Abstract
The hypothesis that germ-line polymorphisms in DNA repair genes influence cancer risk has previously been tested primarily on a cancer site-specific basis. The purpose of this study was to test the hypothesis that DNA repair gene allelic variants contribute to globally elevated cancer risk by measuring associations with risk of all cancers that occurred within a population-based cohort. In the CLUE II cohort study established in 1989 in Washington County, MD, this study was comprised of all 3619 cancer cases ascertained through 2007 compared with a sample of 2296 with no cancer. Associations were measured between 759 DNA repair gene single nucleotide polymorphisms (SNPs) and risk of all cancers. A SNP in O6-methylguanine-DNA methyltransferase, MGMT, (rs2296675) was significantly associated with overall cancer risk [per minor allele odds ratio (OR) 1.30, 95% confidence interval (CI) 1.19–1.43 and P-value: 4.1 × 10−8]. The association between rs2296675 and cancer risk was stronger among those aged ≤54 years old than those who were ≥55 years at baseline (P-for-interaction = 0.021). OR were in the direction of increased risk for all 15 categories of malignancies studied (P < 0.0001), ranging from 1.22 (P = 0.42) for ovarian cancer to 2.01 (P = 0.008) for urinary tract cancers; the smallest P-value was for breast cancer (OR 1.45, P = 0.0002). The results indicate that the minor allele of MGMT SNP rs2296675, a common genetic marker with 37% carriers, was significantly associated with increased risk of cancer across multiple tissues. Replication is needed to more definitively determine the scientific and public health significance of this observed association.
Introduction
Despite the unique etiologic, clinical and prognostic features that distinguish each different type of malignancy, the underlying process of carcinogenesis across human tissues shares common features. Cancer is ultimately a disease of altered DNA structure or function. Hence, most carcinogens cause cancer by damaging DNA. DNA repair mechanisms therefore assume central importance in defending against carcinogenesis. This is demonstrated by the fact that global defects in the DNA repair apparatus can cause inordinately high risks of malignancies in different tissues. Examples are autosomal recessive disorders such as Bloom’s syndrome (1) and xeroderma pigmentosum (2), in which rare but highly penetrant mutations in DNA repair genes lead to early age of onset of malignancies that increase cancer risks in multiple tissues by orders of magnitude greater than seen in the general population. The hypothesis tested in this study was that more common, but less penetrant germ-line allelic variants in DNA repair genes could also introduce inter-individual differences in DNA repair proficiency that could impact on carcinogenesis across multiple tissues, and hence increase overall cancer risk.
DNA repair is accomplished through many interacting biochemical pathways that interact and have overlapping functions. The five principal DNA repair pathways are nucleotide excision repair, base excision repair, nonhomologous end joining, homologous recombinational repair, and mismatch repair (3). Additional DNA repair-related genes include those involved in DNA–DNA crosslink repair, DNA–protein crosslink repair, direct reversal repair, DNA damage signal transduction, poly (ADP ribose) polymerase family genes, hereditary disease genes with deficient DNA repair phenotypes, and genes with putative DNA repair functions (3).
There is a large body of epidemiologic evidence on the associations between DNA repair gene variants and the risk of cancer. For example, a field synopsis of cancer risk in relation to low-penetrant DNA repair gene variants published in 2009 ascertained 361 relevant articles (4). The results identified 31 nominally statistically significant associations; of these, three were rated as providing strong epidemiologic evidence (4). This synopsis was subsequently updated with 25 additional meta-analyses published through 2010 (5). In recent meta-analyses of specific candidate DNA repair gene polymorphisms, studies of different cancer sites were grouped as a way to estimate the associations of genetic variants with overall cancer risk. The results of these meta-analyses have been equivocal, e.g. specific variants in genes such as MLH1 (6), O 6-methylguanine-DNA methyltransferase (MGMT) (7) and ERCC1 (8) have documented evidence supporting associations across cancer types, but in several other instances, the evidence has not supported such cross-cutting associations (9–11).
From this very large body of epidemiologic evidence on DNA repair gene variants in relation to the risk of human cancer, only a few relatively small-scale reports have directly investigated the association of DNA repair gene variants in relation to overall cancer risk. The few previous reports of associations between DNA repair gene variants and multiple malignancies included at most 22 polymorphisms and less than 600 cancer cases (12,13). To more globally test the hypothesis that germ-line DNA repair allelic gene variants contribute to an overall elevated risk of any type of cancer, 759 single nucleotide polymorphisms (SNPs) in 118 DNA repair genes were tested for associations with all pathologically confirmed cancer cases that occurred in a community-based cohort during 18 years of follow-up.
Methods
The methods for this study have been described previously (14) and are thus briefly summarized here. This study uses the same study population and the same genetic markers to address a distinctly different study question, namely the association between DNA repair gene variants and overall cancer risk, whereas the outcome of the previous study was a cancer-prone phenotype defined by the presence of nonmelanoma skin cancer (NMSC) plus another type of cancer.
Study population
This study was embedded within the parent CLUE II (after the campaign slogan ‘Give us a clue to cancer and heart disease’) cohort, established in the Washington County, Maryland, community. In 1989, blood samples were collected from >30 000 adult county residents by drawing 20ml into heparinized Vacutainers. Buffy coats were placed in storage at −70°C. A brief questionnaire was administered that included demographics, cigarette smoking, height and weight. Ascertainment of cancer cases was achieved by linking to the Washington County Cancer Registry, which is also linked with the Maryland Cancer Registry. The study was approved by the Institutional Review Boards of the Johns Hopkins University Bloomberg School of Public Health and the Medical University of South Carolina.
