Abstract
Frequent contact with human waste and liquid manure from intensive livestock breeding, and the increased loads of antibiotic-resistant bacteria that result, are believed to be responsible for the high carriage rates of ESBL-producing E. coli found in birds of prey (raptors) in Central Europe. To test this hypothesis against the influence of avian migration, we initiated a comparative analysis of faecal samples from wild birds found in Saxony-Anhalt in Germany and the Gobi-Desert in Mongolia, regions of dissimilar human and livestock population characteristics and agricultural practices. We sampled a total of 281 wild birds, mostly raptors with primarily north-to-south migration routes. We determined antimicrobial resistance, focusing on ESBL production, and unravelled the phylogenetic and clonal relatedness of identified ESBL-producing E. coli isolates using multi-locus sequence typing (MLST) and macrorestriction analyses. Surprisingly, the overall carriage rates (approximately 5%) and the proportion of ESBL-producers among E. coli (Germany: 13.8%, Mongolia: 10.8%) were similar in both regions. Whereas bla CTX-M-1 predominated among German isolates (100%), bla CTX-M-9 was the most prevalent in Mongolian isolates (75%). We identified sequence types (STs) that are well known in human and veterinary clinical ESBL-producing E. coli (ST12, ST117, ST167, ST648) and observed clonal relatedness between a Mongolian avian ESBL-E. coli (ST167) and a clinical isolate of the same ST that originated in a hospitalised patient in Europe. Our data suggest the influence of avian migratory species in the transmission of ESBL-producing E. coli and challenge the prevailing assumption that reducing human influence alone invariably leads to lower rates of antimicrobial resistance.
Introduction
Previous studies have demonstrated high carriage rates of Extended-spectrum beta-Lactamase- producing E. coli (ESBL-E. coli) in faecal excreta of various wild avian hosts, including birds of prey and waterfowl [1]–[4] Most of these studies conducted in Central Europe, a region with high human and livestock densities [5], [6],facilitating interactions between wild birds and human-influenced habitats like urban environments, wastewater treatment facilities, landfills, and land used for intensive agricultural and livestock farming. It has been suggested that such interactions increase the probability for wildlife to acquire antibiotic-resistant bacteria [7], [8]and preliminary evidence shows that birds of prey carry more of these resistant bacteria when they live in an area with intensive livestock production [9]. Certain ESBL-E. coli isolates from wild avian hosts belong to phylogenetic lineages that are closely related to those found in human and veterinary clinical settings, presumably explaining the frequent observation of ESBL-E. coli in wild birds [10], [11]Thus, an indirect transmission of ESBL-E. coli from humans or domestic animals to wild animals, including wild birds, and vice versa, is plausible. However, the frequencies of such events and the routes of transmission are largely unknown, leaving several unanswered questions; namely whether (a) higher detection rates of ESBL-E. coli in avian hosts could be related to spatially linked higher human density, and/or whether (b) prolonged shedding of ESBL-E. coli in the birds excreta might compensate for infrequent transmission events.
To begin answering these questions, there is a clear need for studies comparing the microbiota of avian hosts in areas with contrasted exposure to human antimicrobial “practice” (use). This study therefore aimed to (i) assess the rate of ESBL-E. coli carriage by birds of prey in remote areas compared to those in Central Europe, (ii) characterize these ESBL-E. coli genotypically, and ultimately, (iii) provide preliminary data that might help assess the possible role of migrating avian hosts in the spread of ESBL-E. coli into remote environments.
The selection of both the sampling areas and the avian species to be sampled was crucial. Beyond enabling factors like legal access to avian samples from remote areas, it was important to select two sampling sites decidedly different in their human and livestock densities and agricultural practices, but still with comparable avian populations; different groups of avian species seem to differ largely in their carriage rates of ESBL-E. coli [11].
We therefore chose sampling spots in semi-desert areas of the South Gobi in Mongolia, among the least densely human populated areas in the world. Besides extensive pasture farming of ruminants, Mongolia features relatively low livestock indices for pigs, cattle and poultry due to the absence of industrial animal breeding. However, as overgrazing by free-ranging livestock, including camels, horses, sheep, goats and cattle, has become a problem in some parts of Mongolia, we were careful not to select these areas. Manure spread on the fields is estimated to be of minor relevance as only a marginal area of the country is arable [5], [6], [12]. By contrast, we selected the sampling area in Saxony-Anhalt, Germany, because it represents typical Central European conditions, e.g. high human densities and industrial animal breeding. High livestock indices for pigs, cattle and poultry presumably lead to increased therapeutic use of antimicrobial substances in livestock farming and intensive liquid manuring [5], [6], [12].
