Abstract
Background
Rectal cancer is one of the most common cancers in the world. Early detection and early therapy are important for the control of death caused by rectal cancer. The present study aims to investigate the genomic alterations in rectal adenoma and carcinoma.
Methods
We detected the genomic changes of 8 rectal adenomas and 8 carcinomas using array CGH. Then 14 genes were selected for analyzing the expression between rectal tumor and paracancerous normal tissues as well as from adenoma to carcinoma by real-time PCR. The expression of GPNMB and DIS3 were further investigated in rectal adenoma and carcinoma tissues by immunohistochemistry.
Results
We indentified ten gains and 22 losses in rectal adenoma, and found 25 gains and 14 losses in carcinoma. Gains of 7p21.3-p15.3, 7q22.3-q32.1, 13q13.1-q14.11, 13q21.1-q32.1, 13q32.2-q34, 20p11.21 and 20q11.23-q12 and losses of 17p13.1-p11.2, 18p11.32-p11.21 and 18q11.1-q11.2 were shared by both rectal adenoma and carcinoma. Gains of 1q, 6p21.33-p21.31 and losses of 10p14-p11.21, 14q12-q21.1, 14q22.1-q24.3, 14q31.3-q32.1, 14q32.2-q32.32, 15q15.1-q21.1, 15q22.31 and 15q25.1-q25.2 were only detected in carcinoma but not in adenoma. Copy number and mRNA expression of EFNA1 increased from rectal adenoma to carcinoma. C13orf27 and PMEPA1 with increased copy number in both adenoma and carcinoma were over expressed in rectal cancer tissues. Protein and mRNA expression of GPNMB was significantly higher in cancer tissues than rectal adenoma tissues.
Conclusion
Our data may help to identify the driving genes involved in the adenoma-carcinoma progression.
Background
Rectal cancer is the 5th leading cause of cancer-related death and its incidence is increasing at a rate of 4.2% per year in China [1]. Early detection and early therapy are important for the control of death caused by rectal cancer.
The majority of epithelial cancers arise through a stepwise progression from normal cells, through dysplasia, into malignant cells that have invasive and metastatic potential. The classic example of this process is the colorectal adenoma to carcinoma progression [2,3]. Genomic aberrations are found frequently in cancers and are believed to contribute to initiation and progression of cancer by deletion-induced down-expression of tumor suppressor genes or amplification and activation of oncogenes. In colorectal cancer the most frequent chromosomal aberrations were gains at 7p, 7q, 8q, 13q, and 20q and losses of 1p, 4p, 4q, 5q, 8p, 14q, 15q, 17p and 18q [4-9]. In particular, 8q, 13q and 20q gains and 8p, 15q and 18q losses are linked with colorectal adenoma to carcinoma progression. However, most of published reports are focused on colon cancer. Little information is available concerning the genomic aberrations of rectal carcinoma, especially DNA copy number changes in the progression from adenoma to tumor.
In the present study, we investigated the genomic aberrations of rectal adenoma and carcinoma by oligonucleotide-based array CGH, and identified common and different alterated chromosome regions between rectal adenoma and carcinoma. Then the expression of 15 genes at selected chromosome regions above was analyzed by real-time PCR or immunohistochemistry.
Methods
Patients and samples
Biopsy tissues from 22 rectal adenoma patients and 36 rectal carcinoma patients were collected by the Department of Endoscopy, Cancer Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China. Biopsy samples were obtained by colonoscopy and stored at −80°C. Definitive pathological result from a biopsy was obtained at a later clinical course. An experienced pathologist confirmed that normal cell content of all the samples was less than 40% by HE staining. All the samples used in this study were residual specimens after diagnosis sampling. And all patients signed separate informed consent forms for sampling and research. The clinicopathological characteristics of the patients in array CGH assay are summarized in Table 1.
Table 1.
Clinical Characteristics of 16 Patients Studied by Array CGH
| Case No. | Sex | Age | Type | Location |
|---|---|---|---|---|
| 1 |
F |
52 |
Adenoma |
Rectum |
| 2 |
F |
49 |
Adenoma |
Rectum |
| 3 |
M |
75 |
Adenoma |
Rectum |
| 4 |
M |
47 |
Adenoma |
Rectum |
| 5 |
M |
57 |
Adenoma |
Rectum |
| 6 |
F |
61 |
Adenoma |
Rectum |
| 7 |
M |
69 |
Adenoma |
Rectum |
| 8 |
F |
75 |
Adenoma |
Rectum |
| 9 |
M |
69 |
Carcinoma |
Rectum |
| 10 |
M |
61 |
Carcinoma |
Rectum |
| 11 |
F |
70 |
Carcinoma |
Rectum |
| 12 |
F |
73 |
Carcinoma |
Rectum |
| 13 |
M |
42 |
Carcinoma |
Rectum |
| 14 |
M |
32 |
Carcinoma |
Rectum |
| 15 |
F |
31 |
Carcinoma |
Rectum |
| 16 | M | 66 | Carcinoma | Rectum |
Genomic DNA extraction and array-based CGH
Genomic DNA was isolated from tumor tissues using the Qiagen DNeasy Blood & Tissue Kit as described by the manufacturer (Qiagen, Hilden, Germany).