Genotyping was attempted for a total of 6277 cohort members. This included all those with a confirmed cancer diagnosis as of September 2007 and a cancer-free comparison group. The cancer-free comparison group was comprised primarily of a 10% age-stratified random sample of adult CLUE II participants. The exception was that 250 controls were added for a substudy focused on lung cancer; these added controls almost all had a positive history of cigarette smoking. Before beginning the data analyses, all those initially selected to be in the cancer-free comparison group in 2007 but who were subsequently diagnosed with cancer by 31 December 2010 were reclassified as cancer cases for the data analyses.
From this total, excluded were those whose DNA sample had <95% of SNPs successfully genotyped (263, 4.2%) and, to guard against population stratification, those of non-Caucasian ethnic ancestry (98, 1.6%). Ethnic ancestry classification was based on principal components analysis comparisons to three HapMap2 populations. After making these exclusions, the final study population was comprised of 3620 cancer cases and 2296 cancer-free controls.
SNP selection.
The SNP selection algorithm was described previously (15). In brief, 118 human DNA repair genes and DNA repair-related genes (3) were assigned to one of the five central DNA repair pathways (nucleotide excision repair, base excision repair, nonhomologous end joining, homologous recombinational repair and mismatch repair) or to one of seven ancillary DNA repair-related pathways (DNA–DNA crosslink repair, DNA–protein crosslink repair, direct reversal repair, poly (ADP ribose) polymerase family, DNA damage signal transduction, hereditary disease genes with deficient DNA repair phenotypes and genes with putative DNA repair functions) based on putative activities, functions, sequence homology or physical associations with other DNA repair proteins. These pathways were prioritized based on evidence of association with NMSC because the primary study question was to address associations with a cancer-prone phenotype defined by the presence of NMSC. Selected for genotyping were nonsynonymous coding SNPs in DNA repair genes and tagging SNPs were selected using a general minor allele frequency (MAF) cutoff of 0.05 with the MAF cutoff relaxed to 0.01 for top priority pathway genes. Despite the inclusion of rare variants in the SNP selection, for the data analyses, all SNPs with MAF < 0.05 were excluded, due to the high prevalence of monomorphic and quasi-monomorphic SNPs in our study population and the lack of statistical precision associated with the low MAF SNPs that were not monomorphic. SNP frequencies and linkage disequilibrium (LD) were estimated using the CEPH population (European ancestry) data from the HapMap (CEU), as the study population for this study was limited to those of European ancestry. R 2 was set to >0.80. Potentially eligible SNPs were excluded if they had a genotyping platform design score <0.60. Tagger™ software was used to access the HapMap data (16). A total of 828 DNA repair gene SNPs with MAF ≥ 0.05 were identified that met these criteria.
Genomic DNA extraction and genotyping.
Buffy coats with no previous DNA extraction were stored at −70°C from collection until DNA extraction. Genomic DNA was extracted from buffy coats using standard phenol/chloroform extraction procedures, followed by ethanol precipitation and re-suspension to concentrations of 150 µg/ml (17). The precipitated DNA was re-suspended in low salt buffer to uniform DNA concentrations (50 µg/ml) for genotyping. DNA was quantified using a NanoDrop spectrophotometer per manufacturer instructions.
When previously extracted DNA was available, that DNA was re-suspended in low salt buffer to the same concentration and utilized for genotyping. Illumina GoldenGate® arrays customized by the manufacturer were used to attempt genotyping on 828 selected DNA repair gene SNPs with MAF ≥ 0.05. After excluding 52 SNPs (6%) with failed genotyping in ≥ 5% of the samples and 17 SNPs (2%) that deviated from Hardy–Weinberg equilibrium at P < 0.001, 759 SNPs were included in the final analyses. A summary of the pathways studied, genes per pathway and number of SNPs per gene was previously published (14); the complete SNP list is provided in Supplementary Table 1, available at Carcinogenesis Online. For quality control purposes, the genotyping results were compared for 24 SNPs that had been previously genotyped in earlier studies. Among CLUE II cohort members who overlapped with this study population (per SNP number of participants ranged from 3943 to 4757), the overall concordance was 98%.
Statistical analyses.
Potential type I error inflation due to genotyping errors and batch effects was assessed using a quantile–quantile (Q–Q) plot of the observed versus expected −log10 P-values. As a SNP screening step, to test the hypothesis that DNA repair gene SNPs contribute to globally elevated cancer risk, the association between each SNP and all cancers combined was tested using the additive genetic model with SNPs coded as having 0, 1 or 2 copies of the minor allele, using logistic regression to estimate odds ratios (OR), confidence intervals (CI) and P-values. Using Bonferroni correction for multiple comparisons, the threshold for statistical significance in this SNP screening step was a two-sided test P-value of 0.05/759 = 6.6 × 10−5.
The lone SNP that exceed this threshold was then assessed in greater detail by using logistic regression to characterize the associations by genotypic, dominant and recessive model and in subgroup analyses based on age and smoking. An interaction with age was tested because true genetically driven risk was hypothesized to be stronger in a younger than older age group. The potential interaction between cigarette smoking and the significant SNP was assessed because it was from a gene MGMT that repairs DNA damage caused by alkylating agents, and cigarette smoke is a source of exposure to alkylating agents. To account for the possibility that any potential effect modification by cigarette smoking could be specific to malignancies established to be caused by cigarette smoking, the smoking-stratified analyses were also performed stratified according to smoking-caused cancer and cancers not currently established to be caused by cigarette smoking. The definition of smoking-caused cancer was taken to be on the basis of judgment of causation in US Surgeon General’s reports (18), which includes the following nine malignancies: lung, oral cavity, esophagus, bladder, pancreas, kidney, uterine cervix, stomach and acute myelogenous leukemia.
Associations between the significant SNP and 14 site-specific cancers with ≥60 cases were assessed; cancer sites with <60 cases were grouped into an ‘other cancer’ category. The nonparametric sign test was used to test the probability of observing by chance alone the number of OR that were in the direction of increased risk out of the total of 15 cancer site-specific OR that were calculated in this step.
