Abstract
Recent clinical studies revealed increased phenylalanine levels and phenylalanine to tyrosine ratios in patients suffering from infection, inflammation and general immune activity. These data implicated down-regulation of activity of phenylalanine hydroxylase by oxidative stress upon in vivo immune activation. Though the structural damage of oxidative stress is expected to be comparably small, a structural rationale for this experimental finding was lacking. Hence, we investigated the impact of side chain oxidation at two vicinal cysteine residues on local conformational flexibility in the protein by comparative molecular dynamics simulations. Analysis of backbone dynamics revealed a highly flexible loop region (Tyr138-loop) in proximity to the active center of phenylalanine hydroxylase. We observed elevated loop dynamics in connection with a loop movement towards the active site in the oxidized state, thereby partially blocking access for the substrate phenylalanine. These findings were confirmed by extensive replica exchange molecular dynamics simulations and serve as a first structural explanation for decreased enzyme turnover in situations of oxidative stress.
Introduction
The mononuclear non-heme iron containing monooxygenase phenylalanine hydroxylase (PAH, EC 1.14.16.1) is a member of the aromatic amino acid hydroxylase family as tyrosine hydroxylase or tryptophan hydroxylases 1, 2. It catalyzes the oxidation of phenylalanine (Phe) to tyrosine (Tyr). This oxidation of the substrate amino acid is achieved by molecular oxygen and the reductive co-factor 5,6,7,8-tetrahydrobiopterin (BH4) [1]. Hydroxylation of Phe to Tyr is the committed step in Phe catabolism and therefore requires strict regulation to ensure homeostasis of the essential amino acid Phe. Dysfunction of PAH causes phenylketonuria (PKU) [2], [3], a common and well-examined genetic disease [4] leading to mental retardation upon accumulation of Phe [5].
The catalytic mechanism of PAH involves the reduction from ferric (III) to ferrous (II) form by the co-factor BH4. Ferrous iron is subsequently oxidized by molecular oxygen to a Fe(IV)O intermediate, which in turn hydroxylates Phe to Tyr [6]–[9]. Eukaryotic PAH is found as a homotetramer, where each subunit contains three distinct domains: an N-terminal autoregulatory domain (residues 1–142), the catalytic domain (residues 143–410) and a tetramerization domain stabilizing the quaternary structure (residues 411–452) [10], [11]. BH4 acts as a negative regulator of PAH activity by stabilizing an inactive form [12], whereas the enzyme is activated by phosphorylation of Ser16 [13] as well as binding of Phe [14]. Activity of PAH increases upon preincubation with Phe up to 100-fold [15]. Allosteric effects are held responsible for the activating effect of Phe by conformational changes in the protein [16]. The positive cooperativity of Phe binding allows for fast responses to increased Phe levels in order to avoid damages to the brain [17]. Recently, a potential binding site of Phe distal to the active site was discovered by a crystallographic study of PAH from Chromobacterium violaceum [18].
Expression of the truncated PAH catalytic domain does not result in a loss in activity suggesting the autoregulatory domain to be a key factor in allosteric regulation [19]. The structural background of PAH activation remains still unclear, as crystal structures are not yet available for the full sequence protein. The only available crystal structures containing the autoregulatory domain of a dimeric rat PAH lack parts of the tetramerization domain and structural insights into residues 1–18 including the phosphorylation site at Ser16 [20], [21]. Nevertheless, the position of the additional residues of the regulatory domain suggest a complete physical blockage of the active site in the inactive enzyme [22]. However, it remains unclear, if activation is due to a localized movement of the regulatory sequence or a global conformational reorganization in the protein.
Current studies found immune activation and inflammation to be associated with an increase in the ratio of Phe to Tyr in blood of patients suffering from trauma, sepsis, carcinoma and infection [23]–[25] suggesting an inhibition of PAH [26]. Recently, treatment with immune modulating interferon-α of patients with hepatitis C virus, resulted in an increase of Phe and the Phe to Tyr ratio [27] consistent with earlier findings concerning patients with multiple melanoma treated with interferon-α [28]. As immune activation of macrophages is paralleled by the release of toxic reactive oxygen species (ROS) and neopterin [29], [30], oxidative stress is discussed as chemical background for PAH deactivation consistent with studies correlating PKU with oxidative stress [31]. This hypothesis is further strengthened, as phenylalanine concentrations in blood samples of cancer patients were shown to correlate to the oxidative stress marker isoprostane-8 [24]. Immune activation marker neopterin was shown to further increase oxidizing capacity of ROS [32] and therefore raise concentrations of protein oxidation products [33] whilst decreasing antioxidant levels [34]. Modulation of PAH activity has also been reported after reaction with various disulfide reagents [35], [36], pointing particular attention to cysteine residues in PAH.
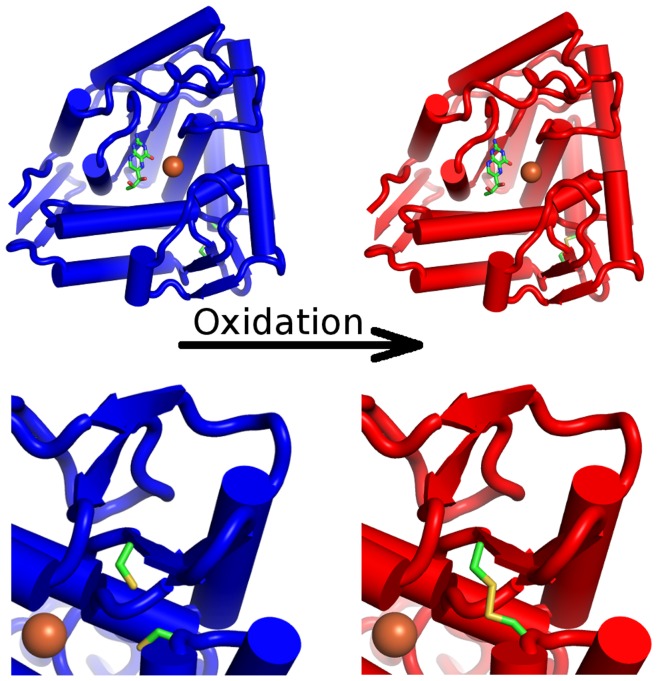
To investigate the impact of oxidative stress on PAH structure and dynamics at atomic level, molecular dynamics (MD) simulations of the catalytic domain were performed. Inspection of 23 available X-ray structures of the catalytic domain of PAH in the PDB [37], [38] revealed one single apparent oxidative site at two free cysteine residues (Cys203, Cys334) in close proximity to each other (4 Å) but distant from the active center of the enzyme (15 Å) (see Figure 1). An intrinsic connection between these cysteine residues and PAH activity in vivo is documented by mutation data from genetic studies of PKU. Point mutations of both residues are associated with mild forms of PKU [39]–[41].
