotide changes. While the majority of induced point mutations are predicted to be functionally silent, other nucleotide changes such as non-sense, mis-sense, RNA splicing defects and regulatory alterations result from chemical mutagenesis. Such changes can have varying effects on gene expression and protein function (Barkley and Wang, 2008; Vaddepalli et al., 2011). Reported densities of induced mutations in sexually propagated species range from 1 mutation per 1 million base pairs (Mb) to 1 in 25 kilobase pairs (kb) (Jankowicz-Cieslak et al., 2011). While densities can be affected by treatment regime and variations in mutation repair pathways, a trend can be observed where higher densities are achieved with increasing ploidy, a phenomenon first observed phenotypically in wheat (Stadler, 1929). An optimal mutation density allows for smaller population sizes and more rapid recovery of desired alleles and therefore is a key to successful breeding and functional genomics approaches.
Little is known with regard to the achievable spectrum and density of mutations in asexually propagated species. Confounding factors include previous reliance on phenotypic observations that can be influenced by environmental conditions and epigenetic variation, the reliance on dominant or hemizygous alleles in the first generation (M1) and the presence of genotypic heterogeneity or chimerism after mutagenesis. Such heterogeneity occurs because at the time of mutagenesis, it is expected that all cells accumulate different EMS-induced mutations randomly throughout the nuclear genome. This is problematic because measured phenotypes and genotypes may represent only a subset of the variation in the material and may not be inherited. Chimerism can be eliminated in sexually propagated crops through the single cell–based mechanisms of meiosis and sexual reproduction (Comai and Henikoff, 2006). While straightforward, this requires the production of a M2 population which necessitates additional resources and time. Further, a structured population typically following single seed descent is often employed to ensure that induced alleles are sampled only once as oversampling increases time and costs and confounds estimations of mutation density. For vegetatively propagated crops, strategies have been developed that employ successive rounds of tissue isolation and bisection aimed at reducing the genotypic complexity of the resulting plantlets (van Harten, 1998). However, methods for rapid dissolution of genotypic heterogeneity, or chimerism, in mitotically propagated tissues have yet to be evaluated at the genomic level.
The genus Musa, consisting of bananas and plantains, represents the fourth most important food crop in some of the world’s least developed countries. Edible varieties tend to be triploid and parthenocarpic. They are seedless and thus must be propagated vegetatively. Although banana is a major export commodity worth over 28 billion US dollars annually, 87% of Musa production is consumed locally (http://faostat.fao.org/site/339/default.aspx 2009 accessed 21 February 2012). Plantations consist of cloned plants that are nearly genetically identical and uniform, which makes the crop particularly susceptible to fungal pathogens causing biotic diseases such as panama and black sigatoka. This has led to speculation that worldwide banana production may cease to be sustainable in the near future, threatening economic stability and food security in developing countries in the tropics and subtropics (Heslop-Harrison and Schwarzacher, 2007). Therefore, we sought to develop a platform for the induction of stably heritable point mutations in mitotically propagated bananas. We observe rapid dissolution of chimeric sectors in EMS-treated meristematic tissues and discuss models for this unexpected result. The findings have potential practical implications for mutagenesis-based functional genomics and breeding of many other vegetatively propagated plants.
Results
Development of an EMS-mutagenized population of vegetatively propagated banana
To establish optimal concentration ranges for EMS mutagenesis of banana shoot tips, cultures were treated with 4 EMS concentrations (0.25%, 0.5%, 1% and 1.5%) and 2 incubation times (2 and 4 h). A total of 25 cuttings were evaluated for survival rate and fresh weight per treatment regime. Regardless of concentration of EMS, all explants incubated for 2 h survived the treatment. After 4 h incubation, only the 1.5% EMS treatment was lethal to the in vitro shoot tip cuttings. The percentage reduction in fresh weight of in vitro plantlets was used to choose mutagenesis conditions for the development of a large population suitable for mutation discovery, evaluation of mutation inheritance and measuring the rate at which plants become genotypically homogeneous. A target range weight reduction of 40%–50% was selected to balance the need for sufficient plant growth while maximizing the density of induced mutations.
For bulk mutagenesis, 4000 in vitro shoot tip cuttings of the triploid ‘Grande Naine’ variety were prepared. Owing to the potentially confounding variables of EMS absorbance time and possible cytotoxic effects on cell function, four different mutagenic treatments were selected to achieve the weight reduction ranges described above. Treatment batches consisted of 1000 banana shoot tips. Mutagenesis was performed at room temperature for 3 h (1% EMS), 6 h (0.5% EMS), 24 h (0.125% EMS) and 48 h (0.06% EMS), respectively. Survival rates ranged from 52% (24 h) to 87% (48 h) with no trend observable between survivability and either EMS dosage or time (see Table S1). A total of 3004 mutagenized samples survived the treatments.
To reduce the chance of chimeric sectors in mutagenized plants, meristems were repeatedly isolated and longitudinally bisected (Figure 1). This results in a reduction in the number and genotypic diversity of meristematic cells that divide to produce progeny. Plants were mitotically propagated through six vegetative cycles to the M1V6 generation. Little is known about the mechanism of chimera dissolution in banana and other vegetatively propagated plants. A previous study in banana relying on the measurement of cytochimeras looking at polyploidy induced by the treatment of plants with colchicine suggested that chimeras are at least partially removed by the third or fourth generation, and so the expectation is that all EMS-mutagenized plants at the M1V6 generation will be genotypically homogenous (Jain et al., 2011; Roux et al., 2001). Seven hundred and sixty-eight plants were chosen at random for the evaluation of mutation density and spectrum (see Table S2). A total of 23 primer pairs specific for putative open reading frames were designed from BAC sequence to amplify gene fragments between ∼755 and 1500 bp. Forty-eight per cent (11/23) of primer pairs produced both high-quality TILLING data and Sanger sequencing reads and were included in this study (Table 1). The remaining primers produced poor-quality Sanger sequencing reads, potentially owing to high levels of natural background polymorphisms driving primer mispriming (Till et al., 2010). Samples were subjected to TILLING screens using the validated gene targets, and a total of 33 putative mutations were identified based on gel analysis. All putative mutations were confirmed by Sanger sequencing to be GC-AT transition changes, the major type of change reported in species treated with EMS (Table 2). As expected, all changes were determined to be heterozygous. Analysis of the mutation spectrum revealed 36% silent, 49% mis-sense and 15% truncation alleles (see Table S3).
Figure 1.