Due to the method of selection of the comparison group, it was on average substantially younger than the cancer cases. In addition to adjusting for age, to further assess for the potential impact of this age difference on the findings, ancillary analyses were restricted to those aged 35 years and older at study baseline, which made the two age distributions much more comparable and resulted in the loss of only a small number of cancer cases from the analyses. Further, there were 250 controls included from a separate study of lung cancer. Of these, 232 had >95% successful genotyping and were included in this study. To address the potential concern that inclusion of a subset of controls selected in a different way may have altered the observed association, a sensitivity analysis with these 232 individuals excluded was performed.
The first three principal components were adjusted for in all analyses to control for potential residual confounding by ethnic ancestry. Variables additionally adjusted for were age, sex, education, smoking status and body mass index because of their associations with cancer risk and because factors such as these have been observed to be associated with DNA repair capacity (19,20). All analyses were carried out in the statistical environment R (http://cran.r- project.org/) and SAS 9.1 (SAS Institute, Cary, NC, USA).
Results
Baseline descriptive characteristics of the cancer cases and controls are summarized in Table I. The cancer-free group was significantly younger than the cancer cases (average age 43.1 versus 58.2 years) and had a significantly high proportion of females (58.1% versus 53.0%). The prevalence of ever-smokers was only slightly higher in the cases than the comparison group (54% versus 49%) but when stratified by smoking-caused malignancies, the expected higher prevalence of former (38% higher) and current (37% higher) smokers was observed. NMSC, breast, prostate, lung and colorectal cancers were the most common cancer diagnoses. The age distribution was much more comparable between the study groups when age was restricted to those aged 35 years and older (Supplementary Table 2, available at Carcinogenesis Online).
Table I.
Baseline characteristics of those with and without cancer, CLUE II Cohort, Washington County, MD (1989–2010)a
| Characteristics | No cancer | Cancer | ||
|---|---|---|---|---|
| Total (row %) | 2296 | 100% | 3620 | 100% |
| Mean age (years), (Q1,Q3) | 43.1 | (30,57) | 58.2 | (49,68) |
| Sex | ||||
| Male | 963 | 41.9% | 1703 | 47.0% |
| Female | 1333 | 58.1% | 1917 | 53.0% |
| Mean body mass index (Q1,Q3) | 25.7 | (22.0,28.4) | 26.6 | (23.4,28.9) |
| Education | ||||
| <12 years | 597 | 26.0% | 1035 | 28.6% |
| 12 years | 998 | 43.5% | 1518 | 42.0% |
| >12 yearss | 701 | 30.5% | 1064 | 29.4% |
| Cigarette smoking | ||||
| Never | 1161 | 50.6% | 1671 | 46.2% |
| Former | 561 | 24.4% | 1298 | 35.9% |
| Current | 574 | 25.0% | 650 | 18.0% |
| Type of cancer | Not applicable | Not applicable | ||
| Non-melanoma skin only | 694 | 19.2% | ||
| Breast | 527 | 14.6% | ||
| Prostate | 483 | 13.3% | ||
| Lung | 332 | 9.2% | ||
| Colorectal | 292 | 8.1% | ||
| Melanoma | 197 | 5.4% | ||
| Bladder | 130 | 3.6% | ||
| Uterus | 113 | 3.1% | ||
| Lymphoma | 89 | 2.5% | ||
| Cervix | 83 | 2.3% | ||
| Ovary | 74 | 2.0% | ||
| Pancreas | 72 | 2.0% | ||
| Leukemias | 69 | 1.9% | ||
| Kidney/urinary tract | 62 | 1.7% | ||
| All other malignancies | 403 | 11.1% | ||
aColumn totals vary due to missing data.
The results of the allelic trend test OR and P-values for each SNP are given in Supplementary Table 1, available at Carcinogenesis Online. A total of 57 SNPs (7.5%) were nominally significant at P ≤ 0.05. These included the following percentage (and number) of SNPs per pathway: 17% nonhomologous end joining (13), 16% base excision repair (9), 11% mismatch repair (11), 10% transduction (6), 7% poly (ADP-ribose) polymerase (1), 5% nucleotide excision repair (10), 4% DNA cross-link repair (2), 4% from CHAF1A, a chromatin gene with putative DNA repair function (1) and 2% homologous recombination (3).
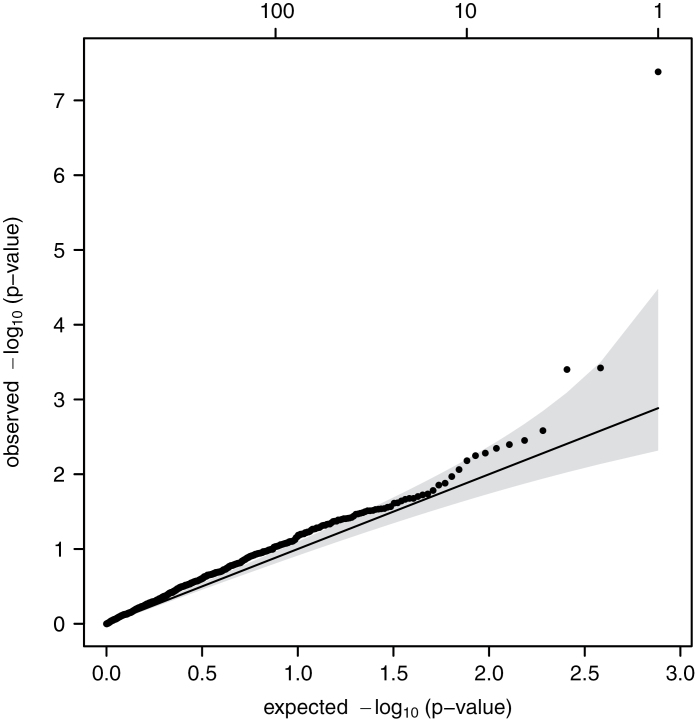
The Q–Q plot of the observed versus expected −log P-values (Figure 1) revealed two key points. First, the slight departure of P-values from the expected line indicated enrichment for high P-values. This pattern could be attributable to (i) the candidate gene association study approach enriching signals for associations between DNA repair SNPs and overall cancer risk and/or (ii) genotyping error rates causing type I error inflation, more commonly seen in studies using custom arrays than those using off-the-shelf platforms. The second key point is that the datum in the top right corner of Figure 1 is for a SNP that was highly statistically significantly associated with overall cancer risk even after correction for multiple comparisons. This was a SNP in the MGMT gene, specifically, MGMT SNP rs2296675, which had a MAF of 0.21 and a P-value from the exact test of Hardy–Weinberg equilibrium of 0.42. It is an intronic SNP located on chromosome 10 at chromosome position 131,454,987. The per minor allele OR was 1.30 (95% CI 1.19–1.43, P = 4.1 × 10−8; Table II). Over one-third (37.2%) of the population were carriers of at least one minor allele, with a genotype distribution of 62.8%, 33.1% and 4.1% homozygous common, heterozygous and homozygous minor, respectively. The ORs were 1.38 (95% CI 1.23–1.55) and 1.43 (95% CI 1.08–1.88) for heterozygous and homozygous minor genotypes, respectively, with an OR of 1.38 (95% CI 1.24–1.55) comparing those with one or two minor alleles to those with two common alleles (dominant model; Table II). The data were much more compatible with a dominant than recessive mode of inheritance for rs2296675 (recessive model: OR 1.28, 95% CI 0.97–1.69 and P = 0.08).