Figure 1. Structural overview of native and oxidized phenylalanine hydroxylase.
The catalytic domain of native phenylalanine hydroxylase in blue cartoon representation (top left). The biopterin co-factor in the active site is shown as sticks, the catalytic iron as brown sphere. Cysteine residues 203 and 334 are shown as sticks to highlight the artificially introduced oxidation site distant from the catalytic center (red, top right). A zoom at the site of oxidation at the back of the top figures is shown at the bottom. Cys203 and Cys334 in proximity to each other (native state, blue, bottom left) are closed to a disulfide bond (oxidized state, red, bottom right).
Comparative MD simulations were performed to analyze conformational differences between native PAH (PAHnat) and PAH in oxidized state (PAHox) containing an artificially introduced disulfide bond between these residues. Findings from these simulations were confirmed by follow-up replica exchange molecular dynamics (REMD) simulations increasing sampling of the observed conformational transitions in the protein.
Methods
Two MD simulations were set up in parallel using the aforementioned starting structures PAHnat and PAHox. In simulation PAHnat of native PAH we treated Cys203 and Cys334 as individual cysteine residues, as reported in all available X-ray structures. In simulation of PAHox these cysteine residues were connected via an introduced disulfide bond to represent an oxidized state of PAH. This introduced disulfide bond between Cys203 and Cys334 is the only difference between the two simulated states PAHnat and PAHox.
As a starting point for simulations we chose the X-ray structure of human PAH with bound co-factor and highest available resolution (PDB: 1J8U [42], resolution:1.50 Å), although the bound co-factor and the iron atom were both found in reduced form. All atom root mean square deviation (RMSD) values to high resolution X-ray structures containing the oxidized species (PDB: 1TG2 [43], 1LTZ [44]) were found to be reasonably low (RMSD <1.13 Å) to set up simulations using template structure 1J8U with both oxidized co-factor 7,8-dihydrobiopterin BH2 and Fe(III).
Both all-atom MD simulations of the PAH catalytic domain containing a Fe(III)-ion and the co-factor BH2 were carried out using the AMBER10 package [45] using the ff99SB parameter set [46] for the protein and GAFF parameters [47] for the co-factor. Partial charges for the co-factor were derived by RESP fitting [48] at HF-6/31G*-level using Gaussian03 [49]. Van der Waals parameters for Fe(III) were derived by distance scanning against a helium atom at HF-6/31G*-level and found to be r = 1.20 Å and e = 0.0140 kcal/(mol*cm) in agreement with other transition metals in ff99SB.
Glutamate as well as aspartate residues were deprotonated, lysine and arginine residues protonated. Histidine (His) protonation was carried out manually according to the respective chemical environment: His200, His201, His208 and His271 at the protein surface were protonated at both imidazole nitrogens, His146, His170, His264 were assigned ε-hydrogen atoms. His285 and His290 coordinating the Fe(III)-ion at the active center were assigned δ-hydrogen atoms. The total charge of the system was found to be +3, which was neutralized by a uniform neutralizing plasma for Particle Mesh Ewald simulations [50].
The solvent was treated explicitly in the simulations including all resolved water molecules from the X-ray structure. The system was soaked with a wall distance of 10 Å by superposing an octahedral box of 7554 TIP3P [51] water molecules, resulting in a box edge of 70.8 Å after NpT equilibration.
After relaxation with harmonic restraints on heavy protein atoms and subsequent minimization, the systems were heated gradually from 100 K to 300 K over 200 ps in NVT ensemble. After heating and 1 ns of pressure and density equilibration, a NpT MD production run of 200 ns was performed to allow for larger conformational rearrangement in the enzyme. Simulations were carried out at 300 K using a Langevin thermostat [52] at 1.0 bar pressure with 8.0 Å nonbonded cutoff. SHAKE algorithm [53] on hydrogen atoms allowed a 2 fs time step. Snapshots were saved to trajectory every 500 steps or equivalent 1 ps for further analysis.
REMD simulations increase conformational sampling compared to standard MD simulations by exchanging replica of the simulated system at different temperatures, if an energy criterion is met. Hence, energy barriers are more likely to be overcome extending the sampling of conformational space [54]. In this study, equilibrated systems of standard MD simulations were used as input coordinates for two REMD simulations (PAHnat, PAHox). Sixteen parallel simulations were performed for both systems in a temperature spacing of 2 K as suggested by Patriksson et al [55] in a temperature range from 300 to 330 K. Exchange probability was set to 0.25 whilst attempting 10 exchanges per nanosecond. Besides a change to NVT ensemble, which is justified after volume adaption during pressure equilibration, identical parameters as for standard MD simulations were applied for REMD simulations. Production runs were performed for the two systems at 16 temperatures for 100 ns, resulting in total REMD trajectory length of 1.6 µs each.
Trajectories were analyzed using ptraj (version 4/2010) from AmberTools [45]. Positional fluctuations were calculated as RMSD of Cα-atoms to assess stability of the simulations. B-factors (temperature factors) were calculated residue-wise as a measure of individual local flexibility. Structures were visualized using pymol [56]. This software package was also used to calculate the accessible surface area of the catalytic Fe(III)-ion over the simulation time by the command ‘get_area’. Binding site volumes were estimated using POVME [57] by creating a 6 Å sphere around the center Fe(III)-ion filling the active site cavity with 1 Å spaced grid points. Subsequently, grid points near protein atoms were removed yielding an estimate of the actual binding site by summing up volume elements of residual grid points.