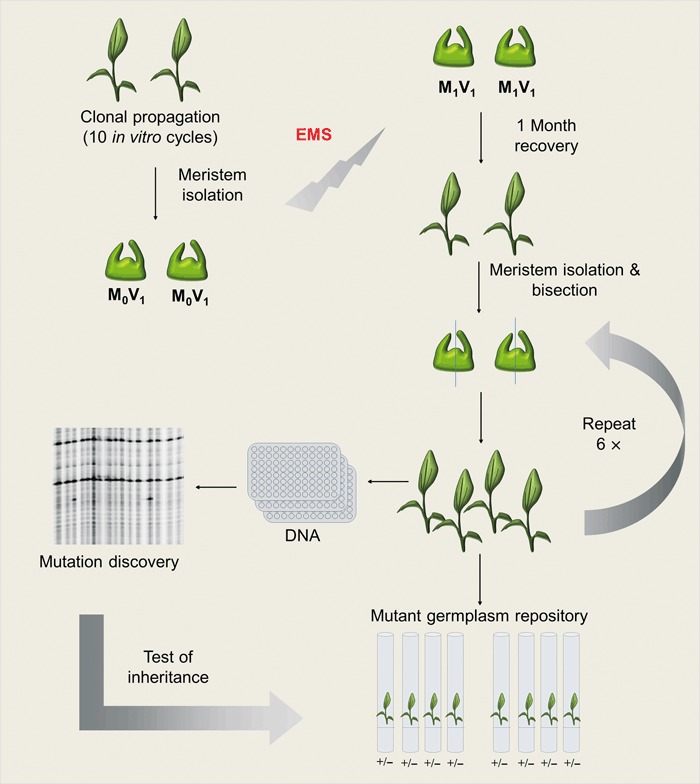
A strategy for meristematic mutagenesis, chimera dissolution and mutation recovery in vegetatively propagated banana. Clonally propagated isogenic plants are prepared through 10 rounds of in vitro multiplication. Shoot apical meristems are isolated and soaked in EMS, then allowed to recover for 1 month. Individuals from the first vegetative generation (M1V1) are propagated through meristem isolation and longitudinal bisection to produce the M1V2 generation. Successive rounds of isolation and division are performed to reduce genotypic heterogeneity, and the number of individuals approximately doubles each generation. Tissue is collected from M1V6 individuals, and DNA is extracted and screened for induced mutations. The inheritance of isolated mutations is evaluated in the M1V6 and subsequent generations, allowing for estimations of the rate of chimera dissolution.
Table 1.
Gene targets and primer sequences used to evaluate induced mutations in banana
| Target name | Annotation | Genbank accession | Target size | Forward primer | Reverse primer |
|---|---|---|---|---|---|
| ACETRANS | N-Acetylglucosaminyltransferase | AC186756 | 1500 | TCGCTCTGGGTTTCAGGAAAGCAGTT | TCAGAGTGTAAACCGGGGTCCCAAAT |
| AMTHLTR | Aminomethyltransferase | AC186747 | 1508 | CGGCATCCAAGTTTCTCATGCCTTTTA | CAACTCGAGCAAAAAGCATCTCACGAT |
| DNAJ | Heatshock protein | AC186747 | 1473 | AGGAGAAGTCAGGGACCAGAACCGAAT | TATAAACCGCCCAAATCTCACCACAGC |
| ELF3 | Eukaryotic translation initiation factor 3 | AC186746 | 1500 | CGACTTCACAATCCCCCACATGTTAGA | GTTGTCCCCTTCAATACCGACGGATG |
| FTSJMT | Ftsj-like methyltransferase family protein | AC186746 | 1479 | ATACAGCAAGGGTGATGCAGCAGACAG | ATTTGGCCTTTATTCTTGCGTCCCTTC |
| GHF17 | Glycoside hydrolase family 17 protein | AC186755 | 1500 | TAGGGCCAAAAGCTCCCCTGAGAAAGA | CGAGAAGGCACATAGCCGTTTCTGAGT |
| MALSYN | Malate synthase | AY484589 | 1500 | CTCACCAGGGATGCCTTGCAGTTC | AGACTTCCATGATAGGCGGGATCAGG |
| NPH3 | Nonphototropic hypocotyl 3 family protein | AC186747 | 1498 | TCGAACCTGCTGCCAAGTTCTGTTATG | GTCCATGCTCACCTTCAAGACCTGGTT |
| PAAL2 | Phenylalanine ammonia lyase | AP009326 | 1500 | AGGAGGACCAAGCAAGGAGGTGCTCTT | GGTCGTCGACGTAGGTGAAGACGTG |
| PUF | Pumilio/Puf RNA binding | AC186756 | 1501 | CGACGGCTTCGATGTCTACGAGTTGAT | TGGGTTGTGAGGAGAAAGTGGCTTCAC |
| RNDR | Ribonucleotide reductase | AY484588 | 1500 | ATGAAGCTCCGGGACTTGCTGATTG | CAGGTTGGAGAATTCCCTGAGCAACAA |
Table 2.
Induced mutations identified in clonally propagated banana
| Gene target | Nucleotide change | Mutant allele* | Effect† | PSSM‡ | SIFT§ |
|---|---|---|---|---|---|
| ACETRANS | M215H | C215T | Intron | ||
| S227B | C227T | Intron | |||
| C653Y | C653T | D107= | |||
| G1127R | G1127A | W265* | |||
| C1312Y | C1312T | S327F | 10.8 | 0.08 | |
| G1346R | G1346A | E338= | |||
| AMTHLTR | C717Y | C717T | Intron | ||
| G1007R | G1007A | E267= | |||
| DNAJ | G239R | G239A | W547* | ||
| ELF3 | G493R | G493A | S342= | ||
| C1040Y | C1040R | G160D | 0.47 | ||
| C1092Y | C1092T | A143T | 5.9 | 0.55 | |
| C1107Y | C1107T | A138T | 8.1 | 0.56 | |
| G1119R | G1119A | R134W | 6.6 | 0.04 | |
| G1126R | G1126R | V131= | |||
| G1138R | G1138A | R127= | |||
| G1148R | G1148A | P124L | 1.00 | ||
| C1150Y | C1150T | L123= | |||
| FTSJMT | G284R | G284A | G485E | −2.8 | 1.00 |
| C623Y | C623T | P560S | 6.3 | 0.27 | |
| C986Y | C986T | Q681* | |||
| GHF17 | C1148Y | C1148T | P538S | 11.5 | 0.04 |
| MALSYN | C182Y | C182T | P88L | 12.8 | 0.07 |
| G685R | G685A | R172Q | 27 | 0 | |
| G1309R | G1309A | Intron | |||
| NPH3 | G270R | G270A | A217T | ||
| G398R | G398A | W259* | |||
| PAAL2 | G240R | G240A | V158I | 2.6 | 0.49 |
| G493R | G493A | G242D | 0.09 | ||
| C696Y | C696T | Q310* | |||
| PUF | C183Y | C183T | T714I | 0.02 | |
| G392R | G392A | G784R | 0.15 | ||
| RNDR | G824R | G824A | R511= |
Owing to natural heterozygosity and triploidy, up to three alleles can be observed at any locus. Nucleotide position is based on amplicon sequence.