Array CGH experiments were performed using standard Agilent protocols (Agilent Technologies, Santa Clara, CA). Commercial human genomic DNA (PROMEGA, Warrington, UK) was used as reference. For each CGH hybridization, 500 ng of reference genomic DNA and the same amount of tumor DNA were digested with Alu I and RSA I restriction enzyme (PROMEGA, Warrington, UK). The digested reference DNA fragments were labeled with cyanine-3 dUTP and the tumor DNA with cyanine-5 dUTP (Agilent Technologies, Santa Clara, CA). After clean-up, reference and tumor DNA probes were mixed and hybridized onto Agilent 44K human genome CGH microarray (Agilent) for 40 h. Washing, scanning and data extraction procedures were performed following standard protocols.
Array CGH data set is available at Gene Expression Omnibus (GEO) http://www.ncbi.nlm.nih.gov/geo/[10], accession number GSE34472.
Microarray data analysis
Microarray data were analyzed using Agilent Genomic Workbench (Agilent Technologies, Santa Clara, CA) and MD-SeeGH (http://www.flintbox.ca). The Aberration Detection Method 2 algorithm with threshold at 6 (Agilent Genomic Workbench) was applied to identify common genomic aberrations. Mean Log2ratio of all probes in a chromosome region between 0.125 and 0.5 was classified as genomic gain, > 0.5 as high-level DNA amplification, < −0.125 as hemizygous loss, and < −0.5 as homozygous deletion. Minimal regions of gains or losses in our study defined as the smallest overlapping aberrant chromosomal regions identified by Agilent Genomic Workbench. Frequency plot comparison method (MD-SeeGH) was used to compare frequency of DNA copy number changes between rectal adenoma and carcinoma.
Total RNA extraction and real-time PCR
Total RNA was isolated from tissues using the RNeasy Mini Kit as described by the manufacturer (Qiagen, Hilden, Germany).
The PCR reactions were performed in a total volume of 20 μl, including 10 μl of 2 X SYBR ® Green PCR Master Mix (Applied Biosystems, Warrington, UK), 2 μl of cDNA (5 ng/μl), 1 μl of primer mix (10 μM each). The PCR amplification and detection were carried out in a 7300 Real Time PCR System (Applied Biosystems) for 45 cycles, each with 15 s at 95 °C, 1 min at 60 °C, and initial denaturation with 10 min at 95 °C. The relative gene expression was calculated using the comparative CT Method [11]. The copy number of the target gene normalized to an endogenous reference (GAPDH), and relative to calibrator was given by the formula 2 − ΔΔCt. ΔCT was calculated by subtracting the average GAPDH CT from the average CT of the gene of interest. The ratio defines the level of relative expression of the target gene to that of GAPDH.
Immunohistochemical staining
Formalin-fixed, paraffin-embedded specimens of rectal adenoma and carcinoma were detected in immunohistochemistry assay. Tissues of each case were repeated for three times. The slides were deparaffinized, rehydrated, immersed in 3% hydrogen peroxide solution for 10 min, heated in citrate buffer (pH 6) for 25 min at 95°C, and cooled for 60 min at room temperature. The slides were blocked by 10% normal goat serum for 30 min at 37°C and then incubated with rabbit polyclonal antibody against DIS3 (PTGLab), rabbit polyclonal antibody against GPNMB (PTGLab) overnight at 4°C. After being washed with PBS, the slides were incubated with biotinylated secondary antibody (diluted 1:100) for 30 min at 37°C, followed by streptavidin-peroxidase (1:100 dilution) incubation for 30 min at 37°C. Immunolabeling was visualized with a mixture of 3,3'-diaminobenzidine solution. Counterstaining was carried out with hematoxylin.
Expression level was determined on the basis of staining intensity and percentage of immunoreactive cells. Negative expression (score = 0) was no or faint staining, or moderate to strong staining in <25% of cells. Weak expression (score = 1) was a moderate or strong staining in 25% to 50% of cells. And strong expression (score = 2) was > 50% of the cells with strong staining. Weak expression and strong expression defined as positive staining.
Statistical analysis
Statistical analyses were conducted using the Student’s t-test and performed with the statistical software SPSS 15.0. The differences were judged as statistically significant when the corresponding two-sided P value were <.05.
Results
Recurrent copy number alterations in rectal adenoma and carcinoma detected by array CGH
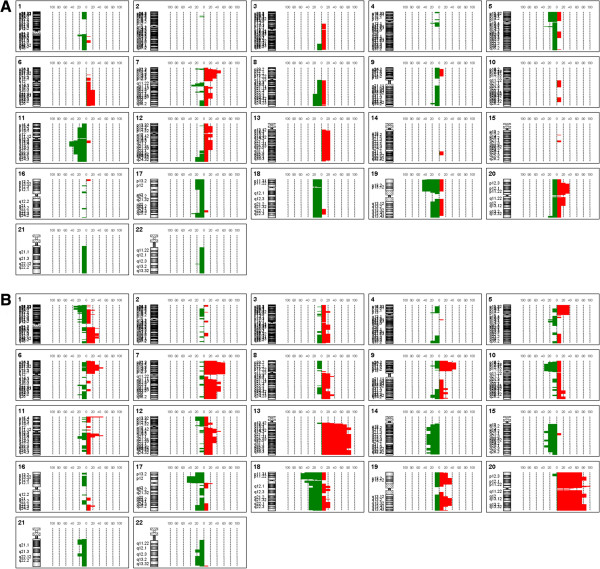
Seven out of eight adenomas and all of carcinomas had genomic aberrations. More alterations were observed in patients of rectal cancer than adenoma, and the numbers of changes were 39.13±20.48 and 14.3±6.164, respectively (Additional file 1: Figure S1). Array CGH results showed that the most frequent copy number alterations in rectal adenoma were gains of 7p21.3-p15.3 and 20p12.3-p11.21 and losses of 5q13.2, 7q11.23, 11q13.1-q14.1, 17q25.1 and 19p13.3-p13.11 (Figure 1A, Tables 2 and 3). And the most common genetic aberrations in rectal carcinoma were gains of 7p21.3-p15.3, 7p15.3-p14.1, 7p14.1-p13, 7p13-p11.2, 13q13.1-q14.11, 13q21.1-q32.1, 13q32.1-q34, 20p11.21, 20q11.23-q12 and 20q13.2-q13.33 and losses of 17p13.1-p11.2, 18p11.32-p11.21 and 18q11.1-q11.2 (Figure 1B, Tables 2 and 3).