Results
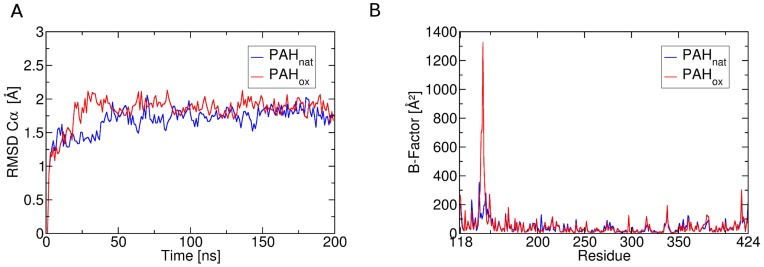
After equilibration, for both simulated systems stable molecular dynamics trajectories over 200 ns with mean backbone Cα RMSD values below 2 Å could be generated (see Figure 2). After 20 ns simulation PAHox shows a sharp increase in backbone dynamics as a loop region around residue 140 distant from the artificially linked cysteine residues starts to rearrange. Despite this structural transition in the so-called Tyr138-loop [58]–[60], simulation PAHox yields a stable trajectory for the whole simulated system resulting in an RMSD plateau below 2 Å at comparable level to simulation PAHnat.
Figure 2. Global dynamic behavior of native and oxidized phenylalanine hydroxylase.
A: RMSD of Cα-atoms over the simulation time of 200 ns standard MD simulations for the two systems PAHnat (blue) and PAHox (red). After an initial phase of loop reorientation for the oxidized system PAHox (around 20 ns simulation time), simulations yield stable trajectories. B: Residue-wise B-factors for the simulated systems highlight residues 130–145 as particularly altered by the introduced cysteine oxidation. Elevated B-factors in this loop region (Tyr138-loop) in PAHox are caused by a reorientation of the loop in an early stage of the simulation. Dynamics of residues around the oxidation sites (Cys-203, Cys334) are similar in both systems.
Calculated B-factors also highlight the Tyr138-loop as region of particular interest with B-factors reaching beyond 1000 Å2 in PAHox compared to 300 Å2 in PAHnat. Other regions show less pronounced flexibility and similar behavior in both MD simulations. This finding also holds true for the vicinities of Cys203 and Cys334, where the artificial oxidation was introduced in PAHox, which are found to be rigid in both simulations.
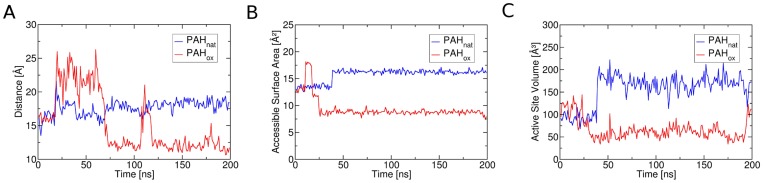
As indicated by the distance plot in Figure 3, the flexible Tyr138-loop performs a movement towards the active site of the protein after an initial phase of elevated dynamics in PAHox (15–70 ns) leading to a contraction of the binding site. This movement is not present in the simulation of the native state PAHnat. This movement in PAHox is paralleled by a decrease in accessibility of the catalytic iron center indicated by lowered accessible surface area of below 10 Å2 in PAHox compared to over 15 Å2 in PAHnat (see Figure 3 and Table 1). Furthermore, the total binding site volume is reduced significantly in the course of this loop rearrangement in PAHox: An average active site volume of over 150 Å3 in PAHnat is reduced to less than half in PAHox by the introduced perturbation.
Figure 3. Dynamics of the binding site region of native and oxidized phenylalanine hydroxylase.
A: Distance between the catalytic iron center and the Cα-atom of Glu141 included in the flexible Tyr138-loop near the active site. Whereas this distance remains mostly stable in the native state PAHnat over the simulation time (blue), simulation of PAHox shows an initial phase of increased loop flexibility that is followed by decrease of that distance indicating a loop movement towards the active site. This finding is confirmed by calculation of the accessible surface area of the iron center (B). After an initial adaption to the perturbation, PAHox shows a constantly reduced accessibility of the catalytic iron center compared to PAHnat. The overall active site volume (C) is similarly reduced over simulation time in PAHox compared to PAHnat, suggesting a reduced enzyme turnover.
Table 1. Averages and standard deviations (SD) of accessible surface area of the catalytic iron as well as the total active site volume for the simulations of PAHnat and PAHox.
| 200 ns MD | 16*100 ns REMD | |||
| Average (SD) | PAHnat | PAHox | PAHnat | PAHox |
| Area [Å2] | 15.74 (1.16) | 9.40 (1.97) | 15.38 (2.33) | 13.68 (3.07) |
| Volume [Å3] | 155.87 (35.11) | 66.58 (22.73) | 116.15 (54.42) | 104.76 (45.82) |
REMD simulations confirm the reduction in both measures for PAHox in comparison to PAHnat for an average over 16 simulations over 100 ns compare to a single observation in a 200 ns standard MD run.
REMD simulations yielded comparably stable trajectories (see Figure S1), confirming trends observed from for standard MD simulations. B-factors in the Tyr138-loop are generally elevated in PAHox compared to PAHnat (see Figure S2). This observation is true for every single direct comparison of 16 temperature-equivalent replicas. Likewise, PAHox shows a lowered average accessibility of the catalytic iron of 13.7 Å2 compared to 15.4 Å2 in PAHnat. The volume of the active site is decreased similarly in the oxidized state from 116.2 Å3 in PAHnat to 104.8 Å3 in PAHox.