Positions of changes on the amino acid sequence.
Mis-sense changes are predicted to be damaging to the encoded protein if the PSSM (PARSESNP) score is >10.
Mis-sense changes are predicted to be damaging to the encoded protein if the SIFT score is <0.05.
Rate of genotypic homogeneity in meristematic cells, mutation density and inheritance
Mutation densities from TILLING screens of sexually propagated species are typically calculated by counting mutations from a single progeny or pooled progeny from a mutation event, thus allowing an estimation by dividing the mutations discovered by the total bases screened (Till et al., 2003). To calculate mutation densities in in vitro mutagenized and mitotically propagated plants, it is first necessary to evaluate at what stage in propagation the pool of meristematic cells became nonchimeric. This is required because, after this point, all sibling progeny plants within a line will be clonally related and inherit the same alleles. Including clonally related plants will result in an inflation of the population size and thereby an underestimation of mutation density when compared to estimations from seed-propagated crops. To calculate the percentage of sibling plants in the M1V6 population inheriting the same mutation, we selected 16 mutant alleles for further testing. Sibling plants were then screened individually for the presence or absence of the mutant allele. A total of 285 plants were tested and mutant alleles were recovered in ∼82% (235/285) of the plants (Table 3). In one case, the same allele (FTSJMT C623T) was identified in a subset of siblings from three different lines. While coincident mutations creating the same allele are expected to occur at a low frequency with random mutagens such as EMS, the recovery of three coincident mutations in a set of 33 discovered alleles is exceedingly unlikely. More likely, the presence of this allele in three unique lines represents human error when labelling plant flasks. Removing these potential errors from the evaluation, 222/243 (91%) of sibling plants within the population shared the same allele. This suggests that the majority of induced mutations are fixed in the treated plant meristem within 1 month after mutagenesis. We used this information to correct for the population size in estimating a mitotic mutation density (MMD) and arrive at one mutation per 57 kb. To further establish that plants in M1V6 are stably inherited, we chose three sequence-confirmed alleles and passaged plants in vitro to the M1V9 generation. One hundred per cent (8/8) of progeny plants carried the induced allele.
Table 3.
Inheritance of induced mutations in sibling individuals
| Gene target | Allele | Line number | Number identified | Number screened |
|---|---|---|---|---|
| ACETRANS | C215T | MT1_5 | 6 | 6 |
| C227T | MT90_83 | 4 | 4 | |
| C653T | MT47_33 | 10 | 10 | |
| G1127A | MT49_43 | 10 | 10 | |
| C1312T | MT73_6 | 9 | 9 | |
| AMTHLTR | C717T | MT80_53 | 73 | 74 |
| G1007A | MT90_83 | 5 | 5 | |
| FTSJMT | G284A | MT57_33 | 10 | 10 |
| C623T | MT82_73 | 8 | 13 | |
| C623T | MT90_23 | 3 | 11 | |
| C623T | MT94_33 | 2 | 18 | |
| C986T | MT82_33 | 9 | 27 | |
| MALSYN | C182T | MT99_83 | 2 | 3 |
| G685A | MT89_53 | 4 | 4 | |
| G1309A | MT81_63 | 7 | 7 | |
| RNDR | G824A | MT80_53 | 73 | 74 |
| Total | 235 | 285 |
Natural mutation rate and evaluation of natural SNPs in the mutant population
We observed no evidence of spontaneous mutations in the 10 vegetative propagation cycles before mutagenesis or the six cycles postmutagenesis. This allows for an estimation of the rate of accumulation of natural mutations in cultured banana. Using the number of generations propagated and the number of base pairs screened, we estimate the spontaneous rate to be <6 × 10−9. This is consistent with a natural mutation rate estimation of 7 × 10−9 calculated for Arabidopsis thaliana (Ossowski et al., 2010). While we observed no evidence of spontaneous mutations arising in tissue culture, high heterozygosity reported for bananas suggests the accumulation of spontaneous mutations over a long evolutionary history (Till et al., 2010). Therefore, natural accumulation of deleterious alleles may increase the utility of using mutagenesis in diploid and polyploid vegetatively propagated plants. To evaluate this, sequence analysis was performed on 8 selected gene targets across the regions screened by enzymatic mismatch cleavage, and polymorphisms compared to the available reference sequence using the PARSESNP and SIFT algorithms (Table 4). Of 132 catalogued polymorphisms common in all plants screened, 99/132 (75%) are silent and 25% are mis-sense changes. Approximately 8% of mis-sense changes are predicted to be deleterious to protein function. Fifty-five per cent of predicted deleterious mis-sense changes are heterozygous and 45% are homozygous compared to the reference sequence. While only a section of coding region was evaluated, heterozygous putatively deleterious alleles were found in 4/8 gene targets, suggesting a high accumulation of potentially deleterious alleles genome-wide.
Table 4.