The host-side sampling of birds of prey were considered since (i) there is growing evidence that these may carry ESBL-E. coli at a high frequency and (ii) in both of the sampling areas some of the same species were present. Furthermore, the German and Mongolian raptor populations were not connected by migration; the main avian migration routes for raptors in both locations are north-south [1]–[4], [13]. Although we sampled different species of raptors in the two areas to reach a minimum acceptable sample size, it should be stressed that raptors demonstrate common feeding behaviours, distinct from other groups of birds.
Methods
Ethics Statement
We carried out the sampling of nestlings in Germany and Mongolia during bird ringing and the animals were released afterward in accordance with the Ornithological Council’s guidelines on the use of wild birds in research [14]. We conducted sampling in Mongolia with the approval and in cooperation with the National University of Mongolia in Ulaan-Baatar, Mongolia. Sampling in Germany was performed under the approval of the State Office of Environmental Protection of Saxony-Anhalt, which also granted M. Stubbe the general permission to ring birds.
According to the IUCN Red List of Threatened species, the conservation status of nearly all animals of this study was of “least concern” (LC), with the exceptions of M. milvus, A. monachus (both nearly threatened; NT) and F. cherrug (endangered, EN).
Sampling of Birds
In spring 2010, while ringing the nestlings of sixteen wild avian species, we obtained cloacal swabs in both Germany and Mongolia; most of the avian hosts sampled were birds of prey. We sampled four non-raptor species in Mongolia, but the isolates originating from these birds were excluded from the calculation of the number of ESBL-E. coli (Tab.1).
We carried out sampling in Central Germany, Saxony-Anhalt, in the Northern region of the Harz-mountains, (around the Hakel-Woodland) in an area of 30 km2 around N 51°56′24.5″; E 11°13′13.2″ (human density: 116 n/km2, livestock densities: cattle/swine 50–100 n/km2, small ruminants 10 n/km2, poultry 1.000–2.500 n/km2) [12] and at several sampling spots in the South-Mongolian semi-desert and in West Mongolia during the Mongolian-German Biological Expedition 2010 by M. and A. Stubbe (detailed geographic origin: Tab. 1, human density: 1–2 n/km2, livestock densities: swine <1 n/km2, cattle 1–5 n/km2, small ruminants 5–10 n/km2, poultry <10 n/km2) [12], [13]. We sampled animals once and shipped cloacal swabs (MASTASWAB containing Amies Medium with charcoal, Mast Diagnostics Reinfeld, Germany) to the lab in Berlin.
Table 1. Wild bird species sampled and number of E. coli and ESBL-producing E. coli isolated from respective host species.
| Origin | Total number of birdsand avian species | No. of E. coliisolated(% avian hosts) | No. ESBL-E. coli(% of all E. coli ) | Strain nameESBL-E. coli | Sampling site ofESBL-E. coli |
| Germany | Total 171 | 65 (38.0) | 9 (13.8) | ||
| 13 Black Kites (Milvus migrans) | 9 (69.2) | 2 (22.0) | IMT21743 IMT21823 | all within 30 km2 aroundN51°56′24.5″, E 11°13′13.2″ | |
| 73 Red Kites (Milvus milvus) | 32 (43.8) | 6 (18.8) | IMT21774 IMT21783 IMT21790 IMT21810 IMT21818 IMT21829 | ||
| 68 Buzzards (Buteo buteo) | 15 (22.0) | 1 (6.6) | IMT21813 | ||
| 2 Sea Eagles (Haliaeetus albicilla) | 1 (50.0) | – | – | – | |
| 1 Spotted Eagle (Aquila pomarina) | 1 (100.0) | – | – | – | |
| 14 Goshawks (Accicepter gentilis) | 7 (50.0) | – | – | – | |
| Mongolia | Total 91 | 37 (40.7) | 4 (10.8) | ||
| 19 Black Kites (Milvus migrans) | 13 (68.4) | 1 (7.6) | IMT23464 | N 44°24′03.6″, E 105°21′17.9″ | |
| 9 Buzzards (Buteo hemilasius) | 3 (33.3) | – | |||
| 30 Black Vultures (Aegypius monachus) | 11 (36.6) | 3 (27.3) | IMT21913 | N 47°40′22.4″, E 105°56′51.9″ | |
| IMT23462 | N 45°48′05.4″, E 107°15′07.9″ | ||||
| IMT23463 | N 45°46′53.6″, E 107°15′23.7″ | ||||
| 4 Steppe Eagles (Aquila nipalensis) | 2 (50.0) | – | – | – | |
| 1 Golden Eagle (Aquila chrysaetos) | 0 (0) | – | – | – | |
| 1 Short-toed Eagle (Circaetus gallicus) | 0 (0) | – | – | – | |
| 3 Eurasian Hobbys (Falco subbuteo) | 0 (0) | – | – | – | |
| 14 Kestrels (Falco tinnunculus) | 4 (28.7) | – | – | – | |
| 8 Saker Falcons (Falco cherrug) | 2 (25.0) | – | – | – | |
| 2 Lesser Kestrels (Falco naumanni) | 2 (100.0) | – | – | – | |
| 15 Demoiselle Cranes (Anthropoides virgo)* | 6 (40.0) | 1 (16.6) | IMT23465 | N 46°41′32.6″, E 106°31′02.0″ | |
| 2 Sandpipers (Actitis hypoleucos)* | 0 (0) | – | – | – | |
| 1 Nightjar (Caprimulgus europaeus)* | 0 (0) | – | – | – | |
| 1 Hoepoe (Upupa epops)* | 0 (0) | – | – | – |
Non-birds of prey species, not included in the calculations;
- = no ESBL identified.