Fig. 1.
Quantile–quantile plot of observed versus expected −log(P values) for association between 759 DNA repair SNPs and risk of any cancer, CLUE II Cohort, Washington County, MD 1989–2007.
Table II.
Association between rs2296675 and risk of all cancers, overall and stratified by age at baseline
| Additive model | Genotype (n)c | Genotypic and dominant models | |||
|---|---|---|---|---|---|
| OR (95% CI)a | OR (95% CI)b | OR (95% CI)a | OR (95% CI)b | ||
| Overall | 1.30 (1.19–1.43) | 1.25 (1.13–1.39) | AA (3633) | 1.0 (referent) | 1.0 (referent) |
| AG (1912) | 1.38 (1.23–1.55) | 1.33 (1.17–1.51) | |||
| GG (235) | 1.43 (1.08–1.88) | 1.28 (0.94–1.74) | |||
| AG + GG | 1.38 (1.24–1.55) | 1.33 (1.17–1.50) | |||
| Stratified by age at baseline; P-for-interaction = 0.021 (dominant model) | |||||
| Age ≤54 years | 1.37 (1.20–1.57) | 1.34 (1.16–1.55) | AA (1889) | 1.0 (referent) | 1.0 (referent) |
| AG (917) | 1.53 (1.31–1.80) | 1.50 (1.26–1.79) | |||
| GG (101) | 1.28 (0.86–1.92) | 1.24 (0.81–1.92) | |||
| AG + GG | 1.51 (1.29–1.76) | 1.47 (1.24–1.74) | |||
| Age ≥55 years | 1.13 (0.97–1.32) | 1.14 (0.98–1.33) | AA (1745) | 1.0 (referent) | 1.0 (referent) |
| AG (995) | 1.13 (0.93–1.36) | 1.13 (0.94–1.37) | |||
| GG (134) | 1.31 (0.84–2.03) | 1.32 (0.84–2.06) | |||
| AG + GG | 1.14 (0.96–1.37) | 1.15 (0.96–1.38) | |||
aAssociations adjusted for first three principal components to control for potential residual confounding by ancestry.
bAssociations adjusted for first three principal components plus age, sex, education, cigarette smoking, and body mass index.
cNumbers may not sum to 5916 due to genotyping failures.
Age was assessed as a potential effect modifier by stratifying by age at study baseline in 1989 (≤54 years versus ≥55 years; median 55 years). The association between rs2296675 and all cancers was stronger in the younger age group (OR 1.51, 95% CI 1.29–1.76) than the older age group (OR 1.14, 95% CI 0.96–1.37), a statistically significant difference (P = 0.021; Table II). Overall, the adjustments for age, sex, education, smoking and body mass index did not appreciably alter the inferences for the results presented in Table III, except for differences in the association for the homozygous minor genotype, the group with the smallest stratum sizes. The associations observed in the overall study population were upheld in the age-restricted study population for the association between rs2296675 and overall cancer risk (Supplementary Table 3, available at Carcinogenesis Online).
Table III.
Association between being a carrier of a minor allele in rs2296675 (dominant model: AG + GG versus AA) and risk of (a) all cancers, (b) smoking-caused cancers, and (c) cancers not currently established to be caused by cigarette smoking, stratified by cigarette smoking status at baseline
| Smoking status | Minimally adjusted | Fully adjusted |
|---|---|---|
| OR (95% CI)a | OR (95% CI)b | |
| Total (P-for-interaction = 0.22) | ||
| Never smokers | 1.53 (1.30–1.79) | 1.41 (1.18–1.70) |
| Former smokers | 1.20 (0.98–1.48) | 1.14 (0.91–1.42) |
| Current smokers | 1.43 (1.13–1.80) | 1.45 (1.13–1.86) |
| Smoking-caused cancers (P-for-interaction = 0.43) | ||
| Never smokers | 1.50 (1.09–2.07) | 1.37 (0.98–1.92) |
| Former smokers | 1.16 (0.86–1.55) | 1.10 (0.81–1.49) |
| Current smokers | 1.47 (1.10–1.82) | 1.49 (1.09–2.03) |
| Cancers not established to be caused by smoking (P-for-interaction = 0.27) | ||
| Never smokers | 1.53 (1.30–1.80) | 1.42 (1.18–1.71) |
| Former smokers | 1.22 (0.98–1.51) | 1.15 (0.91–1.45) |
| Current smokers | 1.39 (1.06–1.82) | 1.41 (1.07–1.88) |
CI, confidence interval; OR, odds ratio.
aAssociations adjusted for first three principal components to control for potential residual confounding by ancestry.
bAssociations adjusted for first three principal components plus age, sex, education, and body mass index.