Natural mutations identified in triploid banana cultivar Grande Naine
| Gene target | Nucleotide change* | Zygosity† | Effect‡ | PARSESNP§ | SIFT¶ |
|---|---|---|---|---|---|
| ACETRANS | T74K | Het | S40A | 10.7 | 0.00 |
| T81Y | Het | F42S | 19.7 | 0.00 | |
| G86K | Het | A44S | 11.5 | 0.00 | |
| C1076T | Hom | Y248= | |||
| G1295A | Hom | S321= | |||
| AMTHLTR | A138M | Het | Intron | ||
| G212A | Hom | Intron | |||
| T220C | Hom | Intron | |||
| A251R | Het | Intron | |||
| C269M | Het | Intron | |||
| G277A | Hom | Intron | |||
| A314G | Hom | Intron | |||
| G347A | Hom | Intron | |||
| C359M | Het | Intron | |||
| A360G | Hom | Intron | |||
| G427R | Het | Intron | |||
| T461A | Hom | Intron | |||
| A522R | Het | Intron | |||
| A530W | Het | Intron | |||
| G533R | Het | Intron | |||
| A585W | Het | Intron | |||
| T589W | Het | Intron | |||
| G591K | Het | Intron | |||
| T608G | Hom | Intron | |||
| A615R | Het | Intron | |||
| A650M | Het | Intron | |||
| A663R | Het | Intron | |||
| T790W | Het | Intron | |||
| G822K | Het | Intron | |||
| C823Y | Het | Intron | |||
| G838R | Het | Intron | |||
| T850K | Het | Intron | |||
| G882R | Het | Intron | |||
| G962R | Het | S252= | |||
| A1256M | Het | P350= | |||
| ELF3 | A73G | Hom | I482= | ||
| G451A | Hom | D356= | |||
| A459T | Hom | S354T | −3.8 | 1 | |
| G649A | Hom | V290= | |||
| C685T | Hom | R278= | |||
| G706C | Hom | S271= | |||
| G716A | Hom | A268V | 10.5 | 0.09 | |
| G778C | Hom | S247= | |||
| A874G | Hom | L215= | |||
| C964G | Hom | T185= | |||
| T1065G | Hom | N152H | 0.24 | ||
| C1070T | Hom | R150H | 0.13 | ||
| T1114C | Hom | K135= | |||
| C1207T | Hom | K104= | |||
| C1275G | Hom | V82L | 0.75 | ||
| G1297A | Hom | R74= | |||
| C1381G | Hom | S46= | |||
| FTSJMT | C252T | Hom | D474= | ||
| A323C | Hom | K498T | 12.6 | 0.47 | |
| A392T | Hom | Intron | |||
| T702A | Hom | V586E | 2.3 | 0.96 | |
| G763T | Hom | E606D | −5.1 | 0.95 | |
| A1038G | Hom | K698R | −3.5 | 1 | |
| T1177A | Hom | S744= | |||
| T1281G | Hom | V779G | 0.7 | 0.58 | |
| T1354G | Hom | G803= | |||
| G1369A | Hom | K808= | |||
| MALSYN | A90R | Het | V57= | ||
| C119Y | Het | P67L | 6.4 | 1 | |
| G163R | Het | A82T | −6.1 | 0.67 | |
| T180C | Hom | P87= | |||
| C222M | Het | V101= | |||
| G313R | Het | Intron | |||
| C331M | Het | Intron | |||
| C386Y | Hom | Intron | |||
| T640W | Het | V157E | 1 | ||
| C737Y | Het | I189= | |||
| T872C | Hom | Intron | |||
| T884K | Het | Intron | |||
| A905G | Hom | Intron | |||
| A915G | Hom | Intron | |||
| G928A | Hom | Intron | |||
| A934C | Hom | Intron | |||
| G959S | Het | V235L | 18 | 0.01 | |
| T990C | Hom | I245T | 18.1 | 0 | |
| C1004A | Hom | L250I | −16.2 | 1 | |
| G1039C | Hom | A261= | |||
| T1153C | Hom | D299= | |||
| G1239R | Het | R328H | −0.6 | 0.1 | |
| C1240T | Hom | R328= | |||
| G1290T | Hom | Intron | |||
| G1320C | Hom | Intron | |||
| C1342T | Hom | Intron | |||
| G1424R | Het | K361= | |||
| NPH3 | G186R | Het | A189T | 16.6 | 0 |
| A256C | Hom | Y212S | −4.9 | 0.86 | |
| T576C | Hom | S319P | 0.3 | ||
| C722Y | Het | S367= | |||
| G860R | Het | E413= | |||
| C1045S | Het | A475G | 9.4 | 0.13 | |
| A1064R | Het | R481= | |||
| PAAL2 | C75Y | Het | Intron | ||
| T132:5 | Hom | Intron | |||
| A138G | Hom | Intron | |||
| G140:5 | Hom | Intron | |||
| A151G | Hom | Intron | |||
| T167Y | Het | N133= | |||
| T179C | Hom | F137= | |||
| A480G | Hom | S238G | −10.7 | 1 | |
| G503R | Het | E245= | |||
| G533R | Het | V255= | |||
| T591C | Hom | L275= | |||
| C602M | Het | L278= | |||
| A759M | Het | K331Q | 0.8 | ||
| G827T | Hom | P353= | |||
| A870R | Het | K368E | 17.4 | 0.04 | |
| T941C | Hom | L391= | |||
| C977Y | Het | V403= | |||
| G1019R | Het | K417= | |||
| C1052Y | Het | N428= | |||
| T1076C | Hom | P436= | |||
| G1094R | Het | G442= | |||
| G1160A | Hom | E464= | |||
| T1286C | Hom | L506= | |||
| A1394G | Hom | R542= | |||
| PUF | C130S | Het | A696= | ||
| C225Y | Het | A728V | 0.06 | ||
| G279A | Hom | R746H | 0.02 | ||
| C408Y | Het | T789I | 0.07 | ||
| A420G | Hom | Q793R | 0.02 | ||
| T436Y | Hom | C798= | |||
| A467M | Het | R809= | |||
| G497K | Het | A819S | 2.4 | 0.16 | |
| C556Y | Het | F838= | |||
| T705Y | Het | Intron | |||
| C719Y | Het | Intron | |||
| T853G | Hom | S908A | 0.36 |
:5, five base pairs deleted.
Het, heterozygous, Hom, homozygous.
Positions of changes on the amino acid sequence.
Mis-sense changes are predicted to be damaging to the encoded protein if the PARSESNP score is >10.
Mis-sense changes are predicted to be damaging to the encoded protein if the SIFT score is <0.05.
Discussion
The use of induced mutations for gene function analysis and crop improvement has been widely employed in angiosperms. Reverse genetic studies of seed-propagated plants have resulted in the production of large data sets on the effects of chemical mutagens on plant genomes. Such studies have involved screening plants after at least one meiotic event postmutagenesis. The work presented here focused on the evaluation of vegetatively propagated banana, whereby mutations were screened prior to meiosis. Chemical mutagens such as EMS have been shown to induce mutations randomly across genomes (Greene et al., 2003). When mutagenizing multicellular tissues such as animal embryos, plant seed or meristems, the treated cells will accumulate different mutations and the resulting organisms will be genotypically heterogeneous or chimeric. Genetic or phenotypic measurement of such individuals is considered inappropriate because measured changes are potentially not heritable due to the fact that only a subset of cells are involved in gametogenesis. This issue has been overcome by performing at least one sexual cross prior to analysis. For plants, the single-cell spore phase ensures that progeny are genotypically homogeneous. In reverse genetic projects such as TILLING, it is often advantageous to measure the first nonchimeric generation (M2) to maximize the number of unique mutations recovered in the least number of samples. Evaluation of mutation segregation from a self-cross of the M2 generation often yields expected Mendelian segregation. However, differences in the genetically effective cell number have been observed in plants, which lead to altered segregation ratios (Greene et al., 2003; Perry et al., 2009).
