Abstract
Populus tomentosa is an economically important tree crop that produces wood for lumber, pulp, paper, and biofuels. Wood quality traits are likely to be strongly affected by the plant hormone gibberellic acid (GA), which regulates growth. GA20Ox encodes one of the major regulatory enzymes of GA biosynthesis and may therefore play a large role in growth and wood quality. Here, linkage disequilibrium (LD) studies were used to identify significant associations between single nucleotide polymorphisms (SNPs) within PtGA20Ox and growth and wood-quality traits of P. tomentosa. We isolated a full-length GA20Ox cDNA from Populus tomentosa by reverse transcription (RT)-PCR; this 1401 bp cDNA clone had an open reading frame of 1158 bp and encoded a protein of 385 amino acids. PtGA20Ox transcripts were maximally expressed in the mature xylem of vascular tissues, suggesting that PtGA20Ox is highly expressed and specifically associated with secondary xylem formation. Resequencing the PtGA20Ox locus of 36 individuals identified 55 SNPs, and the frequency of SNPs was 1/31 bp. The 29 most common SNPs (frequency>0.1) were genotyped in an association population (426 individuals) that was also phenotyped for key growth and wood quality traits. LD did not extend over the entire gene (r 2<0.1, within 500 bp), demonstrating that a candidate-gene-based LD approach may the best way to understand the molecular basis underlying quantitative variation in this species. SNP- and haplotype-based association analyses indicated that four SNPs (false discovery rate Q<0.05) and 14 haplotypes (P<0.05) were significantly associated with growth and wood properties. The phenotypic variance explained by each SNP ranged from 3.44% to 14.47%. The SNP markers identified in this study can be applied to breeding programs for the improvement of growth and wood-property traits by marker-assisted selection.
Introduction
Gibberellins (GAs) are phytohormones that regulate a wide range of growth and developmental processes in plants, including seed germination, leaf expansion, stem elongation, and the development of flowers, seeds, and fruit [1], [2]. GAs also play an important role in promoting the formation of fiber in trees and ultimately in determining the quality of wood [3]. In higher plants, GAs are synthesized through a complex pathway in which GA20-oxidase (GA20Ox) is one of the major regulatory enzymes [4]–[6]. Previous studies have demonstrated that the expression of GA20-oxidase genes from various species can modify endogenous GA levels and in turn enhance plant growth [5], [7], [8]. Consequently, it is crucial to enhance our understanding of the role of GA20Ox in regulating growth and wood fiber properties to effectively manipulate wood biosynthesis in trees.
One of the main goals of forest tree breeding programs is to increase the quantity and quality of wood products. Marker-assisted selection (MAS) is a useful tool in tree breeding for reducing breeding cycles and increasing selection accuracy, particularly for wood properties [9]. Before MAS can be applied to tree breeding, markers that are significantly associated with target traits need to be identified. Association studies are powerful methods for identifying markers that are significantly linked to traits in natural or breeding populations [10]. DNA markers that are commonly used in association studies include single nucleotide polymorphisms (SNPs) and insertions or deletions (InDels) of the DNA sequence [11]. SNP-based linkage disequilibrium (LD) mapping provides another strategy for MAS in forest trees [12]. In contrast to traditional linkage analysis, LD mapping can be readily applied to natural or breeding populations of unrelated individuals to identify marker-trait associations. For example, Thumma et al. [13] discovered polymorphisms in the cinnamoyl-CoA reductase (CCR) gene that were associated with microfibril angle in Eucalyptus nitens. Similarly, 13 SNPs of five xylem genes associated with microfibril angle, cellulose, pulp yield, and total lignin were identified in E. nitens [14]. In addition, Wegrzyn et al. [15] found 27 highly significant, unique, single-marker associations (false discovery rate Q<0.10) across 40 candidate genes in three composite traits including the lignin content, syringyl to guaiacyl ratio, and C6 sugars in black cottonwood (Populus trichocarpa). Previous studies have demonstrated that LD mapping can be used to identify alleles associated with quantitative traits, suggesting that this new approach could be particularly useful in forest tree breeding programs.
Chinese white poplar (P. tomentosa Carr.) belongs to the section Populus in the genus Populus. This species is one of the main commercial trees for timber production in China and plays an important role in ecological and environmental protection along the Yellow River [16]. With the development of targeted cultivation of industrial commercial forests, new varieties of poplar must be fast-growing and produce high-quality wood. Therefore, methods to improve the growth and wood quality of P. tomentosa are essential. To this end, association studies of SNPs associated with growth and wood properties of P. tomentosa are important to improve the growth and wood quality of P. tomentosa by MAS breeding programs. Population structure is the leading cause of false positives in genetic association studies [17]. The presence of population structure leads to bias because subgroups of relatives tend to share more markers and gene alleles genome-wide than a pair of individuals drawn at random from the population [18]. Therefore, correction for the confounding effects of population structure present in plant populations is essential in association mapping. Huang [19] was the first to provide climatic regionalization in the distribution zones of P. tomentosa and showed that the three climatic zones can be treated as genetic regions. In addition, the structure of the natural populations has also been explored by Du et al [20]; we used this information on population structure in our association analysis in this study. Because the gene encoding GA20-oxidase, one of the major regulatory enzymes in GA synthesis, likely serves a key role in the regulation of growth and development [5], [7], [8], it can be used as a model to address the significance of allelic variation linked to growth and wood-quality traits in trees. Here, we report the identification and characterization of the gene encoding GA20Ox, PtGA20Ox, from a mature xylem cDNA library of P. tomentosa. Real-time PCR analysis revealed that this gene may be involved in wood formation and may be up-regulated by GA treatment in trees. Several common SNPs and their haplotypes in PtGA20Ox were identified for associations with growth and wood properties while accounting for population structure. The selected SNPs were being investigated further to test whether RNA transcript accumulation varies among the different genotypes that showed significant associations.
Results
Phenotypic Data Distribution and Correlations
To quantify traits for association mapping, we measured growth and wood-quality properties in our association population of 426 individuals (see Methods). In the association population, all 10 wood-quality and growth traits have abundant phenotypic variation; for example, fiber length, fiber width and volume values ranged from 0.866 to 1.512 mm (mean value of 1.170 mm), from 1.6984 to 33.503 µm (average of 23.160 µm), and from 0.037 to 3.022 m3 (mean 0.602 m3), respectively. The maximum tree height (22.50 m) was approximately eight times higher than that of the minimum tree (2.90 m), and the average height was 14.61 m. Descriptive statistics of the trait distributions are presented in Table S1. As expected, the frequency distributions for each trait measured in the association population followed an approximately normal distribution (data not shown).
The growth and wood-quality traits in the association population showed significant correlations (Table S2). For example, fiber length was positively correlated with fiber width (P<0.01). In addition, significant positive pairwise correlation was observed between tree diameter and volume (P<0.01). The details of the phenotypic correlations among these traits in the association population are shown in Table S2.
