Abstract
Purpose
To identify the mutations in the pre-mRNA processing factor 31 homolog (PRPF31) gene in Chinese families with autosomal dominant retinitis pigmentosa (adRP) and to characterize the clinical features of those patients who were found to have mutations in the PRPF31 gene.
Methods
Detailed ocular examinations were performed, and genomic DNA was isolated by standard methods for genetic diagnosis. Probands from each family were screened for mutations in the PRPF31 gene that was known to cause adRP. Cosegregation analysis was performed in the available family members. Linkage analysis was performed for one missense mutation to calculate the likelihood of its pathogenicity. Two hundred unrelated, healthy Chinese subjects were screened to exclude nonpathogenic polymorphisms.
Results
Forty Chinese families with adRP were selected by an analysis of pedigrees. We identified four mutations (c.196_197delAA, c.544_618del75bp, c.615delC, and c.895T>C) in total, and three deletions were novel. Cosegregation analysis of the available family members (20 patients and 17 unaffected family members) revealed that each index patient and all affected family members showed a heterozygous mutation in the PRPF31 gene. In two families, incomplete penetrance was observed. Linkage analysis achieved the maximum LOD score of c.895T>C is 2.09, achieved at θ=0. The four probands with PRPF31 mutations showed classical signs of RP, with relatively preserved central vision and severe visual field constriction.
Conclusions
Our studies extended the mutation spectrum of PRPF31, and mutations in PRPF31 were found at a relatively high frequency (10%, 4 of 40 adRP families) in our cohort.
Introduction
Retinitis pigmentosa (RP), broadly defined to include nonsyndromic and syndromic forms, is a group of clinically and genetically heterogeneous, inherited retinal diseases characterized by the progressive degeneration of the photoreceptors, which eventually leads to blindness. Nonsyndromic RP is the most common inherited form of severe retinal degeneration, with a prevalence of approximately 1/4,000 RP cases worldwide [1]. Nonsyndromic cases can be inherited as an autosomal-dominant (20%–25%), autosomal-recessive (15%–20%), X-linked recessive (10%–15%), or sporadic/simplex (30%) trait [2]. The typical clinical manifestations of nonsyndromic RP include the presence of night blindness, peripheral visual field defects, lesions in the fundus (bone spicule-like retinal pigmentary deposits and chorioretinal atrophy), and a severely reduced or nondetectable electroretinogram (ERG) [1].
To date, mutations in 23 different genes have been associated with autosomal dominant retinitis pigmentosa (adRP; RetNet) genes. One of these is the gene pre-mRNA processing factor 31 homolog (PRPF31), which is localized on chromosome 19q13.4 (RP11). Interestingly, PRPF31 is one of six pre-mRNA splicing factors identified as causing adRP. The PRPF31 gene comprises 14 exons spanning approximately 18 kb of genomic DNA and encodes a protein of 499 amino acids. The PRPF31 gene is ubiquitously expressed, including in the neural tissues, brain, and retina [3]. This 61-kDa protein is required for the formation of the U4/U6·U5 tri-small nuclear ribonucleoprotein spliceosome complex, which serves to ensure the accurate and efficient splicing of pre-mRNA [4].
According to previous reports, the prevalence of PRPF31 mutations ranges from 2% to 10% in adRP cohorts from various geographical origins [5-10]. To date, more than 50 adRP-associated PRPF31 mutations have been identified, including missense/nonsense mutations, splicing mutations, deletions/insertions, small indels, and complex mutations (HGMD). Most of the previous genetic studies on PRPF31 were performed in the western world, and only limited data from Chinese patients are available [9,11-13]. The purpose of this study is to assess the genotypes and phenotypes of Chinese adRP patients with PRPF31 mutations.