The potential interaction between cigarette smoking and rs2296675 was assessed because MGMT repairs DNA damage caused by alkylating agents and cigarette smoke is a source of exposure to alkylating agents. There was no interaction between cigarette smoking and rs2296675 on the risk of overall cancer (likelihood ratio test P-value = 0.22; Table III). To account for the possibility that cigarette smoking may act as an effect modifier specifically for smoking-caused cancers but not for cancers not caused by smoking, these analyses were further stratified by the outcome categories of smoking-caused cancers and cancers not currently established as caused by smoking. The associations between rs2296675 and these cancer endpoints was consistent within the different categories of smoking status, reinforcing the absence of evidence that cigarette smoking was an effect modifier for the association between rs2296675 and cancer risk (Table III). The smoking-stratified analyses yielded very similar results to those described above when analyses were restricted to those aged 35 years and older at study baseline (Supplementary Table 3, available at Carcinogenesis Online).
A total of 21 MGMT SNPs were genotyped. Other than rs2296675, none was significantly associated with risk of all cancers (allelic trend test P > 0.05 for all) (Supplementary Table 4, available at Carcinogenesis Online). None of these 21 MGMT SNPs were in LD with rs2296675 (all R 2 < 0.04).
OR for rs2296675 in relation to site-specific cancers were in the direction of increased risk for all 15 groupings of malignancies (nonparametric sign test P-value <0.0001; Table IV). In minimally adjusted analyses, the OR ranged from 1.22 (P = 0.42) for ovarian cancer to 2.01 (P = 0.008) for kidney/urinary tract cancers; the smallest P-value was for female breast cancer (OR 1.45, P = 0.0002). P-values <0.05 were observed for cancers of the skin, breast, prostate, lung, colorectum, uterus, bladder, kidney, as well as leukemias, lymphomas and the ‘other cancers’ group. After additionally adjusting for age, sex, education, cigarette smoking and body mass index, the OR ranged from 1.04 (P = 0.80) for prostate cancer to 1.74 (P = 0.04) for kidney/urinary tract cancer; the smallest P-value was for breast cancer (OR 1.58, P = 6.6 ×10−5). After these additional adjustments, the OR for all but uterine and ovarian cancer were closer to the null, and for some of these, such as prostate (1.28 to 1.04) and bladder (1.47 to 1.08), the attenuation was substantial. Only the associations for breast and kidney cancer remained statistically significant. Although the more fully adjusted associations tended to not be statistically significant and to be closer to the null, all 15 OR were still greater than the null value of 1.0 (sign test P-value <0.0001). Similar results were observed in the age-restricted analyses (Supplementary Table 5, available at Carcinogenesis Online).
Table IV.
Associations between rs2296675 and risk of specific malignancies
| Types of cancer | # cases | Minimally adjusted | Fully adjusted | ||
|---|---|---|---|---|---|
| OR (95% CI)a | P valuea | OR (95% CI)b | P valueb | ||
| Nonmelanoma skinc | 694 | 1.23 (1.03–1.48) | 0.02 | 1.15 (0.94–1.40) | 0.17 |
| Breast | 527 | 1.45 (1.19–1.76) | 0.0002 | 1.58 (1.27–1.97) | 6.6×10−5 |
| Prostate | 483 | 1.28 (1.01–1.61) | 0.04 | 1.04 (0.79–1.37) | 0.80 |
| Lung | 332 | 1.48 (1.16–1.86) | 0.002 | 1.16 (0.88–1.51) | 0.29 |
| Colorectal | 292 | 1.54 (1.18–2.00) | 0.002 | 1.33 (1.00–1.78) | 0.054 |
| Melanoma | 197 | 1.36 (1.00–1.85) | 0.048 | 1.31 (0.96–1.80) | 0.09 |
| Bladder | 130 | 1.47 (1.03–2.11) | 0.04 | 1.08 (0.73–1.59) | 0.72 |
| Uterus | 113 | 1.47 (0.99–2.18) | 0.057 | 1.51 (0.99–2.30) | 0.055 |
| Lymphoma | 89 | 1.41 (0.92–2.18) | 0.12 | 1.31 (0.84–2.05) | 0.23 |
| Cervix | 83 | 1.32 (0.84–2.08) | 0.23 | 1.33 (0.83–2.13) | 0.23 |
| Ovary | 74 | 1.22 (0.75–2.00) | 0.42 | 1.28 (0.76–2.14) | 0.35 |
| Pancreas | 72 | 1.57 (0.98–2.53) | 0.06 | 1.39 (0.85–2.27) | 0.19 |
| Leukemias | 69 | 1.80 (1.11–2.91) | 0.02 | 1.59 (0.97–2.61) | 0.06 |
| Kidney/urinary tract | 62 | 2.01 (1.20–3.34) | 0.008 | 1.74 (1.03–2.94) | 0.04 |
| All other malignanciesd | 403 | 1.28 (1.03–1.59) | 0.03 | 1.13 (0.90–1.43) | 0.30 |
CI, confidence interval; OD, odds ratio.
aResults for dominant model (AG + GG versus AA genotypes). All associations adjusted for first three principal components to control for potential residual confounding by ancestry.
bAssociations adjusted for first three principal components plus age, sex, education, cigarette smoking, and body mass index.
cThis group is comprised of those whose only cancer diagnosis was non-melanoma skin cancer.
dThis group includes all remaining cancer cases not included in the above categories.