Discussion
As a structural explanation of the impairment of PAH caused by oxidative stress has been lacking by now, we propose a computational model of an oxidized PAH specie. This protein structure shows an altered behavior in MD simulations in the Tyr138-loop in close proximity to the active site [61] leading to a stable structure showing a contracted binding site. As the loop rearrangement occurs as a single event in the 200 ns MD trajectory, the statistical significance of this observation cannot be inferred from standard MD simulations of that time scale [62]. This intramolecular movement hence appears on a time scale beyond sampling of standard state-of-the-art MD simulations.
To statistically substantiate this proposed structural rationale for PAH down-regulation, REMD simulations providing improved conformational sampling were performed. These computationally demanding simulations agreed with trends seen in standard MD simulations, namely elevated loop dynamics paralleled with a decrease in accessibility of the catalytic iron center and binding site contraction. This contraction of the binding pocket would obviously lead to reduced substrate binding and hence enzyme down-regulation or even dysfunction. The enhanced sampling by REMD simulations decreased the observed differences between the simulations of PAHnat and PAHox, but still supported findings from standard MD simulations, as 16 additional parallel simulations confirmed the trends observed from a single standard MD run.
Findings from MD simulations linking dynamics of the Tyr138-loop region with catalytic activity are in best agreement with experimental data on allosteric regulation of PAH. Li and co-workers showed by an increase in exchange kinetics of hydrogen/deuterium exchange mass spectrometry, that region 129–143 is indeed a solvent-accessible and highly flexible region in PAH that is influenced by presence of Phe [63]. This observation is further confirmed, as this region is not resolved in several crystal structures of PAH in absence of a substrate stabilizing the active site. Furthermore, modulation of the dynamics of region 129–143 by incubation with phenylalanine, an activator of PAH, indicates a mechanistic role in allosteric regulation [63].
The critical regulatory role of surface loops has been discussed in a recent review on allosteric regulation of PAH by Fitzpatrick [60]. He describes the Tyr138-loop as undergoing the most drastic change upon ligand binding in accordance with crystal structure analyses of Andersen et al [58], [59]. Tyr138 included in that loop closes down on the active site leading to a major contraction restricting further access similar to the movement observed in our simulations (see Figure 4). A similar movement of a surface loop has been proposed by Sura et al. for the homologous enzyme tyrosine hydroxylase [64].
Figure 4. Changes in the active site of phenylalanine hydroxylase introduced by side chain oxidation.
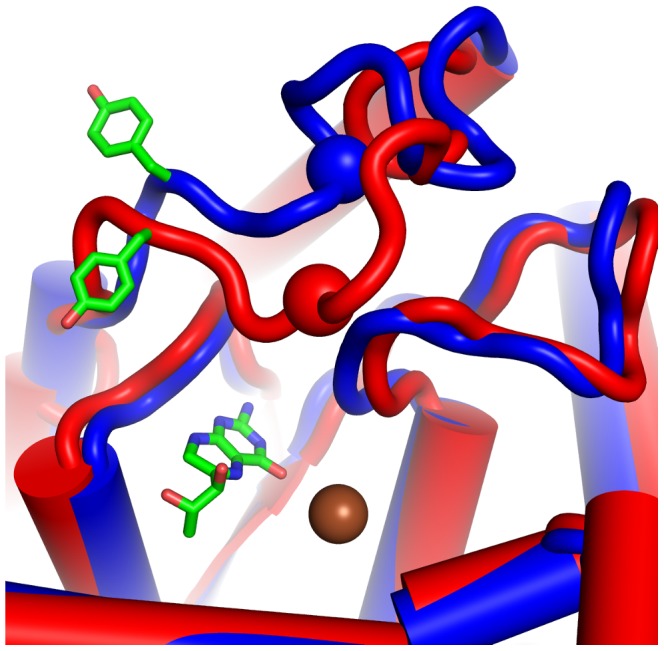
Starting coordinates of both simulations (blue cartoon) overlaid with an average structure of the time frames 150–160 ns of standard MD simulation PAHox (red cartoon). The pronounced loop movement towards the catalytic center (iron ion as brown sphere, co-factor as sticks) was measured as distance of the Cα-atom of Glu141 (red and blue sphere respectively) to the catalytic iron (brown). This rearrangement is paralleled by a reorientation of Tyr138 (shown as sticks, left), in turn concertedly blocking access to the active site of PAHox.
This dynamic behavior of the loop region could be seen as an analogy to the behaviour of the autoregulatory sequence of PAH. This domain of the protein physically blocks entrance to the active site of PAH in the inactive form. A smaller but similar impact of the surface loop can be inferred from presented molecular dynamics simulations directly explaining the observed PAH impairment upon oxidative stress.
The importance of conformational flexibility for PAH activity has been revealed in several studies: Cerreto et al and Gersting et al showed correlations between PAH conformational stability and PAH down-regulation by PKU-causing mutations [65], [66]. Changes in stability were shown by MD simulations upon phosphorylation of Ser-16 [67]. This computational technique was also used to infer factors for specificity and affinity of PAH ligand binding [68], [69].