One major difference in asexually propagated plants is the lack of passage of genetic material through the male and female haploid gametic phases. Continual mitotic propagation typically occurs in multicellular tissues. Making plants nonchimeric is therefore potentially more difficult. Chimerism in asexually propagated plants has long been observed and exploited for the generation of commercially important traits such as leaf variegation and flower colour (Marcotrigiano and Gradziel, 1997). Models for mechanisms of chimera disassociation and mosaic inheritance have been informed by observable phenotypic traits such as leaf mottling, but the underlying genetic mechanisms are often not well defined. Possible causes for such phenotypic abnormalities include transposable genetic elements, plastid mutations and mutations in nuclear genes. Phenotypic evaluations can also be influenced by somaclonal variation, a phenomenon whereby extended tissue culture can generate phenotypes, potentially influenced by epigenetic changes in cultured plants (Kaeppler et al., 2000). The work presented here specifically focuses on the effect of EMS treatment on genomic DNA sequence and therefore allows for a less-biased investigation of the inheritance of genetic variants under asexual propagation. One advantage of using EMS as a mutagen is that it produces primarily GC-AT transition mutations because of the alkylation of the guanine residue and subsequent fixation of the base change during replication. For plants like Arabidopsis thaliana and Triticum aestivum, 100% transition changes have been reported. In some species such as Drosophila melanogaster, the recovery of fewer transition mutations may be driven by selective pressures from maintaining living populations (Cooper et al., 2008). In the work presented here, screening an EMS-mutagenized banana population resulted in the recovery of 100% transition mutations. This suggests that there is limited meiotic selection against transversion mutations or small indels in previously tested TILLING populations. Another important metric in evaluating the effect of mutagenesis and the usability of mutant population is mutation density. To make an accurate estimation of mutation density in mitotically propagated plants, it is necessary to consider the genetic relationship of the siblings screened. Siblings in the M1V6 were chosen because a previous study in banana suggested that most chimeras should be dissolved before this generation, likely in the M1V3 or M1V4 (Roux et al., 2001). However, after mutations are fixed and plants are genotypically homogenous, all subsequent siblings will be genetic clones and this must be corrected for when estimating mutation density. For example, if chimeras were dissolved in the M1V3 generation, it is expected that 25% of siblings in the M1V6 generation would be clonally related and share the same mutations. Upon investigation of inheritance of mutations in siblings, we observed that, in most lines, all siblings inherited the same mutations and on average approximately 90% of siblings inherit the same allele. Such a measurement reflects the genetic composition of the totipotent shoot apical meristem cells that divide to produce all above-ground tissues in the banana plant. Thus, the measurement of inheritance in the M1V6 shows that most totipotent cells were genetically homogeneous in the M1V1 1 month after mutagenesis just prior to meristem isolation and bisection to generate the M1V2 plants.
This intriguing result was not expected based on observations that cells in the central meristematic zone (CZ) maintain their position and competency over time (Carles and Fletcher, 2003). Studies of CLAVATA3 expression in Arabidopsis thaliana, a marker for the CZ, suggest that there are approximately 35 stem cells in the shoot apical meristem (Yadav et al., 2009). Efforts are ongoing to accurately map and computer model the behaviour and gene expression of the shoot apical meristem (SAM). Past studies suggest that tissues are derived from at least 2–3 stem cells in CZ (Gross-Hardt and Laux, 2003). Based on our observations, we propose the following model of meristematic cell behaviour after treatment with EMS. In this model, all cells in the treated tissue accumulate transition mutations randomly throughout their nuclear genomes. Resulting cells are therefore genotypically distinct from one another, and each cell harbours a different fitness for cell division. This would result in a competitive environment where the most rapidly dividing cell eventually populates the apical meristematic region. Competitive cell divisions have been termed ‘diplontic drift’ or ‘diplontic selection’ to explain the instability of some phenotypes in vegetatively propagated plants (Balkema, 1972; Gaul, 1958; Klekowski and Kazarinovafukshansky, 1984). While previous work evoked nonmutagenized cells outcompeting cells that have naturally accumulated one or more deleterious alleles, our study allows monitoring of induced mutations, including silent mutations, accumulating in all cells without the need of observable phenotypic variation. Likely in addition to differences in cell division, the EMS treatment results in some level of cell death. This was postulated in EMS studies in Arabidopsis thaliana (Irish and Sussex, 1992). While we have no measure of cell death in mutagenized banana, this is not incompatible with our proposed model. A mechanism of competitive division or cell death would not be necessary if only one cell in the central zone were contributing to differentiated leaf tissue. However, if only one cell were involved, previously reported chimerism in banana would not have been observable. This therefore supports multiple cells contributing to developing primordia in Musa acuminata. Owing to the wide reproducibility of the effects of EMS mutagenesis in seed-propagated plants, we postulate that phenomena similar to that observed in banana may be observable in other vegetatively propagated plants. Furthermore, the same process may occur in any mutagenized meristem, including those in seed. This has practical applications when developing TILLING projects because considerable resources are required to generate a structured M2 population in seed-propagated plants. If rapid selection is occurring after seed mutagenesis, cells may become genotypically homogenous in the M1, allowing the development of DNA libraries from later leaf tissues of the first generation. Rapid mutation discovery would allow the propagation of only those M2 seed-harbouring desired alleles. We are currently testing this in seed-propagated crops.
We consider several mechanisms for the observed rapid genotypic homogeneity in the M1V1 generation. First, cytotoxic effects of alkylating agents may affect the competency of cells to divide (Britt, 1996). Differential competencies could be driven by variations in cytotoxicity because of cell cycle stage, activity of repair mechanisms, or by variations in EMS absorbance influenced by cellular position. Secondly, we consider genotoxic effects. While dramatic genetic changes such as chromosomal deletions and aneuploidy could be envisioned to explain variations in cellular division competencies, the large body of data from prokaryotic, eukaryotic and transgene reporter assays suggest that EMS is inducing almost exclusively point alleles (Kurowska et al., 2011; Ohta et al., 2000; Yoshihara et al., 2006). When evaluating natural SNPs in the mutagenized banana cultivar, we observed that many gene targets harboured at least one potentially deleterious allele in the relatively small region of sequence interrogated. Obligate vegetatively propagated species have no mechanism to remove deleterious alleles and therefore will continue to accumulate them until some external pressure is applied, such as human selection. Plants may be maintained so long as at least one functional copy of essential genes is present. Such functional haploidy would be revealed in the M1 generation via mutagenesis where induced heterozygous point mutations could result in expressed phenotypes in otherwise recessive traits. Therefore, it is possible that genetic factors contribute to the observed rapid clearing of chimeric sectors. Interestingly, we observed 99/109 plants from 27 lines showing some phenotypic deviation from wild type in the M1V10 generation (see Table S4). Most differences were in leaf pigmentation and were variable during glasshouse growth. This could be due to the effect of induced mutations, environmental factors or somaclonal variation. However, one possible mechanism for somaclonal variation, induction of spontaneous mutation, was not observed in this population. Leaf morphology differences did not change during extended glasshouse growth. For example, a narrow leaf phenotype was observed in line MT90-83 in 26/28 (93%) of siblings assayed. This is consistent with genotypic segregation of 100% measured in this line in the M1V6 (Table 3). On average, phenotypic inheritance was observed in 91% of siblings (see Table S4), similar to genotypic measurements, supporting the model of rapid fixation of meristematic cells after mutagenesis. A larger-scale genomic analysis is required to determine whether mutations affecting gene functions are driving genotypic homogeneity in mutagenized banana cultures. This will be aided by the recent release of the banana genome sequence (D’Hont et al., 2012).