Isolation of a PtGA20Ox Gene from P. tomentosa
To identify GA20Ox SNPs, we first isolated the GA20Ox locus. A full-length cDNA encoding GA20Ox was isolated from a cDNA library prepared from the mature xylem zone of P. tomentosa by reverse transcription (RT)-PCR. The cDNA clone is 1401 bp in length and has an open reading frame of 1158 bp, including 126 bp of 5′ untranslated region (UTR) and 117 bp of 3′ UTR. Alignment of the full-length cDNA sequence with the genomic sequence showed that PtGA20Ox is composed of two introns and three exons (Figure 1). The deduced protein sequence of PtGA20Ox revealed a protein of 385 amino acids with an estimated molecular mass of 44.0 kD and a pI of 8.38. A BLASTP search with PtGA20Ox as the query sequence revealed that the PtGA20Ox protein shares 65.8% identity with the Arabidopsis GA20Ox.
Figure 1. Genomic organization of PtGA20Ox.
Exons are shown as boxes and introns as lines. Positions of common SNP markers are shown as vertical lines.
Expression Analysis of PtGA20Ox
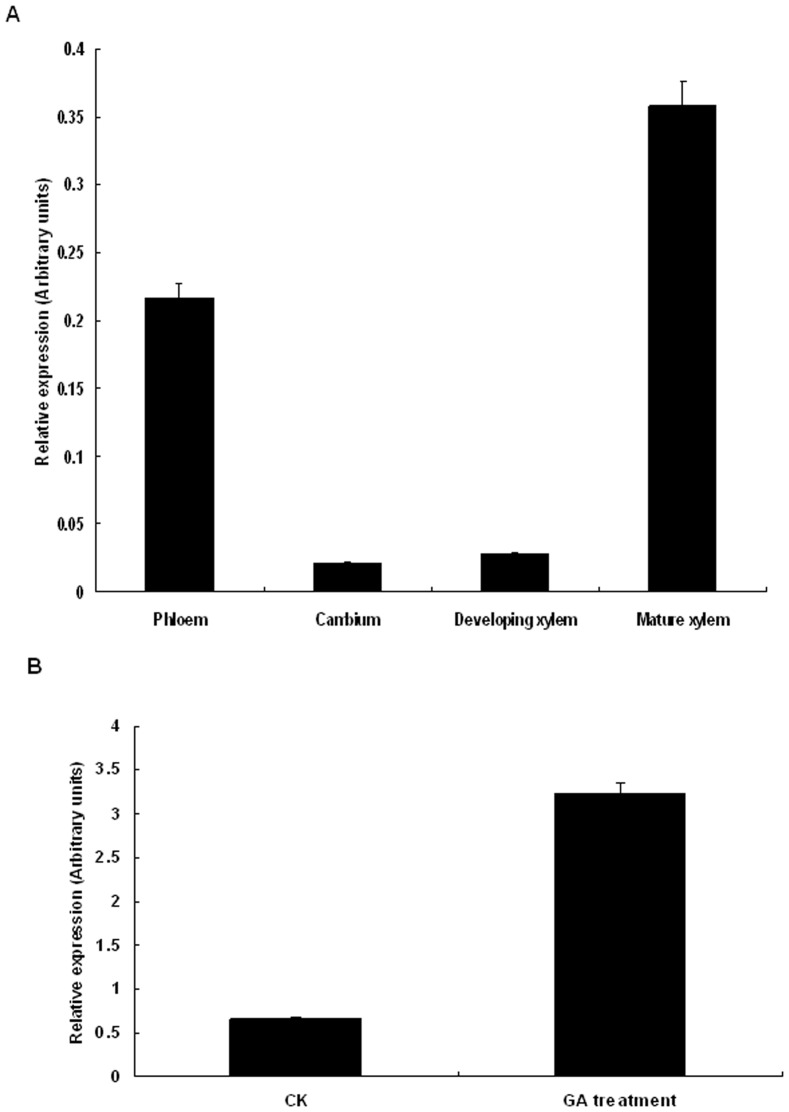
Using gene-specific primer and Actin as an internal control, real-time quantitative PCR was used to perform transcript profiling of PtGA20Ox mRNA in various poplar vascular tissues: phloem, cambium, developing xylem, and mature xylem. PtGA20Ox was preferentially expressed in mature xylem (Figure 2A); consistent with that its full-length cDNA was originally isolated from a cDNA library prepared from the mature xylem zone of P. tomentosa. By contrast, relatively lower levels of PtGA20Ox mRNA were detectable in the primary tissues of the cambium. Thus, PtGA20Ox appears to be a highly expressed gene specifically associated with secondary xylem formation. We further tested whether PtGA20Ox was inducible by treatment with GA (Figure 2B). The expression of PtGA20Ox after GA treatment was about five times higher than the expression of the control, suggesting that PtGA20Ox is up-regulated by GA.
Figure 2. Relative transcript levels of PtGA20Ox.
(A) Relative transcript levels of PtGA20Ox in various poplar vascular tissues. (B) Relative transcript levels of PtGA20Ox before and after GA treatment.
SNP Diversity and Genotyping
To identify polymorphisms for association mapping, we re-sequenced the PtGA20Ox region in 36 unrelated individuals from the association population. An approximately 1693 bp genomic region of PtGA20Ox, including 126 bp of 5′ UTR, 1158 bp of coding regions, 292 bp of intron, and 117 bp of 3′ UTR, was amplified and sequenced from 36 unrelated individuals, representing almost the entire natural range of P. tomentosa. Table 1 summarizes the statistical analysis of nucleotide polymorphisms (excluding indels) over different regions of PtGA20Ox. Across the samples, 55 SNPs were detected in the entire gene at a frequency of approximately one SNP every 31 bp (Table 1). Thirty of these SNPs were found in exons, of which 20, 9, and 1 variants were categorized as silent, missense, and nonsense mutations, respectively (Table 1). Altogether, 29 of 55 SNPs (53%) were considered common (frequency>0.10). In general, the PtGA20Ox locus has high nucleotide diversity (πT), where πT = 0.00988 and θw = 0.00797 (Table 1). More specifically, estimates of nucleotide diversity for the different gene regions ranged from 0.00594 (exon 2) to 0.03478 (intron 1), and θw varied between 0.00442 (5′ UTR) and 0.02173 (intron 1). Within coding regions, the value of non-synonymous nucleotide substitutions (πnonsyn) was markedly lower than πsyn, with a πnonsyn/πsyn ratio of 0.27, suggesting that diversity at the non-synonymous sites of exon regions resulted from strong purifying selection (Table 1). The 29 common SNPs were successfully genotyped across 426 individuals in the association population using locked nucleic acid (LNA) technology.