Isolation of E. coli
Cloacal swabs were streaked out on CHROMagar orientation (with and without 4 µg/ml cefotaxime; Mast Diagnostica, Reinfeld, Germany) and incubated overnight to isolate E. coli and to preselect for cefotaxime-resistant E. coli. One colony per sample with coliform appearance on CHROMagar was processed further and bacterial species identification was carried out using the automated VITEK®2 system (BioMérieux, Germany).
Phenotypic Characterization of ESBL-E. coli
E. coli isolates showing growth on CHROMagar containing cefotaxime were confirmed as ESBL producers using the phenotypic confirmatory test for ESBL production, performed and interpreted according to CLSI guideline M31-A3 [15].[Using the VITEK®2 system (BioMérieux, Germany) minimal inhibitory concentration testing for antimicrobials was performed according to the guidelines given by the CLSI.
Genotypic Characterization of ESBL-E. coli
The genomic make-up of the confirmed ESBL-E. coli was characterised using established PCR protocols with amplification and subsequent sequencing for the most prevalent beta lactam resistance genes (bla CTX-M, bla SHV, bla TEM and bla OXA) and non-beta-lactam resistance genes (tet(A), tet(B), tet(C), sul1, sul2, sul3, strA, strB, aadA1-like, aacC4, acc(6′)-Ib qnrA, qnrB, and qnrS) [16]–[26] The presence of the intI1 and intI2 genes, encoding class 1 and 2 integrases, was also determined by PCR [23].
MLST and Phylogenetic Grouping by Structure Analysis
Multi-locus sequence type (MLST) determination was carried out as described previously [27], [28]. Gene amplification and sequencing were performed by using primers specified on the E. coli MLST web site (University of Cork website, http://mlst.ucc.ie/mlst/mlst/dbs/Ecoli, accessed 2012 November 29th). Sequences were analysed by the software package Ridom SeqSphere 0.9.39 (Ridom website, http://www3.ridom.de/seqsphere, accessed 2012 November 29th) and sequence types were computed automatically. The phylogenetic group of the E. coli strains was determined using the software Structure 2.3.X based on the concatenated sequences of the seven housekeeping genes used for MLST (University of Chicago website http://pritch.bsd.uchicago.edu/structure.html, accessed 2012 November 29th).
Macrorestriction Analyses by Pulsed Field Gel Electrophoresis (PFGE)
To asses possible clonal relatedness of ESBL-producing E. coli isolates, macrorestriction analysis was performed as previously described using a CHEF DRIII System (BioRad, Munich, Germany) [27]. PFGE profiles generated by restriction of chromosomal DNA with XbaI were compared digitally using BioNumerics software (Version 6.6, Applied Maths, Belgium). Cluster analysis of Dice similarity indices based on the unweighted pair group method with arithmetic mean (UPGMA) was applied to generate dendrograms depicting the relationships among PFGE profiles. Isolates were considered to belong to a group of clonally related strains if the Dice similarity index of the PFGE pattern was ≥85% [29].
