Abstract
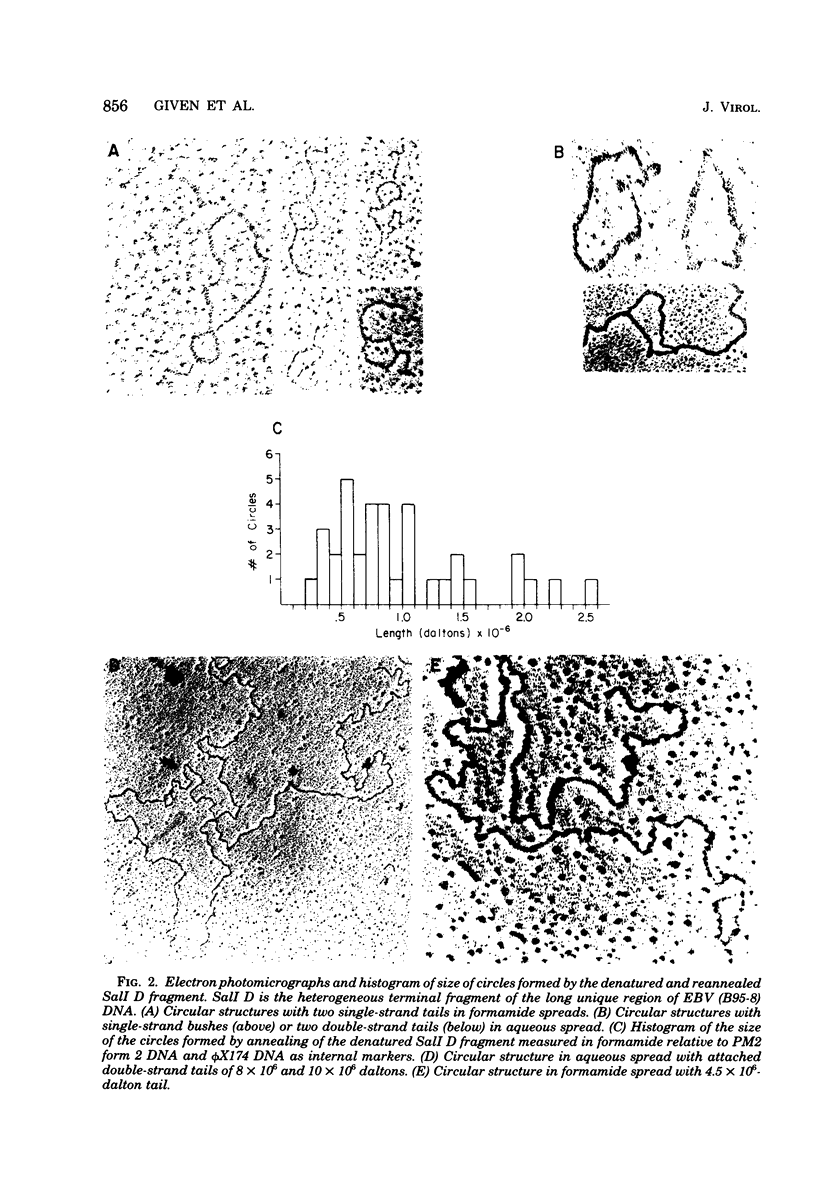
Previous data indicated that Epstein-Barr virus DNA is terminated at both ends by direct or inverted repeats of from 1 to 12 copies of a 3 X 10(5)-dalton sequence. Thus, restriction endonuclease fragments which include either terminus vary in size by 3 X 10(5)-dalton increments (D. Given and E. Kieff, J. Virol. 28:524--542, 1978; S. D. Hayward and E. Kieff, J. Virol. 23:421--429, 1977). Furthermore, defined fragments containing either terminus hybridize to each other (Given and Kieff, J. Virol. 28:524--542, 1978). The 5' ends of the DNA are susceptible to lambda exonuclease digestion (Hayward and Kieff, J. Virol. 23:421--429, 1977). To determine whether the terminal DNA is a direct or inverted repeat, the structures formed after denaturation and reannealing of the DNA from one terminus and after annealing of lambda exonuclease-treated DNA were examined in the electron microscope. The data were as follows. (i) No inverted repeats were detected within the SalI D or EcoRI D terminal fragments of Epstein-Barr virus DNA. The absence of "hairpin- or pan-handle-like" structures in denatured and partially reannealed preparations of the SalI D or EcoRI D fragment and the absence of repetitive hairpin- or pan-handle-like structures in the free 5' tails of DNA treated with lambda exonuclease indicate that there is no inverted repeat within the 3 X 10(5)-dalton terminal reiteration. (ii) Denatured SalI D or EcoRI D fragments reanneal to form circles ranging in size from 3 X 10(5) to 2.5 X 1O(6) daltons, indicating the presence of multiple direct repeats within this terminus. (iii) Lambda exonuclease treatment of the DNA extracted from virus that had accumulated in the extracellular fluid resulted in asynchronous digestion of ends and extensive internal digestion, probably a consequence of nicks and gaps in the DNA. Most full-length molecules, after 5 min of lambda exonuclease digestion, annealed to form circles, indicating that there exists a direct repeat at both ends of the DNA. (iv) The finding of several circularized molecules with small, largely double-strand circles at the juncture of the ends indicates that the direct repeat at both ends is directly repeated within each end. Hybridization between the direct repeats at the termini is likely to be the mechanism by which Epstein-Barr virus DNA circularizes within infected cells (T. Lindahl, A. Adams, G. Bjursell, G. W. Bornkamm, C. Kaschka-Dierich, and U. Jehn, J. Mol. Biol. 102:511-530, 1976).
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A., Bjursell G., Kaschka-Dierich C., Lindahl T. Circular Epstein-Barr virus genomes of reduced size in a human lymphoid cell line of infectious mononucleosis origin. J Virol. 1977 May;22(2):373–380. doi: 10.1128/jvi.22.2.373-380.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A., Lindahl T. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1477–1481. doi: 10.1073/pnas.72.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D. M., Radding C. M. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J Biol Chem. 1971 Apr 25;246(8):2502–2512. [PubMed] [Google Scholar]