Further, a comparison of the results with the 232 controls selected for a lung cancer case–control study excluded showed that the overall inferences were unchanged (data not shown), and in fact, the association between MGMT rs2296675 and all cancer actually increased, from an OR of 1.30 (95% CI 1.19–1.43, P = 4.1 × 10−8) to 1.38 (95% CI 1.25–1.52, P = 1.25 × 10−10). For most individual cancer sites, the associations were not materially changed, but there was some fluctuation in the associations, e.g. the association for melanoma went from being nominally statistically significant (OR 1.36, 95% CI 1.00–1.85 and P-value = 0.048) to nonsignificant (OR 1.26, 95% CI 0.97–1.65 and P-value = 0.08).
The second and third ranked SNPs, with additive model P-values of 0.004, were two base excision repair SNPs in XRCC1, rs1799782 and rs3213344 that were in high LD (R 2 = 0.99). Although not exceeding the Bonferroni-defined level of significance, these associations can be considered significant in the context of the overall P-value distribution. The MAF for both of these SNPs was 6%, and carriers of a minor allele had >25% lower risk of cancer (rs1799782: OR 0.74, 95% CI 0.63–0.87; rs3213344: OR 0.73, 95% CI 0.62–0.86). For both SNPs, associations in the direction of decreased risk were observed for carriers of the minor allele for 12 of the 15 cancer groupings (nonparametric sign test P-value = 0.04).
Discussion
A large-scale, population-based study with extensive coverage of common allelic DNA repair gene variants revealed a robust association between the minor allele of MGMT SNP rs2296675 and increased overall cancer risk. Carriers of the rs2296675 minor allele had 38% increased risk of developing any type of cancer and comprised 37% of the study population (33% heterozygotes and 4% homozygous rare genotype). Consistent with a genetically driven cancer risk, the association of rs2296675 with cancer risk was significantly stronger in younger individuals. The strength of the associations varied by cancer site, but all of the cancer site-specific associations were in the direction of increased risk.
MGMT directly repairs alkyl adducts that arise in the O 6 position of guanine. The O 6 position is important because O 6-methylguanine tends to be read as adenine during replication, resulting in G:C to A:T transition mutations (21). Germ-line MGMT polymorphisms could potentially affect carcinogenesis via at least three mechanisms: (i) directly diminishing MGMT-mediated DNA repair via a functional coding change; (ii) the sequence variation could increase MGMT’s susceptibility to epigenetic silencing of its transcription and (iii) other potential biologic pathways, such as, if this region of MGMT was a target for RNA interference (RNAi), which can silence gene expression by degrading mRNA (22,23).
Concerning the first, there is some evidence that at least one functional MGMT SNP may be associated with overall cancer risk. For example, several functional MGMT polymorphisms (not including rs2296675) were assessed for associations with overall cancer risk by using meta-analytic techniques that combined the results of site-specific cancer studies. The results indicated Leu84Phe (rs12917; but not Ile143Val, rs2308321) was associated with overall cancer risk, particularly among populations of European ancestry (7). Although not directly relevant to rs2296675, the results of the meta-analysis lend credence to the notion that MGMT functional polymorphisms could potentially contribute to human carcinogenesis in different tissues via putative effects on protein activity. On a cautionary note, however, the results of this study were null for rs12917, emphasizing the need for replication of our study findings with respect to rs2296675. The association observed for rs2296675 could be a consequence of being in high LD with a functional SNP. MGMT SNP rs2296675 is an intronic SNP located on the 5ʹ side of exon 5, within 66 base pairs of rs2308321, a functional SNP in exon 5.
Hypermethylation of CpG islands in the MGMT promoter blocks gene transcription (24). MGMT promoter hypermethylation is found in >20% of colon, lung, testicular, head-neck, retinoblastoma, cervical, lymphoma and brain tumors and 10–20% of esophageal, stomach, pancreatic and melanoma tumors (25–27). The epigenetic silencing of MGMT transcription renders cells unable to remove O 6 alkylguanine adducts, a defect that leads to increased mutations in key tumor suppressor genes and oncogenes, such p53 and K-ras (27).
Thus, MGMT variants could plausibly impact susceptibility to a broad spectrum of cancers by enhancing MGMT promoter hypermethylation, thereby inducing gene silencing, and creating a more permissive, procarcinogenic environment across multiple tissues. In support of this line of reasoning, germ-line MGMT polymorphisms have been linked to both MGMT promoter hypermethylation and gene silencing in colorectal cancer (28). In a study of 182 colorectal tumors, the results for rs2296675 were not statistically significant but compared with those with two common alleles, MGMT promoter hypermethylation in colorectal tumor tissue was 1.7 and 4.0 times more prevalent in those with one and two minor alleles, respectively (28). Further, the rs2296675 minor allele was associated with loss of MGMT expression, which increased from 28% to 38% to 50% in those with zero, one, and two minor alleles, respectively (28). These are the only previously published data we know of for rs2296675; they raise the possibility that the rs2296675 minor allele could be a marker of susceptibility to MGMT promoter hypermethylation and loss of gene expression, at least in the colorectum.
The levels of methylation in tumor compared with normal tissue may provide evidence to assess the likelihood that rs2296675 variants may be associated with cancer risk via a pathway that includes MGMT promoter methylation. In healthy individuals without colorectal neoplasia, similar patterns of MGMT promoter methylation in normal mucosa were seen as in tumor tissue from patients with colorectal cancer (29), suggesting that promoter methylation may be involved early in the carcinogenic pathway in the colon and rectum. MGMT activity is inversely correlated with promoter methylation (30), and there is more evidence available for MGMT activity in tumor compared with normal tissue (31). Comparisons of MGMT activity in malignant tissue to normal tissue showed that for cancers relevant to this study’s findings (breast, colon and rectum and lung), MGMT activity was uniformly greater in the tumor tissue than the normal tissue. To the extent that MGMT activity serves as a useful proxy for promoter methylation, this differential between tumor and normal tissue does not support the hypothesis that germ-line variants alone determine enhanced susceptibility to promoter methylation.