Results and Discussion
We isolated comparable rates of E. coli from birds sampled in the two sampling areas; 38% of the German and 41% of the Mongolian birds carried E. coli (Tab. 1). We confirmed ESBL-production in 13.8% (n = 9) of the sixty-five German and in 10.8% (n = 4) of the thirty-seven Mongolian E. coli isolates. Although we detected an ESBL-E. coli, originating from a Demoiselle Crane, the strain was excluded from the calculations as it represented a non-raptor bird, but we have provided the typing results of the strain in the manuscript. All ESBL-E. coli from this study originated from different individual raptors in different nests, thus precluding a possible bias caused by inter-sibling transfer in the nest. By including all birds of the study –even those who did not carry E. coli, 5.2% of the wild birds from Germany and 4.5% of those from Mongolia carried ESBL-E. coli.
The detection rate of ESBL-E. coli observed for the German isolates (13.8%) correlates with data from other studies on raptors from Central Europe where detection rates from 10–20% have been observed [11]. Although based on a limited number of studies available on raptors, high carriage rates seem to be present in Europe independently from the origin of the birds, whether from natural preserves or land used for agricultural production [11]. There is no data on the occurrence of ESBL-E. coli in raptors from remote areas, but the detection rates of ESBL-E. coli for other avian species (Glaucus Winged Gull) were only about 1%. In this regard, the carriage rates of ESBL-E. coli detected in this study among Mongolian raptors (10.8%) is surprisingly high. Furthermore, the ESBL-E. coli detected by Hernandez et al. (2010) [30] was typed as O25b:H4-ST131-CTX-15, presenting an anthropogenic clinically important zoonotic pathogen and no commensal E. coli [31]. The mere detection of antimicrobial resistant commensal E. coli in the Mongolian samples, which has been described for wild birds or rodents in remote areas [32], [33] could have been anticipated, but high rates of clinically important multi-resistant ESBL-E. coli like ST648 and ST167 have, to the best of our knowledge, not been yet detected in remote areas. As described in detail in the following, large proportions of the Mongolian ESBL-E. coli in this study displayed a high similarity to relevant clinical pathogenic strains from Europe, clearly indicating their zoonotic potential. Thus our data underline that the previous finding of O25b:H4-ST131-CTX-15 in a wild bird was not by chance [30], but that besides commensal E. coli displaying antimicrobial resistance, clinical zoonotic pathogens have also reached remote areas in significant numbers.
Genotypic characterisation of the isolates revealed that all ESBL-E. coli harboured bla CTX-M genes, with bla CTX-M-1 predominating among German (100%) and bla CTX-M-9 amongst Mongolian isolates (80%) (Tab. 2). bla CTX-M-1 represents the predominant ESBL type in poultry, pigs and cattle in Europe, but not in human clinical samples where bla CTX-M-1 counts for only about 7%, as revealed by a meta-analysis recently published by our group [26]. The high prevalence of bla CTX-M-1 in the German birds suggests transmission from livestock breeding into the environment, and subsequently, to wild birds, rather than from human sources. To our knowledge, clinical data on ESBL-E. coli from Mongolia are not available, but as for the rest of Asia, the major type found in the Mongolian birds (bla CTX-M-9) only plays a minor role in human clinical samples [34]. bla CTX-M-9 has also been found less frequently in livestock in Asia, underlining the highly complex spread of ESBL-E. coli [34].
Table 2. Molecular characteristics of ESBL-producing E. coli obtained from wild avian hosts according to phylogenetic background and resistance profile.
| Strain | Host | Origin | Ances-tralgroup | ST | STC | ESBL type | Non extended spectrum beta-lactam resistancegenes and integron cassettes |
| IMT21743 | Milvus migrans | Ger | A | 744 | 10 | bla CTX-M-1 | blaTEM-1-like, tet(B), sul2, strA, strB, aac(6′)-IB-cr, integron class I |
| IMT21774 | Milvus milvus | Ger | A | 744 | 10 | bla CTX-M-1 | blaTEM-1-like, tet(B), sul2, strA, strB, aac(6′)-IB-cr, integron class I |