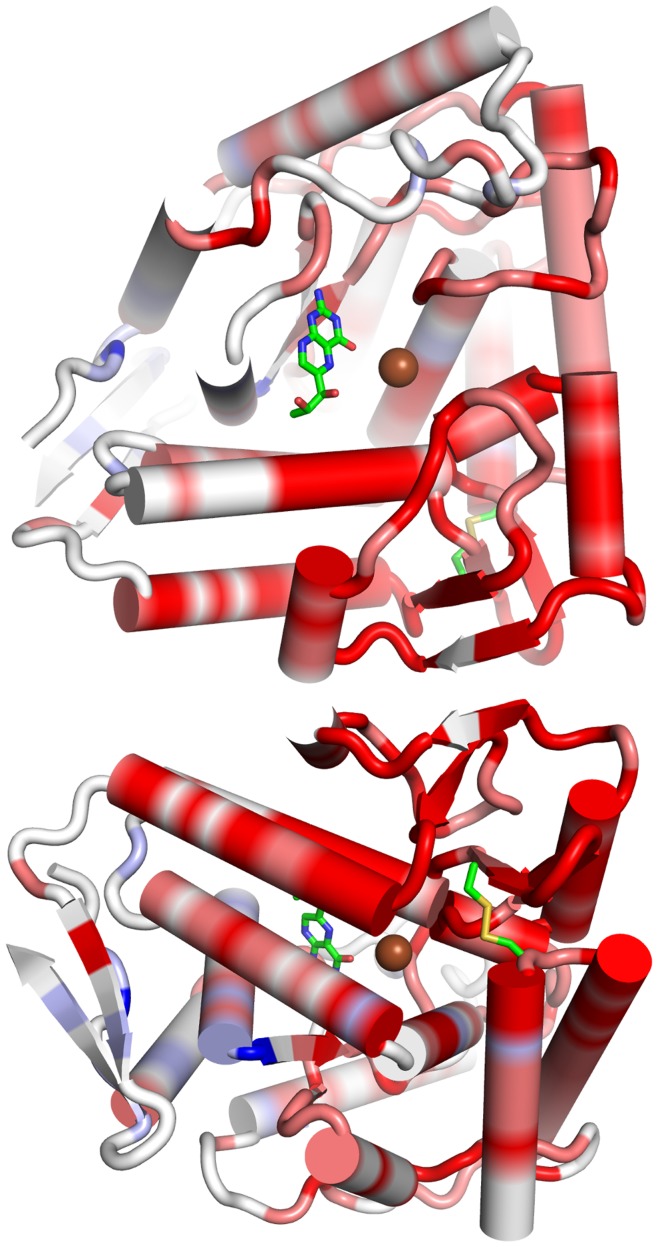
As presented MD simulations allow to keep track of alterations in molecular motions within the protein, a pathway for the inferred allosteric signaling between Cys203/Cys334 and the active site could be anticipated. An analysis of hydrogen bonding within the protein did not highlight a particular alteration in molecular interactions within the protein but rather suggested a concerted flow of dynamic behavior throughout the protein backbone of PAH (see Figure 5). The differences in molecular flexibility between PAHox and PAHnat REMD simulations are highlighted by a color-coding from blue for regions more flexible in PAHnat over white to red for regions more flexible in PAHox. The dominating red coloring of PAHox starting from the artificial oxidation site propagates over the protein backbone towards the surface loops in close proximity to the active site, whereas only minor blue regions are found in a small part of the binding site next to the biopterin co-factor. This highlights a putative interaction path for the allosteric effect of disulfide formation on the active center via the central helices directly to the Tyr138-loop near the active site. A loop movement near the active site in turn restricts accessibility of the catalytic site, hence explaining reduced enzyme turnover.
Figure 5. Alteration in local dynamics in phenylalanine hydroxylase upon side chain oxidation.
Residue-wise B-factors for the 2*16 REMD trajectories were calculated for Cα-atoms and compared between simulations of equal temperatures. Rank-based differences were mapped on the protein structure, where blue regions indicate regions more flexible in more of the 16 simulations of PAHnat. Unaffected regions are shown in white, whereas red regions indicate regions showing elevated backbone dynamics in more of the 16 simulations of PAHox. Elevated B-factors in the central helices suggest an allosteric signal transduction over this path from the introduced oxidation site (Cys203, Cys334) to the Tyr138-loop near the catalytic center. The bottom picture shows the same structure rotated by 220° to highlight local effects on the oxidation site in the back of the top picture.
Our study offers a new example in the emerging field of redox signaling in enzyme activity, that is facilitated by novel findings concerning molecular mechanisms of redox switches in proteins [70]. Schroeder et al for example recently highlighted how the reduction of disulfide bonds activates an antimicrobial protein [71]. Tanner et al. recently reviewed the redox regulation of protein tyrosine phosphatases, where an active site cysteine takes several oxidation states and is inactivated by redox signaling [72]. A regulatory mechanism similar to the proposed signal propagation in PAH has been found by Wang et al. in C-reactive protein, where a single disulfide bond serves as a switch for the function of the protein [73].
An obvious limitation of the presented study is the underlying simplistic model of PAHox. Although reduced Cys203 and Cys334 in close proximity are a clear weak-point for oxidation in the protein, this model might not cover all effects of oxidative stress. Cysteine residues might undergo further oxidation processes besides simple disulfide formation. On the other hand, also the active site of PAH is susceptible to oxidative species, as the enzyme co-factor is readily oxidizable under oxidizing conditions. These latter two effects could not be covered in the presented simulations and hence might be starting points for further investigations from theoretical and experimental side.
In conclusion, our study provides a structural basis for the down-regulation of PAH activity upon oxidative stress. MD simulations highlight a flexible loop region around residue 140 (Tyr138-loop) as susceptible to side chain oxidation in PAH. A complex movement over the whole protein leads to a contraction of the binding site leading to the experimentally observed reduced enzyme activity.
Supporting Information
Dynamics of REMD simulations. A: RMSD of Cα-atoms over the simulation time of 16 REMD simulations a 100 ns of PAHnat (blue). Stable trajectories showing comparable deviations to the starting structure as in standard molecular dynamics runs were obtained. The same holds true for 16 REMD simulations of PAHox (red, B), where deviations are slightly elevated due to the initial perturbation by disulfide formation.
(TIFF)
Positional fluctuations in REMD simulations. A: Residue-wise B-factors for 16 REMD simulations a 100 ns of PAHnat (blue). The flexible Tyr138-loop shows several conformational transitions, whereas other parts of the protein remain stable. The Tyr138-loop shows even higher elevated B-factors in PAHox (red, B) indicating a major rearrangement in this loop region.
(TIFF)
Acknowledgments
The authors thank Knut Teigen and Aurora Martinez for fruitful discussions about the project. The authors acknowledge the platform High Performance Computing at Leopold Franzens University of Innsbruck for providing access to the Leo II computer cluster.