Methods
Recruitment of subjects
All participants were identified at Peking Union Medical College Hospital. The diagnosis of RP was based on the presence of night blindness, typical fundus findings (characteristic retinal pigmentation, vessel attenuation, and various degrees of retinal atrophy), the severe loss of peripheral visual field, and abnormal electroretinogram findings (dramatic diminution in amplitudes or the complete absence of response). To be included in our adRP cohort, the patients had to have at least three generations of affected individuals or two generations with evidence of male-to-male transmission. Written informed consent was obtained from all participating individuals. This study was approved by the Institutional Review Board of Peking Union Medical College Hospital and adhered to the tenets of the Declaration of Helsinki and the Guidance on Sample Collection of Human Genetic Diseases by the Ministry of Public Health of China.
Clinical evaluations
For each patient, a full medical and family history was taken and an ophthalmological examination was performed. Each underwent a standard ophthalmic examination: best correct visual acuity according to Snellen charts, slit-lamp biomicroscopy, dilated indirect ophthalmoscopy, fundus photography if possible, and visual field tests (Octopus; Interzeag, Schlieren, Switzerland). Retinal structure was examined by using optical coherence tomography (OCT; Topcon, Tokyo, Japan). ERGs were performed (RetiPort ERG system; Roland Consult, Wiesbaden, Germany) using corneal “ERGjet” contact lens electrodes. The ERG protocol complied with the standards published by the International Society for Clinical Electrophysiology of Vision.
Genetic studies
Genomic DNA was extracted from the peripheral white blood cells. We used 2ml blood which collected in EDTA vacutainer tubes. Genomic DNA was isolated from the peripheral leukocytes by using a QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. All 14 coding exons, including the intron–exon boundaries of the PRPF31 gene, were amplified by PCR, using primers published previously [14]. After purification, amplicons were sequenced using both forward and reverse primers on an ABI 3730 Genetic Analyzer (ABI, Foster City, CA). Sequences were assembled and analyzed using Lasergene SeqMan software (DNASTAR, Madison, WI). The results were compared with the PRPF31 reference sequence (GenBank accession number: NM_015629.3). The presence of novel variants was also investigated in 200 unrelated healthy Chinese control subjects by using direct sequencing to exclude nonpathogenic polymorphisms. Cosegregation analysis was performed in the available family members.
Mutation identification
The pathogenicity of c.895T>C was measured by calculating the LOD score using the Linkage software package (version 5.2) with parameters during calculation were set as below. Allele frequencies of affection status were set at 0.9997 and 0.0003, according to the prevalence of RP; allele frequencies of the candidate mutation site (c.895T>C) were set at 0.995 and 0.005, according to Sullivan’s counting. The penetrance was set at 95%. MLINK program in Linkage software package was used in calculating the LOD score [15].
Results
Mutation analysis
Upon complete sequence analysis of the coding and adjacent intronic regions of PRPF31, four mutations were detected in four out of 40 families, including three novel deletions and one previously reported missense mutation (Figure 1, Figure 2, Table 1). Two variants (c.895T>C and c.615delC) were not found in the 200 unrelated healthy Chinese controls. The three deletions were predicted to lead to premature stop codons. Cosegregation analysis revealed two families with incomplete penetrance (Table 1).
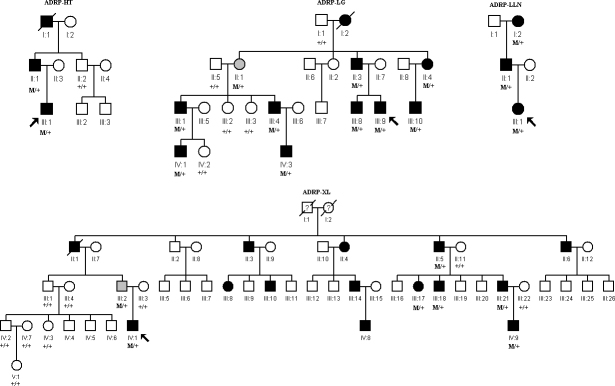
Figure 1.
Pedigrees of four Chinese families with autosomal dominant retinitis pigmentosa associated with mutations in the PRPF31 gene. Genotypes are shown beneath the symbols. Affected individuals are represented by black symbols, unaffected ones by unfilled, asymptomatic ones by gray; squares signify males, circles females. Arrows mark the index patients. M refers to the mutant allele, and + means normal allele.
Figure 2.