| IMT21783 | Milvus milvus | Ger | A | 744 | 10 | bla CTX-M-1 | blaTEM-1-like, tet(B), sul2, strA, strB, aac(6′)-IB-cr, integron class I |
| IMT21790 | Milvus milvus | Ger | B2 | 12 | 12 | bla CTX-M-1 | bla TEM-1-like, tet(A), integron class I |
| IMT21810 | Milvus milvus | Ger | B1 | 847 | none | bla CTX-M-1 | bla TEM-1-like, bla OXA-1, tet(A), sul2, strA, strB, integron class I |
| IMT21813 | Buteo buteo | Ger | A | 744 | 10 | bla CTX-M-1 | blaTEM-1-like, tet(B), sul2, strA, strB, aac(6′)-IB-cr, integron class I |
| IMT21818 | Milvus milvus | Ger | A | 2199 | 155 | bla CTX-M-1 | tet(B), sul2, strA, strB, integron class I |
| IMT21823 | Milvus migrans | Ger | D | 2198 | none | bla CTX-M-1 | bla TEM-1-like, sul2, strA, strB, integron class I |
| IMT21829 | Milvus milvus | Ger | AxB1 | 1640 | 350 | bla CTX-M-1 | sul2, strA, strB, aac(6′)-IB-cr, integron class I |
| IMT21913 | Aegypius monachus | Mon | ABD | 117 | 117 | bla CTX-M-55 | blaTEM-1-like, sul2, strA, strB, aac(3)-IV |
| IMT23462 | Aegypius monachus | Mon | A | 167 | 10 | bla CTX-M-9 | sul2, strA, strB, integron class I |
| IMT23463 | Aegypius monachus | Mon | ABD | 648 | 648 | bla CTX-M-9 | blaTEM-1-like, blaOXA-1, tet(A), sul2, strA,strB, aac (6′)-IB-cr, integron class I |
| IMT23464 | Milvus migrans | Mon | ABD | 648 | 648 | bla CTX-M-9 | blaTEM-1-like, blaOXA-1, tet(A), sul2,strA, strB, aac (6′)-IB-cr, integron class I |
| IMT23465 | Anthropoides virgo* | Mon | B2 | 2346 | none | bla CTX-M-9 | bla TEM-1-like, bla OXA-1, tet(A), sul1, integron class I |
Abbreviations: ST = sequence type; STC = ST complex; Ger = Germany; Mon = Mongolia,
Non-birds of prey species.
Moreover, we detected several other resistance determinants for both sampling areas along with the beta-lactamase encoding genes (Tab. 2); the phenotypic resistance patterns these strains displayed confirmed these results (Tab. S1). Besides the production of ESBLs we also found concomitant resistance to tetracycline, sulphonamide/trimethoprim, and, to a lesser extent, fluoroquinolones. These results confirm recent data on common resistance in wild birds [35]. Ancestral group B2 strains (Sequence type ST12, ST2346), which are believed to be of high extra-intestinal virulence (ExPEC; extra-intestinal pathogenic E. coli), were present in one bird from Mongolia and one from Germany. Hybrid group ABD ESBL-E. coli strains (ST117, ST648) which are also believed to be of extra-intestinal virulence, predominated in Mongolian birds (60%). Thus several isolates from both sampling areas combined multiresistance with a certain extra-intestinal virulence potential resembling a trend that has been observed in ESBL isolates worldwide and is highlighted by the intercontinental spread of O25b:H4-ST131-CTX-15 which represents an ESBL-producing ExPEC [31].
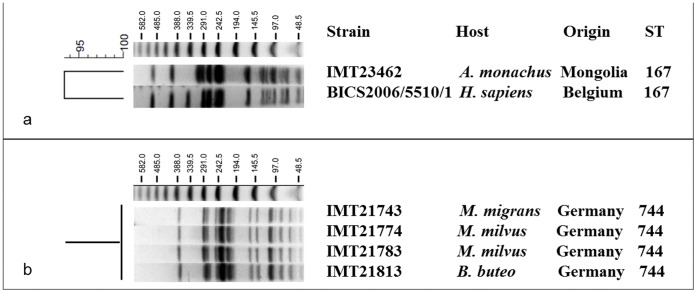
Overall, ten different STs were detected among the avian ESBL-E. coli; several of these, including ST12 (ancestral group B2), ST847 (B1), ST167 (A), and ST117 (ABD) have already been reported from human and veterinary clinical ESBL-producing isolates [34], [36]–[39]. Interestingly, ST167 belongs to STs that have been associated with the global carriage of ESBL-E. coli in humans [34], [38]–[41]. Recently, we found that this phylogenetic lineage was highly prevalent in ESBL-E. coli from companion animals (own unpublished data), while to the best of our knowledge, this is the first report of ST167 ESBL-E. coli in wildlife. In a previously published study on the epidemiology of ESBL-producing Enterobacteriaceae in Belgian hospitals [37] an ESBL-producing ST167 E. coli isolate (BICS2006/5510/1) was detected in a clinical sample from a 67-yr old patient with urinary tract infection. Indeed, a comparative macrorestriction analysis of this strain with the Mongolian ST167 Black vulture ESBL-E. coli isolate IMT23462 confirmed the clonal relatedness of both isolates (dice similarity index >90%) (Fig. 1 A). This finding is in line with recent data indicating that wildlife principally carries strains from the same phylogenetic background as clinical strains or even identical ESBL strains [11].