Funding Statement
The research of the manuscript was supported by funding of the Austrian Science Fund FWF: project “Targeting Influenza Neuraminidase” (P23051). Julian E. Fuchs is a recipient of a DOC-fellowship of the Austrian Academy of Sciences at the Institute of General, Inorganic and Theoretical Chemistry at University of Innsbruck. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Werner ER, Blau N, Thöny B (2011) Tetrahydrobiopterin: Biochemistry and Pathophysiology. Biochem J 438: 397–414. [DOI] [PubMed] [Google Scholar]
- 2. Kaufman S, Holtzman NA, Milstien S, Butler IJ, Krumholz A (1975) Phenylketonuria Due to a Deficiency of Dihydrobiopteridine Reductase. N Engl J Med 293: 785–790. [DOI] [PubMed] [Google Scholar]
- 3. Blau N, Van Spronsen FJ, Levy HL (2010) Phenylketonuria. Lancet 376: 1417–1427. [DOI] [PubMed] [Google Scholar]
- 4. Scriver CR, Hurtubise M, Konecki D, Phommorinh M, Prevost L, et al. (2003) PAHdb 2003: What a Locus-Specific Knowledgebase Can Do. Hum Mutat 21: 333–344. [DOI] [PubMed] [Google Scholar]
- 5. Folling A (1934) Ueber Ausscheidung von Phenylbrenztraubensaeure in den Harn als Stoffwechselanomalie in Verbindung mit Imbezillitaet. Ztschr Physiol Chem 227: 169–176. [Google Scholar]
- 6. Fitzpatrick PF (2003) Mechanism of Aromatic Amino Acid Hydroxylation. Biochemistry 42: 14083–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olsson E, Martinez A, Teigen K, Jensen VR (2011) Formation of the Iron-Oxo Hydroxylating Species in the Catalytic Cycle of Aromatic Amino Acid Hydroxylases. Chem Eur J 17: 3746–3758. [DOI] [PubMed] [Google Scholar]
- 8. Olsson E, Martinez A, Teigen K, Jensen VR (2011) Substrate hydroxylation by the Oxido-Iron Intermediate in Aromatic Amino Acid Hydroxylases: A DFT Mechanistic Study. Eur J Inorg Chem 17: 2720–2732. [Google Scholar]
- 9. Panay AJ, Lee M, Krebs C, Bollinger JM, Fitzpatrick PF (2011) Evidence for a High-Spin Fe(IV) Species in the Catalytic Cycle of a Bacterial Phenylalanine Hydroxylase. Biochemistry 50: 1928–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Erlandsen H, Fusetti F, Martinez A, Hough E, Flatmark T, et al. (1997) Crystal Structure of the Catalytic Domain of Human Phenylalanine Hydroxylase Reveals the Structural Basis for Phenylketonuria. Nat Struct Biol 4: 995–1000. [DOI] [PubMed] [Google Scholar]
- 11. Fusetti F, Erlandsen H, Flatmark T, Stevens RC (1998) Structure of Tetrameric Human Phenylalanine Hydroxylase and Its Implications for Phenylketonuria. J Biol Chem 273: 16962–16967. [DOI] [PubMed] [Google Scholar]
- 12. Mitnaul LJ, Shiman R (1995) Coordinate Regulation of Tetrahydrobiopterin Turnover and Phenylalanine Hydroxylase Activity in Rat Liver Cells. Proc Nastl Acad Sci USA 92: 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abita JP, Milstien S, Chang N, Kaufman S (1976) In Vitro Activation of Rat Liver Phenylalanine Hydroxylase by Phosphorylation. J Biol Chem 17: 5310–5314. [PubMed] [Google Scholar]
- 14. Shiman R, Mortimore GE, Schworer CM, Gray DW (1982) Regulation of Phenylalanine Hydroxylase Activity by Phenylalanine in Vivo, in Vitro, and in Perfused Rat Liver. J Biol Chem 257: 11213–11216. [PubMed] [Google Scholar]
- 15. Shiman R, Gray DW (1980) Substrate Activation of Phenylalanine Hydroxylase. J Biol Chem 255: 4793–4800. [PubMed] [Google Scholar]
- 16. Fitzpatrick PF (1999) Tetrahydropterin-Dependent Amino Acid Hydroxylases. Annu Rev Biochem 68: 355–381. [DOI] [PubMed] [Google Scholar]
- 17.Scriver CR, Kaufman S (2001) Hyperphenylalaninemia: phenylalanine hydroxylase deficiency. In: Scriver CR, Beaudet AL, Valle D, Sly WS, editors. The metabolic and molecular basis of inherited disease, 8th edition. 1667–1724.
- 18.Ronau JA, Abu-Omar MM, Das C (2012) Crystallographic characterization of a substrate-bindin site distinct from the active site in a bacterial phenylalanine hydroxylase. To be published, PDB: 3TCY.