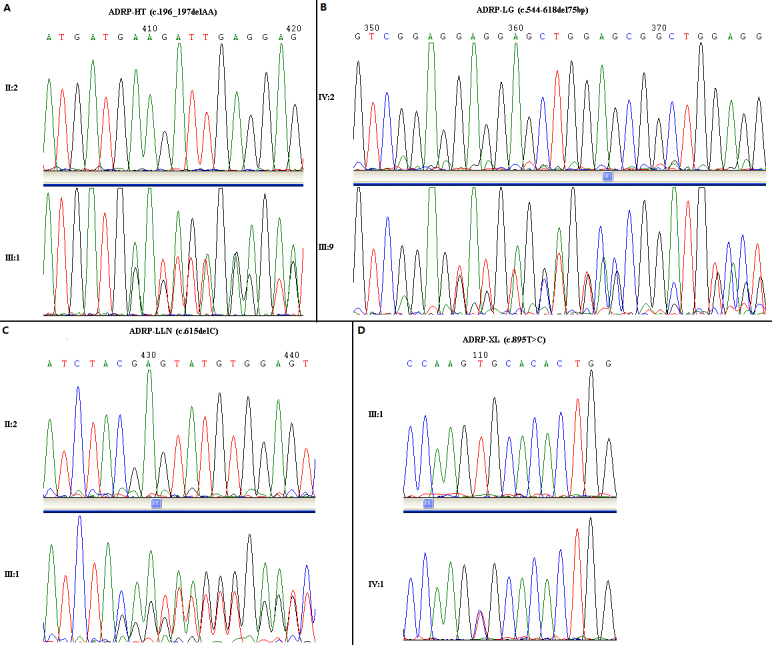
Sequencing results of the PRPF31 mutations in the four families. A: Family ADRP-HT; B: Family ADRP-LG; C: Family ADRP-LLN; D: Family ADRP-XL. The sequences of control subjects are shown in each top panel (A-D). The corresponding sequences of four probands are shown in the lower panel (A-D).
Table 1. Four PRPF31 mutations identified in a Chinese adRP cohort.
| Families | Exon | Nucleotide exchange | Protein effect | Information about penetrance |
|---|---|---|---|---|
| ADRP-HT |
3 |
c.196_197delAA# |
K66DfsX2 |
segregates |
| ADRP-LG |
7 |
c.544_618del75 bp# |
E182_E206del |
incomplete |
| ADRP-LLN |
7 |
c.615delC# |
p.Y205* |
segregates |
| ADRP-XL | 9 | c.895T>C [7] | p.C299R | incomplete |
#: novel mutation.
Mutation identification
Under the given parameters, the maximum LOD score of c.895T>C was 2.09, achieved at θ=0.
Clinical assessment of four probands
The clinical features of the four index patients with PRPF31 mutations are summarized in Table 2 and Table 3. Three of them were males and one was female, with ages ranging from 26 to 29 years. Ages at the time of testing ranged from 24 to 27 years. The symptoms that led to the diagnosis were dominated by night blindness in all patients. Central visual acuity was relatively preserved. Anterior segment examinations showed a remarkable posterior subcapsular cataract in only one patient (IV:1, ADRP-XL). All patients showed severe visual-field constriction. Fundus examination with indirect ophthalmoscopy after full dilatation showed relatively normal disc color, varying amounts of pigment deposits, and different degrees of retinal pigment-layer atrophy (Figure 3). OCT examinations showed relatively preserved foveal lamination and the disappearance of the IS/OS layer, as well as the retinal pigment epithelium layer in the periphery (Figure 3). Full-field ERGs were nonrecordable under scotopic conditions (rods), and the cone responses were markedly hypovolted or nonrecordable in all these patients.