Figure 1.
Genome-wide frequency plot of rectal adenoma (A) and adenocarcinoma (B) in array CGH assay. Line on the right of 0%-axis: gain; Line on the left of 0%-axis: loss.
Table 2.
Genomic Gains in Rectal Adenoma and Adenocarcinoma
|
Chromosome Region |
Rectal adenoma |
Rectal adenocarcinoma |
||||||
|---|---|---|---|---|---|---|---|---|
| Start | End | No. of probes | No. of cases | Start | End | No. of probes | No. of cases | |
| 1q21.3 |
|
|
|
|
150819451 |
150852905 |
3 |
3 |
| 1q25.3-q31.3 |
|
|
|
|
183720174 |
197184608 |
157 |
3 |
| 1q32.1-q41 |
|
|
|
|
204180950 |
214439909 |
173 |
3 |
| 5p13.3-p12 |
|
|
|
|
33503866 |
45681293 |
165 |
3 |
| 6p21.33-p21.31 |
|
|
|
|
30737615 |
33655570 |
151 |
4 |
| 6q16.3-q27 |
100547312 |
168205989 |
848 |
2 |
|
|
|
|
| 7p21.3-p15.3 |
11041844 |
23202043 |
119 |
4 |
7671318 |
23172047 |
142 |
5 |
| 7p15.3-p14.1 |
|
|
|
|
23821348 |
39813908 |
231 |
5 |
| 7p14.1-p13 |
|
|
|
|
40099046 |
44497196 |
64 |
5 |
| 7p13-p11.2 |
|
|
|
|
44890654 |
55242365 |
111 |
5 |
| 7q21.11-q21.12 |
81196827 |
86205180 |
42 |
2 |
|
|
|
|
| 7q21.12-q21.3 |
|
|
|
|
87207024 |
97321855 |
144 |
4 |
| 7q22.3-q32.1 |
106191096 |
127234809 |
245 |
2 |
105253205 |
127519635 |
260 |
4 |
| 8q12.1 |
|
|
|
|
59565778 |
61340797 |
21 |
3 |
| 8q24.21-q24.22 |
|
|
|
|
128816904 |
133653633 |
42 |
3 |
| 9p24.1-p21.1 |
|
|
|
|
7058096 |
31463899 |
251 |
4 |
| 11p15.5 |
|
|
|
|
192958 |
2278596 |
76 |
4 |
| 11q13.2 |
|
|
|
|
66917525 |
67689856 |
30 |
4 |
| 12p13.31-p11.21 |
9053548 |
30700931 |
337 |
2 |
|
|
|
|
| 12q12-q13.11 |
37052371 |
47174877 |
139 |
2 |
|
|
|
|
| 12q13.13 |
|
|
|
|
50568352 |
51486634 |
34 |
4 |
| 12q14.1-q22 |
57350276 |
91428773 |
354 |
2 |
|
|
|
|
| 13q13.1-q14.11 13q21.1-q32.1 13q32.3-q34 |
21038984 |
109780488 |
909 |
2 |
32490193 |
39679219 |
79 |
7 |
| |
|
|
|
|
52774228 |
94079000 |
275 |
7 |
| |
|
|
|
|
100091512 |
114022929 |
148 |
7 |
| 19p13.2-p13.11 |
|
|
|
|
9800520 |
19631574 |
473 |
3 |
| 19q13.13-q13.33 |
|
|
|
|
43396893 |
55615310 |
550 |
3 |
| 20p11.21 |
7296794 |
23132344 |
189 |
3 |
22510206 |
23380542 |
15 |
8 |
| 20q11.23-q12 |
29592072 |
42681834 |
275 |
2 |
35467169 |
41087006 |
78 |
7 |
| 20q13.2-q13.33 | 52017030 | 62323759 | 215 | 7 | ||||
Note: The number of rectal adenoma and adenocarcinoma in Array CGH study are both 8 cases.
Table 3.