Table 1. Nucleotide polymorphisms at the PtGA20Ox locus.
| Region | No. of bp | No. of polymorphic sites | Percentage polymorphism | Nucleotide diversity | |
| π | θw | ||||
| 5′ UTR | 126 | 2 | 1.59 | 0.00936 | 0.00442 |
| Exon 1 | 569 | 15 | 2.64 | 0.00865 | 0.00636 |
| Synonymous | 122.30 | 9 | 7.36 | 0.01901 | 0.01901 |
| Non-synonymous | 444.70 | 6 | 1.35 | 0.00583 | 0.00325 |
| Intron 1 | 111 | 10 | 9.01 | 0.03478 | 0.02173 |
| Exon 2 | 322 | 9 | 2.80 | 0.00594 | 0.00674 |
| Synonymous | 69.70 | 8 | 11.48 | 0.02587 | 0.02768 |
| Non-synonymous | 251.30 | 1 | 0.40 | 0.00043 | 0.00096 |
| Intron 2 | 181 | 6 | 3.31 | 0.01012 | 0.00851 |
| Exon 3 | 267 | 6 | 2.25 | 0.00655 | 0.00542 |
| Synonymous | 59.19 | 3 | 5.07 | 0.00458 | 0.01222 |
| Non-synonymous | 204.81 | 3 | 1.46 | 0.00721 | 0.00353 |
| 3′ UTR | 117 | 7 | 5.98 | 0.01081 | 0.01455 |
| Total silenta | 761.19 | 45 | 5.91 | 0.01610 | 0.01426 |
| Synonymous | 252.19 | 20 | 7.93 | 0.01745 | 0.01912 |
| Non-synonymous | 902.81 | 10 | 1.11 | 0.00463 | 0.00267 |
| Total PtGA20Ox b | 1693 | 55 | 3.25 | 0.00988 | 0.00797 |
Total silent = synonymous plus silent sites.
Total PtGA20Ox = silent sites plus non-synonymous sites.
Regions containing indels were excluded from the calculations.
Linkage Disequilibrium
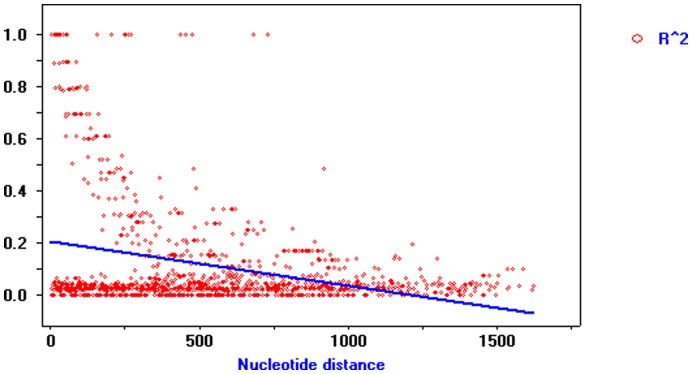
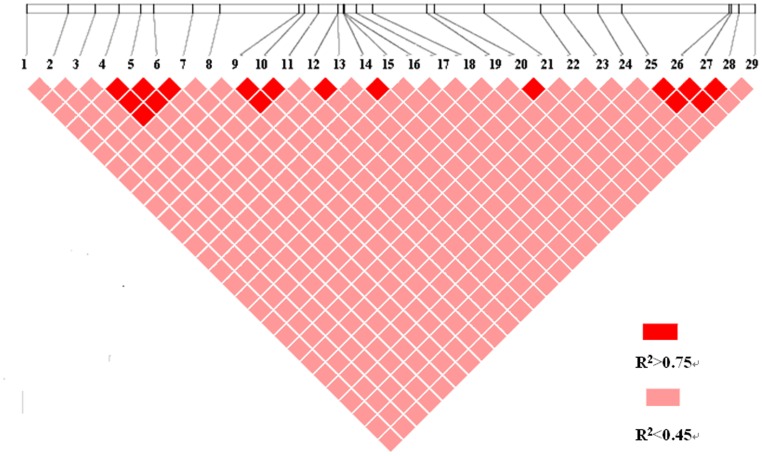
The pattern of the squared allelic correlation coefficient (r 2) with base-pair distance within the PtGA20Ox gene illustrated rapid LD decay in the P. tomentosa population (Figure 3), with r 2 values dropping to 0.1 within 500 bp, indicating that the LD of the SNPs within this gene did not extend over the entire gene region. LD analysis using genotype data of 29 SNPs from 426 individuals in the association population (Figure 4) revealed three distinct haplotype blocks within the PtGA20Ox gene, from SNP4 to 7, SNP9 to 11, and SNP25 to 28. Within each block, LD between the SNPs was high (r 2>0.75), whereas LD was low between the three haplotype blocks (r 2<0.3) (Figure 4).
Figure 3. Decay of linkage disequilibrium within PtGA20Ox.
Pairwise correlations between SNPs are plotted against the physical distance between the SNPs in base pairs. The straight line describes the least-squares fit of r 2 (Er2) to its expectation. Linkage disequilibrium decays drastically within 500 bp.
Figure 4. Pairwise linkage disequilibrium (r2) between SNP markers.
The common genotyped SNPs are shown on a schematic of PtGA20Ox and the pairwise r2 values are shown by color coding in the matrix below.
Marker-trait Association and Haplotype Analysis
Associations between 29 SNPs and 10 growth and wood-quality traits were tested by comparing results from a general linear model (GLM) and a mixed linear model (MLM) in TASSEL version 2.1 software. The number of significant markers (P<0.05) was 15 using GLM but fell to 13 with MLM (Table S3). After a qFDR (false discovery rate) test, the number of significant associations of SNPs (Q<0.05) with growth and wood-property traits was reduced to seven. This analysis revealed that four SNP markers (SNP10, SNP19, SNP22, and SNP29) were significantly associated with five traits, including fiber length, fiber width, microfibril angle, holocellulose content, and tree height (Table 2). These associations were identified in exonic, intronic, and 3′ UTR regions of PtGA20Ox. Of these markers, SNP10, a missense mutation in exon 1 resulting in an encoded amino acid change from Asn to Lys, was significantly associated with multiple traits, i.e., fiber length, fiber width, holocellulose content, and tree height; SNP19 in exon 2, a synonymous mutation, was associated with fiber length, SNP22 in intron 2 was associated with fiber width, and SNP29 from 3′ UTR was closely linked to microfibril angle. Altogether, these SNP associations explained a small proportion of the phenotypic variance, with the individual effects ranging from 3.44% to 14.47%.
Table 2. SNP markers significantly associated with growth and wood-property traits using the mixed linear model (MLM).
| Trait | Marker | Position | P- value | Q-value | R2 | FST |
| Fiber length | SNP10 | Exon 1 | 8.72×10−14 | 1.26×10−11 | 14.47% | 0.0023 |
| SNP19 | Exon 2 | 9.94×10−5 | 0.0048 | 4.66% | 0.0001 | |
| Fiber width | SNP10 | Exon 1 | 4.17×10−9 | 3.02×10−7 | 10.20% | 0.0023 |
| SNP22 | Intron 2 | 0.0005 | 0.0160 | 4.13% | 0.0016 | |
| Microfibril angle | SNP29 | 3′UTR | 0.0003 | 0.0091 | 4.36% | 0.0002 |
| Holocellulose content | SNP10 | Exon 1 | 5.61×10−5 | 0.0033 | 5.78% | 0.0023 |
| Tree height | SNP10 | Exon 1 | 0.0010 | 0.0287 | 3.44% | 0.0023 |
P-value = the significant level for association (the significance is P≤0.05), R2 = percentage of the phenotypic variance explained, Q-value = a correction for multiple testing [false discovery rate FDR (Q) ≤0.05], FST = variation due to differentiation among subpopulations.