Table 2. Clinical data of probands from families with adRP due to PRPF31 mutations.
| Family/patient | Age at time of testing | Sex | Symptoms at time of diagnosis | BCVA OD/OS | Lens | Fundus examination |
|---|---|---|---|---|---|---|
| ADRP-HT/III:1 |
27 |
M |
Night blindness |
20/20
20/20 |
Clear |
Normal disc color, multiple bone spicules in the mid- periphery retina |
| ADRP-LG/III:9 |
24 |
M |
Night blindness |
20/20
20/20 |
Clear |
RPE atrophy in the temporal area of optic disc |
| ADRP-LLN/III:1 |
27 |
F |
Night blindness |
20/40
20/50 |
Clear |
Multiple bone spicules in the mid- periphery retina |
| ADRP-XL/IV:1 | 27 | M | Night blindness | 20/25 20/25 | Posterior subcapsular cataracts (OU) | Multiple bone spicules in the mid- periphery retina |
The following abbreviation was used: BCVA: best corrected visual acuity.
Table 3. Functional data.
| Family/patient | OCT | Visual field | Full field ERG (photopic and scotopic) |
|---|---|---|---|
| ADRP-HT/III:1 |
Preserved foveal lamination, disappearance of IS/OS layer in periphery |
10° both horizontally and vertically |
Non-recordable |
| ADRP-LG/III:9 |
Preserved foveal lamination, disappearance of IS/OS layer in periphery |
10° both horizontally and vertically |
Only residual cone responses |
| ADRP-LLN/III:1 |
Preserved foveal lamination, disappearance of IS/OS layer in periphery |
Tunnel vision, <5° both horizontally and vertically |
non-recordable in scotopic ERG and the cone responses were markedly reduced |
| ADRP-XL/IV:1 | Preserved foveal lamination, disappearance of IS/OS layer in periphery | Tunnel vision, <5° both horizontally and vertically, more severe in the left eye | Non-recordable |
Figure 3.
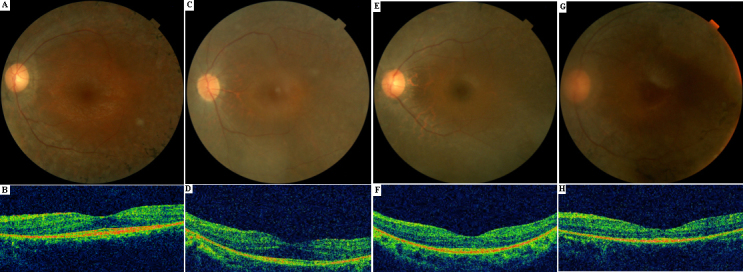
Fundus photographs(top panel) and optical coherence tomography images (lower panel) of four probands with mutations in the PRPF31 gene: A and B: III:1 (ADRP-HT); C and D: III:9 (ADRP-LG); E and F: III:1 (ADRP-LLN); G and H: IV:1 (ADRP-XL). Typical retinitis pigmentosa appearance of the fundus can be seen (A, C, E, and G). Optical coherence tomography images (B, D, F, and H) reveal relatively preserved foveal lamination.
The phenotype of two obligate carriers
In family ADRP-XL, an asymptomatic carrier (III:2) of the missense mutation (c.895T>C) in the PRPF31 gene was identified, suggesting incomplete penetrance in this pedigree. The best corrected visual acuity of this asymptomatic carrier was 20/20 (in both eyes). The fundus and OCT exam did not reveal any changes typical of RP (Figure 4). Another family (ADRP-LG) with a relatively large deletion in the PRPF31 gene (c.544_618del75bp) also exhibited incomplete penetrance. One of the proband’s paternal aunts (II:1, 57 years old, ADRP-LG) did not have any history of night blindness or visual field defects. Her best corrected visual acuity was 20/40 (right eye) and 20/25 (left eye). The refractions were −7.00 DS–2.25 DC × 153° (OD) and −5.00 DS − 1.25 DC × 50° (OS). Dilated fundus examination showed typical bilateral high myopic fundus changes (Figure 5). Detailed ophthalmic examination revealed relatively normal visual field and ERG tests (Figure 6), and therefore, a diagnosis of RP was ruled out for this person.
Figure 4.