Genomic Losses in Rectal Adenoma and Adenocarcinoma
|
Chromosome Region |
Rectal adenoma |
Rectal adenocarcinoma |
||||||
|---|---|---|---|---|---|---|---|---|
| Start | End | No. of probes | No. of cases | Start | End | No. of probes | No. of cases | |
| 1p36.23-p36.22 |
|
|
|
|
7804415 |
11633739 |
82 |
3 |
| 1p36.22-p36.13 |
|
|
|
|
12600054 |
16167534 |
41 |
3 |
| 1p36.12-p35.3 |
|
|
|
|
21802142 |
29525663 |
226 |
3 |
| 1q21.2-q21.3 |
148163183 |
149505863 |
60 |
2 |
|
|
|
|
| 1q21.3-q23.1 |
151880217 |
155031244 |
154 |
2 |
|
|
|
|
| 4q12 |
55913547 |
57653302 |
38 |
2 |
|
|
|
|
| 5p15.33-p12 |
260981 |
45865412 |
433 |
2 |
|
|
|
|
| 5q13.2 |
68434643 |
68900029 |
18 |
3 |
|
|
|
|
| 7p22.2-p22.1 |
4298590 |
6547570 |
42 |
2 |
|
|
|
|
| 7q11.23 |
72003839 |
75977276 |
77 |
3 |
|
|
|
|
| 7q22.1 |
99538250 |
101895994 |
79 |
2 |
|
|
|
|
| 8q22.2-q24.3 |
100781187 |
143914353 |
448 |
2 |
|
|
|
|
| 8q24.3 |
143914353 |
146250824 |
75 |
2 |
|
|
|
|
| 9q34.11 |
130111425 |
132321365 |
64 |
2 |
|
|
|
|
| 10p14-p11.21 |
|
|
|
|
11825924 |
35645512 |
315 |
3 |
| 11p15.2-p11.12 |
14750051 |
50638829 |
468 |
2 |
|
|
|
|
| 11q13.1-q14.1 |
63802950 |
80046693 |
442 |
4 |
|
|
|
|
| 12q24.23-q24.33 |
116956235 |
132193660 |
257 |
2 |
|
|
|
|
| 14q12-q21.1 |
|
|
|
|
30209271 |
38927323 |
130 |
3 |
| 14q22.1-q24.3 |
|
|
|
|
48874529 |
77750644 |
544 |
3 |
| 14q31.3-q32.1 |
|
|
|
|
87763614 |
93260389 |
110 |
3 |
| 14q32.2-q32.32 |
|
|
|
|
99254905 |
102592287 |
70 |
3 |
| 15q15.1-q21.1 |
|
|
|
|
38653893 |
42843706 |
119 |
3 |
| 15q22.31 |
|
|
|
|
61519869 |
64628895 |
74 |
3 |
| 15q25.1-q25.2 |
|
|
|
|
76206143 |
79967204 |
77 |
3 |
| 17p13.1-p11.2 |
84287 |
21386319 |
606 |
2 |
8327645 |
20974722 |
266 |
4 |
| 17q25.1 |
70528777 |
71603516 |
61 |
3 |
|
|
|
|
| 18p11.32-p11.21 |
170229 |
13875315 |
173 |
2 |
2580000 |
13752309 |
137 |
5 |
| 18q11.1-q11.2 |
16904187 |
76018409 |
684 |
2 |
16976046 |
20313378 |
51 |
4 |
| 19p13.3-p13.11 |
1432408 |
19699544 |
795 |
4 |
|
|
|
|
| 19q13.11-q13.43 |
37554715 |
63672832 |
1114 |
2 |
|
|
|
|
| 20q13.33 |
60039825 |
62320720 |
85 |
2 |
|
|
|
|
| 22q13.1 | 37689058 | 37715431 | 3 | 2 | ||||
Note: The number of rectal adenoma and adenocarcinoma in Array CGH study are both 8 cases.
Common and distinct genomic events in rectal adenoma and carcinoma
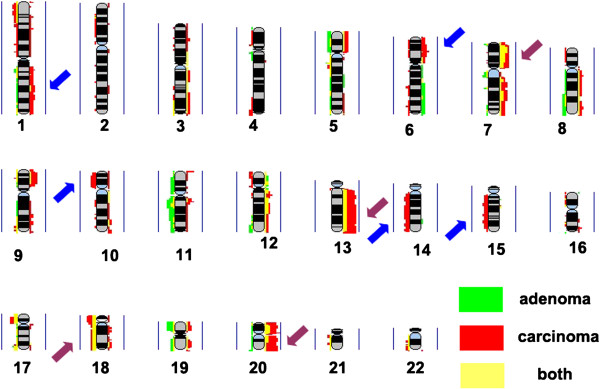
By comparing the genomic aberrations of rectal adenoma and carcinoma, we found that gains of 7p21.3-p15.3, 7q22.3-q32.1, 13q13.1-q14.11, 13q21.1-q32.1, 13q32.3-q34, 20p11.21 and 20q11.23-q12 and losses of 17p13.1-p11.2, 18p11.32-p11.21, and 18q11.1-q11.2 were shared by rectal adenoma and carcinoma. However, gains of 1q, 6p21.33-p21.31 and losses of 10p14-p11.21, 14q12-q21.1, 14q22.1-q24.3, 14q31.3-q32.1, 14q32.2-q32.32, 15q15.1-q21.1, 15q22.31 and 15q25.1-q25.2 were detected in carcinoma but not in adenoma (Figure 2, Tables 2 and 3).
Figure 2.
Frequency plot comparison of rectal adenoma and carcinoma. Red: carcinoma; green: adenoma; yellow: shared by both. The presentation is per array probe; gains and losses are represented by the colors on the right and left, respectively. Vertical blue line represents 100% of the samples. Brown and blue arrows highlight the changed chromosomal areas that were common or distinct between rectal adenoma and carcinoma, respectively.