Most of the associations were consistent with modes of gene action other than codominance (Table 3). For example, heterozygotes for SNP10 had shorter fiber length on average than either homozygote class (1.2804 µm for AA, 1.1688 µm for AG, 1.2167 µm for GG). Three of the seven marker-trait pairs for which dominance and additive effects could be calculated were consistent with over- or underdominance (|d/a|>1.25). The remaining four marker-trait pairs were split between modes of gene action that were partially to fully dominant (0.50<|d/a|<1.25, n = 2) or codominant (|d/a|≤0.5, n = 2). Differences in microfibril angle among the three genotypes of SNP29 were significant, indicating that the pattern of gene action was consistent with additive effects.
Table 3. List of marker effects for significant marker-trait pairs.
| Trait | SNP | 2a1 | d2 | d/a | 2a/sp 3 | Frequency4 | a5 | |
| Fiber length | SNP 10 | 0.0637 | −0.0797 | −2.5024 | 0.7611 | 0.48 | (G) | 0.0192 |
| SNP19 | 0.0392 | −0.0114 | −0.5816 | 0.4683 | 0.49 | (T) | 0.0152 | |
| Fiber width | SNP10 | 3.6247 | −0.8306 | −0.4583 | 1.8285 | 0.48 | (G) | −0.5954 |
| SNP22 | 3.9557 | −2.5851 | −1.3070 | 1.9955 | 0.49 | (T) | 0.2697 | |
| Microfiber angle | SNP29 | 8.2694 | 1.3075 | 0.3162 | 1.8279 | 0.46 | (C) | 6.2997 |
| Holocellulose | SNP10 | 7.7360 | 2.0023 | 0.5177 | 0.7192 | 0.48 | (G) | −2.5755 |
| Tree height | SNP10 | 1.4100 | −2.0458 | −2.9018 | 0.4905 | 0.48 | (G) | 0.5098 |
Calculated as the difference between the phenotypic means observed within each homozygous class (2a = |GBB–Gbb|, where Gij is the trait mean in the ijth genotypic class).
Calculated as the difference between the phenotypic mean observed within the heterozygous class and the average phenotypic mean across both homozygous classes [d = GBb−0.5(GBB+Gbb), where Gij is the trait mean in the ijth genotypic class].
sp, standard deviation of the phenotypic trait under consideration.
Allele frequency of either the derived or minor allele. Single nucleotide polymorphism (SNP) alleles corresponding to the frequency listed are given in parentheses.
The additive effect was calculated as a = pB(GBB)+pb(GBb)–G, where G is the overall trait mean, Gij is the trait mean in the ijth genotypic class, and pi is the frequency of the ith marker allele. These values were always calculated with respect to the minor allele.
We used haplotype trend regression (HTR) to identify significant haplotypes associated with growth and wood-quality traits from the regions surrounding the significant SNPs. We found 14 common haplotypes (frequency>1%) associated with growth and wood-quality traits (Table 4). Among these, three haplotypes from SNP9–11 were associated with fiber length, two haplotypes from SNP20–22 and three haplotypes from SNP10–12 were associated with fiber width, and six haplotypes from SNP8–10 were associated with tree height. The proportion of phenotypic variation explained by these haplotypes, which originated from exon 1, exon 2, intron 2, and 3′UTR, ranged from 3.98% to 8.37%.
Table 4. Haplotypes significantly associated with growth and wood-property traits.
| Trait | P-value | R2 | Haplotype | Frequency |
| Fiber length | 7.93×10−6 | 8.37% | SNPs 9–11 | |
| C-A-C | 0.492 | |||
| A-G-T | 0.469 | |||
| A-A-T | 0.025 | |||
| Fiber width | 0.0018 | 4.05% | SNPs 20–22 | |
| T-G-A | 0.495 | |||
| C-A-T | 0.495 | |||
| 6.61×10−6 | 7.62% | SNPs 10–12 | ||
| A-A-C | 0.492 | |||
| G-T-A | 0.491 | |||
| A-A-A | 0.014 | |||
| Tree height | 0.003 | 3.98% | SNPs 8–10 | |
| T-C-A | 0.247 | |||
| C-C-A | 0.247 | |||
| T-A-G | 0.237 | |||
| C-A-G | 0.237 | |||
| T-A-A | 0.014 | |||
| C-A-A | 0.014 |
P = the significance level for haplotype-based association (P≤0.05); R2 = percentage of the phenotypic variance explained.
Transcript Analysis of SNP Genotypes
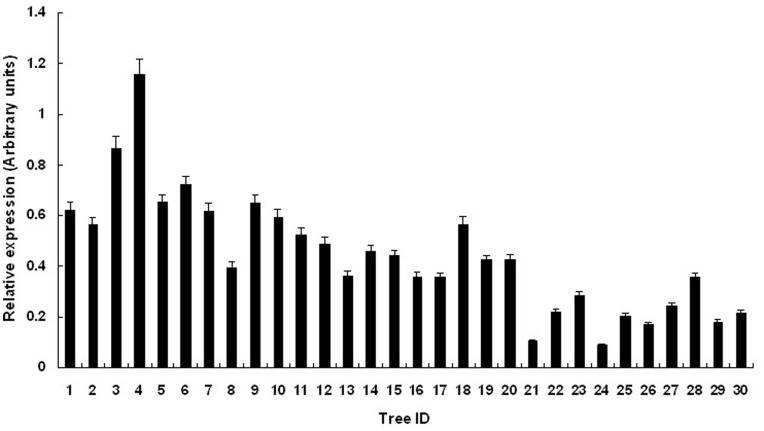
To determine whether PtGA20Ox expression was altered in the different genotypic classes, transcript levels of the four significantly associated SNPs were compared using real-time quantitative PCR with gene-specific primers. The assays used secondary xylem from 20-year-old trees to quantify mRNA levels in 30 trees (10 trees for each genotype) of the association population. Among the four SNPs tested, only SNP10 exhibited significant differences in the transcript levels among the three genotypes (Figure 5). The highest PtGA20Ox mRNA levels were found in the AA group, followed by the AG group, and the transcript levels of the GG group were lowest. The mean relative expression levels of mRNA products for the AA, AG, and GG groups were 0.6832, 0.4407, and 0.2067, respectively. These results demonstrated that the transcript level of the AA group was about 1.55 times that of the AG group and 3.31 times that of the GG group.
Figure 5. Expression levels of three genotypic classes for SNP10.
1–10 represent the AA group, 11–20 represent the AG group, and 21–30 represent the GG group.
Discussion
Analysis of Statistical Models for Association
An appropriate statistical model is necessary for phenotype-genotype associations to avoid false positive or spurious associations that arise from population and family structure [21]. One widely used approach, the general linear model (GLM) was first implemented in TASSEL (Trait Analysis by aSSociation, Evolution and Linkage) to reduce the risk of false positives arising from population structure. However, Q-values alone are not adequate because the Q-matrix only provides a rough dissection of population differentiation [22]. More recently, a unified mixed model method, Q+K, that combines information from population structure and relatedness (kinship), has been shown to be superior to these former methods [22], [23]. In the present study, these two statistical models are available for testing associations, the number of significant markers (P<0.05) was 15 using GLM but reduced to 13 using MLM (data not shown). This result was consistent with previous studies reporting that MLM was better than GLM at reducing false positive associations. Therefore, we ultimately used MLM to test our phenotype-genotype associations. Similarly, Ehrenreich et al. [24] used MLM to conduct candidate gene association mapping of Arabidopsis flowering time. In addition, Sexton et al. [11] used MLM for association studies in Eucalyptus pilularis Smith. Taken together, choosing of the proper statistical models can help to reduce the number of false positive associations.