Analysis of inheritance of induced mutations in siblings allowed a correction for population size and an estimation of mutation density analogous to those reported in sexually propagated plants and animals. We calculate a density of 1 mutation per 57 kb. With a genome size of approximately 520 Mb (D’Hont et al., 2012), we estimate that on average each plant carries approximately 10 000 induced alleles. While DNA sequence-based mutation densities have not previously been reported for triploids, the density is higher than that reported for diploids but lower than tetraploid and hexaploid wheat that was screened using the same mutation discovery protocol. This controls for variations in ascertainment biases in different mutation discovery methods (Jankowicz-Cieslak et al., 2011). The calculated estimate is expected as increasing copy number provides redundancy that protects against mutagens, thus allowing higher mutation densities to be achieved with increasing ploidy (Slade et al., 2005; Stadler, 1929; Uauy et al., 2009). That the expected density was recovered in mitotically propagated tissues suggests that treatment of isolated meristems in liquid EMS was near optimal. The expected triploid mutation density is based on sexually propagated species, suggesting that most of the mutations induced in vegetatively propagated tissues are meiotically heritable. This has practical implications for applications in facultative vegetatively propagated plants. Finally, we showed that induced alleles remain stable in culture conditions through ten cycles of propagation.
The work presented suggests strategies that may be applicable for a range of vegetatively propagated plant species, including those of commercial importance (potato, hops, grapevine, citrus) and those of importance for food security in developing countries (cassava, sweet potato, banana). Further experiments are ongoing to test our models in other asexually propagated species. The utility of induced mutations and reverse genetics may potentially be of greatest applicability in facultative vegetatively propagated crops where meiosis is possible, but either inefficient or seldom used. Here, tissue culture mutagenesis could be employed to produce nonchimeric M1 populations rapidly for genotypic mutation screening at a core facility. Only plants containing desired mutants need to be selected and delivered for self-crossing and field evaluation. Thus, the effective population size compared to forward genetic mutation breeding approaches could be reduced by several orders of magnitude, and bottlenecks in sexual propagation, such as low fecundity, can be overcome by clonal propagation. Because standardized in vitro propagation protocols can be applied to many species, and vegetative material can easily be shipped as micropropagules, a single facility could service a wide geographical range. The utility of point mutations for trait development in obligate vegetatively propagated plants and the prevalence of functional haploidy remain to be further investigated. Alternative mutagenesis approaches, such as inducing aneuploidy, may provide a means to generate more observable phenotypes in plants where meiotic propagation is not possible (Henry et al., 2010). Tissue culture has been established for many plant species, and the methods described here, where a mutant population can be rapidly produced, may also prove highly advantageous for seed-propagated species, as these approaches uncouple the dependency on field propagation and shorten the time until desired alleles are recovered.
Experimental procedures
Plant material
In vitro plantlets of banana (Musa acuminata) ecotype Grande Naine (Cavendish AAA, ITC accession 0180) were obtained from the Bioversity’s International Transit Centre (http://www.crop-diversity.org/banana/#AvailableITCAccessions, accessed 8 March 2012). Plants were maintained in vitro through aseptic shoot tip cultures in liquid S27 media supplemented with 20 μm 6-benzylaminopurine, 1 mg/L thiamine, 40 mg/L cystein HCl and 40 g/L sucrose, pH 5.8. The liquid cultures were maintained under constant horizontal rotation at 60/min with a continuous light (65 μmol/m2/s; Cool White fluorescent tubes, Philips TLP 36/86, Philips, Amsterdam, the Netherlands) at 22° ± 2°. Shoot multiplication was induced by cutting a single isolated shoot tip longitudinally through the apex using a scalpel blade (Cronauer and Krikorian, 1984). The resulting two shoot halves were placed in freshly prepared liquid media for 3–4 weeks to allow shoot regeneration. At this point, outer leaves produced from the dividing shoot tip and the shoot bases were trimmed away. Shoot isolation and division was then carried out. This process was repeated for a total of 10 generative cycles until the number of clonal propagules reached 4400.
Optimization of chemical mutagenesis
To determine a dosage range for the treatment of banana with ethyl methanesulphonate (EMS), shoot tips were incubated for either 2 or 4 h in 0.25%, 0.5%, 1% and 1.5% concentrations of mutagen. For each treatment combination, 25 in vitro cuttings were soaked in EMS solution of the selected percentage in sterilized water plus 2% dimethyl sulphoxide (v/v, DMSO). Control material was treated in parallel under similar conditions minus addition of EMS. After the treatment, shoot tips were rinsed a minimum of five times in distilled water and subsequently placed into culture media. After 24 h, mutagenized tissue was transferred into fresh media and incubated for an additional 30 days as described above. After this period, the fresh weight of mutagenized and nonmutagenized tissue was recorded and survivability measured. This was used as a qualitative measure of response to mutagenic treatment. To evaluate the effect of mutagenesis on sample weight, analysis of variance (ANOVA) was conducted using Statistica 9.0 software (StatSoft®, Tulsa, OK). Significant differences were observed between treated and control samples (P = 0.05). This was used to select parameters for bulk mutagenesis.
Bulk mutagenesis
To balance mutation density and survivability, a series of treatments was applied. A total of 4000 shoot tips were treated with EMS in 1000 sample batches for 3 h (1% EMS), 6 h (0.5% EMS), 24 h (0.125% EMS) and 48 h (0.06% EMS). After mutagenic treatment, groups of 10 isolated shoot tips were placed in individual culture flasks. Each shoot tip cutting was defined as the progenitor material for a subsequent set of related sibling progeny (referred to as a line). Cultures were transferred weekly into fresh liquid media to reduce possible accumulation of phenolic compounds because of the stress of mutagenesis. Thirty days after the treatment, survival rates of the mutagenized population were recorded. At this point, each progenitor in vitro plantlet was transferred into a separate flask with fresh media and assigned a unique number that represents the line number for siblings that will be produced from this individual. The progenitor material was labelled M1V1. V1 signifies the first vegetative generation after mutagenic treatment. Increasing numbers following V represent successive vegetative generations, and increasing numbers after M indicate meiotic propagation. This allows tracking of generations in both facultative and obligate vegetatively propagated species. To remove potential chimeric sectors caused by the random mutagenesis of different cells in the shoot tip, repeated propagation was carried out through the successive division of apical shoot tip meristems as described above. At each division, material was transferred into fresh culture media and allowed to recover for 4–5 weeks. This process was repeated five times to make a population of M1V6 individuals (Figure 1). From this, a total of 1000 plants were randomly selected for DNA extraction and mutation screening. These plants were transferred to solid media containing culture media plus 2.2% Gelrite and maintained at 22° ± 2° under stationary incubation and 12 h light cycle for the duration of the study.