Candidate target genes of interesting gains and losses
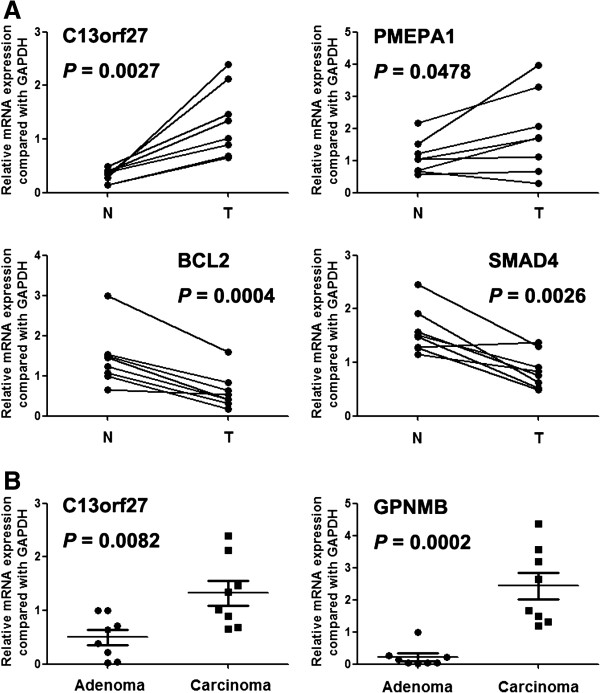
Further, we selected 14 genes of 1q, 6p, 7p, 13q, 18q and 20q to analyze the mRNA expression by real-time PCR (Table 4). Array CGH found that copy number increase of GPNMB (7p15.2), OXGR1 (13q32.1), C13orf27 (13q32.2-q34), PMEPA1 (20q13.31), PHACTR3 (20q13.32) and decrease of SMAD4 (18q21.2), BCL2 (18q21.33) occurred in both rectal adenoma and carcinoma. Our real-time PCR results showed that C13orf27 and PMEPA1 were overexpressed in rectal cancer tissues comparing with paracancerous normal tissues. BCL2 and SMAD4 were underexpressed in tumor tissue (Figure 3A). And the expression level of C13orf27 and GPNMB was significantly higher in cancer tissues than rectal adenoma tissues (Figure 3B).
Table 4.
Primers of genes in Real-time PCR assay
| Gene | Forward primer | Backward primer | Size (bp) |
|---|---|---|---|
| GAPDH |
GGTCGTATTGGGCGCCTGGTC |
TGACGGTGCCATGGAATTTGCCA |
148 |
| KIFC1 |
TCTCTGGGTGGTAGTGCTAAGA |
TAAGTCACTTCCTGTTGGCCTG |
148 |
| SOX4 |
GACCGGGACCTGGATTTTAACT |
TGAAAACCAGGTTGGAGATGCT |
133 |
| PBX2 |
AAGTTCCAAGAGGAGGCAAACA |
TCCTGAGAGATTGAAAGAGCCG |
132 |
| ESRRG |
GCTATCCTGCAGCTGGTAAAGA |
GCTATCCTGCAGCTGGTAAAGA |
133 |
| KDM5B |
CCCTCAGACACATCCTATTCCG |
CAGTCCACCTCATCTCCTTCTG |
101 |
| PTGS2 |
TGTATCCTGCCCTTCTGGTAGA |
AAGGAGAATGGTGCTCCAACTT |
85 |
| EFNA1 |
GTGGCAAAATCACTCACAGTCC |
CTATGTAGAACCCGCACCTCTG |
91 |
| BCL2 |
AGGATTGTGGCCTTCTTTGAGT |
CGGTTCAGGTACTCAGTCATCC |
113 |
| SMAD4 |
TGTTGATGACCTTCGTCGCTTA |
ATGCTCTGTCTTGGGTAATCCG |
81 |
| PHACTR3 |
TATGACAGGAGGGCAGACAAAC |
GCTTGCTTGATGCATGTACCTC |
118 |
| C13orf27 |
TCAGGCTCAGCAGATGAAATGT |
TCCAGTGGATTTTATGGGGAGC |
85 |
| PMEPA |
CTGAGCCACTACAAGCTGTCTG |
CTTCTGAGGACAGGGCATCTTC |
85 |
| OXGR1 |
ATCTTGAGGGTCATTCGGATCG |
TGTCGCTGACCACCACATATAG |
148 |
| GPNMB | GTCACTGTGATCTCCCTCTTGG | TTTGCACGGTTGAGAAAGACAC | 116 |
Figure 3.
Expression of genes which were located on the common aberrant chromosomal regions in rectal adenoma and carcinoma. N: paracancerous normal tissues; T: rectal cancer tissues.
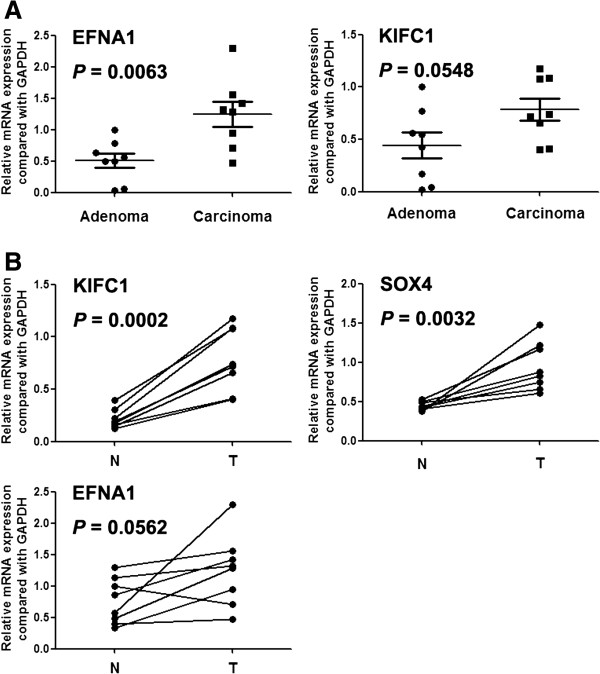
Copy number increase of EFNA1 (1q22), PTGS2 (1q31.1), KDM5B (1q32.1), ESRRG (1q41), KIFC1 (6p21.32), PBX2 (6p21.32) and SOX4 (6p22.3) were only detected in rectal cancer in array CGH. Among them, EFNA1 had increased expression in carcinoma compared with adenoma, and KIFC1 had an upward trend but not significant in statistical analysis (Figure 4A). Of these genes KIFC1 and SOX4 were also significantly overexpressed in rectal tumor tissues than paracancerous tissues (Figure 4B).