- Dolyniuk M., Pritchett R., Kieff E. Proteins of Epstein-Barr virus. I. Analysis of the polypeptides of purified enveloped Epstein-Barr virus. J Virol. 1976 Mar;17(3):935–949. doi: 10.1128/jvi.17.3.935-949.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D. Molecular weights of coliphages and coliphage DNA. IV. Molecular weights of DNA from bacteriophages T4, T5 and T7 and the general problem of determination of M. J Mol Biol. 1970 Dec 28;54(3):567–577. doi: 10.1016/0022-2836(70)90127-0. [DOI] [PubMed] [Google Scholar]
- Gerber P., Nkrumah F. K., Pritchett R., Kieff E. Comparative studies of Epstein-Barr virus strains from Ghana and the United States. Int J Cancer. 1976 Jan 15;17(1):71–81. doi: 10.1002/ijc.2910170111. [DOI] [PubMed] [Google Scholar]
- Given D., Kieff E. DNA of Epstein-Barr virus. IV. Linkage map of restriction enzyme fragments of the B95-8 and W91 strains of Epstein-Barr Virus. J Virol. 1978 Nov;28(2):524–542. doi: 10.1128/jvi.28.2.524-542.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HYman R. W., Oakes J. E., Kudler L. In vitro repair of the preexisting nicks and gaps in herpes simplex virus DNA. Virology. 1977 Jan;76(1):286–294. doi: 10.1016/0042-6822(77)90303-8. [DOI] [PubMed] [Google Scholar]
- Hayward S. D., Kieff E. DNA of Epstein-Barr virus. II. Comparison of the molecular weights of restriction endonuclease fragments of the DNA of Epstein-Barr virus strains and identification of end fragments of the B95-8 strain. J Virol. 1977 Aug;23(2):421–429. doi: 10.1128/jvi.23.2.421-429.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson E., Miller G., Robinson J., Heston L. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology. 1977 Jan;76(1):152–163. doi: 10.1016/0042-6822(77)90292-6. [DOI] [PubMed] [Google Scholar]
- Jehn U., Lindahl T., Klein C. Fate of virus DNA in the abortive infection of human lymphoid cell lines by Epstein-Barr virus. J Gen Virol. 1972 Sep;16(3):409–412. doi: 10.1099/0022-1317-16-3-409. [DOI] [PubMed] [Google Scholar]
- Jondal M., Klein G. Surface markers on human B and T lymphocytes. II. Presence of Epstein-Barr virus receptors on B lymphocytes. J Exp Med. 1973 Dec 1;138(6):1365–1378. doi: 10.1084/jem.138.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Adams A., Bjursell G., Bornkamm G. W., Kaschka-Dierich C., Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976 Apr 15;102(3):511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- Masamune Y., Fleischman R. A., Richardson C. C. Enzymatic removal and replacement of nucleotides at single strand breaks in deoxyribonucleic acid. J Biol Chem. 1971 Apr 25;246(8):2680–2691. [PubMed] [Google Scholar]