Validation under either different environmental conditions and/or genetic backgrounds has become the gold standard for assessing statistical results from association studies, even though this replication requirement may cause real genetic effects to be missed [25]. The importance of validation has been well established in candidate gene-based association studies, and several examples in forest trees have recently been published [11], [13], [26]–[28]. The current lack of validation across different association and field-testing environments in this study, and the varying numbers of significant markers (P<0.05) using GLM and MLM (data not shown), suggest that these estimates should be considered with caution. To compensate for the lack of a validation population, we conducted functional validation of SNP associations via gene expression analyses to identify whether these significant associations affect gene expression at the mRNA level. And the functional support for significant associations in LD mapping strategy lends strength to their proposed effects.
Linkage Disequilibrium Test in Trees
Linkage disequilibrium (LD) refers to the nonrandom association of alleles at different loci [29] and plays an important role in association studies for identifying significant markers or haplotypes. Therefore, understanding the patterns of LD in the species is the important prerequisite for association mapping, whether genome-wide associations are feasible or whether a candidate gene-based approach has to be considered. In this study, a rapid LD decay was observed in PtGA20Ox (r 2<0.1, within 500 bp; Figure 3). It is consistent with previous studies that suggested limited LD in trees. For example, Brown et al. [30] found a rapid decline in LD within several kilobases in loblolly pine. Similar results of limited LD were reported for candidate genes in other species [13], [26], [31]–[33]. In Populus, previous studies based on SNP markers have indicated that a rapid decay of LD occurs within just 300–1,700 bp in candidate genes among related species of Populus [15], [34]–[36], suggesting high recombination rates in this outcrossing species. However, Slavov et al [37] found that the decay of linkage disequilibrium with physical distance was slower than expected from previous studies in the P. trichocarpa genome, with r2 dropping below 0.2 within 3–6 kb.
The reasons for the low LD in forest trees may be a large effective population size, trees’ outcrossing habit and long history of recombination [38]. The low LD usually seen in forest trees suggests that candidate-gene-based LD mapping should be the ideal approach to understand the molecular basis underlying quantitative variation in trees. However, genome-wide association mapping strategies may be inefficient because a large number of markers are needed.
Detection of Phenotype-genotype Associations in P. tomentosa
Candidate gene-based LD mapping approaches have been used to dissect complex growth and wood-property traits in forest trees [13], [15], [26]–[28], [39], [40]. Generally, the power of a single-marker association test is often limited because LD information contained in flanking markers is ignored. Intuitively, haplotypes (a block of linked ordered markers) may be more powerful than individual markers [15]. In this study, a comparison of single-marker and haplotype-based association demonstrates that the single-marker-based association analyses have either similar or greater power than haplotype-based tests (Table 2 and 4). These results suggest that single-marker-based tests were preferred to haplotype-based tests to avoid uncertainty in haplotype determination from diploid SNP data sets. Therefore, combining these two methods may provide a better potential to detect functional allelic variance underlying quantitative traits in association populations.
In our study, the fact that significant associations were found with both growth (e.g., tree height) and wood quality (e.g., fiber length, fiber width, microfibril angle and holocellulose content) traits is encouraging (see Table 2), as we are usually interested in selecting for both traits simultaneously. In total, seven significant associations were found and the phenotypic variance explained by a single SNP association ranges from 3.44% to 14.47%. Most of the associations explained a small proportion of the phenotypic variance. This was consistent with other association studies in which between 1.5% and 6.5% of the total phenotypic variation was accounted for by SNPs [13], [26]–[28], [40]. These small SNP effects are in accordance with polygenic quantitative models of wood traits [31]. Among the significant associations, SNP10 explained 14.47% and 10.20% of phenotypic variance in fiber length and width traits, respectively. The effects which were high compared to other studies of quantitative traits in trees may be due to the underlying biological mechanism or other factors. These estimates need to be further validated.
Our discovery of a non-synonymous exonic SNP (SNP10) is somewhat surprising. SNP10 was significantly associated with three wood-quality traits and one growth trait, which may represent pleiotropic effects of the PtGA20Ox gene [41]. A similar phenomenon has been identified in previous studies. For example, Beaulieu et al. [40] found some SNPs to be significantly associated with more than one trait whereas others were positive with both the additive and the dominant effects models. Similarly, Southerton et al. [14] identified two SNPs in Eni-CAD1 and four SNPs in Eni-HB1 that were associated with multiple wood traits. SNP10 was located in exon 1 of PtGA20Ox and represented a missense mutation. This mutation leads to an amino acid change from Asn to Ser. The traits that associated with SNP10 were fiber length, fiber width, holocellulose content and tree height. The phenomenon that SNP10 was significantly associated with both fiber length and fiber width was consistent with the strong positive phenotypic correlation between these two traits. The association between SNP10 and fiber length was consistent with previous studies that GA alone can affect the length of xylem fibers, as injecting GA inhibitors into woody stems leads to reductions in fiber length [42]. Also, GA-induced increase in both the number and length of xylem fibers in transgenic GA-overproducing trees [43], suggests that GA is mainly active during xylem fiber development. Three significant common haplotypes surrounding SNP10 were also significantly associated with fiber length, and had significant differences (1.2220 mm for C-A-C, 1.1765 mm for A-A-T and 1.1601 mm for A-G-T). Holocellulose is the total of the polysaccharide components of the walls of the secondary xylem cells, which are almost entirely composed of cellulose and hemicelluloses, accounting for nearly 80% of secondary xylem tissue [44]. In the present study, differences in holocellulose content among the three genotypes of SNP10 were significant (74.4160% for AA, 72.5503% for AG and 66.6800% for GG), illustrating that patterns of gene action are consistent with dominant effects. Besides SNP10, six common haplotypes surrounding SNP10 were significantly associated with tree height. The associations with tree height provided evidence for the theory that GAs can accelerate the growth and development of plants. Characterization of GA-deficient mutants has established that GA has a role in promoting shoot growth and stem elongation, as GA mutants are associated with severe dwarfism [45]. GA first promotes cell elongation, and cell division could be stimulated as a result of cell growth [46], [47]. In addition, quantitative Real-time PCR also showed significant expression differences among the three genotypes of SNP10. Moreover, most of the common haplotypes significantly associated with wood traits surround SNP10. Taken together, these results strongly suggest that SNP10 may be a functional polymorphism that is in or near a locus involved in the control of wood traits of P. tomentosa. In addition, the four traits (fiber width, fiber length, holocellulose content, and tree height) associated with SNP10 are not negatively correlated (Table S2). Thus, SNP10 could serve as an important marker in breeding for improved growth and wood-property traits of P. tomentosa.