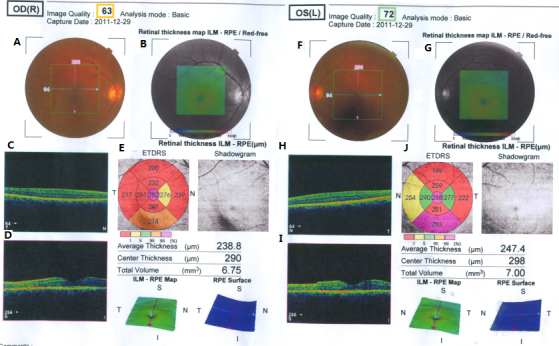
Fundus photographs (A and F) and optical coherence tomography images (C, D, H, and I) of both eyes for III:2 (ADRP-XL). Panel B and G indicated red-free fundus images. Panel E and J indicated the calculating results of macular thickness and total volume in the given areas. Right eye: panel A-E. Left eye: panel F-J. No signs of RP can be seen.
Figure 5.
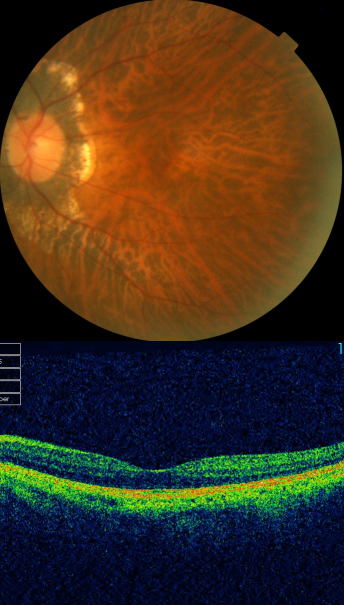
Fundus photograph and optical coherence tomography image of the right eye for II:1 (ADRP-LG). Fundus examination showed typical high myopic fundus changes including tilting of the optic disc, myopic conus, and tessellated fundus. Optical coherence tomography image revealed relatively normal macular lamination.
Figure 6.
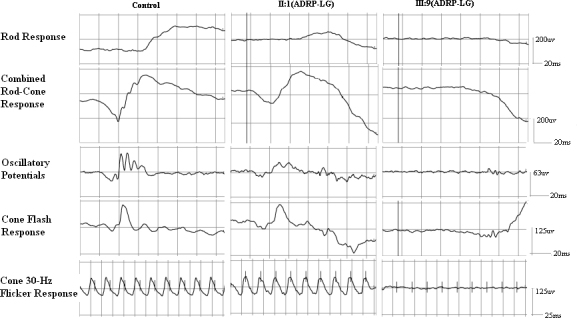
The comparison of full-field electroretinography between the proband (III:9) and the obligate carrier (II:1) in family ADRP-LG. Scotopic electroretinographs are nonrecordable, and other ERGs are severely reduced in III:9. Although the amplitudes of a-waves and b-waves of bright flash ERGs are slightly reduced, the other ERGs are within the normal range in the asymptomatic carrier II:1. The ERG protocol we used in this study complied with the standards published by the International Society for Clinical Electrophysiology of Vision (ISCEV). The light levels used to stimulate the eyes are available on the ISCEV Internet site.
Discussion
To determine the types and frequencies of disease-causing mutations in the PRPF31 gene, we tested 40 Chinese families with adRP for mutations. We identified four different disease-causing mutations in four of 40 unrelated adRP families, including three novel deletions and one known missense mutation. To date, only few PRPF31 variations have been reported to be missense mutations [10]. This also holds true for our study. As far as we know, this study is the largest series to date performed in Chinese adRP patients, and our findings expand the spectrum of PRPF31 mutations. However, the mutation-screening protocols adopted in this study (PCR-based methods) cannot detect gross genomic rearrangements such as the large deletions that were found—using multiplex ligation-dependent probe amplification—to account for 2.5% of adRP families [16]. Thus, the 10% (four in 40 families) frequency of PRPF31 mutations in our cohort is likely to be an underestimate. According to a previous study by Lim et al. [9], the variant frequency for the PRPF31 gene (detected by direct sequencing) in a small cohort of Chinese adRP patients was estimated to be 11.1% (1/9 adRP families), indicating that PRPF31-associated adRP is relatively common in Chinese populations.