- Menezes J., Jondal M., Leibold W., Dorval G. Epstein-Barr virus interactions with human lymphocyte subpopulations: virus adsorption, kinetics of expression of Epstein-Barr virus-associated nuclear antigen, and lymphocyte transformation. Infect Immun. 1976 Feb;13(2):303–310. doi: 10.1128/iai.13.2.303-310.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Lipman M. Comparison of the yield of infectious virus from clones of human and simian lymphoblastoid lines transformed by Epstein-Barr virus. J Exp Med. 1973 Dec 1;138(6):1398–1412. doi: 10.1084/jem.138.6.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci U S A. 1973 Jan;70(1):190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson K., Klein G., Henle W., Henle G. The establishment of lymphoblastoid lines from adult and fetal human lymphoid tissue and its dependence on EBV. Int J Cancer. 1971 Nov 15;8(3):443–450. doi: 10.1002/ijc.2910080312. [DOI] [PubMed] [Google Scholar]
- Pattengale P. K., Smith R. W., Gerber P. Selective transformation of B lymphocytes by E.B. virus. Lancet. 1973 Jul 14;2(7820):93–94. doi: 10.1016/s0140-6736(73)93286-8. [DOI] [PubMed] [Google Scholar]
- Pizzo P. A., Magrath I. T., Chattopadhyay S. K., Biggar R. J., Gerber P. A new tumour-derived transforming strain of Epstein-Barr virus. Nature. 1978 Apr 13;272(5654):629–631. doi: 10.1038/272629a0. [DOI] [PubMed] [Google Scholar]
- Pritchett R. F., Hayward S. D., Kieff E. D. DNA of Epstein-Barr virus. I. Comparative studies of the DNA of Epstein-Barr virus from HR-1 and B95-8 cells: size, structure, and relatedness. J Virol. 1975 Mar;15(3):556–559. doi: 10.1128/jvi.15.3.556-559.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Traub N., Pritchett R., Kieff E. DNA of Epstein-Barr virus. III. Identification of restriction enzyme fragments that contain DNA sequences which differ among strains of Epstein-Barr virus. J Virol. 1978 Aug;27(2):388–398. doi: 10.1128/jvi.27.2.388-398.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekosh D. M., Russell W. C., Bellet A. J., Robinson A. J. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977 Jun;11(2):283–295. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- Rymo L., Forsblom S. Cleavage of Epstein-Barr virus DNA by restriction endonucleases EcoRI, HindIII and BamI. Nucleic Acids Res. 1978 Apr;5(4):1387–1402. doi: 10.1093/nar/5.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Sriprakash K. S., Lundh N., Huh MM-O, Radding C. M. The specificity of lambda exonuclease. Interactions with single-stranded DNA. J Biol Chem. 1975 Jul 25;250(14):5438–5445. [PubMed] [Google Scholar]
- Sugden B., Summers W. C., Klein G. Nucleic acid renaturation and restriction endonuclease cleavage analyses show that the DNAs of a transforming and a nontransforming strain of Epstein-Barr virus share approximately 90% of their nucleotide sequences. J Virol. 1976 May;18(2):765–775. doi: 10.1128/jvi.18.2.765-775.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]