Figure 4.
Expression of genes which were located on the distinct aberrant chromosomal regions in rectal adenoma and carcinoma. N: paracancerous normal tissues; T: rectal cancer tissues.
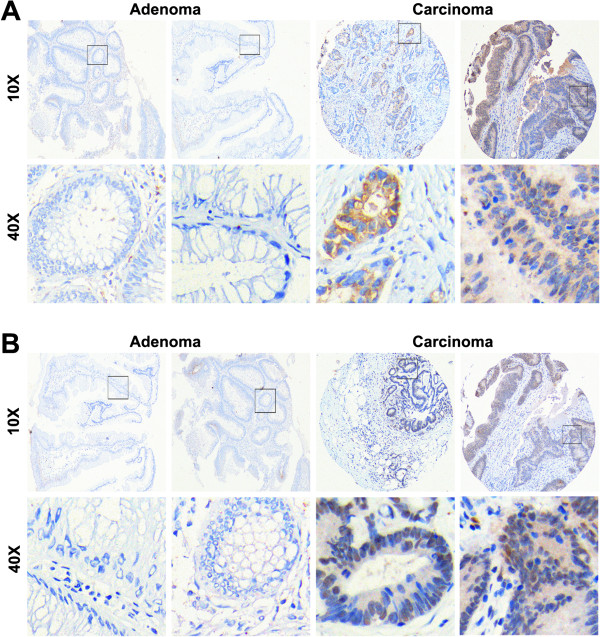
We also analyzed the protein expression of GPNMB (7p15.2) and DIS3 (13q22.1) by immunohistochemistry. Of all six detected rectal adenoma tissues, GPNMB and DIS3 had no expression. In twenty rectal cancer tissues, GPNMB and DIS3 were positively stained in six and five cases, respectively (Figure 5).
Figure 5.
Expression of GPNMB and DIS3 by immunohistochemistry assay.
Discussion
In the past decades, a number of genomic changes were found in colorectal adenoma and carcinoma, but the target genes are limited and molecular mechanism of adenoma to carcinoma progression is still unknown.
Previous studies found that 8q, 13q and 20q gains and 8p, 15q and 18q losses are linked with colorectal adenoma to carcinoma progression [4-9]. Our study narrowed down the gain regions to 13q13.1-q14.11, 13q21.1-q32.1, 13q32.2-q34 and 20q11.23-q12 and the loss regions to 18q11.2. Furthermore, gains of 7p21.3-p15.3 and 7q22.3-q32.1 and losses of 17p13.1-p11.2, 18p11.32-p11.21 were also found in both rectal adenoma and carcinoma.
Our study also showed that some genomic aberrations were present in rectal tumor but not in adenoma. They are gains of 1q and 6p21.33 and losses of 10p14-p11.21, 14q12-q21.1, 14q22.1-q24.3, 14q31.3-q32.1, 14q32.2-q32.32, 15q15.1-q21.1, 15q22.31 and 15q25.1-q25.2. These aberrations occurred at the later stages of rectal carcinogenesis, and may contribute the progression from adenoma to carcinoma.
Identifying the candidate targets underlying the genomic aberrations was important for understanding the mechanism of carcinogenesis. Carvalho et al. found that the overexpressions of C20orf24, AURKA, RNPC1, TH1L, ADRM1, C20orf20 and TCRL5 in carcinomas compared with adenomas were correlated with 20q gain [4]. Habermann et al. showed that copy number changes of 7q, 8p, 8q, 13q, 18p, 18q, 20p and 20q deregulated the average expression levels of the genes on these chromosome arms [12]. However, most of samples detected in these reports were colon cancer which had some different genomic aberrations compared with rectal cancer [13], expression-dysregulated genes in the carcinogenesis of rectum were still limited. By literature analyses, we selected 14 genes to compare their expression between in tumor and paracancerous tissues or between in rectal adenoma and carcinoma tissues. Of them, copy number and mRNA expression of EFNA1 increased from rectal adenoma to carcinoma, and C13orf27 and PMEPA1 with gains in both adenoma and carcinoma were overexpressed in rectal cancer tissues. These results revealed that copy number increase maybe the reason of expression up-regulation. Interestingly, both mRNA and protein expression of GPNMB was higher in cancer tissues than rectal adenoma tissues.