It is a common phenomenon that adRP patients with PRPF31 mutations show “all or none” of the forms of incomplete penetrance, making it a high-priority candidate gene in such cases [6,7,10-12]. Consistent with previous reports, incomplete penetrance was observed in two of our four pedigrees, suggesting that most of the PRPF31 mutations are indeed associated with incomplete penetrance. The mechanism presumed to explain this phenomenon is an allelic imbalance with the overexpression of the wild-type allele, compensating for the nonfunctional mutant allele in asymptomatic carriers [17,18]; however, the exact mechanism remains to be determined. Although one of the obligate carriers with a relatively large deletion of the PRPF31 gene exhibited clear evidence of high myopia, given the high prevalence of myopia in East Asia, this finding could simply be a coincidence [19].
The missense mutation c.895T>C (p.C299R) was previously reported by Sullivan et al. in 2006: they predicted that it was probably a pathogenic mutation [7]. We identified this known mutation in a three-generation adRP family with 13 affected patients. Six patients and five unaffected family members took part in the genetic study. The family exhibited incomplete penetrance, as the proband’s father was an asymptomatic mutation carrier. We calculated the LOD score in MLINK, and the result was 2.09. In most cases this would not be sufficient to prove linkage, but combined with prior evidence regarding PRPF31 and this specific mutation, it is highly convincing. What is more, the change was absent in 200 unrelated, healthy Chinese controls, which together suggest that c.895T>C is a true mutation.
Patients carrying mutations in the PRPF31 gene in this study demonstrated classic RP with night blindness as the initial symptom, followed by the gradual constriction of the visual field and a decline in visual acuity later in life. The fundi of the four probands were remarkably similar, with some variations in the amount of bone spicule pigmentation. The three deletions described in this study have a truncating effect on the protein and are likely to result in the degradation of the mRNA. Patients carrying a missense mutation, which would be expected to result in the mildest phenotype, are not necessarily affected more mildly. Our data also show that it is not always true that patients carrying a missense change will have the mildest phenotype. This strongly suggests that disease severity is not determined solely by PRPF31 genotype; other genetic modifiers and/or environmental factors might influence phenotype expression. Therefore, the correlation between the genotype and the severity of the phenotype cannot be determined.
In conclusion, this study described three novel and one known mutation in the PRPF31 gene in Chinese adRP patients. The identification of the genetic cause allows more precise genetic counseling, and in the future, this will be of importance when gene-specific or mutation-specific therapies become available.
Acknowledgments
We thank the patients and their families for taking part in this research. Our work was supported by the Ministry of Science and Technology of the People’s Republic of China (Grant No.: 2010DFB33430), the Ministry of Human Resource and Social Security of the People’s Republic of China and the Foundation Fighting Blindness, USA. (Grant No.: CD-CL-0808–0470-PUMCH).
References
- 1.Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari S, Di Iorio E, Barbaro V, Ponzin D, Sorrentino FS, Parmeggiani F. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics. 2011;12:238–49. doi: 10.2174/138920211795860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer ND, Willis C, Moore AT, Bird AC, Hunt DM, Bhattacharya SS. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell. 2001;8:375–81. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 4.Makarova OV, Makarov EM, Liu S, Vornlocher HP, Luhrmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–57. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-Gimeno M, Gamundi MJ, Hernan I, Maseras M, Millá E, Ayuso C, García-Sandoval B, Beneyto M, Vilela C, Baiget M, Antiñolo G, Carballo M. Mutations in the pre-mRNA splicing-factor genes PRPF3, PRPF8, and PRPF31 in Spanish families with autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2003;44:2171–7. doi: 10.1167/iovs.02-0871. [DOI] [PubMed] [Google Scholar]