- 19. Daubner SC, Hillas PJ, Fitzpatrick PF (1997) Expression and Characterization of the Catalytic Domain of Human Phenylalanine Hydroxylase. Arch Biochem Biophys 348: 295–302. [DOI] [PubMed] [Google Scholar]
- 20. Kobe B, Jennings IG, House CM, Michaell BJ, Goodwill KE, et al. (1999) Structural basis of autoregulation of phenylalanine hydroxylase. Nat Struct Biol 6: 442–448. [DOI] [PubMed] [Google Scholar]
- 21. Miranda FF, Teigen K, Thorolfsson M, Svebak RM, Knappskog PM, et al. (2002) Phosphorylation and Mutations of Ser16 in Human Phenylalanine Hydroxylase. J Biol Chem 277: 40937–40943. [DOI] [PubMed] [Google Scholar]
- 22. Jennings IG, Teh T, Kobe B (2001) Essential Role of the N-terminal Autoregulatory Sequence in the Regulation of Phenylalanine Hydroxylase. FEBS Lett 488: 196–200. [DOI] [PubMed] [Google Scholar]
- 23. Ploder M, Neurauter G, Spittler A, Schroecksnadel K, Roth E, et al. (2008) Serum phenylalanine in patients post trauma and with sepsis correlate to neopterin concentrations. Amino Acids 35: 303–307. [DOI] [PubMed] [Google Scholar]
- 24. Neurauter G, Grahmann AV, Klieber M, Zeimet A, Ledochowski M, et al. (2008) Serum phenylalanine concentrations in patients with ovarian carcinoma correlate with concentrations of immune activation markers and of isoprostane-8. Cancer Lett 272: 141–147. [DOI] [PubMed] [Google Scholar]
- 25. Zangerle R, Kurz K, Neurauter G, Kitchen M, Sarcletti M, et al. (2010) Increased blood phenylalanine to tyrosine ratio in HIV-1 infection and correction following effective antiretroviral therapy. Brain Behav Immun 24: 403–408. [DOI] [PubMed] [Google Scholar]
- 26. Rosenblatt D, Scriver CR (1968) Heterogeneity in Genetic Control of Phenylalanine Metabolism in Man. Nature 218: 677–678. [DOI] [PubMed] [Google Scholar]
- 27. Zoller H, Schloegel A, Schroecksnadel S, Vogel W, Fuchs D, et al. (2012) Interferon-Alpha Therapy in Patients with HCV Infection Increases Plasma Phenylalanine and the Phenylalanine to Tyrosine Ratio. J Interf Cytok Res 32: 216–220. [DOI] [PubMed] [Google Scholar]
- 28. Van Gool AR, Van Oijk HH, Kruit WHJ, Bannink M, Mulder PGH, et al. (2004) Pegylated interferon-α2b treatment in melanoma patients: influence on amino acids, 5-hydroxyindolacetic acid and pterine plasma concentrations. Ant-Cancer Drug 15: 587–591. [DOI] [PubMed] [Google Scholar]
- 29. Nathan CF, Murray HW, Wiebe ME, Rubin BY (1983) Identification of interferon-γ as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med 158: 670–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murr C, Widner B, Wirleitner B, Fuchs D (2002) Neopterin as a Marker for Immune System Activation. Curr Drug Metab 3: 175–187. Anticancer Res 19: 1721–1728. [DOI] [PubMed] [Google Scholar]
- 31. Ribas GS, Sitta A, Wajner M, Vargas CR (2011) Oxidative Stress in Phenylketonuria: What is the Evidence? Cell Mol Neurobiol 31: 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffmann G, Wirleitner B, Fuchs D (2003) Potential Role of Immune System Activation-Associated Production of Neopterin Derivatives in Humans. Inflamm Res 52: 313–321. [DOI] [PubMed] [Google Scholar]
- 33. Witko-Sarsat V, Friedlander M, Capeillere-Blandin C, Nguyen-Khoa T, Nguyen NT, et al. (1996) Advanced oxidation protein products as a novel marker of oxidative stress in uremia. Kidney Int 49: 1304–1313. [DOI] [PubMed] [Google Scholar]
- 34.Murr C, Fuith LC, Widner B, Wirleitner B, Baier-Bitterlich G, et al. (1999) Increased neopterin concentrations in patients with cancer: Indicator of oxidative stress? [PubMed]
- 35. Parniak MA, Kaufman S (1981) Rat Liver Phenylalanine Hydroxylase. J Biol Chem 256: 6876–6882. [PubMed] [Google Scholar]
- 36. Koizumi S, Suzuki T, Takahashi S, Satake K, Takeuchi T, et al. (1987) Sulfhydryl Modification and Activation of Phenylalanine Hydroxylase by Dinitrophenyl Alkyl Disulfide. Biochemistry 26: 6461–6465. [DOI] [PubMed] [Google Scholar]
- 37. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kirchmair J, Patrick M, Distinto S, Schuster D, Spitzer GM, et al. (2008) The Protein Data Bank (PDB), Its Related Services and Software Tools as Key Components for in Silico Drug Discovery. J Med Chem 51: 7021–7040. [DOI] [PubMed] [Google Scholar]
- 39. Eisensmith RC, Martinez DR, Kuzmin AI, Goltsov AA, Brown A, et al. (1996) Molecular Basis of Phenylketonuria and a Correlation Between Genotype and Phenotype in a Heterogeneous Southeastern US Population. Pediatrics 97: 512–516. [PubMed] [Google Scholar]
- 40. Guldberg P, Rey F, Zschocke J, Romano V, Francois B, et al. (1998) European Multicenter Study of Phenylalanine Hydroxylase Deficiency: Classification of 105 Mutations and a General System for Genotype-Based Prediction of Metabolic Phenotype. Am J Hum Genet 63: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Erlandsen H, Patch MG, Gamez A, Straub M, Stevens RC (2003) Structural Studies on Phenylalanine Hydroxylase and Implications Toward Understanding and Treating Phenylketonuria. Pediatrics 112: 1557–1565. [PubMed] [Google Scholar]
- 42. Andersen OA, Flatmark T, Hough E (2001) High Resolution Crystal Structure of the Catalytic Domain of Human Phenylalanine Hydroxylase in its Catalytically Active Fe(II) Form and Binary Complex with Tetrahydrobiopterin. J Mol Biol 314: 279–291. [DOI] [PubMed] [Google Scholar]
- 43. Erlandsen H, Pey AL, Gamez A, Perez B, Desviat LR, et al. (2004) Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations. Proc Natl Acad Sci USA 101: 16903–16908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Erlandsen H, Kim JY, Patch MG, Han A, Volner A, et al. (2002) Structural Comparison of Bacterial and Human Iron-dependent Phenylalanine Hydroxylases: Similar Fold, Different Stability and Reaction Rates. J Mol Biol 320: 645–661. [DOI] [PubMed] [Google Scholar]
- 45.Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, et al. (2008) AMBER 10. University of California, San Francisco.
- 46. Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, et al. (2006) Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins 65: 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and Testing of a General Amber Force Field. J Comput Chem 25: 1157–1174. [DOI] [PubMed] [Google Scholar]
- 48. Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges: The RESP Model. J Phys Chem 97: 10269–10280. [Google Scholar]
- 49.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al. (2004) Gaussian 03, Revision C.02. Gaussian Inc, Wallingfort CT.