GPNMB is a type I transmembrane protein and overexpressed in several malignant human tissues relative to the corresponding normal tissues. Ectopic overexpression of GPNMB/osteoactivin can promote the metastasis and invasion of glioma, breast and hepatocellular carcinoma [14-17]. EFNA1 was overexpressed in hepatocellular carcinoma and can inhibit growth of malignant mesothelioma by phosphorylating EPHA2 [18,19]. C13orf27 was overexpressed in rectal tumor in our study, but the function of C13orf27 was unknown. PMEPA1 was also identified in our study, which is mapped to 20q13.3 is a TGF-beta inducible gene and encodes a NEDD4 E3 ubiguitin ligase binding protein. PMEPA1 is over-expressed in prostate, breast, renal cell, stomach and rectal carcinomas [20-22]. But little is known about the function of PMEPA1, Further study should be conducted to investigate the roles of the above genes in human colorectal cancer.
Loss of 18q is a common event in colorectal cancer, and 18q deletion and loss of SMAD4 expression are associated with liver metastasis. In colorectal cancer, patients with reduced SMAD4 expression frequently presented an unfavorable survival because of liver metastasis [23-26]. High expression level of SMAD4 reflected significantly longer overall and disease-free survival time than low expression level [27]. Bixiang et al. found that transgenic expression of SMAD4 can significantly reduce the oncogenic potential of MC38 and SW620 cells [28]. Our study confirmed the decreased expression of SMAD4 in rectal cancer.
In summary, we identified EFNA1 (1q), C13orf27 (13q), PMEPA1 (20q), GPNMB (7q) as candidate driving genes of genomic aberrations in rectal cancer. Further study was needed to reveal the mechanisms by which these genes may be involved in the carcinogenesis of the rectum.
Conclusions
Our data provide detailed information on genomic aberrations present in rectal adenoma or carcinoma, especially both in two groups or only in rectal cancer. Real-time PCR and immunohistochemistry assay selected EFNA1, C13orf27, PMEPA1 and GPNMB as candidate amplification targets. Our results may help to identify the driving genes involved in the adenoma-carcinoma progression.
Competing interests
The authors declared that they have no competing interest.
Authors’ contributions
ZYM participated in the collection of specimens. HJJ prepared the genomic DNA and total RNA. SZZ carried out the array CGH study. WBS performed microarray data analysis, LJW, CX and ZY carried out the real-time PCR assay. SL and ZTT performed immunohistochemistry assay. SZZ, WMR, WGQ and ZY participated in the design of the study and performed the statistical analysis. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Figure S1. Comparison of rectal adenoma and carcinoma in number of genomic aberrations.
Contributor Information
Zhi-Zhou Shi, Email: zhizhoushi@gmail.com.
Yue-Ming Zhang, Email: zhangyueming1028@126.com.
Li Shang, Email: 274271130@qq.com.
Jia-Jie Hao, Email: hjj8173@126.com.
Tong-Tong Zhang, Email: 163zttong@163.com.
Bo-Shi Wang, Email: wbs137@126.com.
Jian-Wei Liang, Email: liangjianwei0@yahoo.com.cn.
Xi Chen, Email: dejan11@sohu.com.
Ying Zhang, Email: tilamisu-jj@163.com.
Gui-Qi Wang, Email: wangguiq@126.com.
Ming-Rong Wang, Email: wangmr2015@cicams.ac.cn.
Yu Zhang, Email: zhangyu909@126.com.
Acknowledgements
The authors would like to thank Kai-Tai Zhang, Department of Etiology and Carcinogenesis of Peking Union Medical College as the help of array CGH experiment.
Funding
Supported by: This work was supported by Special Public Health Fund of China (200902002-5) and Chinese Hi-Tech R&D Program Grant (2011AA022706).
References
- Li M, Gu J. Changing patterns of colorectal cancer in China over a period of 20 years. World J Gastroenterol. 2005;11:4685–4688. doi: 10.3748/wjg.v11.i30.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- Muto T, Bussey HJ, Morson BC. The evolution of cancer of the colon and rectum. Cancer. 1975;36:2251–2270. doi: 10.1002/cncr.2820360944. [DOI] [PubMed] [Google Scholar]
- Carvalho C, Postma S, Mongera E, Hopmans S, Diskin MA, van de Wiel, Van Criekinge W, Thas O, Matthai A, Cuesta MA, Terhaar Sive Droste JS, Craanen M, Schrock E, Ylstra B, Meijer GA. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- Diep CB, Kleivi K, Ribeiro FR, Teixeira MR, Lindgjaerde OC, Lothe RA. The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer. 2006;45:31–41. doi: 10.1002/gcc.20261. [DOI] [PubMed] [Google Scholar]
- Hoglund M, Gisselsson D, Hansen GB, Sall T, Mitelman F, Nilbert M. Dissecting karyotypic patterns in colorectal tumors: two distinct but overlapping pathways in the adenoma-carcinoma transition. Cancer Res. 2002;62:5939–5946. [PubMed] [Google Scholar]
- Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, Tomlinson IP, Carter NP. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–4825. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- Nakao K, Mehta KR, Fridlyand J, Moore DH, Jain AN, Lafuente A, Wiencke JW, Terdiman JP, Waldman FM. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis. 2004;25:1345–1357. doi: 10.1093/carcin/bgh134. [DOI] [PubMed] [Google Scholar]
- Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Brennan C, Rook M, Wolfe JL, Leo C, Chin L, Pan H, Liu WH, Price B, Makrigiorgos GM. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 2004;32:e76. doi: 10.1093/nar/gnh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann JK, Paulsen U, Roblick UJ, Upender MB, McShane LM, Korn EL, Wangsa D, Kruger S, Duchrow M, Bruch HP, Auer G, Ried T. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes Chromosomes Cancer. 2007;46:10–26. doi: 10.1002/gcc.20382. [DOI] [PubMed] [Google Scholar]
- He QJ, Zeng WF, Sham JS, Xie D, Yang XW, Lin HL, Zhan WH, Lin F, Zeng SD, Nie D, Ma LF, Li CJ, Lu S, Guan XY. Recurrent genetic alterations in 26 colorectal carcinomas and 21 adenomas from Chinese patients. Cancer Genet Cytogenet. 2003;144:112–118. doi: 10.1016/S0165-4608(02)00959-7. [DOI] [PubMed] [Google Scholar]
- Tse KF, Jeffers M, Pollack VA, McCabe DA, Shadish ML, Khramtsov NV, Hackett CS, Shenoy SG, Kuang B, Boldog FL, MacDougall JR, Rastelli L, Herrmann J, Gallo M, Gazit-Bornstein G, Senter PD, Meyer DL, Lichenstein HS, LaRochelle WJ. CR011, a fully human monoclonal antibody-auristatin E conjugate, for the treatment of melanoma. Clin Cancer Res. 2006;12:1373–1382. doi: 10.1158/1078-0432.CCR-05-2018. [DOI] [PubMed] [Google Scholar]
- Onaga M, Ido A, Hasuike S, Uto H, Moriuchi A, Nagata K, Hori T, Hayash K, Tsubouchi H. Osteoactivin expressed during cirrhosis development in rats fed a choline-deficient, L-amino acid-defined diet, accelerates motility of hepatoma cells. J Hepatol. 2003;39:779–785. doi: 10.1016/S0168-8278(03)00361-1. [DOI] [PubMed] [Google Scholar]
- Rich JN, Shi Q, Hjelmeland M, Cummings TJ, Kuan CT, Bigner DD, Counter CM, Wang XF. Bone-related genes expressed in advanced malignancies induce invasion and metastasis in a genetically defined human cancer model. J Biol Chem. 2003;278:15951–15957. doi: 10.1074/jbc.M211498200. [DOI] [PubMed] [Google Scholar]
- Rose AA, Pepin F, Russo C, Abou Khalil JE, Hallett M, Siegel PM. Osteoactivin promotes breast cancer metastasis to bone. Mol Cancer Res. 2007;5:1001–1014. doi: 10.1158/1541-7786.MCR-07-0119. [DOI] [PubMed] [Google Scholar]
- Nasreen N, Mohammed KA, Lai Y, Antony VB. Receptor EphA2 activation with ephrinA1 suppresses growth of malignant mesothelioma (MM) Cancer Lett. 2007;258:215–222. doi: 10.1016/j.canlet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Cui XD, Lee MJ, Yu GR, Kim IH, Yu HC, Song EY, Kim DG. EFNA1 ligand and its receptor EphA2: potential biomarkers for hepatocellular carcinoma. Int J Cancer. 2010;126:940–949. doi: 10.1002/ijc.24798. [DOI] [PubMed] [Google Scholar]
- Giannini G, Ambrosini MI, Di Marcotullio L, Cerignoli F, Zani M, MacKay AR, Screpanti I, Frati L, Gulino A. EGF- and cell-cycle-regulated STAG1/PMEPA1/ERG1.2 belongs to a conserved gene family and is overexpressed and amplified in breast and ovarian cancer. Mol Carcinog. 2003;38:188–200. doi: 10.1002/mc.10162. [DOI] [PubMed] [Google Scholar]
- Rae FK, Hooper JD, Nicol DL, Clements JA. Characterization of a novel gene, STAG1/PMEPA1, upregulated in renal cell carcinoma and other solid tumors. Mol Carcinog. 2001;32:44–53. doi: 10.1002/mc.1063. [DOI] [PubMed] [Google Scholar]
- Xu LL, Shanmugam N, Segawa T, Sesterhenn IA, McLeod DG, Moul JW, Srivastava S. A novel androgen-regulated gene, PMEPA1, located on chromosome 20q13 exhibits high level expression in prostate. Genomics. 2000;66:257–263. doi: 10.1006/geno.2000.6214. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, Utsunomiya J, Kuroki T, Mori T. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- Losi L, Bouzourene H, Benhattar J. Loss of Smad4 expression predicts liver metastasis in human colorectal cancer. Oncol Rep. 2007;17:1095–1099. [PubMed] [Google Scholar]
- Kawakami M, Yamaguchi T, Takahashi K, Matsumoto H, Yasutome M, Horiguchi S, Hayashi Y, Funata N, Mori T. Assessment of SMAD4, p53, and Ki-67 alterations as a predictor of liver metastasis in human colorectal cancer. Surg Today. 2010;40:245–250. doi: 10.1007/s00595-009-4028-3. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Watanabe T, Kitayama J, Kanazawa T, Kazama Y, Tanaka J, Kazama S, Nagawa H. Chromosome 18q deletion as a novel molecular predictor for colorectal cancer with simultaneous hepatic metastasis. Diagn Mol Pathol. 2009;18:219–225. doi: 10.1097/PDM.0b013e3181910f17. [DOI] [PubMed] [Google Scholar]
- Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Jarvinen H, Mecklin JP, Hemminki A, Schwartz S Jr, Aaltonen LA, Arango D. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005;11:2606–2611. doi: 10.1158/1078-0432.CCR-04-1458. [DOI] [PubMed] [Google Scholar]
- Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL, Datta PK. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138:969–980. doi: 10.1053/j.gastro.2009.11.004. e961-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of rectal adenoma and carcinoma in number of genomic aberrations.