- 6.Sato H, Wada Y, Itabashi T, Nakamura M, Kawamura M, Tamai M. Mutations in the pre-mRNA splicing gene, PRPF31, in Japanese families with autosomal dominant retinitis pigmentosa. Am J Ophthalmol. 2005;140:537–40. doi: 10.1016/j.ajo.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, Garcia CA, Ruiz RS, Blanton SH, Northrup H, Gire AI, Seaman R, Duzkale H, Spellicy CJ, Zhu J, Shankar SP, Daiger SP. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47:3052–64. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandra M, Anandula V, Authiappan V, Sundaramurthy S, Raman R, Bhattacharya S, Govindasamy K. Retinitis pigmentosa: mutation analysis of RHO, PRPF31, RP1, and IMPDH1 genes in patients from India. Mol Vis. 2008;14:1105–13. [PMC free article] [PubMed] [Google Scholar]
- 9.Lim KP, Yip SP, Cheung SC, Leung KW, Lam ST, To CH. Novel PRPF31 and PRPH2 mutations and co-occurrence of PRPF31 and RHO mutations in Chinese patients with retinitis pigmentosa. Arch Ophthalmol. 2009;127:784–90. doi: 10.1001/archophthalmol.2009.112. [DOI] [PubMed] [Google Scholar]
- 10.Audo I, Bujakowska K, Mohand-Saïd S, Lancelot ME, Moskova-Doumanova V, Waseem NH, Antonio A, Sahel JA, Bhattacharya SS, Zeitz C. Prevalence and novelty of PRPF31 mutations in French autosomal dominant rod-cone dystrophy patients and a review of published reports. BMC Med Genet. 2010;11:145. doi: 10.1186/1471-2350-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Ribaudo M, Zhao K, Yu N, Chen Q, Sun Q, Wang L, Wang Q. Novel deletion in the pre-mRNA splicing gene PRPF31 causes autosomal dominant retinitis pigmentosa in a large Chinese family. Am J Med Genet A. 2003;121A:235–9. doi: 10.1002/ajmg.a.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia K, Zheng D, Pan Q, Liu Z, Xi X, Hu Z, Deng H, Liu X, Jiang D, Deng H, Xia J. A novel PRPF31 splice-site mutation in a Chinese family with autosomal dominant retinitis pigmentosa. Mol Vis. 2004;10:361–5. [PubMed] [Google Scholar]
- 13.Lu SS, Zhao C, Cui Y, Li ND, Zhang XM, Zhao KX. Novel splice-site mutation in the pre-mRNA splicing gene PRPF31 in a Chinese family with autosomal dominant retinitis pigmentosa. Zhonghua Yan Ke Za Zhi. 2005;41:305–11. [PubMed] [Google Scholar]
- 14.Taira K, Nakazawa M, Sato M. Mutation c. 1142 del G in the PRPF31 gene in a family with autosomal dominant retinitis pigmentosa (RP11) and its implications. Jpn J Ophthalmol. 2007;51:45–8. doi: 10.1007/s10384-006-0394-1. [DOI] [PubMed] [Google Scholar]
- 15.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36:460–5. [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan LS, Bowne SJ, Seaman CR, Blanton SH, Lewis RA, Heckenlively JR, Birch DG, Hughbanks-Wheaton D, Daiger SP. Genomic rearrangements of the PRPF31 gene account for 2.5% of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006;47:4579–88. doi: 10.1167/iovs.06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vithana EN, Abu-Safieh L, Pelosini L, Winchester E, Hornan D, Bird AC, Hunt DM, Bustin SA, Bhattacharya SS. Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: a molecular clue for incomplete penetrance? Invest Ophthalmol Vis Sci. 2003;44:4204–9. doi: 10.1167/iovs.03-0253. [DOI] [PubMed] [Google Scholar]
- 18.Rivolta C, McGee TL, Rio Frio T, Jensen RV, Berson EL, Dryja TP. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat. 2006;27:644–53. doi: 10.1002/humu.20325. [DOI] [PubMed] [Google Scholar]
- 19.Lu B, Jiang D, Wang P, Gao Y, Sun W, Xiao X, Li S, Jia X, Guo X, Zhang Q. Replication study supports CTNND2 as a susceptibility gene for high myopia. Invest Ophthalmol Vis Sci. 2011;52:8258–61. doi: 10.1167/iovs.11-7914. [DOI] [PubMed] [Google Scholar]