- 50. Darden T, York D, Pedersen L (1993) Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Phys 98: 10089–10092. [Google Scholar]
- 51. Jorgensen WL, Chandrasekhar J, Madura J, Impey RW, Klein ML (1983) Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys 79: 926–935. [Google Scholar]
- 52. Adelman SA, Doll JD (1976) Generalized Langevin Equation Approach for Atom-Solid-Surface Scattering – General Formulation for Classical Scattering Off Harmonic Solids. J Chem Phys 64: 2375–2388. [Google Scholar]
- 53. Ciccotti G, Ryckaert JP (1986) Molecular Dynamics Simulation of Rigid Molecules. Comput Phys Rep 4: 345–392. [Google Scholar]
- 54. Sugita Y, Okamoto Y (1999) Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett 314: 141–151. [Google Scholar]
- 55. Patriksson A, Van der Spoel D (2008) A temperature predictor for parallel tempering simulations. Phys Chem Chem Phys 10: 2073–2077. [DOI] [PubMed] [Google Scholar]
- 56.The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.
- 57. Durrant JD, De Oliveira CAF, McCammon JA (2011) POVME: An Algorithm for Measuring Binding-Pocket Volumes. J Mol Graph Model 29: 773–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Andersen OA, Flatmark T, Hough E (2002) Crystal Structure of the Ternary Complex of the Catalytic Domain of Human Phenylalanine Hydroxylase with Tetrahydrobiopterin and 3-(2-Thienyl)-L-alanine, and its Implications for the Mechanism of Catalysis and Substrate Activation. J Mol Biol 320: 1095–1108. [DOI] [PubMed] [Google Scholar]
- 59. Andersen OA, Stokka AJ, Glatmark T, Hough E (2003) 2.0 A Resolution Crystal Structures of the Ternary Complexes of Human Phenylalanine Hydroxylase Catalytic Domain with Tetrahydrobiopterin and 3-(2-Thienyl)-L-alanine or L-Norleucine: Substrate Specificity and Molecular Motions Related to Substrate Binding. J Mol Biol 333: 747–757. [DOI] [PubMed] [Google Scholar]
- 60. Fitzpatrick PF (2012) Allosteric Regulation of Phenylalanine Hydroxylase. Arch Biochem Biophys 519: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fuchs JE, Huber RG, Wallnoefer HG, Von Grafenstein S, Spitzer GM, et al. (2011) Side Chain Oxidation Modulates Local Dynamics of Phenylalanine Hydroxylase. Pteridines 22: 31–32. [Google Scholar]
- 62. Fuchs JE, Huber RG, Wallnoefer HG, Von Grafenstein S, Spitzer GM, et al. (2012) Conformational Changes of Phenylalanine Hydroxylase Upon Simulated Oxidative Stress. Pteridines 23: 57–58. [Google Scholar]
- 63. Li J, Dangott LJ, Fitzpatrick PF (2010) Regulation of Phenylalanine Hydroxylase: Conformational Changes Upon Phenylalanine Binding Detected by Hydrogen/Deuterium Exchange and Mass Spectrometry. Biochemistry 49: 3327–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sura GR, Lasagna M, Gawandi V, Reinhart GD, Fitzpatrick PF (2006) Effects of Ligands on the Mobility of an Active-Site Loop in Tyrosine Hydroxylase as Monitored by Flourescence Anisotropy. Biochemistry 45: 9632–9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cerreto M, Cavaliere P, Carluccio C, Amato F, Zagari A, et al. (2011) Natural phenylalanine hydroxylase variants that confer a mild phenotype affect the enzyme’s conformational stability and oligomerization equilibrium. Biochim Biophys Acta 1812: 1435–1445. [DOI] [PubMed] [Google Scholar]
- 66. Gersting SW, Femter KF, Staudig M, Messing DD, Danecka MK, et al. (2008) Loss of Function in Phenylketonuria Is Caused by Impaired Molecular Motions and Conformational Instability. Am J Hum Genet 83: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miranda FF, Thorolfsson M, Teigen K, Sanchez-Ruiz JM, Martinez A (2004) Structural and stability effects of phosphorylation: Localized structural changes in phenylalanine hydroxylase. Protein Sci 13: 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Teigen K, Martinez A (2003) Probing Cofactor Specificity in Phenylalanine Hydroxylase by Molecular Dynamics Simulations. J Biomol Struct Dyn 20: 733–740. [DOI] [PubMed] [Google Scholar]
- 69. Teigen K, McKinney JA, Haavik J, Martinez A (2007) Selectivity and Affinity Determinants for Ligand Binding to the Aromatic Amino Acid Hydroxylases. Curr Med Chem 14: 455–467. [DOI] [PubMed] [Google Scholar]
- 70. Kemp M, Go YM, Jones DP (2008) Non-equilibrium thermodynamics of thiol/disulfide redox systems: A perspective on redox systems biology. Free Radic Biol Med 44: 921–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, et al. (2011) Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 469: 419–425. [DOI] [PubMed] [Google Scholar]
- 72. Tanner JJ, Parsons ZD, Cummings AH, Zhou H, Gates KS (2011) Redox Regulation of Protein Tyrosine Phosphatases: Structural and Chemical Aspects. Antioxid Redox Signaling 15: 77–97. [DOI] [PubMed] [Google Scholar]
- 73. Wang MY, Ji SR, Bai CJ, El Kebir D, Li HY, et al. (2011) A redox switch in C-reactive protein modulates activation of endothelial cells. FASEB J 25: 3186–3196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dynamics of REMD simulations. A: RMSD of Cα-atoms over the simulation time of 16 REMD simulations a 100 ns of PAHnat (blue). Stable trajectories showing comparable deviations to the starting structure as in standard molecular dynamics runs were obtained. The same holds true for 16 REMD simulations of PAHox (red, B), where deviations are slightly elevated due to the initial perturbation by disulfide formation.
(TIFF)
Positional fluctuations in REMD simulations. A: Residue-wise B-factors for 16 REMD simulations a 100 ns of PAHnat (blue). The flexible Tyr138-loop shows several conformational transitions, whereas other parts of the protein remain stable. The Tyr138-loop shows even higher elevated B-factors in PAHox (red, B) indicating a major rearrangement in this loop region.
(TIFF)