Figure 1. Dendrogram showing (A) the relationship of one avian ESBL-E. coli isolate and a human clinical isolate [39], both of ST167, and (B) PFGE profiles of four avian ST744 ESBL-E. coli isolates based on XbaI restriction calculated with Bionumerics 6.6 (Applied Maths, Belgium).
ST = sequence type; A. = Aegypius; B. = Buteo; H. = Homo; M. = Milvus, A size marker (Lambda Ladder PFG Marker; New England Biolabs GmbH, Frankfurt a. M., Germany) with respective fragment sizes (kb) is given on top of the agarose gel.
Interestingly, four of the isolates collected from wild birds in Germany were all assigned to ST744 and were subsequently found to belong to a single and identical PFGE clone (Fig. 1 B), although they originated from individuals (belonging to three avian species) sampled at different locations within an area of 30 square kilometres. A possible explanation for this could be either the existence of one single environmental origin or a yet unidentified non-point source, thus implying the general occurrence of this particular clone in that area to that time point.
The low-level prevalence of ESBL-E. coli that has been reported previously in wild birds from remote areas [30] contrasts with the findings in the present study. This is surprising since it has been recently shown that proximity to human-influenced settings was associated with an increase in antimicrobial-resistant bacteria [42]. The occurrence of ESBL-E. coli seems to be species dependent and this may have influenced the high rates obtained in this study [11]. Nevertheless the southward migration of Mongolian Birds to areas with higher human influence and the possible spill-over of multi-resistant bacteria from a spatially segregated, polluted environment might be the reason for the frequent occurrence of ESBL-E. coli in the avian hosts in remote areas. This would also contradict previous assumptions that multi-resistant bacterial contaminations should be low in remote environments that lack constant antibiotic pressure [43].
Conclusions
The possible contribution of avian migration to the transmission of multi-resistant bacteria has been discussed previously [11], [44]. In this study, we found that ESBL-E. coli from wild birds originating from Germany and from remote regions of Mongolia differed in their resistance genes and phylogenetic background. This is not unexpected, since the examined avian species do not migrate between Mongolia and Central Europe. Nonetheless, all Mongolian avian hosts sampled in this study undergo southward migration, namely on the Korean Peninsula (Black vulture), to China (Buzzards), and to India (Demoiselle Crane), connecting remote areas to the globalised world with high frequencies of ESBL-E. coli in human and livestock [13], [45]–[47].
Although we acknowledge that the presented data are based on a limited number of samples, they clearly suggest the equation “no man, no resistance” is an over simplification [48], as clinically relevant ESBL-E. coli are present in remote environments as well. The contribution of international human travel to the spread of multi-resistant E. coli has only recently garnered attentions [49] and avian migration follows essentially the same principles. Its importance is often neglected although the number of migrating birds worldwide has been estimated to be five billion a year [50]. We would encourage additional studies focusing on carriage rates, persistence and duration of shedding/excretion of ESBL-E. coli in migratory birds on a larger scale. Studies on the necessary frequency of antibiotic exposure to generate significant resistance in wildlife are needed. Such data could help to estimate the potential influence of avian hosts on the pandemic spread of ESBL-E. coli into the environment, the community and ultimately, human and veterinary clinical settings.
Supporting Information
Results of minimal inhibitory concentration testing of avian ESBL-producing E. coli (mg/L).
(DOC)
Acknowledgments
We would like to thank Michael F.C. Moreland for careful proofreading of the manuscript.
Funding Statement
This work was supported by the Federal Ministry of Education (BMBF) and Research Network Zoonosis (FBI-Zoo, Grant no. 01KI1012A) and the German Research Foundation (DFG) funded Indo-German Research Training Group (Grant GRK1673). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pinto L, Radhouani H, Coelho C, Martins da Costa P, Simoes R, et al. (2010) Genetic detection of extended-spectrum beta-lactamase-containing Escherichia coli isolates from birds of prey from Serra da Estrela Natural Reserve in Portugal. Appl Environ Microbiol 76: 4118–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Costa D, Poeta P, Saenz Y, Vinue L, Rojo-Bezares B, et al. (2006) Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J Antimicrob Chemother 58: 1311–1312. [DOI] [PubMed] [Google Scholar]
- 3. Radhouani H, Pinto L, Coelho C, Goncalves A, Sargo R, et al. (2010) Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M classes in faecal samples of common buzzards (Buteo buteo). J Antimicrob Chemother 65: 171–173. [DOI] [PubMed] [Google Scholar]
- 4. Literak I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, et al. (2010) Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl Environ Microbiol 76: 8126–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bloom DE (2011) 7 billion and counting. Science 333: 562–569. [DOI] [PubMed] [Google Scholar]
- 6.Eds. Steinfeld H, Mooney HA, Schneider F, Neville LE (2010) Livestock in a Changing Landscape, Drivers, Consequences and Responses. Island Press.