SNP19, SNP22 and SNP29 were also significant markers. They were located in different regions of PtGA20Ox, including exon, intron, and 3′UTR locations. Silent SNPs should not be considered a priori as potential false positives because they may affect transcript levels and codon usage [48], [49]. In this study, SNP19 represented a synonymous substitution that was significantly associated with fiber width. A similar phenomenon has been identified in previous studies. For example, Thumma et al. [26] discovered a synonymous exonic SNP of EniCOBL4A associated with cellulose content and kraft pulp yield. Similarly, Southerton et al. [50] confirmed that the CO07 SNP in Eni-COBL4A was a synonymous substitution but was significantly associated with cellulose content. Both SNP22 located in intron 2 and SNP29 located in 3′UTR occurred in non-coding regions of PtGA20Ox. SNPs in introns could affect phenotypic traits because those particular introns may play an important role in regulating gene expression and exon splicing. Although the mutation of the 3′ flanking region did not result in an amino acid change, it may regulate expression of the gene. Previous studies have observed that sequences in the 3′ flanking region can affect the mechanisms of mRNA deadenylation and degradation [51], [52]. Significant SNP markers in non-coding regions of a gene, such as introns and 3′UTR, have also been reported elsewhere. Gonzalez-Martinez et al. [33] analyzed association genetics in Pinus taeda and found a strong association between SNP M10 located in intron 1 and earlywood microfibril angle. Fang et al. [53] detected a novel SNP in the 3′ flanking region of the goat BMP-2 gene associated with growth traits.
These association results suggest that PtGA20Ox can affect the growth and development, fiber formation, and wood quality of P. tomentosa; these findings are consistent with previous studies. In terms of tree breeding, GA20Ox has been found to affect the growth and wood quality of trees. Eriksson et al. [43] transferred GA20Ox of Arabidopsis thaliana (AtGA20Ox) to P. tremula × P. tremuloides and discovered higher levels of endogenous GA and improved biomass with bigger leaves, a taller stem, and more fiber cells in the xylem. This suggested that PtGA20Ox may have a similar function considering that the PtGA20Ox protein shares 65.8% identity with AtGA20Ox. Similarly, Deng et al. [54] transferred cotton GA20Ox to P. tomentosa and found that stem growth and biomass were enhanced. Thus, PtGA20Ox is an important candidate gene for future tree-breeding programs. The SNP markers identified in this study can be applied to breeding programs. However, a combination of these significant markers may be required because the percentage of trait variation explained by each individual SNP was small.
Materials and Methods
Association Population
The association population consisted of 426 unrelated P. tomentosa individuals growing in Guan Xian County, Shandong Province, where root segments of 1047 native individuals collected from the entire natural distribution range of P. tomentosa were used to establish a clonal arboretum in 1982 using a randomized complete block design with three replications. On the basis of principal component analysis and ISODATA fuzzy clustering of 16 meteorological factors [19], the total climatic zone covered by these individuals can be divided into three large climatic regions: southern, northwestern, and northeastern. In the present study, 426 unrelated individuals representing almost the entire geographic distribution of P. tomentosa (180 from the southern region, 86 from the northwestern region, and 160 from the northeastern region) were used for association analysis. In addition, 36 P. tomentosa individuals were sequenced to identify SNPs within PtGA20Ox.
This study was carried out in strict accordance with the recommendations in the Guide for Observational and field studies. All necessary permits were obtained for the described field studies. The sampling of all individuals of P. tomentosa was approved by the Youhui Zhang, director of National Garden of P. tomentosa in Guan Xian County, Shandong Province.
Phenotypic Data
Ten traits were measured: lignose content, holocellulose content, alpha-cellulose content, fiber length, fiber width, microfibril angle, tree height, tree diameter, volume of wood, and tree height/tree diameter. In this study, wood sample materials containing bark and pith were cored from each of the 426 poplars in the association population at breast height using increment borers. These sample materials (15 cm long × 10 mm in diameter) were collected in 2010.
Four referential standard procedures (GB/T2677.8–1994, GB/T2677.10–1995, GB/T 744–2004, and FZ/T50010.4–1998) were consulted to test for contents of lignose, holocellulose, and alpha-cellulose, with consideration of the experimental conditions. Fiber length and width were measured using a Colour CCTV Camera (Panasonic SDII), and microfibril angle was measured using an X-ray powder diffractometer (Philips). Data for tree height and diameter at breast height were collected during field surveys in 2011; these data were used to determine the volume of wood. SAS for Windows ver. 8.2 (SAS Institute, Cary, NC, USA) was used to conduct analysis of variance (ANOVA) and correlations for the above phenotypic traits.
Isolation of PtGA20Ox cDNA
The P. tomentosa stem mature xylem cDNA library was constructed using the Superscript λ System following the manufacturer’s suggestions (Life Technologies, Rockville, MD, USA). The constructed cDNA library consisted of 5.0 × 106 pfu with an insert size of 1.0–4.0 kb. Random end sequencing of 500 cDNA clones and comparison with all available Arabidopsis GA20Ox sequences identified a full-length cDNA with high similarity to AtGA20Ox; this was named PtGA20Ox.
SNP Identification
To identify SNPs, 36 unrelated individuals from the association population were used to sequence GA20Ox. First, genomic DNA (20 ng per reaction) isolated separately from the 36 unrelated individuals of the association population was used as an amplification template to clone the GA20Ox gene using gene specific primers. The primer pairs for amplification were designed using Primer 3 software (primer3.sourceforge.net). All PCR products were resolved by agarose gel electrophoresis, excised, and purified using Ultrafree®-DA (Millipore, Billerica, MA, USA) centrifugal filter units. The purified DNA was then ligated into the pGEM®-T Easy Vector and transformed into JM109 competent cells (Promega). Plasmid DNA was extracted from overnight cultures using the QIAprep Spin Miniprep protocol and was sequenced on both strands with conservative primers using the Big Dye Terminator version 3.1 Cycle Sequencing kit (Applied Biosystems, Beijing, China) and a LI-COR 4300 genetic analyzer. Finally, the 36 genomic clones were sequenced and analyzed using the software MEGA4.0 and DnaSP4.50.4.
SNP Genotyping
Twenty-nine common SNPs were genotyped in 426 trees in the association population by amplification using locked nucleic acid (LNA) technology [55], [56]. Amplification was performed in a final reaction volume of 25 µl containing 20 ng genomic DNA, 0.8 U Taq DNA polymerase (Promega), 50 ng forward primer, 50 ng reverse primer, 10×PCR buffer (Promega), and 0.2 mM dNTPs (Promega). The PCR conditions were as follows: 94°C for 3 min, 30 cycles of 94°C denaturation for 30 s, annealing at 54–58°C (depending on the primers) for 15 s, and extension at 72°C for 1 min, with a final extension at 72°C for 5 min.
Real-time Quantitative PCR
Real-time quantitative PCR was performed on a DNA Engine Opticon 2 machine (MJ Research) using the LightCycler-FastStar DNA master SYBR Green I kit (Roche). The PCR program included an initial denaturation at 94°C for 5 min; 40 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C; and a final melt-curve of 70–95°C. The specificity of the amplified fragments was checked using the generated melting curve. All reactions were conducted in triplicate, and the generated real-time data were analyzed using the Opticon Monitor Analysis Software 3.1 tool.