Alkylating agents are potent carcinogens that originate from a variety of endogenous and exogenous sources (32,33). Many endogenous alkylating agents (or precursors) have been identified, including S-adenosyl-methionine, a common methyl donor in biochemical reactions, nitrosated amines or bile acids produced enzymatically by Escherichia coli, and nitrosated alkaloids possibly produced by endogenous nitrosating agents such as Nox (33). With respect to exogenous sources of exposure to alkylating agents, N-nitroso compounds are common environmental alkylating agents found in tobacco products and prepared foods, or formed during natural and industrial processes (33). Active cigarette smoking is a major environmental source of exposure to alkylating agents (33). Cigarette smoke contains many N-nitroso compounds that have been shown to produce O 6-meG or other methylated bases after metabolic activation (34). Other than smoking status, a limitation of this study was the lack of information on potential sources of alkylating agent exposure.
The results observed in this study, indicating that carrying the minor rs2296675 allele was robustly associated with increased risk of all cancers, as well as several specific cancer sites, raises the question as to why this SNP, or others in LD with it, have not previously been reported to be associated with cancer in prior genome-wide association studies (GWAS) of specific cancer sites. One factor contributing to this apparent discrepancy may be a loss of power in GWAS due to corrections for multiple comparisons. That is, SNPs in high LD with rs2296675 may have been associated with risk of specific cancer sites, but these associations were not statistically significant after correcting for multiple comparisons. For some cancer sites, the associations in this study were large enough to be detected in GWAS. Differences in defining the cancer phenotype may have contributed to important differences in the magnitude of the observed associations between this study and GWAS. Typically, individuals with a history of NMSC are not excluded from the control group. However, it is now becoming well established that NMSC is associated with increased risk of malignancy in virtually all other tissues (35). In this study, rs2296675 was shown to be weakly associated with NMSC risk; this association would have been masked had NMSC patients been classified as controls. Including NMSC cases in the control group could therefore have introduced misclassification that biased associations toward the null. This bias could be substantial because the prevalence of NMSC among Caucasian populations is very high. Misclassification of NMSC patients is an issue that needs to be carefully addressed in future validation studies.
A consideration of these issues emphasizes the need for replication of our study finding before strong inferences can be made about the observed association between rs2296675 and overall cancer risk. In the absence of replication, an important limitation is the possibility that the results of this study could be false-positive findings. Concerns about the possibility of a false-positive finding could be heightened by the fact that the P-value distribution was slightly enriched for lower P-values, possibly due to genotyping error rates leading to type I error inflation. However, as is evident in Figure 1, this deviation from the expected null distribution is not severe and cannot reasonably explain the P-values of the most significant findings, a marker with an appreciable MAF of 0.21. As previously discussed, the lack of any published supportive data from GWAS nonetheless adds to the index of suspicion of a false-positive finding. False-positive findings from epidemiology studies have been a source of growing concern (36,37). Replication studies could come in the form of taking a similar cohort study approach as in this study, or alternatively pooling existing data across different cancer sites from case–control or nested case–control studies. The approach of pooling across case–control studies has the advantage of more cancer site-specific matching for potential confounding variables, whereas the cohort study approach has the advantage of being inclusive of all malignancies occurring in a population, in keeping with the goal of this study. Even though replication is lacking, this study does contain the appropriate safeguards of accounting for multiple comparisons and appropriately cautious inferences, taking into account previous findings and biologic plausibility (37).
In summary, the hypothesis that germ-line SNPs in DNA repair genes were associated with risk of all cancers combined was tested. Compared with those with two common alleles, carriers of the minor allele of MGMT SNP rs2296675 had 38% greater risk of any cancer, an association that was highly statistically significant even after accounting for multiple comparisons. This overall increased cancer risk was not due to associations with a few malignancies but rather associations in the direction of increased risk were observed for all of the specific cancer sites studied. This implies that the association, if it is in actual fact genuine, may be relevant to a broad spectrum of malignancies. Replication of this finding in other study populations is needed before more definitive inferences concerning the potential scientific and public health importance of this finding can be made.
Supplementary material
Supplementary Tables 1–5 can be found at http://carcin.oxfordjournals.org/
Funding
US National Cancer Institute (R01 CA105069, HHSN26120080001E) and the Intramural Research Program of the NCI, Center for Cancer Research. This publication does not necessarily reflect the views or policies of the National Cancer Institute, National Institute of Mental Health, National Institutes of Health, US Department of Health and Human Services, the US government, or the Maryland Cancer Registry, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Supplementary Material
Acknowledgements
Cancer incidence data were provided by the Maryland Cancer Registry, Center for Cancer Surveillance and Control, Department of Health and Mental Hygiene, which is funded by the State of Maryland, the Maryland Cigarette Restitution Fund, and the National Program of Cancer Registries of the Centers for Disease Control and Prevention.
Conflicts of Interest Statement: None declared.
Glossary
Abbreviations:
- CI
confidence interval
- GWAS
genome-wide association studies
- LD, linkage disequilibrium; MAF
minor allele frequency
- MGMT
O6-methylguanine-DNA methyltransferase
- NMSC
nonmelanoma skin cancer
- OR
odds ratio
- SNP
single nucleotide polymorphisms.