- 7. Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, et al. (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8: 251–259. [DOI] [PubMed] [Google Scholar]
- 8. Nelson M, Jones SH, Edwards C, Ellis JC (2008) Characterization of Escherichia coli populations from gulls, landfill trash, and wastewater using ribotyping. Dis Aquat Organ 81: 53–63. [DOI] [PubMed] [Google Scholar]
- 9. Blanco G, Lemus JA, Grande J, Gangoso L, Grande JM, et al. (2007) Geographical variation in cloacal microflora and bacterial antibiotic resistance in a threatened avian scavenger in relation to diet and livestock farming practices. Environ Microbiol 9: 1738–1749. [DOI] [PubMed] [Google Scholar]
- 10. Wieler LH, Ewers C, Guenther S, Walther B, Lubke-Becker A (2011) Methicillin-resistant staphylococci (MRS) and extended-spectrum beta-lactamases (ESBL)-producing Enterobacteriaceae in companion animals: nosocomial infections as one reason for the rising prevalence of these potential zoonotic pathogens in clinical samples. Int J Med Microbiol 301: 635–641. [DOI] [PubMed] [Google Scholar]
- 11. Guenther S, Ewers C, Wieler LH (2011) Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbio 2: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wint W, Robinson T (2007) Gridded Livestock of the World 2007. FAO, Rome: 131.
- 13. Stubbe M, Stubbe A, Batsajchan N (2010) Grid mapping and breeding ecology of raptors in Mongolia. Erforsch biol Ress Mongolei (Halle/Saale) 11: 23–175. [Google Scholar]
- 14.The Ornithological Counsil website. Available: http://oacu.od.nih.gov/WildBirdGuide.pdf. Accessed 2012 Nov 28.
- 15.CLSI (2008) Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard - Third Edition CLSI document M31-A3. Wayne, PA, U.S.A.: Clinical and laboratory standards institute, 2008.
- 16. Rodriguez I, Barownick W, Helmuth R, Mendoza MC, Rodicio MR, et al. (2009) Extended-spectrum beta-lactamases and AmpC beta-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J Antimicrob Chemother 64: 301–309. [DOI] [PubMed] [Google Scholar]
- 17. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63: 219–228. [DOI] [PubMed] [Google Scholar]
- 18. Bertrand S, Weill FX, Cloeckaert A, Vrints M, Mairiaux E, et al. (2006) Clonal emergence of extended-spectrum beta-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J Clin Microbiol 44: 2897–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, et al. (2006) Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nature medicine 12: 83–88. [DOI] [PubMed] [Google Scholar]
- 20. Park YJ, Yu JK, Lee S, Oh EJ, Woo GJ (2007) Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: a multicentre study from Korea. J Antimicrob Chemother 60: 868–871. [DOI] [PubMed] [Google Scholar]
- 21. Grimm V, Ezaki S, Susa M, Knabbe C, Schmid RD, et al. (2004) Use of DNA microarrays for rapid genotyping of TEM beta-lactamases that confer resistance. J Clin Microbiol 42: 3766–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jouini A, Vinué L, Slama KB, Sáenz Y, Klibi N, et al. (2007) Characterization of CTX-M and SHV extended-spectrum beta-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J Antimicrob Chemother 60: 1137–1141. [DOI] [PubMed] [Google Scholar]
- 23. Skurnik D, Le Menac’h A, Zurakowski D, Mazel D, Courvalin P, et al. (2005) Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrobial agents and chemotherapy 49: 3062–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maguire AJ, Brown DF, Gray JJ, Desselberger U (2001) Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob Agents Chemother 45: 1022–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinez-Martinez L, Pascual A, Jacoby GA (1998) Quinolone resistance from a transferable plasmid. Lancet 351: 797–799. [DOI] [PubMed] [Google Scholar]
- 26. Ewers C, Grobbel M, Stamm I, Kopp PA, Diehl I, et al. (2010) Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum beta-lactamase-producing Escherichia coli among companion animals. J Antimicrob Chemother 65: 651–660. [DOI] [PubMed] [Google Scholar]
- 27. Ewers C, Janssen T, Kiessling S, Philipp HC, Wieler LH (2004) Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet Microbiol 104: 91–101. [DOI] [PubMed] [Google Scholar]