Data Analysis
Linkage disequilibrium analysis: We used “SAS genetics” to test Hardy-Weinberg equilibrium (HWE) of the SNPs and estimated the relationship of linkage disequilibrium with physical distance by using the linear regression analysis approach which is built into the DnaSP software, version 4.0 [57], [58]. The software package HAPLOVIEW (http://www.broad.mit.edu/mpg/haploview.html) [59] was used to evaluate LD among 29 SNPs in P. tomentosa. The r2 (squared allele frequency) is the parameter most frequently used to estimate LD [60]–[62]. The interval value of the parameter varies from 0 to 1. The significance (P-values) of r2 for each SNP locus was calculated using 100,000 permutations.
Association testing: The phenotype-genotype associations in this study were identified by comparing results from a general linear model (GLM) and a mixed linear model (MLM) in the software package TASSEL2.1 (http://www2.maizegenetics.net/index.php?page=bioinformatics/tassel/index.html) [21], [63]. The values of estimated membership probability (Q) and pairwise kinship (K) were used to evaluate the effects of population structure and relatedness among individuals for marker-trait associations. The GLM only uses the Q value, whereas MLM uses both Q and K values. The Q matrix was identified based on the significant subpopulations (K = 11) [20] by 20 neutral genomic SSR markers, as assessed according to the statistical model described by [64], using the software package STRUCTURE VERISON 2.3 (http://pritch.bsd.uchicago.edu/structure.html) [65]. The K matrix was calculated on the basis of 20 SSR loci using the method proposed by Ritland [66], which is built into the program SPAGeDi, version 1.3 [67]. The K matrix was calculated as described by Yu et al. [21] and all negative values between individuals were set to 0. The false positive discovery rate (FDR) method was additionally applied to correct for multiple testing using QVALUE software [68], [69].
Haplotype analysis: Haplotype frequencies from genotype data were estimated, and haplotype association tests were conducted on a three-marker sliding window using haplotype trend regression software [70]. The significance of the haplotype associations was based on 1000 permutation tests.
Modes of gene action: The modes of gene action were quantified using the ratio of dominance (d) to additive (a) effects estimated from least-square means for each genotypic class. Partial or complete dominance was defined as values in the range 0.50<|d/a|<1.25, whereas additive effects were defined as values in the range |d/a|≤0.5. Values of |d/a|>1.25 were equated with under- or overdominance [15], [71].
Supporting Information
The minimum, and maximum values, mean, standard error (SE) and coefficient of phenotypic variation [CV (%)] for each growth and wood property trait measured in P. tomentosa association population.
(DOC)
Estimates of phenotypic correlations (R) for these ten phenotypic traits in the association population.
(DOC)
All the association results. a. All the association results using the general linear model (GLM). b. All the association results using the mixed linear model (MLM).
(DOC)
Acknowledgments
Sequence data from this article have been deposited in the GenBank Data Library under the accession Nos. JX305425-JX305461.
Funding Statement
This work was supported by the State Key Basic Research Program of China (No. 2012CB114506) and as the Projects of the National Natural Science Foundation of China (Nos. 31170622, 30872042). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223. [DOI] [PubMed] [Google Scholar]
- 2. Swain SM, Singh DP (2005) Tall tales from sly dwarves: Novel functions of gibberellins in plant development. Trends Plant Sci 10: 123–129. [DOI] [PubMed] [Google Scholar]
- 3. Shen HJ (1996) Plant hormone and wood formation. Scientla Silwae Sinicae 32(2): 165–170. [Google Scholar]
- 4. Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460. [DOI] [PubMed] [Google Scholar]
- 5. Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530. [DOI] [PubMed] [Google Scholar]
- 6. Olszewski N, Sun T, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14 supp.: 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Niki T, Nishijima T, Nakayama M, Hisamatsu T, Oyama-Okuba N, et al. (2001) Production of dwarf lettuce by overexpressing a pumpkin gibberellin 20-oxidase gene. Plant Physiol 126: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Biemelt S, Tschiersch H, Sonnewald U (2004) Impact of altered gibberellin metabolism on biomass accumulation, lignin, biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol 135: 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ribaut JM, Hoisington DA (1998) Marker-assisted selection: new tools and strategies. Trends Plant Sci 3: 236–239. [Google Scholar]
- 10. Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12: 111–122. [DOI] [PubMed] [Google Scholar]
- 11. Sexon TR, Henry RJ, McManus LJ, Henson M, Thomas DS, et al. (2010) Genetic association studies in Eucalyptus pilularis Smith (blackbutt). Austral For 73(4): 254–258. [Google Scholar]
- 12. Zhang DQ, Zhang ZY (2005) Single nucleotide polymorphisms discovery and linkage disequilibrium. For Studies China 7: 1–14. [Google Scholar]
- 13. Thumma BR, Nolan MF, Evans R, Moran GF (2005) Polymorphisms in Cinnamoyl CoA Reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171: 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Southerton SG, MacMillan CP, Bell JC, Bhuiyan N, Downes G, et al. (2010) Association of allelic variation in xylem genes with wood properties in Eucalyptus nitens . Austral For 73(4): 259–264. [Google Scholar]
- 15. Wegrzyn JL, Eckert AJ, Choi M, Lee JM, Stanton BJ, et al. (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol 188: 515–532. [DOI] [PubMed] [Google Scholar]
- 16. Zhang DQ, Zhang ZY, Yang K, Li BL (2004) Genetic mapping in (Populus tomentosa × P. bolleana) and P. tomentosa Carr. using AFLP markers. Theor Appl Genet 108 (4): 657–662. [DOI] [PubMed] [Google Scholar]
- 17. Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6: 45–61. [DOI] [PubMed] [Google Scholar]
- 18. Breseghello F, Sorrells ME (2006) Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci 46: 1323–1330. [Google Scholar]
- 19. Huang ZH (1992) The study on the climatic regionalization of the distributional region of Populus tomentosa . J Beijing For Univ 14: 26–32. [Google Scholar]
- 20.Du QZ, Wang BW, Wei ZZ, Zhang DQ, Li BL (2012) Genetic diversity and population structure of Populus tomentosa (Chinese white poplar) revealed by SSR markers. J Hered 2012, 103. doi: 10.1093/jhered/ess061. [DOI] [PubMed]
- 21. Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208. [DOI] [PubMed] [Google Scholar]
- 22. Yang XH, Yan JB, Shah T, Warburton ML, Li Q, et al. (2010) Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor Appl Genet 121: 417–431. [DOI] [PubMed] [Google Scholar]
- 23. Shao YF, Jin L, Zhang G, Lu Y, Shen Y, et al. (2011) Association mapping of grain color, phenolic content, avonoid content and antioxidant capacity in dehulled rice. Theor Appl Genet 122: 1005–1016. [DOI] [PubMed] [Google Scholar]
- 24. Ehrenreich IM, Hanzawa Y, Chou L, Roe JL, Kover PX, et al. (2009) Candidate gene association mapping of Arabidopsis flowering time. Genetics 183: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Greene CS, Penrod NM, Williams SM, Moore JH (2009) Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS ONE 4: e5639 doi:10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thumma BR, Matheson BA, Zhang DQ, Meeske C, Meder R, et al. (2009) Identification of a Cis-acting regulatory polymorphism in a Eucalypt COBRA-like gene affecting cellulose content. Genetics 183: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dillon SK, Nolan M, Li W, Bell C, Wu HX, et al. (2010) Allelic variation in cell wall candidate genes affecting solid wood properties in association populations and land races of Pinus radiata . Genetics 185: 1477–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dillon SK, Brawner JT, Meder R, Lee DJ, Southerton SG (2012) Association genetics in Corymbia citriodora subsp. Variegate identifies single nucleotide polymorphisms affecting wood growth and cellulosic pulp yield. New Phytol 195: 596–608. [DOI] [PubMed] [Google Scholar]