Genomic DNA isolation
Approximately 50 mg of fresh leaf tissue was harvested from the randomly selected M1V6 plants, and genomic DNA was extracted using the DNeasy 96 Plant Kit (Qiagen, Valencia, CA). Sample quality and quantity was evaluated using standard agarose gel assays and normalized to a concentration of 3 ng/μL. Seven hundred and sixty-eight DNA samples were arrayed and pooled eightfold using a two-dimensional pooling strategy (Till et al., 2006). Samples were diluted to 0.1 ng/μL in Tris–EDTA (TE) buffer prior to PCR amplification.
Primer design, TILLING and evaluation of induced and natural mutations
Primers were designed for 23 genic target regions from nine publically available BAC clones using the CODDLe and Primer 3 software with default parameters for TILLING assays (Till et al., 2006). Melting temperatures ranged between 67° and 73° (Table 1). PCR amplification, enzymatic mismatch cleavage using a crude celery juice extract, mutation detection on a LI-COR DNA analyzer, data analysis using GelBuddy software and sequence validation of identified mutations was performed as previously described (Till et al., 2006). The effect of induced and natural mutations on protein sequence was evaluated using the PARSESNP programme that incorporates the SIFT algorithm (Ng and Henikoff, 2006; Taylor and Greene, 2003). Nucleotide positions for alleles were derived from annotated regions of reference sequences listed in Table 1. Sequences of induced and natural alleles are deposited in GenBank (http://www.ncbi.nlm.nih.gov/) with accession numbers JX123279 through JX123314.
Estimation of mutation density
Population mutation density was calculated by dividing the total number of sequence-validated mutations by the total number of bases screened (sum of amplicon sizes multiplied by total number of samples screened). This was corrected for the terminal 100 base pairs on each fragment end where mutation discovery is hindered (Greene et al., 2003). Because sibling plants derived from a single meristem may be clonally related and harbouring the same mutations, mutation density was adjusted by estimating the number of genotypically unique individuals in the screened population. This was carried out by screening multiple sibling plants of the same lineage to evaluate the segregation of induced mutations (Table 3). The resulting population size of unique individuals was used in the calculation of mutation density. A further correction factor for mutation density was applied because mutation screening was performed simultaneously on all three genomes rather than screening only one homeologue in a polyploid species as previously described (Slade et al., 2005). Accurate recovery of single nucleotide variations by simultaneous screening of homeologous sequences was previously shown in studies evaluating natural nucleotide variation in Musa (Till et al., 2010). The calculation used for mutation density is as follows:
 |
To differentiate between mutation densities reported in TILLING literature from meiotically propagated plants and animals, we refer to this adjusted mutation density as MMD (mitotic mutation density).
Heritability of induced mutations
Plants harbouring three confirmed mutant alleles (MALSYN G1309A, FTSJMT C623T and FTSJMT C986T) were selected to evaluate the heritability of induced mutations in culture conditions. In vitro plantlets were propagated and maintained in solid media for a period of 12 months up to the M1V9 generation. Presence or absence of the induced allele was evaluated using Sanger sequencing.
Phenotypic evaluation of mutagenized plants
One hundred and nine randomly selected plantlets representing 27 mutant lines were propagated to the M1V10 generation as described above and transferred to the glasshouse in pots containing Torboton I soil (Gartenhilfe Ges.m.b.H, Linz, Austria). Plants were acclimatized to standardized glasshouse conditions with average temperature of 25° and 40%–50% air humidity for 24 months before measurements were recorded. Phenotypical observations were taken following standardized banana descriptors (http://cropgenebank.sgrp.cgiar.org/images/file/learning_space/descriptors_banana.pdf, accessed 8 August 2012).
Acknowledgments
The authors would like to thank Andreas Draganitsch and Guenter Berthold for general support in the laboratory and glasshouse, Solotiana Harivelo Andrianaivo for assistance in postmutagenesis micropropagation of banana culture, Marilyn Alforque for assistance in plant phenotyping, Mirta Matijevic for assistance with sequencing reactions and Rachel Howard-Till and Brian Forster for critical comments on the manuscript.
Conflict of interest
The authors have no conflict of interest related to the work described in this manuscript.
Supporting information
Additional supporting information may be found in the online version of this article:
Table S1 Plant survival after treatment with EMS.
Table S2 Mutagenesis treatment and line numbers of banana samples included in this study.
Table S3 Expected and observed mutations for tested amplicons.
Table S4 Frequencies of observed phenotypesafter 24 months greenhouse growth.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ahloowalia BS, Maluszynski M, Nichterlein K. Global impact of mutation-derived varieties. Euphytica. 2004;135:187–204. [Google Scholar]
- Balkema GH. Diplontic drift in chimeric plants. Radiat. Bot. 1972;12:51–55. [Google Scholar]
- Barkley NA, Wang ML. Application of TILLING and EcoTILLING as reverse genetic approaches to elucidate the function of genes in plants and animals. Curr. Genomics. 2008;9:212–226. doi: 10.2174/138920208784533656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt AB. DNA damage and repair in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:75–100. doi: 10.1146/annurev.arplant.47.1.75. [DOI] [PubMed] [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R. A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.) Plant J. 2004;40:143–150. doi: 10.1111/j.1365-313X.2004.02190.x. [DOI] [PubMed] [Google Scholar]
- Carles CC, Fletcher JC. Shoot apical meristem maintenance: the art of a dynamic balance. Trends Plant Sci. 2003;8:394–401. doi: 10.1016/S1360-1385(03)00164-X. [DOI] [PubMed] [Google Scholar]
- Comai L, Henikoff S. TILLING: practical single-nucleotide mutation discovery. Plant J. 2006;45:684–694. doi: 10.1111/j.1365-313X.2006.02670.x. [DOI] [PubMed] [Google Scholar]