- 28. Wirth T, Falush D, Lan R, Colles F, Mensa P, et al. (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60: 1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carrico JA, Pinto FR, Simas C, Nunes S, Sousa NG, et al. (2005) Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J Clin Microbiol 43: 5483–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hernandez J, Bonnedahl J, Eliasson I, Wallensten A, Comstedt P, et al. (2010) Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring blaCTX-M-15, found in Glaucous-winged gull at remote Commander Islands, Russia. Environ Microbiol Rep 2: 329–332. [DOI] [PubMed] [Google Scholar]
- 31. Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, et al. (2008) Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61: 273–281. [DOI] [PubMed] [Google Scholar]
- 32. Gilliver MA, Bennett M, Begon M, Hazel SM, Hart CA (1999) Antibiotic resistance found in wild rodents. Nature 401: 233–234. [DOI] [PubMed] [Google Scholar]
- 33.Williams NJ, Sherlock C, Jones TR, Clough HE, Telfer SE, et al. (2011) The prevalence of antimicrobial-resistant Escherichia coli in sympatric wild rodents varies by season and host. J Appl Microbiol. [DOI] [PubMed]
- 34. Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH (2012) Extended-spectrum ß-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clinical Microbiology and Infection 18: 646–655. [DOI] [PubMed] [Google Scholar]
- 35. Guenther S, Grobbel M, Lubke-Becker A, Goedecke A, Friedrich ND, et al. (2010) Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Vet Microbiol 144: 219–225. [DOI] [PubMed] [Google Scholar]
- 36. Nemoy LL, Kotetishvili M, Tigno J, Keefer-Norris A, Harris AD, et al. (2005) Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates. J Clin Microbiol 43: 1776–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez-Villalobos H, Bogaerts P, Berhin C, Bauraing C, Deplano A, et al. (2011) Trends in production of extended-spectrum beta-lactamases among Enterobacteriaceae of clinical interest: results of a nationwide survey in Belgian hospitals. J Antimicrob Chemother 66: 37–47. [DOI] [PubMed] [Google Scholar]
- 38. Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, et al. (2011) Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect 17: 873–880. [DOI] [PubMed] [Google Scholar]
- 39. Oteo J, Diestra K, Juan C, Bautista V, Novais A, et al. (2009) Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int J Antimicrob Agents 34: 173–176. [DOI] [PubMed] [Google Scholar]
- 40. Naseer U, Haldorsen B, Tofteland S, Hegstad K, Scheutz F, et al. (2009) Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS 117: 526–536. [DOI] [PubMed] [Google Scholar]
- 41. Valverde A, Canton R, Garcillan-Barcia MP, Novais A, Galan JC, et al. (2009) Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother 53: 5204–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skurnik D, Ruimy R, Andremont A, Amorin C, Rouquet P, et al. (2006) Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. . J Antimicrob Chemother 57: 1215–1219. [DOI] [PubMed] [Google Scholar]
- 43. Sjölund M, Bonnedahl J, Hernandez J, Bengtsson S, Cederbrant G, et al. (2008) Dissemination of multidrug-resistant bacteria into the Arctic. Emerg Infect Dis 14: 70–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonnedahl J (2011) Antibiotic resistance of Enterobacteriaceae from Wild birds. Acta Universitatis Upsaliensis Uppsala.
- 45. Woodford N, Turton JF, Livermore DM (2011) Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS microbiology reviews 35: 736–755. [DOI] [PubMed] [Google Scholar]
- 46. Naseer U, Sundsfjord A (2011) The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist 17: 83–97. [DOI] [PubMed] [Google Scholar]
- 47. Peirano G, Pitout JD (2010) Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents 35: 316–321. [DOI] [PubMed] [Google Scholar]
- 48. Thaller MC, Migliore L, Marquez C, Tapia W, Cedeno V, et al. (2010) Tracking acquired antibiotic resistance in commensal bacteria of Galapagos land iguanas: no man, no resistance. PloS one 5: e8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peirano G, Laupland KB, Gregson DB, Pitout JDD (2011) Colonization of Returning Travelers With CTX-M-Producing Escherichia coli . J Travel Med 18: 299–303. [DOI] [PubMed] [Google Scholar]
- 50.Berthold P (2001) Bird migration. A general survey. Oxford ornithological series 2nd ed. Oxford Univ. Press Oxford, Great Britain.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of minimal inhibitory concentration testing of avian ESBL-producing E. coli (mg/L).
(DOC)