References
- 1. German J. (1997). Bloom’s syndrome. XX. The first 100 cancers. Cancer Genet. Cytogenet. 93 100–106 [DOI] [PubMed] [Google Scholar]
- 2. Cleaver J.E. (2005). Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer 5 564–573 [DOI] [PubMed] [Google Scholar]
- 3. Wood R.D., et al. (2005). Human DNA repair genes, 2005. Mutat. Res. 577 275–283 [DOI] [PubMed] [Google Scholar]
- 4. Vineis P., et al. (2009). A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J. Natl. Cancer Inst. 101 24–36 [DOI] [PubMed] [Google Scholar]
- 5. Ricceri F., et al. (2011). Is there evidence of involvement of DNA repair polymorphisms in human cancer? Mutat. Res. 736 117–121 [DOI] [PubMed] [Google Scholar]
- 6. Pan X.M., et al. (2011). The association between MLH1 -93 G>A polymorphism of DNA mismatch repair and cancer susceptibility: a meta-analysis. Mutagenesis 26 667–673 [DOI] [PubMed] [Google Scholar]
- 7. Zhong Y., et al. (2010). Effects of O6-methylguanine-DNA methyltransferase (MGMT) polymorphisms on cancer: a meta-analysis. Mutagenesis 25 83–95 [DOI] [PubMed] [Google Scholar]
- 8. Zhang L., et al. (2012). Nucleotide excision repair gene ERCC1 polymorphisms contribute to cancer susceptibility: a meta-analysis. Mutagenesis 27 67–76 [DOI] [PubMed] [Google Scholar]
- 9. Yu H., et al. (2012). Association between PARP-1 V762A polymorphism and cancer susceptibility: a meta-analysis. Genet. Epidemiol. 36 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding D., et al. (2012). Genetic variation of XPA gene and risk of cancer: a systematic review and pooled analysis. Int. J. Cancer 131 488–496 [DOI] [PubMed] [Google Scholar]
- 11. Liu J., et al. (2012). XPA A23G polymorphism and susceptibility to cancer: a meta-analysis. Mol. Biol. Rep. 39 6791–6799 [DOI] [PubMed] [Google Scholar]
- 12. Matullo G., et al. (2006). DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis 27 997–1007 [DOI] [PubMed] [Google Scholar]
- 13. Neasham D., et al. (2009). Double-strand break DNA repair genotype predictive of later mortality and cancer incidence in a cohort of non-smokers. DNA Repair (Amst.) 8 60–71 [DOI] [PubMed] [Google Scholar]
- 14. Ruczinski I., et al. (2012). A population-based study of DNA repair gene variants in relation to non-melanoma skin cancer as a marker of a cancer-prone phenotype. Carcinogenesis 33 1692–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jorgensen T.J., et al. (2009). Hypothesis-driven candidate gene association studies: practical design and analytical considerations. Am. J. Epidemiol. 170 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Bakker P.I., et al. (2005). Efficiency and power in genetic association studies. Nat. Genet. 37 1217–1223 [DOI] [PubMed] [Google Scholar]
- 17. Yuhki N., et al. (1990). DNA variation of the mammalian major histocompatibility complex reflects genomic diversity and population history. Proc. Natl. Acad. Sci. U.S.A. 87 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.US Department of Health and Human Services (US DHHS) The health effects of active smoking: A report of the Surgeon General. U.S. Government Printing Office; Washington, DC: (2004). [Google Scholar]
- 19. Berwick M., et al. (2000). Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J. Natl. Cancer Inst. 92 874–897 [DOI] [PubMed] [Google Scholar]
- 20. Trzeciak A.R., et al. (2008). Age, sex, and race influence single-strand break repair capacity in a human population. Free Radic. Biol. Med. 45 1631–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tubbs J.L., et al. (2007). DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst.) 6 1100–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Westbrook T.F., et al. (2005). Dissecting cancer pathways and vulnerabilities with RNAi. Cold Spring Harb. Symp. Quant. Biol. 70 435–444 [DOI] [PubMed] [Google Scholar]
- 23. Mahmood-ur-Rahman A.I., et al. (2008). RNA interference: The story of gene silencing in plants and humans. Biotech. Adv. 26 202–209 [DOI] [PubMed] [Google Scholar]
- 24. Jacinto F.V., et al. (2007). MGMT hypermethylation: a prognostic foe, a predictive friend. DNA Repair (Amst.) 6 1155–1160 [DOI] [PubMed] [Google Scholar]
- 25. Esteller M., et al. (1999). Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 59 793–797 [PubMed] [Google Scholar]
- 26. Esteller M., et al. (2001). A gene hypermethylation profile of human cancer. Cancer Res. 61 3225–3229 [PubMed] [Google Scholar]
- 27. Esteller M., et al. (2004). Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene 23 1–8 [DOI] [PubMed] [Google Scholar]
- 28. Ogino S., et al. (2007). MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis 28 1985–1990 [DOI] [PubMed] [Google Scholar]
- 29. Menigatti M., et al. (2009). Normal colorectal mucosa exhibits sex- and segment-specific susceptibility to DNA methylation at the hMLH1 and MGMT promoters. Oncogene 28 899–909 [DOI] [PubMed] [Google Scholar]
- 30. Christmann M., et al. (2012). O(6)-methylguanine-DNA methyltransferase (MGMT): impact on cancer risk in response to tobacco smoke. Mutat. Res. 736 64–74 [DOI] [PubMed] [Google Scholar]
- 31. Christmann M., et al. (2011). O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: enzyme activity, promoter methylation and immunohistochemistry. Biochim. Biophys. Acta 1816 179–190 [DOI] [PubMed] [Google Scholar]
- 32. Fu D., et al. (2012). Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12 104–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drabløs F., et al. (2004). Alkylation damage in DNA and RNA–repair mechanisms and medical significance. DNA Repair (Amst.) 3 1389–1407 [DOI] [PubMed] [Google Scholar]
- 34. Hecht S.S. (1999). DNA adduct formation from tobacco-specific N-nitrosamines. Mutat. Res. 424 127–142 [DOI] [PubMed] [Google Scholar]
- 35. Wheless L., et al. (2010). Nonmelanoma skin cancer and the risk of second primary cancers: a systematic review. Cancer Epidemiol. Biomarkers Prev. 19 1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ioannidis J.P., et al. (2011). The false-positive to false-negative ratio in epidemiologic studies. Epidemiology 22 450–456 [DOI] [PubMed] [Google Scholar]
- 37. Wacholder S., et al. (2004). Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 96 434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.