- Cooper JL, Greene EA, Till BJ, Codomo CA, Wakimoto BT, Henikoff S. Retention of induced mutations in a Drosophila reverse-genetic resource. Genetics. 2008;180:661–667. doi: 10.1534/genetics.108.092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronauer SS, Krikorian AD. Multiplication of Musa from excised stem tips. Ann. Bot. 1984;53:321–328. doi: 10.1093/oxfordjournals.aob.a086696. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C, Jabbari K, Cardi C, Poulain J, Souquet M, Labadie K, Jourda C, Lengelle J, Rodier-Goud M, Alberti A, Bernard M, Correa M, Ayyampalayam S, McKain MR, Leebens-Mack J, Burgess D, Freeling M, Mbeguie AMD, Chabannes M, Wicker T, Panaud O, Barbosa J, Hribova E, Heslop-Harrison P, Habas R, Rivallan R, Francois P, Poiron C, Kilian A, Burthia D, Jenny C, Bakry F, Brown S, Guignon V, Kema G, Dita M, Waalwijk C, Joseph S, Dievart A, Jaillon O, Leclercq J, Argout X, Lyons E, Almeida A, Jeridi M, Dolezel J, Roux N, Risterucci AM, Weissenbach J, Ruiz M, Glaszmann JC, Quetier F, Yahiaoui N, Wincker P. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul H. Present aspects of induced mutations in plant breeding. Euphytica. 1958;7:275–289. [Google Scholar]
- Gilchrist EJ, O’Neil NJ, Rose AM, Zetka MC, Haughn GW. TILLING is an effective reverse genetics technique for Caenorhabditis elegans. BMC Genomics. 2006;7:262. doi: 10.1186/1471-2164-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR, Comai L, Henikoff S. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics. 2003;164:731–740. doi: 10.1093/genetics/164.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Laux T. Stem cell regulation in the shoot meristem. J. Cell Sci. 2003;116:1659–1666. doi: 10.1242/jcs.00406. [DOI] [PubMed] [Google Scholar]
- van Harten AM. Mutation Breeding. Theory and Practical Applications. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- Henikoff S, Till BJ, Comai L. TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 2004;135:630–636. doi: 10.1104/pp.104.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Miller ES, Burkart-Waco D, Comai L. Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics. 2010;186:1231–1245. doi: 10.1534/genetics.110.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T. Domestication, genomics and the future for banana. Ann. Bot. (Lond) 2007;100:1073–1084. doi: 10.1093/aob/mcm191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. A fate map of the Arabidopsis embryonic shoot apical meristem. Development. 1992;115:745–753. [Google Scholar]
- Jain SM, Till B, Suprasanna P, Roux N. Mutations and cultivar development of banana. In: Pillay M, Tenkouano A, editors. Banana Breeding, Progress and Challenges. Boca Raton, FL: CRC Press; 2011. pp. 203–217. [Google Scholar]
- Jankowicz-Cieslak J, Huynh OA, Bado S, Matijevic M, Till BJ. Reverse-genetics by TILLING expands through the plant kingdom. Emir. J. Food Agric. 2011;23:290–300. [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y. Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 2000;43:179–188. doi: 10.1023/a:1006423110134. [DOI] [PubMed] [Google Scholar]
- Klekowski EJ, Kazarinovafukshansky N. Shoot apical meristems and mutation – selective loss of disadvantageous cell genotypes. Am. J. Bot. 1984;71:28–34. [Google Scholar]
- Kurowska M, Daszkowska-Golec A, Gruszka D, Marzec M, Szurman M, Szarejko I, Maluszynski M. TILLING – a shortcut in functional genomics. J. Appl. Genet. 2011;52:371–390. doi: 10.1007/s13353-011-0061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano M, Gradziel TM. Genetic mosaics and plant improvement. In: Janick J, editor. Plant Breeding Reviews. New York: John Wiley& Sons, Inc; 1997. pp. 43–77. [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeted screening for induced mutations. Nat. Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Artificial transmutation of the gene. Science. 1927;66:84–87. doi: 10.1126/science.66.1699.84. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Some genetic aspects of sex. Am. Nat. 1932;66:118–138. [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat. Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Murashig T. Plant propagation through tissue-cultures. Annu. Rev. Plant Physiol. 1974;25:135–166. [Google Scholar]
- Muth J, Hartje S, Twyman RM, Hofferbert HR, Tacke E, Prufer D. Precision breeding for novel starch variants in potato. Plant Biotechnol. J. 2008;6:576–584. doi: 10.1111/j.1467-7652.2008.00340.x. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Ohta T, Watanabe-Akanuma M, Yamagata H. A comparison of mutation spectra detected by the Escherichia coli lac(+) reversion assay and the Salmonella typhimurium his(+) reversion assay. Mutagenesis. 2000;15:317–323. doi: 10.1093/mutage/15.4.317. [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Brachmann A, Welham T, Binder A, Charpentier M, Groth M, Haage K, Markmann K, Wang TL, Parniske M. TILLING in lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiol. 2009;151:1281–1291. doi: 10.1104/pp.109.142190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley JH. Vegetative propagation from the standpoint of plant anatomy. In: Agriculture U.S.D.o., editor. Technical Bulletin No. 151. Washington, DC: United States Department of Agriculture; 1929. pp. 1–89. [Google Scholar]
- Roux N, Dolezel J, Swennen R, Zapata-Arias FJ. Effectiveness of three micropropagation techniques to dissociate cytochimeras in Musa spp. Plant Cell Tiss Org. 2001;66:189–197. [Google Scholar]
- Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 2005;23:75–81. doi: 10.1038/nbt1043. [DOI] [PubMed] [Google Scholar]
- Stadler LJ. Mutations in Barley induced by X-Rays and radium. Science. 1928;68:186–187. doi: 10.1126/science.68.1756.186. [DOI] [PubMed] [Google Scholar]
- Stadler LJ. Chromosome number and the mutation rate in Avena and Triticum. Proc. Natl. Acad. Sci. USA. 1929;15:876–881. doi: 10.1073/pnas.15.12.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NE, Greene EA. PARSESNP: a tool for the analysis of nucleotide polymorphisms. Nucleic Acids Res. 2003;31:3808–3811. doi: 10.1093/nar/gkg574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, Henikoff JG, Comai L, Henikoff S. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Zerr T, Comai L, Henikoff S. A protocol for TILLING and Ecotilling in plants and animals. Nat. Protoc. 2006;1:2465–2477. doi: 10.1038/nprot.2006.329. [DOI] [PubMed] [Google Scholar]
- Till BJ, Cooper J, Tai TH, Colowit P, Greene EA, Henikoff S, Comai L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 2007;7:19. doi: 10.1186/1471-2229-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Jankowicz-Cieslak J, Sagi L, Huynh OA, Utsushi H, Swennen R, Terauchi R, Mba C. Discovery of nucleotide polymorphisms in the Musa gene pool by Ecotilling. Theor. Appl. Genet. 2010;121:1381–1389. doi: 10.1007/s00122-010-1395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, Comai L, Dubcovsky J. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepalli P, Fulton L, Batoux M, Yadav RK, Schneitz K. Structure-function analysis of STRUBBELIG, an Arabidopsis atypical receptor-like kinase involved in tissue morphogenesis. PLoS ONE. 2011;6:e19730. doi: 10.1371/journal.pone.0019730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara R, Nakane C, Takimoto K. A new system for detecting mutations in arabidopsis thaliana and the mutational spectra resulting from ethylmethanesulfonate treatment. J. Radiat. Res. (Tokyo) 2006;47:223–228. doi: 10.1269/jrr.0623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