- 29. Lewontin RC (1964) The interaction of selection and linkage. I. general considerations; heterotic models. Genetics 49(1): 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown GR, Gill GP, Kuntz RJ, Langley CH, Neale DB (2004) Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc Natl Acad Sci USA 101: 15255–15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neale DB, Savolainen O (2004) Association genetics of complex traits in conifers. Trends in Plant Science 9: 325–330. [DOI] [PubMed] [Google Scholar]
- 32. Krutovsky KV, Neale DB (2005) Nucleotide diversity and linkage disequilibrium in cold-hardiness and wood quality-related candidate genes in Douglas-fir. Genetics 171: 2029–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gonzalez-Martinez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB (2007) Association genetics in Pinus taeda L. I. wood property traits. Genetics 175: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ingvarsson PK (2005) Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European aspen (Populus tremula L., Salicaceae). Genetics 169: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ingvarsson PK, Garcia MV, Luquez V, Hall D, Jansson S (2008) Nucleotide polymorphism and phenotypic associations within and around the phytochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178: 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ismail M, Soolanayakanahally RY, Ingvarsson PK, Guy RD, Jansson S, et al. (2012) Comparative nucleotide diversity across north American and European Populus species. J Mol Evol 74: 257–272. [DOI] [PubMed] [Google Scholar]
- 37. Slavo GT, DiFazio1 SP, Martin J, Schackwitz W, Muchero W, et al (2012) Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa . New Phytol 196: 713–725. [DOI] [PubMed] [Google Scholar]
- 38. Abdurakhmonov IY, Abdukarimov A (2008) Application of association mapping to understanding the genetic diversity of plan germplasm resources. International Journal of Plant Genomics 2008: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sexton TR, Henry RJ, Harwood CE, Thomas DS, McManus LJ, et al. (2011) Pectin methyltransferase genes influence solid wood properties of Eucalyptus pilularis . Plant Physiol 158: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beaulieu J, Doerksen T, Boyle B, Clement S, Deslauriers M, et al. (2011) Association genetics of wood physical traits in the conifer White Spruce and relationships with gene expression. Genetics 188: 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sari-Gorla M, Krajewski P, Di Fonzo N, Villa M, Frova C (1999) Genetic analysis of drought tolerance in maize by molecular markers. II. Plant height and flowering. Theor Appl Genet 99: 289–295. [Google Scholar]
- 42. Ridoutt BG, Pharis RP, Sands R (1996) Fibre length and gibberellins A1 and A20 are decreased in Eucalyptus globulus by acylcyclohexanedione injected into stem. Physiol Plant 96: 559–566. [Google Scholar]
- 43. Eriksson ME, Israelsson M, Olsson O, Moritz T (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotech 18: 784–788. [DOI] [PubMed] [Google Scholar]
- 44.Higuchi T (1997) Biochemistry and molecular biology of wood. London: Springer Verlag.
- 45. Ross JJ, Murfet IC, Reid JB (1997) Gibberellin mutants. Physiol Plant 100: 550–560. [Google Scholar]
- 46. Sauter M, Kende H (1992) Gibberellin-induced growth and regulation of the cell division cycle in deepwater rice. Planta 188: 362–368. [DOI] [PubMed] [Google Scholar]
- 47. Daykin A, Scott IM, Francis D, Causton DR (1997) Effects of gibberellin on the cellular dynamics of dwarf pea internode development. Planta 203: 526–535. [Google Scholar]
- 48. Kimchi-Sarfati C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, et al. (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528. [DOI] [PubMed] [Google Scholar]
- 49. Chamary HV, Hurst LD (2009) The price of silent mutations. Sci Am 300(6): 46–53. [DOI] [PubMed] [Google Scholar]
- 50. Southerton SG, MacMillan CP, Bell JC, Bhuiyan N, Downes G, et al. (2010) Association of allelic variation in xylem genes with wood properties in Eucalyptus nitens . Austral For 73(4): 259–264. [Google Scholar]
- 51. Xu N, Chen CY, Shyu AB (1997) Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol 17: 4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu N, Loflin P, Chen CY, Shyu AB (1998) A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res 26: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fang XT, Xu HX, Zhang CL, Zhang JM, Lan CW, et al. (2010) Polymorphisms in BMP-2 gene and their associations with growth traits in goats. Genes Genom 32: 29–35. [Google Scholar]
- 54. Deng W, Lu LT, Luo KM, Li Y (2008) Transformation of gibberellin 20-oxidase gene of cotton into Chinese white poplar. Acta Bot. Boreal. –Occident. Sin 28(6): 1095–1100. [Google Scholar]
- 55. Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, et al. (1998) LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligo merisation, and unprecendented nucleic acid recognition. Tetrahedron 54: 3607–3630. [Google Scholar]
- 56. Koshkin AA, Rajwanshi VK, Wengel J (1998) Novel convenient syntheses of LNA [2.2.1] bicyclonucleosides. Tetrahedron Lett 39: 4381–4384. [Google Scholar]
- 57. Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) Bioinformatics. 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- 58.Sokal RR, Rohlf FJ (1981) Biometry, Second Edition. New York: W. H. Freeman and Company.
- 59. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 60. Hedrick PW (1987) Gametic disequilibrium measures: Proceed with caution. Genetics 117: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weir BS (1996) Genetic data analysis II: methods for discrete population genetic data. Sunderland, MA: Sinauer Associates Inc.
- 62. Gupta P, Rustgi S, Kulwal P (2005) Linkage disequilibrium and association studies in higher plants: Present status and future prospects. Plant Mol Biol 57: 461–485. [DOI] [PubMed] [Google Scholar]
- 63. Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, et al. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- 64. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 65. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ritland K (1996) Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67: 175–185. [Google Scholar]
- 67. Hardy OJ, Vekemans X (2002) spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 618–620. [Google Scholar]
- 68. Storey JD (2002) A direct approach to false discovery rates. J Roy Stat Soc Ser B 64: 479–498. [Google Scholar]
- 69. Storey JD, Taylor JE, Siegmund D (2004) Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J Roy Stat Soc Ser B 66: 187–205. [Google Scholar]
- 70. Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, et al. (2002) Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 53: 79–91. [DOI] [PubMed] [Google Scholar]
- 71. Eckert AJ, Bower AD, Wegrzyn JL, Pande B, Jermstad KD, et al. (2009) Association genetics of coastal Douglas fir (Pseudotsuga menziesii var. menziesii, Pinaceae). I. Cold hardiness related traits. Genetics 182: 1289–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The minimum, and maximum values, mean, standard error (SE) and coefficient of phenotypic variation [CV (%)] for each growth and wood property trait measured in P. tomentosa association population.
(DOC)
Estimates of phenotypic correlations (R) for these ten phenotypic traits in the association population.
(DOC)
All the association results. a. All the association results using the general linear model (GLM). b. All the association results using the mixed linear model (MLM).
(DOC)